Abstract
Background
Nosocomial infections caused by multi-drug resistant Enterobacteriaceae are a global public health threat that ought to be promptly identified, reported, and addressed accurately. Many carbapenem-resistant Enterobacteriaceae-associated genes have been identified in Saudi Arabia but not the endemic Klebsiella pneumoniae carbapenemases (KPCs), which are encoded by blaKPC-type genes. KPCs are known for their exceptional spreading potential.
Methods
We collected n = 286 multi-drug resistant (MDR) Klebsiella spp. isolates as part of screening for resistant patterns from a tertiary hospital in Saudi Arabia between 2014 and 2018. Antimicrobial susceptibility testing was carried out using both VITEK II and the broth microdilution of all collected isolates. Detection of resistance-conferring genes was carried out using Illumina whole-genome shotgun sequencing and PacBio SMRT sequencing protocols.
Results
A Carbapenem-resistant Enterobacteriaceae (CRE) Klebsiella quasipneumoniae subsp. similipneumoniae strain was identified as a novel ST-3510 carrying a blaKPC-2 carbapenemase encoding gene. The isolate, designated as NGKPC-421, was obtained from shotgun Whole Genome Sequencing (WGS) surveillance of 286 MDR Klebsiella spp. clinical isolates. The NGKPC-421 isolate was collected from a septic patient in late 2017 and was initially misidentified as K. pneumoniae. The sequencing and assembly of the NGKPC-421 genome resulted in the identification of a putative ~ 39.4 kb IncX6 plasmid harboring a blaKPC-2 gene, flanked by transposable elements (ISKpn6-blaKPC-2–ISKpn27).
Conclusion
This is the first identification of a KPC-2-producing CRE in the Gulf region. The impact on this finding is of major concern to the public health in Saudi Arabia, considering that it is the religious epicenter with a continuous mass influx of pilgrims from across the world. Our study strongly highlights the importance of implementing rapid sequencing-based technologies in clinical microbiology for precise taxonomic classification and monitoring of antimicrobial resistance patterns.
Keywords: MDR, Klebsiella quasipneumoniae, Carbapenemases, KPC-2, blaKPC-2, Tn3
Background
Beta-lactam antibiotics such as penicillins, cephalosporins, and carbapenems represent up to 60% of the available treatment options for antibiotic-resistant Gram-negative bacteria [1]. These antibiotics can be hydrolyzed by the production of extended-spectrum beta-lactamases (ESBL) and carbapenemases [2]. Carbapenem-resistant Enterobacteriaceae (CRE) is known to cause a variety of nosocomial infections that are associated with high mortality rates [3, 4]. Many CRE-associated genes have been identified, including the endemic Klebsiella pneumoniae carbapenemases (KPCs) and the New Delhi Metallo-beta-lactamases (NDMs) [3, 5]. Rapid identification and reporting of such markers could aid in minimizing the horizontal transfer of these genes to other bacterial species and ultimately preventing the spread of antibiotic resistance [6, 7]. The KPC-type enzymes, which are encoded by blaKPC-type genes, are hard to detect by routine susceptibility screening and are known for their exceptional dissemination potential in various CREs [8]. To date, 24 variants of the blaKPC-type genes have been reported from different countries, including the USA, Poland, South Korea, Malaysia, and Thailand [9–12]. These variants are typically identified within a Tn3-family of transposable elements capable of transferring resistance genes at high frequency [13].
Klebsiella pneumoniae was initially classified into three genetically closely-related phylogroups, and more recently, they have been classified into three distinct species: K. pneumoniae [KPI], K. quasipneumoniae [KpII], and K. variicola [KpIII] [14]. This distinction was possible through the comparison of their core genomes and not through conventional Multilocus Sequence Typing (MLST) and capsule genotyping [13, 14]. Due to significant overlap in their biochemical profiles, phenotypic testing using traditional clinical microbiological assays is incapable of accurately differentiating between Klebsiella spp. leading to false reporting of clinically isolated K. quasipneumoniae to be K. pneumoniae [13].
K. quasipneumoniae was initially known as a commensal intestinal colonizer. However, recent genomics-driven studies have documented it as an etiologic agent in a number of clinical Klebsiella-related infection cases [13, 15–17]. Clearly, such findings would underline the importance and usefulness of comparative genomics in defining the relatedness and differences between such strains, which could ultimately aid in reaching an accurate and prompt diagnosis.
Here, we report the first blaKPC-2 harboring plasmid in a K. quasipneumoniae strain isolated from a tertiary care hospital in the Gulf states (GCC) region, identified by a whole-genome sequencing (WGS) approach to detect multi-drug resistant (MDR) markers in a large collection of Klebsiella spp. isolates.
Materials and methods
Clinical setting and the case
As part of a larger study aiming to characterize Klebsiella spp. in a tertiary hospital in King Abdulaziz Medical City (KAMC) in Jeddah, Saudi Arabia, we collected MDR strains (n = 286) between 2014 and 2018. All strains were subjected to routine identification by using VITEK MS and VITEK II systems (bioMérieux Inc., Durham, NC) and were recorded in the laboratory information system as K. pneumoniae. Following the initial analysis of shotgun sequencing of the 286 MDR Klebsiella clinical isolates by Illumina sequencing technology (Illumina, USA) (data not shown), we identified a single isolate (NGKPC-421) as K. quasipneumoniae, which previously was falsely identified as Klebsiella pneumoniae by the clinical microbiology laboratory in the hospital. This isolate was obtained from a male patient in his mid-60s with chronic comorbidities admitted due to trauma-associated airway obstruction in November 2017, which promptly required a Percutaneous Tracheostomy. The patient developed nosocomial infection one-week post-surgery, and his health deteriorated gradually with symptoms mainly indicative of pneumonia. The first positive culture from the patient’s tracheostomy wound was identified as MDR K. penumoniae in December 2017 using VITEK II and VITEK MS. The infection persisted for a couple of months without improvement despite the antibiotic treatment and eventually caused septic shock and subsequent death. Antibiotic susceptibility testing (AST) and Minimum Inhibitory Concentration (MIC) of the NGKPC-421 isolate was conducted using the automated MICRONAUT system (Merlin Diagnostika, Germany). The AST result was interpreted using the resistance breakpoints defined by the EUCAST [18]. The resistance phenotype reported by both VITEK II and MICRONAUT methods were in complete agreement (Table 1).
Table 1.
The AST profile and MIC of the NGKPC-421 isolate
| Antimicrobial agent | MIC (mg/L) | VITEK IIa | Micronauta |
|---|---|---|---|
| Amikacin | 4 | S | S |
| Amoxicillin-clavulanate | 32 | R | – |
| Ampicillin | 32 | R | R |
| Cefazolin | 30 | R | R |
| Cefepime | 4 | R | R |
| Cefoxitin | 30 | – | R |
| Cefotaxime | 2 | R | R |
| Ceftazidime | 64 | R | R |
| Ceftriaxone | 6 | R | – |
| Chloramphenicol | 4 | – | S |
| Ciprofloxacin | 0.5 | R | R |
| Colistin | 1 | – | S |
| Fosfomycin | < 32 | – | S |
| Gentamicin | > 16 | R | – |
| Imipenem | 4 | R | R |
| Levofloxacin | 1 | – | R |
| Meropenem | 8 | R | R |
| Piperacillin | 16 | R | R |
| Piperacillin-tazobactam | 64/4 | – | R |
| Temocillin | < 32 | – | S |
| Tigecycline | 0.25 | S | S |
| Trimethoprim-sulfamethoxazole | 4/76 | R | R |
aVITEK II and MICRONAUT assays were repeated (n = 2) and (n = 3), respectively
R indicates resistant and S indicates susceptible to a given antibiotic based on the EUCAST guidelines [18] and (−) indicates the undetermined antimicrobial sensitivity test for a given antimicrobial agent
Genome sequencing, assembly, and analyses
Genomic DNA of the NGKPC-421 strain was extracted from a single colony, and DNA library was prepared using the NEBNext Ultra II kit for Illumina (New England BioLabs, UK). Paired-end shotgun WGS was performed on Illumina HiSeq 4000 platform (Illumina, CA, USA). To obtain long sequence reads for assembling the complete genome, we also sequenced the high molecular-weight genomic DNA on a Pacbio RS II platform (Pacific Biosciences, CA, USA). The raw PacBio reads were first converted to fastq format by using bamtools v2.3.0 [19] and then assembled on Canu v1.6 [20] by using the default assembly settings (Additional file 1). The resulting assembly file was further polished with Pilon v1.20 [21] by supplying the sorted bam file that was generated from bwa v0.7.17 [22] mapping of Illumina reads to the initial Canu assembly (Additional file 1). The gfa output file from the Canu-assembly of NGKPC-421 was further loaded on Bandage [23] to visualize the connections between contigs so as to identify the putative chromosome and plasmid sequence-containing contigs. All contig sequences that showed no connections to the chromosome (potentially extrachromosomal in nature) were probed for the presence of signature genes that are commonly found associated with bacterial plasmids i.e., genes that encode for plasmid replication (Rep) or conjugative transfer (Tra) proteins. These identified putative plasmids that were then further analyzed on pMLST v1.4 and PlasmidFinder v2.1 web-tools [24] web-tools. Antibiotic resistance gene identification was done with the Kleborate tool v0.3.0 [25] and Resfinder [26] and confirmation of the gene annotations was made using the PATRIC online database [27].
Phylogenetic analysis and annotation
The final assembly was used to assign core genome MLST (cgMLST) by using the K. quasipneumoniae BIGSdb database (http://bigsdb.pasteur.fr/Klebsiella/Klebsiella.html) [28, 29]. Available assembled complete genomes of [KPII-A], [KPII-B] K. quasipneumoniae reference sequences were downloaded from the NCBI database (November 2018) and aligned with identified K. quasipneumoniae (NGKPC-421) genome using progressiveMauve [30]. Alignment-based phylogenetic tree was constructed of K. quasipneumoniae subspecies (CP014154/HKUOPA4, CP034136/G747, CP034129/G4584, CP031257/L22, CP030171/A708, CP029597/ATCC 700603, CP023480/KPC-142 CP029443/CAV1947, CP029432/CAV2018, CP014156/HKUOPL4, CP014155/HKUOPJ4, CP012300/HKUOPLC, CP012252/HKUOPLA, and CP0023478/HKUOPA4) as well as outgroup strains of [KPI] K. pneumoniae (CP013322/CAV1193) and [KPIII] K. variicola (CP000964/342). Locally collinear blocks (LCBs) larger than 500 bp were selected from the full alignment file using a Perl script stripSubsetLCBs [30]. The core alignment file in .xmfa format was then converted into the .fasta format using the Perl script xmfa2fasta (https://github.com/kjolley/seq_scripts/blob/master/xmfa2fasta.pl). The core alignment (4.25 Mb) was further inspected visually for any misaligned regions. To ensure the bacterial strain type, a phylogenetic tree was generated using RAxML v8.2.3 [31] with the GTRGAMMA model, bootstrapping (1,000 replicates), and the best maximum likelihood tree inference. Additionally, a core-genome-SNP-based tree was constructed on Parsnp v1.2 from the Harvest suite [32] by using the recombination filtration (−x) and curated genome directory (−c) parameters.
Results
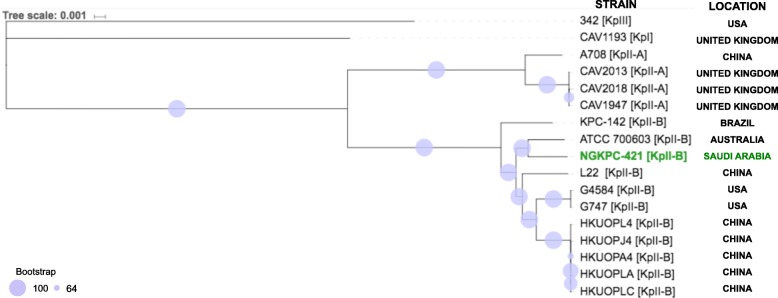
Analysis of the WGS data from the K. pneumoniae surveillance study identified one isolate (NGKPC-421) as K. quasipneumoniae subsp. similipneumoniae [KpII-B], that was originally misidentified as K. pneumoniae in the hospital laboratory. As shown in Fig. 1, the phylogenetic comparison of the NGKPC-421 isolate with 16 publicly available strains of K. quasipneumoniae 10 [KpII-B], and 4 [KpII-A], as well as K. pneumoniae [KpI], and K. variicola [KpIII], revealed a close clustering of NGKPC-421 isolate with the ATCC strain 700,603 [KpII-B], originally isolated from Australia in 2016. A separate core-genome-SNP-based tree that was constructed with Parsnp also showed identical topology (supplementary-Fig. 1). Our cgMLST analysis defined NGKPC-421 isolate as a novel K. quasipneumoniae subsp. similipneumoniae assigned as ST-3510.
Fig. 1.
The phylogenetic tree based on segmental alignments of chromosomal sequences. The phylogenetic tree was constructed using RAxML (maximum likelihood) using reference sequences downloaded from NCBI database, including K. quasipneumoniae subspecies as well as strains of K. pneumoniae [KPI] (CAV1193) and K. variicola [KPIII] and viewed with the iTOL online tool v 4.4.2 [49]. Bootstrap values are represented by the size of the circles on each node. The NGKPC-421 [KPII-B] is most closely related to the ATCC strain 700,603 [KpII-B], originally isolated from Australia. K. quasipneumoniae subspecies (CP014154/HKUOPA4, CP034136/G747, CP034129/G4584, CP031257/L22, CP030171/A708, CP029597/ATCC 700603, CP023480/KPC-142 CP029443/CAV1947, CP029432/CAV2018, CP014156/HKUOPL4, CP014155/HKUOPJ4, CP012300/HKUOPLC, CP012252/HKUOPLA, and CP0023478/HKUOPA4) as well as outgroup strains of [KPI] K. pneumoniae (CP013322/CAV1193) and [KPIII] K. variicola (CP000964/342)
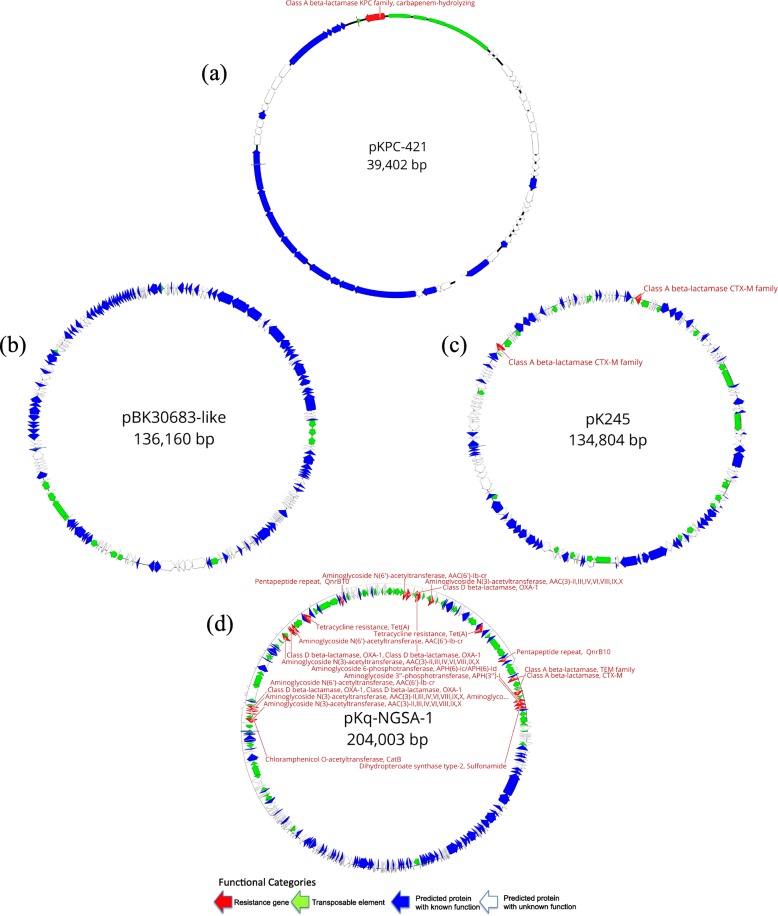
Searching for genetic markers encoding antibiotic resistance showed the presence of KPC-2 gene in the NGKPC-421 isolate. The presence and location of KPC-2 gene were identified and confirmed as a result of the assembly of 6 contigs comprised of one large chromosome (~ 5.26 Mb) and four putative circular plasmids of varying sizes (Table 2). Further analysis of the complete genome also showed the presence of other antibiotic resistance genes and virulence factors (Table 2).
Table 2.
Antibiotic resistance genes on the assembled chromosome and plasmid sequences in K. quasipneumoniae NGKPC-421 strain (ENA ERS3013985)
| Namea | MLST/pMLST | Size (Kb) | CDS/GC content | Resistance genes | Predicted phenotype |
|---|---|---|---|---|---|
| Chromosome | ST3510 | 5255.70 | 5816/57.35 | blaOKPB-3 | β-lactam |
| oqxA | Fluoroquinolone | ||||
| oqxB | Fluoroquinolone | ||||
| pKPC-421 plasmid | IncX6 | 39.40 | 58/47.32 | blaKPC-2 | β-lactam |
| pBK30683-like plasmid | IncFII | 136.16 | 199/55.48 | n/a | n/a |
| pKq-NGSA-1 plasmid | IncFII(k) | 204.00 | 303/53.31 | aac(3)lla | Aminoglycoside |
| aac(3″)lb | Aminoglycoside | ||||
| aac(6′)lb-cr | Fluoroquinolone | ||||
| aac(6)ld | β-lactam | ||||
| blaOXA-1 | Phenicol | ||||
| blaCTX-M-15 | |||||
| blaTEM-1B | |||||
| qnrB1 | Fluoroquinolone | ||||
| catB3 Sul2 | Sulphonamide | ||||
| tet(A) | Tetracycline | ||||
| drA14 | Trimethoprim | ||||
| pK245 plasmid | IncR | 134.81 | 188/53.68 | blaCTX-M-14b | β-lactam |
aAssembly of K. quasipneumoniae NGKPC-421 sequences resulted in 6 contigs comprising of 1 chromosome and 4 putative circular plasmids (Note: 2 contigs represent the pKq-NGSA-1 plasmid)
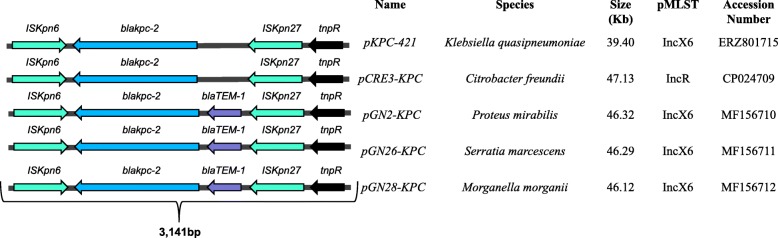
Four putative plasmids identified in the NGKPC-421 isolate were annotated as pKPC-421, pBK30683-like, pK245, and a potentially novel plasmid pKq-NGSA-1 (Fig. 2). The plasmid pKPC-421 (~ 39.4 Kb) belongs to the Inc6X group, where the blaKPC-2 gene is flanked by genes belonging to the Tn3-based transposon family (ISKpn6 and ISKpn27) (Fig. 3). Notably, a public database search of the ISKpn6-blaKPC-2-ISKpn27 harboring sequences showed its presence on several plasmids across numerous bacterial species previously reported from many countries but not from the Gulf region (Fig. 3). The largest plasmid pKq-NGSA-1 (~ 204 Kb), represented by two contigs, was found to be an IncFII(k), harboring the majority of resistance genes, including blaOXA-1, blaCTX-M-15, and blaTEM-1B in multiple copies (Table 2). The second-largest plasmid pBK30683-like (~ 136 Kb), found as a single contig, had no known antibiotic resistance genes present. The fourth plasmid pK245 (~ 135 Kb) belonged to the IncR group and was found to harbor blaCTX-M-14b (Table 2).
Fig. 2.
Schematic gene organization of all 4 putative circular plasmids in NGKPC-421. a pKPC-421 plasmid, b pKq-NGSA-1 plasmid, c pK245 plasmid, and d pBK30683 plasmid
Fig. 3.
Schematic of gene arrangements of the blaKPC-2 flanked by Tn3-based transposon in plasmids across different species. IS481 and IS1182 family of genes harboring transposase and resolvase genes and insertion sequences, i.e., ISKpn6, and ISKpn27 in 5 plasmids compared to the pKPC-421 of K. quasipneumoniae. This fragment has been previously identified in various plasmids in several Gram-negative species from China. The cassette of SKpn6-blaKPC-2-ISKpn27 can be found in similar IncX6 plasmids (included in Fig. 2) as well as multiple types of plasmids across various bacterial species
Discussion
Although K. quasipneumoniae was initially considered as a benign human gut commensal bacteria, recent reports suggest its involvement in potentially fatal infections [15, 16, 33]. Despite being rarely documented, K. quasipneumoniae appears to be capable of undergoing homologous recombination with other Klebsiella spp., allowing it to exchange both chromosomal and plasmid-borne antibiotic resistance genes [14]. Additionally, KPC-positive isolates co-producing Metallo-β-lactamase enzymes are known to be associated with diverse mobile genetic elements causing large outbreaks in China, Europe and the USA [34–37]. Our data demonstrate that blaKPC-2, in addition to other carbapenemase resistance genes, is present in the Gulf region and that K. quasipneumoniae subsp. similipneumoniae could be misidentified as K. pneumoniae in clinical laboratories [38].
This finding should encourage further surveillance within Saudi Arabia and neighboring countries for additional occurrences of such resistance markers. This is particularly of epidemiological significance because the blaKPC-2 was found in an IncX6 plasmid (pKPC-421) that is known to circulate among Enterobacteriaceae species [39]. The blaKPC-2 was found to be flanked by transposons, which could have major implications on the spread of antibiotic resistance within the region. Furthermore, the isolate carried other plasmids (pKq-NGSA-1, pBK30683, and pK245) containing various other genes that encode for antibiotic resistance. While such genes are known to be present in clinical CRE-associated K. pneumoniae strains [5, 40, 41], none of the plasmids included here were reported nor genetically characterized prior to this study in the Gulf region [42–45].
Although the presence of various β-lactamase genes among Enterobacteriaceae such as blaSHV-1, blaTEM-1, blaCTX-M-1, blaNDM-1 and blaOXA-48 in Saudi Arabia and the Gulf region has been previously reported [42–45], though this is the first report of a KPC-type producing K. quasipneumoniae. The discovery of a misidentified MDR K. quasipneumoniae isolate is worrisome, suggesting the need to update identification methods and the consideration of the implementation of rigorous molecular surveillance [46]. The data reported in this study support the need to investigate the emergence and spread of KPC-producing strains from the region. Furthermore, the inability of conventional laboratory techniques to detect KPC-2 gene in this CRE isolate increases the need for routine reconnaissance using WGS. Considering the importance of Saudi Arabia as a host of one of the largest recurring mass gathering events (i.e. Hajj and Umra pilgrimage) with 7 million pilgrims from more than 180 countries [47], infection control efforts are essential so as to monitor the potential spread of such KPC-producing Enterobacteriaceae. Current conventional molecular microbiology laboratory diagnostic methods are sufficient for diagnosing infectious diseases, and WGS may be used as an additional confirmatory tool [48]. Although performing WGS techniques would essentially require access to expensive equipment and reagents in addition to computational infrastructures and relevant expertise; our study re-emphasizes that the introduction of clinical genomics-driven surveillance methods in the GCC region could aid in accurate surveillance and confirmed diagnoses of etiologic agents and the determination of their antibiotic resistance markers. This is of particular public health relevance for monitoring the potential spread of KPC-producing Enterobacteriaceae during mass gathering events such as Hajj and Umrah in Saudi Arabia.
Supplementary information
Additional file 1: Command-lines for the tools
Acknowledgments
We thank the infectious disease department and the pathology laboratory at King Khaled hospital in the Ministry of the National Guards – Health Affairs western region for their support of surveillance research. We also would like to thank the Bioscience core lab in KAUST for their help with the sequencing. We also thank Fathia Ben Rached, Malak Haidar, Qingtian Guan, and Sara Mfarrej from KAUST Microbial Genomics Laboratory for their technical help. We thank the team of curators at the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles and/or isolates at http://bigsdb.pasteur.fr.
Abbreviations
- CRE
Carbapenem Resistant Enterobacteriaceae
- ESBL
Extended-spectrum beta-lactamase
- KPC
Klebsiella pneumonia Carbapenemase
- MIC
Minimum Inhibitory Concentration
- WGS
Whole Genome Shotgun Sequencing
Authors’ contribution
SH, AS, AT, BH, and MA collected the clinical data and clinical interpretation; SH, AM, MK, and MA performed the biochemical experiments; SH performed the wetlab characterization experiments; SH and CPA performed the bioinformatics analysis; SH, AA, and AP conceived the study; SH, HZ, CPA and AP prepared figures and wrote or revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by baseline funding (BAS/1/1020-01-01) from KAUST to AP.
Availability of data and materials
The raw reads and/or assembly files from this study are publicly available at the European Nucleotide Archive (ENA) under the study accession number PRJEB30599 http://www.ebi.ac.uk/ena/data/view/PRJEB30599 and at the Zenodo open-access repository- 10.5281/zenodo.2583210
Ethics approval and consent to participate
The project was approved by the IRB committees at NGHA (RJ17–023-J) and KAUST (17IBEC38_Pain).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sharif Hala, Email: sharif.hala@kaust.edu.sa.
Chakkiath Paul Antony, Email: chakkiat.antony@kaust.edu.sa.
Mohammed Alshehri, Email: ashehrim@gmail.com.
Abdulhakeem O. Althaqafi, Email: ahthaqafi@yahoo.com
Asim Alsaedi, Email: alsaedias@ngha.med.sa.
Areej Mufti, Email: areejmufti@gmail.com.
Mai Kaaki, Email: KaakiMM@ngha.med.sa.
Baraa T. Alhaj-Hussein, Email: Alhajhusseinbt@ngha.med.sa
Hosam M. Zowawi, Email: h.zowawi@uq.edu.au
Abdulfattah Al-Amri, Email: MowalladAW@ngha.med.sa.
Arnab Pain, Email: arnab.pain@kaust.edu.sa.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13756-019-0653-9.
References
- 1.Wong DR, van Duin D. Novel Beta-lactamase inhibitors: unlocking their potential in therapy. Drugs. 2017;77(6):615–628. doi: 10.1007/s40265-017-0725-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zowawi HM, Balkhy HH, Walsh TR, Paterson DL. β-Lactamase production in key gram-negative pathogen isolates from the Arabian peninsula. Clin Microbiol Rev. 2013;26(3):361–380. doi: 10.1128/CMR.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Yoseph Haggai, Cohen Nadav, Korytny Alexander, Andrawus Elias R., Even Dar Razi, Geffen Yuval, Hussein Khetam, Paul Mical. Risk factors for mortality among carbapenem-resistant enterobacteriaceae carriers with focus on immunosuppression. Journal of Infection. 2019;78(2):101–105. doi: 10.1016/j.jinf.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Rees CA, Nasir M, Smolinska A, Lewis AE, Kane KR, Kossmann SE, et al. Detection of high-risk carbapenem-resistant Klebsiella pneumoniae and Enterobacter cloacae isolates using volatile molecular profiles. Sci Rep. 2018;8(1):13297. doi: 10.1038/s41598-018-31543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krapp F, Ozer EA, Qi C, Hauser AR, editors. Case Report of an Extensively Drug-Resistant Klebsiella pneumoniae Infection With Genomic Characterization of the Strain and Review of Similar Cases in the United States. Open forum infectious diseases 2018;Vol. 5, No. 5, p. ofy074. US: Oxford University Press. [DOI] [PMC free article] [PubMed]
- 6.Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg Infect Dis. 2010;16(6):1014. doi: 10.3201/eid1606.091671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zowawi HM. Antimicrobial resistance in Saudi Arabia: an urgent call for an immediate action. Saudi Med J. 2016;37(9):935. doi: 10.15537/smj.2016.9.16139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaidya VK. Horizontal transfer of antimicrobial resistance by extended-spectrum β lactamase-producing enterobacteriaceae. J Lab Physicians. 2011;3(1):37. doi: 10.4103/0974-2727.78563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baraniak A., Izdebski R., Fiett J., Sadowy E., Adler A., Kazma M., Salomon J., Lawrence C., Rossini A., Salvia A., Vidal Samso J., Fierro J., Paul M., Lerman Y., Malhotra-Kumar S., Lammens C., Goossens H., Hryniewicz W., Brun-Buisson C., Carmeli Y., Gniadkowski M. Comparative Population Analysis of Klebsiella pneumoniae Strains with Extended-Spectrum β-Lactamases Colonizing Patients in Rehabilitation Centers in Four Countries. Antimicrobial Agents and Chemotherapy. 2013;57(4):1992–1997. doi: 10.1128/AAC.02571-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machulska M, Baraniak A, Zak I, Bojarska K, Zabicka D, Sowa-Sierant I, et al. KPC-2-producing Klebsiella pneumoniae ST11 in a Children's Hospital in Poland. Pol J Microbiol. 2017;66(3):401–405. doi: 10.5604/01.3001.0010.4884. [DOI] [PubMed] [Google Scholar]
- 11.Lee MY, Ko KS, Kang C-I, Chung DR, Peck KR, Song J-H. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. Int J Antimicrob Agents. 2011;38(2):160–163. doi: 10.1016/j.ijantimicag.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Ramos AC, Gales AC, Monteiro J, Silbert S, Chagas-Neto T, Machado AM, et al. Evaluation of a rapid immunochromatographic test for detection of distinct variants of Klebsiella pneumoniae carbapenemase (KPC) in Enterobacteriaceae. J Microbiol Methods. 2017;142:1–3. doi: 10.1016/j.mimet.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Long SW, Linson SE, Saavedra MO, Cantu C, Davis JJ, Brettin T, et al. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. Msphere. 2017;2(4):e00290–e00217. doi: 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci. 2015;112(27):E3574–E3E81. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breurec S, Melot B, Hoen B, Passet V, Schepers K, Bastian S, et al. Liver abscess caused by infection with community-acquired Klebsiella quasipneumoniae subsp. quasipneumoniae. Emerg Infect Dis. 2016;22(3):529. doi: 10.3201/eid2203.151466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brinkac LM, White R, D'Souza R, Nguyen K, Obaro SK, Fouts DE. Emergence of New Delhi Metallo-ß-lactamase (NDM-5) in Klebsiella quasipneumoniae from neonates in a Nigerian hospital. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolás MF, PIP R, Marques de Carvalho F, Camargo DR, de Fátima Morais Alves C, Loss de Morais G, et al. Comparative genomic analysis of a clinical isolate of Klebsiella quasipneumoniae subsp. similipneumoniae, a KPC-2 and OKP-B-6 beta-lactamases producer harboring two drug-resistance plasmids from southeast Brazil. Front Microbiol. 2018;9:220. doi: 10.3389/fmicb.2018.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden.
- 19.Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27(12):1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren Sergey, Walenz Brian P., Berlin Konstantin, Miller Jason R., Bergman Nicholas H., Phillippy Adam M. Canu: scalable and accurate long-read assembly via adaptivek-mer weighting and repeat separation. Genome Research. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31(20):3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carattoli A, Zankari E, Garcìa-Fernandez A, Larsen MV, Lund O, Villa L, et al. PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrobial Agents and Chemotherapy. 2014:AAC.02412–14. [DOI] [PMC free article] [PubMed]
- 25.Wick RR, Heinz E, Holt KE, Wyres KL. Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. Journal of clinical microbiology. 2018;56(6):e00197–18. [DOI] [PMC free article] [PubMed]
- 26.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2013;42(D1):D581–DD91. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, et al. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of listeria monocytogenes. J Clin Microbiol. 2015;53(9):2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling AE, Mau B, Perna NT. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. 2018;18(1):6. doi: 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maltezou H, Giakkoupi P, Maragos A, Bolikas M, Raftopoulos V, Papahatzaki H, et al. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece) J Infect. 2009;58(3):213–219. doi: 10.1016/j.jinf.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Steinmann J, Kaase M, Gatermann S, Popp W, Steinmann E, Damman M, et al. Outbreak due to a Klebsiella pneumoniae strain harbouring KPC-2 and VIM-1 in a German university hospital, July 2010 to January 2011. Eurosurveillance. 2011;16(33):19944. [PubMed] [Google Scholar]
- 36.Yang J, Ye L, Guo L, Zhao Q, Chen R, Luo Y, et al. A nosocomial outbreak of KPC-2-producing K lebsiella pneumoniae in a C hinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect. 2013;19(11):E509–EE15. doi: 10.1111/1469-0691.12275. [DOI] [PubMed] [Google Scholar]
- 37.Woodford N, Tierno PM, Young K, Tysall L, Palepou M-FI, Ward E, et al. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class a β-lactamase, KPC-3, in a New York medical center. Antimicrob Agents Chemother. 2004;48(12):4793–4799. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, et al. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother. 2014;58(6):3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Feng J, Zhan Z, Yin Z, Jiang Q, Wei P, et al. Dissemination of KPC-2-encoding IncX6 plasmids among multiple Enterobacteriaceae species in a single Chinese hospital. Front Microbiol. 2018;9:478. doi: 10.3389/fmicb.2018.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curiao T, Morosini MI, Ruiz-Garbajosa P, Robustillo A, Baquero F, Coque TM, et al. Emergence of Bla KPC-3-Tn 4401 a associated with a pKPN3/4-like plasmid within ST384 and ST388 Klebsiella pneumoniae clones in Spain. J Antimicrob Chemother. 2010;65(8):1608–1614. doi: 10.1093/jac/dkq174. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y-T, Shu H-Y, Li L-H, Liao T-L, Wu K-M, Shiau Y-R, et al. Complete nucleotide sequence of pK245, a 98-kilobase plasmid conferring quinolone resistance and extended-spectrum-β-lactamase activity in a clinical Klebsiella pneumoniae isolate. Antimicrob Agents Chemother. 2006;50(11):3861–3866. doi: 10.1128/AAC.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.uz Zaman T, Aldrees M, Al Johani SM, Alrodayyan M, Aldughashem FA, Balkhy HH. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Infect Dis. 2014;28:186–192. doi: 10.1016/j.ijid.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 43.Memish ZA, Assiri A, Almasri M, Roshdy H, Hathout H, Kaase M, et al. Molecular characterization of carbapenemase production among gram-negative bacteria in Saudi Arabia. Microb Drug Resist. 2015;21(3):307–314. doi: 10.1089/mdr.2014.0121. [DOI] [PubMed] [Google Scholar]
- 44.Zaman TU, Albladi M, Siddique MI, Aljohani SM, Balkhy HH. Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect Drug Resist. 2018;11:1183. doi: 10.2147/IDR.S161146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdalhamid B, Elhadi N, Albunayan S, Alsamman K, Aljindan R. First description of methyltransferases in extensively drug-resistant Klebsiella pneumoniae isolates from Saudi Arabia. J Med Microbiol. 2017;66(7):859–863. doi: 10.1099/jmm.0.000480. [DOI] [PubMed] [Google Scholar]
- 46.Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24(12):944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Elachola H, Gozzer E, Zhuo J, Memish ZA. A crucial time for public health preparedness: Zika virus and the 2016 Olympics, Umrah, and hajj. Lancet. 2016;387(10019):630–632. doi: 10.1016/S0140-6736(16)00274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunne W, Westblade L, Ford B. Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis. 2012;31(8):1719–1726. doi: 10.1007/s10096-012-1641-7. [DOI] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W2W5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Command-lines for the tools
Data Availability Statement
The raw reads and/or assembly files from this study are publicly available at the European Nucleotide Archive (ENA) under the study accession number PRJEB30599 http://www.ebi.ac.uk/ena/data/view/PRJEB30599 and at the Zenodo open-access repository- 10.5281/zenodo.2583210