Abstract
The aim of the present study was to identify a vascular invasion-associated gene signature for predicting prognosis in patients with hepatocellular carcinoma (HCC). Using RNA-sequencing data of 292 HCC samples from The Cancer Genome Atlas (TCGA), the present study screened differentially expressed genes (DEGs) between patients with and without vascular invasion. Feature genes were selected from the DEGs by support vector machine (SVM)-based recursive feature elimination (RFE-SVM) algorithm to build a classifier. A multi-gene signature was selected by L1 penalized (LASSO) Cox proportional hazards (PH) regression model from the feature genes selected by the RFE-SVM to develop a prognostic scoring model. TCGA set was defined as the training set and was divided by the gene signature into a high-risk group and a low-risk group. Involvement of the DEGs between the two risk groups in pathways was also investigated. The presence and absence of vascular invasion between patients of training set was 175 DEGs. A classification model of 42 genes performed well in differentiating patients with and without vascular invasion on the training set and the validation set. A 14-gene prognostic model was built that could divide the training set or the validation set into two risk groups with significantly different survival outcomes. A total of 762 DEGs in the two risk groups of the training set were revealed to be significantly associated with a number of signaling pathways. The present study provided a 42-gene classifier for predicting vascular invasion, and identified a vascular invasion-associated 14-gene signature for predicting prognosis in patients with HCC. Several genes and pathways in HCC development are characterized and may be potential therapeutic targets for this type of cancer.
Keywords: gene signature, prognosis, vascular invasion, hepatocellular carcinoma, overall survival
Introduction
Hepatocellular carcinoma (HCC) is a major type of primary liver cancer (1). The mortality rate is increasing, and patients with the tumor present with a poor prognosis (2). An increasing number of studies have demonstrated that vascular invasion is an adverse prognostic factor in HCC (3–5). Furthermore, vascular invasion is an independent predictive factor of long-term survival in patients with early-stage HCC, and is significantly associated with intrahepatic metastasis (6). Hence, it is extremely necessary to differentiate patients with HCC that present with vascular invasion from those patients with HCC that do not present with vascular invasion, so as to improve survival time.
A risk classification model of micro-vascular invasion based on histopathological features has been introduced for predicting the prognosis of patients with HCC (7). Differentially expressed genes (DEGs) in HCC tissue samples in the presence or absence of vascular invasion have been studied in order to extract multi-gene signatures for detecting vascular invasion (8,9). High-throughput technologies allow for the development of a classification model, wherein vascular invasion information can be derived from molecular features. The Cancer Genome Atlas (TCGA) provides comprehensive maps of genomic alterations in various types of cancer (https://portal.gdc.cancer.gov/). A recent study derived a 16-miRNA-based classifier from the analysis of micro (mi)RNA and mRNA expression data derived from TCGA, which could effectively identify vascular invasion and predict overall survival (OS) (10). These studies indicated the feasibility of these multi-gene signatures for prediction of cancer prognosis. Nevertheless, more efforts should be made in order to generate more reliable and accurate prognostic models based on feature genes of vascular invasion.
The present study analyzed HCC RNA-sequencing data from TCGA in order to identify feature genes using a recursive feature elimination (RFE) method (11), thus constructing a support vector machine (SVM) classifier for separating patients with vascular invasion from patients without vascular invasion. Furthermore, L1 penalized (LASSO) Cox proportional hazards (PH) regression model was used to determine prognostic genes from the identified feature genes of vascular invasion so as to develop a prognostic scoring model. The performance of the classifier and the prognostic model was tested on an independent set. In addition, a function analysis was performed in order to provide further insights into the molecular mechanisms underlying HCC.
Materials and methods
Data resource
The present study obtained the RNA- sequencing data of 373 HCC samples from TCGA portal based on Illumina HiSeq 2000 RNA Sequencing platform (Download date: 18th, October, 2018). Among these samples, 292 had clinical information of vascular invasion and survival information, including survival time and survival status, and were therefore selected as the training set (TCGA set). Furthermore, the GSE10141 (12) dataset was downloaded from Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) based on the GPL5474 Human 6k Transcriptionally Informative Gene Panel platform, including the microarray gene expression data of 80 HCC tissue samples with survival information. Only 62 HCC samples had vascular invasion, and these were selected as the validation set.
The present study performed uni- and multivariate Cox regression analyses in order to analyze the associations between clinical factors and OS in the training set using survival package v2.44-1.1 (13) of R language (http://bioconductor.org/packages/survivalr/). The significant clinical factors (log-rank P<0.05) were selected as the cut-off to classify the training set.
Differential expression analysis
Data from the TCGA and GEO databases were normalized using R software (version 3.4.1; http://www.r-project.org/). Following data normalization, the present study performed a differential gene expression analysis using HCC samples with and without vascular invasion in the training set using the limma (14) package (version 3.34.7; http://bioconductor.org/packages/release/bioc/html/limma.html) of R software. The genes with false discovery rate (FDR) <0.05 and |log2 FC|>0.263 were selected and subsequently underwent a two-way hierarchical clustering analysis based on centered pearson correlation (15) algorithm using pheatmap package (16) (version 1.0.8) of R language (version 3.34.7). The results were presented in a heatmap.
Development of an SVM classifier
The present study initially performed a Cox regression analysis to investigate the associations between the identified DEGs and OS. From the significant DEGs with log-rank P<0.05, the present study then identified the optimal combination of feature genes using an RFE (17) algorithm in the caret (18) package (version 6.0–79; https://cran.r-project.org/web/packages/caret) of R language, which was then used to develop an SVM classifier using the SVM (19) function with a sigmoid kernel.
In both the training set and the validation set, the robustness of the established SVM classifier was evaluated using concordance index (C-index) (20), Brier score (21), log-rank P-value of cox-PH regression, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and area under receiver operating characteristic curve (AUROC). C-index and Brier score was calculated using the survcomp version 3.9 (22) package (http://www.bioconductor.org/packages/release/bioc/html/survcomp.html) of R language (version 3.4.1), which are two metrics for assessing accuracy. Kaplan-Meier estimate was applied to depict survival time using the survival package in R language. The Log-rank P-value for the difference in OS time between the two groups was calculated. AUROC ranged from 0.5 to 1, with a higher value implying better performance. Sensitivity, specificity, PPV and NPV of ROC curves were computed using pROC v1.15.3 (23) package of R language (https://cran.r-project.org/web/packages/pROC/index.html).
Development and validation of a prognostic scoring model
The present study further utilized the feature genes to fit a LASSO Cox-PH regression model (24) in order to determine the optimal panel of genes for prognosis using the penalized package (v0.9-51) of R language. Based on Cox-PH regression coefficients and expression levels of the identified optimal genes, a prognostic scoring model was built using the following formula:
CoefDEGs represents Cox-PH regression coefficients of DEGs; ExpDEGs represents expression levels of DEGs.
Risk score was calculated for each sample in the training set. Samples in the training set were then split into a high-risk group and a low-risk group according to median risk score (0.0663803). Kaplan-Meier survival curves were plotted for both risk groups using survival package (version 2.41-1) of R language, and OS of the two groups was compared by log-rank test. Similarly, samples in the validation set were divided into a high-risk group and a low-risk group using the median risk score of the validation set (0.132434) so as to test the prognostic ability of the prognostic scoring model in this set. The present study further validated the results by using SurvExpress, which is an online biomarker validation tool for cancer gene expression data (25). A total of four datasets, including GSE10143 (12), GSE10186 (26), TCGA-Liver-cancer and LIHC-TCGA-Liver HCC were included into SurvExpress.
Stratified analysis
In both the high and low-risk groups of the training set, the present study investigated the associations between clinical factors and OS by performing a Cox regression analysis with the survival package in R language (version 2.41-1).
Functional analysis
The cases in the training set were divided into high- and low-risk groups according to the risk score of the gene signature. The present study then screened for DEGs in the two risk groups using a strict cut-off at FDR<0.05 and |log2FC|>0.263. The signficant DEGs were selected for the pathway enrichment analysis using Gene Set Enrichment Analysis (27) (GSEA, version 3.0; http://software.broadinstitute.org/gsea/index.jsp). P<0.05 was considered to indicate a statistically significant result.
Results
Vascular invasion is an independent predictor of prognosis
The present study performed uni- and multivariate Cox regression analyses in order to analyze the associations between clinical factors and OS in the training set using the survival package in R language. As presented in Table I, vascular invasion and pathological M stage (28) were identified as independent predictors of prognosis in the univariate and multivariate analysis (P<0.05). However, there were only three samples at pathological M1 stage, which was an insufficient amount for accurately assessing prognostic value of pathological M stage. Therefore, the present study classified all samples of the training set into two groups according to vascular invasion. Patients without vascular invasion (n=190) had significantly better survival time compared with patients with vascular invasion (n=102; P=8.609×10−3; Fig. 1).
Table I.
Uni-and multivariate Cox regression analysis of the training set.
| Uni-variable cox | Multi-variable cox | ||||
|---|---|---|---|---|---|
| Clinical characteristics | TCGA (n=292) | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, years, mean ± SD | 59.85±12.92 | 1.017 (0.999–1.035) | 0.051 | – | – |
| sex (male/female) | 194/98 | 0.731 (0.474–1.127) | 0.154 | – | – |
| Pathological M (M0/M1/-) (28) | 220/3/69 | 4.91 (1.523–15.84) | 0.003 | 3.848 (1.089–13.588) | 0.036 |
| Pathological N (N0/N1/-) | 210/2/80 | 1.602 (0.221–2.610) | 0.638 | – | – |
| Pathological T (T1/T2/T3/T4/-) | 160/75/48/8/1 | 1.538 (1.23–1.923) | <0.001 | 0.607 (0.217–1.699) | 0.342 |
| Pathological stage (I/II/III/IV/-) | 150/72/48/4/18 | 1.473 (1.153–1.881) | 0.003 | 2.177 (0.797–5.944) | 0.129 |
| Histological grade (G1/G2/G3/G4/-) | 36/141/101/12/2 | 1.19 (0.889–1.593) | 0.243 | – | – |
| Virus infection (HBV/HCV/Mixed/-) | 50/10/35/197 | 1.167 (0.801–1.702) | 0.420 | – | – |
| Vascular invasion (yes/no) | 102/190 | 1.353 (1.087–2.098) | 0.009 | 1.678 (1.195–2.962) | 0.037 |
| Recurrence (yes/no/-) | 119/156/17 | 1.343 (0.843–2.141) | 0.213 | – | – |
| Status (dead/alive) | 87/205 | – | – | – | – |
| Overall survival time, months, mean ± SD | 26.52±24.43 | – | – | – | – |
TCGA, The Cancer Genome Atlas; SD, standard deviation; M, metastasis; N, node; T, tumor; HBV, hepatitis B virus; HCV, hepatitis C virus; HR, hazard ratio; CI, confidence interval; -, information unavailable.
Figure 1.
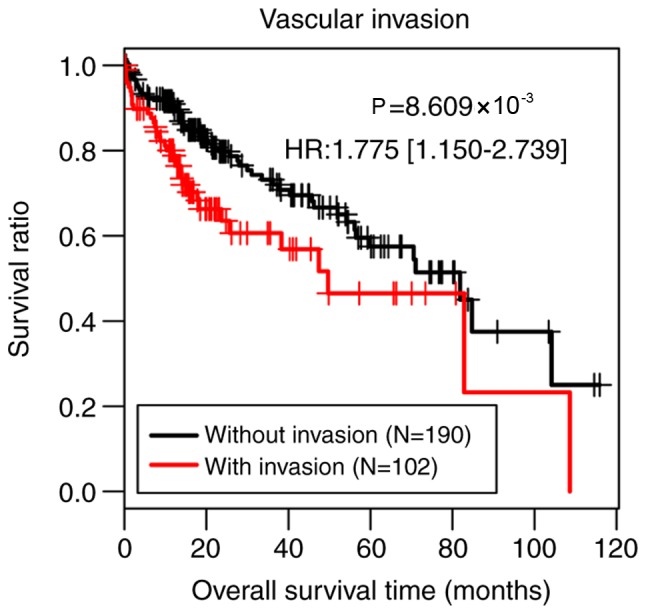
Kaplan-Meier curves of overall survival time of patients with or without vascular invasion of The Cancer Genome Atlas set. HR, hazard ratio.
DEGs were screened between patients with and without vascular invasion
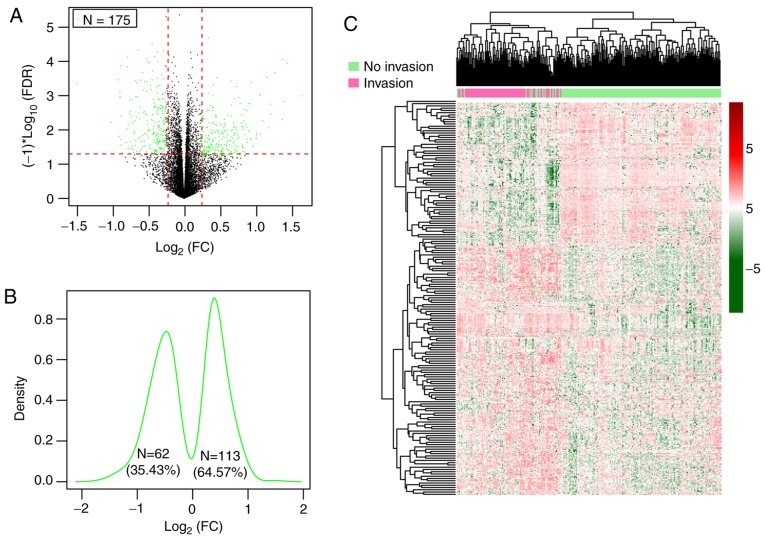
Following the removal of genes with a median expression level of 0, a total of 13,812 genes were inputted into the Limma package and among them, 175 significant DEGs in patients both with and without vascular invasion in the training set that satisfied the cut-off threshold (FDR<0.05 and |log2FC|>0.263) were identified (Table SI), consisting of 62 (35.43%) downregulated genes and 113 (64.57%) upregulated genes in the HCC samples with vascular invasion (Fig. 2A-C).
Figure 2.
DEGs of patients with presence and absence of vascular invasion of the training set. (A) Volcano plot of 175 DEGs. Green spots represent DEGs; red horizontal dash line implies FDR <0.05; two red vertical dash lines indicate |logFC|>0.263. (B) Kernel density plot of log2 (FC) of 175 DEGs. (C) Heatmap for two-way hierarchical clustering of samples based on expression of DEGs. The red and green represent upregulated and downregulated genes, respectively. DEGs, differentially expressed genes; FDR, false discovery rate; FC, fold change.
SVM analysis
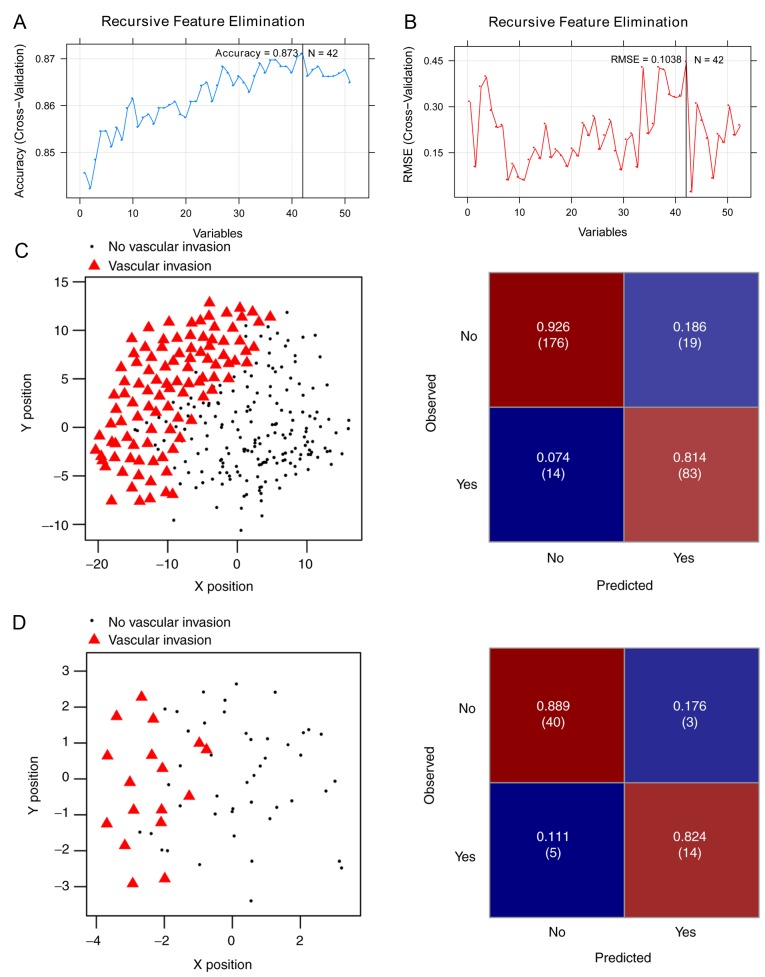
Of the aforementioned 175 DEGs, 51 were significantly associated with OS (log-rank P<0.05) in the Cox regression analysis (Table SII). For the purpose of obtaining the optimal feature genes for predicting vascular invasion in HCC, the present study utilized an SVM-RFE algorithm based on the 51 prognosis-associated genes. Maximal prediction accuracy (0.873) (Fig. 3A) and minimal root-mean-square error (0.1038) (Fig. 3B) were reached when using a 42-gene combination (Table II).
Figure 3.
The (A) Accuracy and (B) RMSE curves of the optimal gene combination screened by recursive feature elimination algorithm. The horizontal axis represents the number of gene variables, and the vertical axis represents cross-validation accuracy and RMSE, and the marked position is the number of genes corresponding to the optimal value. Performance of the 42-gene classifier on (C) the training set and (D) the validation set. Left images: Scatter plots presenting the prediction results by the 42-gene classifier. Black round spots represent samples from patients without vascular invasion; red triangles represent samples from patients with vascular invasion. Right images: Confusion matrix for the classification results. The X and Y axes represent the coordinates corresponding to the position in a two-dimensional plane generated by SVM. The top-left corner represents true positive rate (number), the top-right corner represents false negative rate (number), the left bottom represents false positive rate (number) and the right bottom represents true negative rate (number). SVM, support vector machine; RMSE, root-mean-square error.
Table II.
Combination of 42-genes.
| Gene | logFC | P-value | FDR |
|---|---|---|---|
| DNMT3L | −0.457857972 | 4.250×10−05 | 0.000344393 |
| WNT1 | −0.440653373 | 0.00229823 | 0.018624233 |
| AVPR2 | −0.337010196 | 0.000130506 | 0.001057586 |
| CRYAA | −0.327349605 | 5.220×10−05 | 0.000423239 |
| ADRA1A | −0.323457976 | 0.000132 | 0.001069691 |
| RERGL | −0.307031974 | 0.00027205 | 0.002204622 |
| HSD17B13 | −0.303883897 | 4.350×10−05 | 0.00035246 |
| CRHBP | −0.282544406 | 0.000378787 | 0.003069588 |
| GPR17 | −0.27487125 | 0.001557011 | 0.012617592 |
| AP1M2 | 0.265012128 | 0.002298097 | 0.018623151 |
| CCDC74B | 0.26607111 | 0.005538491 | 0.04488242 |
| EPHX4 | 0.273106635 | 0.001616394 | 0.013098814 |
| MYLK2 | 0.277944797 | 0.001898397 | 0.015384094 |
| S100P | 0.280211942 | 0.000796024 | 0.006450761 |
| SCIN | 0.286745667 | 0.001401359 | 0.011356228 |
| GULP1 | 0.293405465 | 0.002064432 | 0.016729591 |
| TMC5 | 0.304348871 | 0.001717824 | 0.013920779 |
| HOXD9 | 0.327961519 | 4.660×10−05 | 0.000377344 |
| DHDH | 0.331147822 | 0.001303337 | 0.01056189 |
| RUNDC3A | 0.344356975 | 0.001049184 | 0.0085023 |
| FXYD3 | 0.347111205 | 0.002610568 | 0.021155333 |
| FAM90A1 | 0.349492054 | 0.001789546 | 0.014501995 |
| POF1B | 0.353413663 | 0.00098377 | 0.007972208 |
| FAM163A | 0.357671188 | 0.001474436 | 0.01194843 |
| KCNN1 | 0.365217375 | 0.001202203 | 0.009742322 |
| TFAP2A | 0.365567331 | 6.750×10−05 | 0.000547399 |
| COL24A1 | 0.382367663 | 0.002049211 | 0.016606245 |
| DIRAS2 | 0.405965625 | 0.000995196 | 0.0080648 |
| FRMD1 | 0.411164402 | 0.004146525 | 0.033602313 |
| EPO | 0.413544952 | 0.000992878 | 0.008046009 |
| USH1C | 0.417142972 | 0.000668281 | 0.005415564 |
| CA9 | 0.422098465 | 0.001719337 | 0.013933041 |
| ART5 | 0.423955728 | 0.005437747 | 0.044066018 |
| MMP12 | 0.43064025 | 0.000852896 | 0.006911633 |
| TRIM54 | 0.438512907 | 0.001013864 | 0.008216081 |
| PPFIA4 | 0.467076549 | 5.000×10−05 | 0.000405366 |
| SLC35F3 | 0.503285506 | 0.002228769 | 0.018061337 |
| ELOVL3 | 0.524990121 | 0.000117912 | 0.00095553 |
| NPTX1 | 0.532157704 | 0.001637864 | 0.013272803 |
| ZNF695 | 0.601449278 | 0.000219446 | 0.001778327 |
| HOXD10 | 0.633083055 | 2.580×10−05 | 0.000209174 |
| PPP2R2C | 0.685073697 | 1.090×10−05 | 8.810×10−05 |
FC, fold change; FDR, false discovery rate.
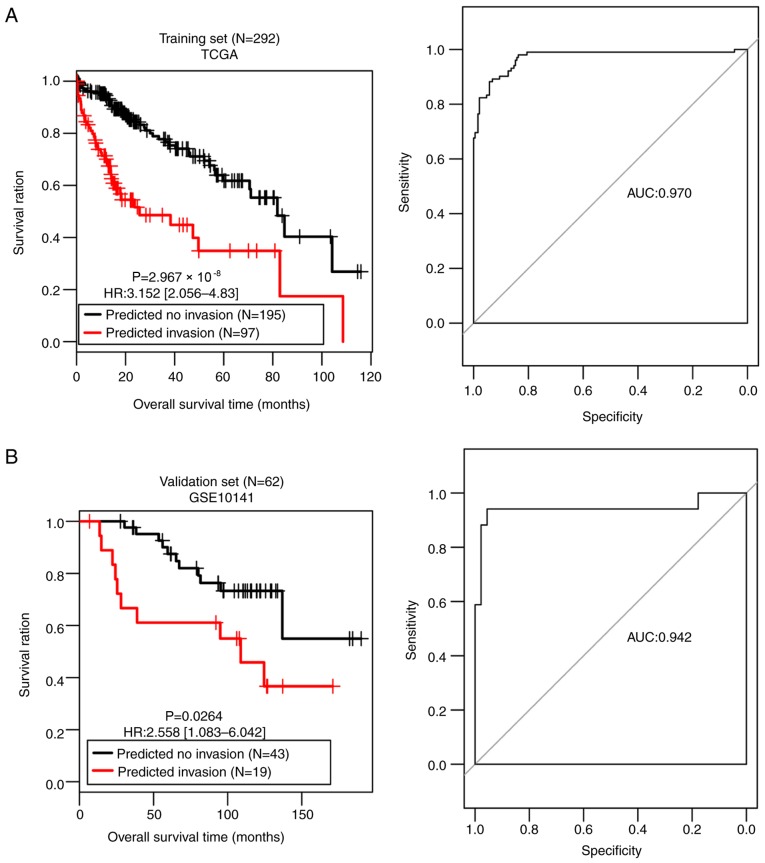
The SVM classifier was built with the 42-gene combination and its performance was assessed in both the training set and the validation set. A scatter plot and confusion matrix for the training set or the validation set classified by the classifier are presented in Fig. 3C and D. Table III demonstrates that both sets generated high C-index scores (>0.75), low Brier scores (<0.1) and significant average log-rank P-values (2.97×10−08; 0.0264) in OS difference between the patients with and without vascular invasion (Fig. 4). AUROC of the two sets were 0.970 and 0.942, respectively (Table III; Fig. 4). The sensitivity, specificity, PPV and NPV values are presented in Table III. These results suggest that the SVM classifier was able to classify the samples effectively.
Table III.
Performances of the SVM classifier on the training and validation sets.
| Overall survival | ROC curve | |||||||
|---|---|---|---|---|---|---|---|---|
| Datasets | C-index | Brier score | Log-rank P-value | AUROC | Sensitivity | Specificity | PPV | NPV |
| Training set (TCGA, N=292) | 0.814 | 0.0394 | <0.0001 | 0.970 | 0.814 | 0.926 | 0.856 | 0.903 |
| Validation set (GSE10141, N=62) | 0.757 | 0.0884 | 0.0264 | 0.942 | 0.824 | 0.889 | 0.737 | 0.930 |
SVM, support vector machine; TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic curve; AUROC, area under receiver operating characteristic curve; PPV, positive predictive value; NPV, negative predictive value.
Figure 4.
Kaplan-Meier and receiver operating characteristic curves for (A) Training set and (B) the validation set classified by the 42-gene classifier. TCGA, The Cancer Genome Atlas; HR, hazard ratio; AUC, area under the curve.
Prognostic model based on a 14-gene signature
The present study also used the 42 feature genes to create a LASSO Cox-PH regression model. When the maximal value of cross-validation likelihood (−498.517) was achieved, the optimal lambda value was 13.049, and the optimal panel of 14 genes was obtained (Table IV), including Wnt family member 1 (WNT1), crystallin α A (CRYAA), RAS like estrogen regulated growth inhibitor like (RERGL), hydroxysteroid 17-Beta dehydrogenase 13 (HSD17B13), scinderin (SCIN), premature ovarian failure (POF)1B, erythropoietin (EPO), USH1 protein network component harmonin (USH1C), ADP-ribosyltransferase 5 (ART5), matrix metalloproteinase (MMP)12, tripartite motif containing 54 (TRIM54), solute carrier family 35 member F3 (SLC35F3), homeobox D (HOXD)10 and protein phosphatase 2 regulatory subunit Bgamma (PPP2R2C). The following results were obtained using the risk score formula:
Table IV.
Prognostic signature with 14 genes.
| Gene | Coefficient | Hazard ratio (95%CI) | P-value |
|---|---|---|---|
| WNT1 | −0.2500 | 0.602 (0.459–0.789) | 2.400×10−04 |
| CRYAA | −0.0002 | 0.108 (0.0092–0.493) | 4.963×10−02 |
| RERGL | −0.0263 | 0.463 (0.244–0.854) | 4.533×10−02 |
| HSD17B13 | −0.0153 | 0.586 (0.176–0.906) | 4.688×10−02 |
| SCIN | 0.0852 | 1.115 (1.086–1.267) | 4.939×10−02 |
| POF1B | 0.0756 | 1.085 (1.001–1.178) | 1.513×10−02 |
| EPO | 0.0616 | 1.068 (1.013–1.152) | 4.897×10−02 |
| USH1C | 0.0106 | 1.043 (1.001–1.071) | 4.897×10−02 |
| ART5 | 0.0134 | 1.047 (1.035–1.171) | 4.231×10−02 |
| MMP12 | 0.0236 | 1.051 (1.048–1.165) | 3.410×10−02 |
| TRIM54 | 0.0454 | 1.059 (1.028–1.164) | 2.392×10−02 |
| SLC35F3 | 0.0124 | 1.057 (1.029–1.203) | 3.974×10−02 |
| HOXD10 | 0.1010 | 1.448 (1.127–1.924) | 3.069×10−03 |
| PPP2R2C | 0.0047 | 1.004 (1.002–1.085) | 4.926×10−01 |
CI, confidence interval.
Risk score=(−0.2500) × ExpWNT1 + (−0.0002) × ExpCRYAA + (−0.0263) × ExpRERGL + (−0.0153) × ExpHSD17B13 + (0.0852) × ExpSCIN + (0.0756) × ExpPOF1B + (0.0616) × ExpEPO + (0.0106) × ExpUSH1C + (0.0134) × ExpART5 + (0.0236) × ExpMMP12 + (0.0454) × ExpTRIM54+ (0.0124) × ExpSLC35F3 + (0.1010) × ExpHOXD10 + (0.0047) × ExpPPP2R2C.
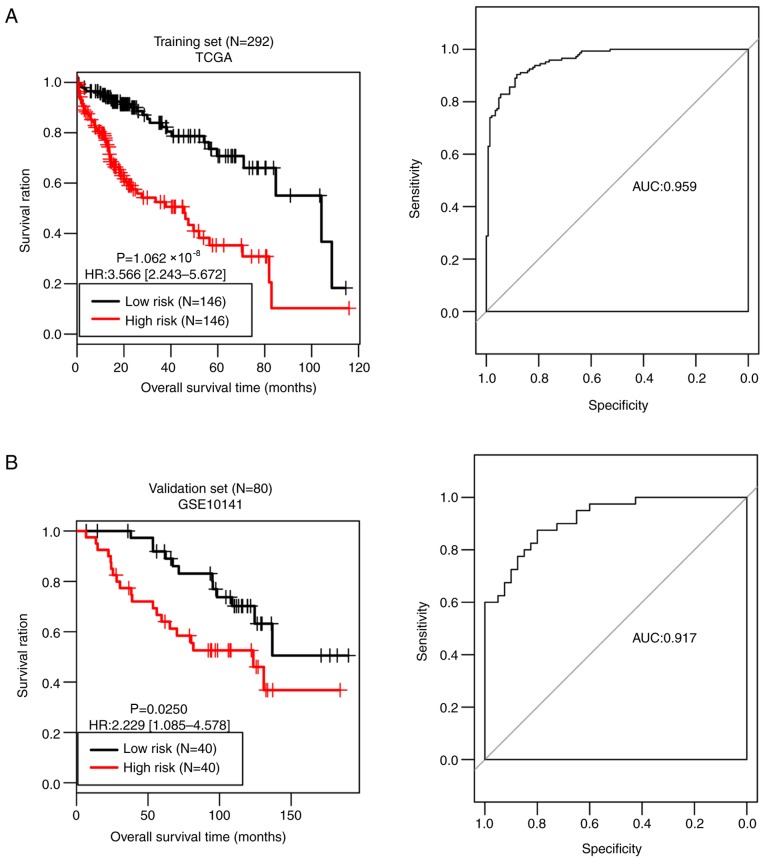
Based on the median risk score, all samples of the training set were divided into a high-risk group (n=146) and a low-risk group (n=146). As presented in Fig. 5A, the OS time was significantly different between the two risk groups (P=1.062×10−08), with an AUC value of 0.959. OS time was significantly different between the high-risk group (n=40) and the low-risk group (n=40) in the validation set (P=0.0250), with an AUC value of 0.917 (Fig. 5B). These observations prove the predictive robustness of the 14-gene signature.
Figure 5.
Kaplan-Meier and receiver operating characteristic curves for the training set (A) and the validation set (B) divided by the 14-gene signature. TCGA, The Cancer Genome Atlas; HR, hazard ratio; AUC, area under the curve.
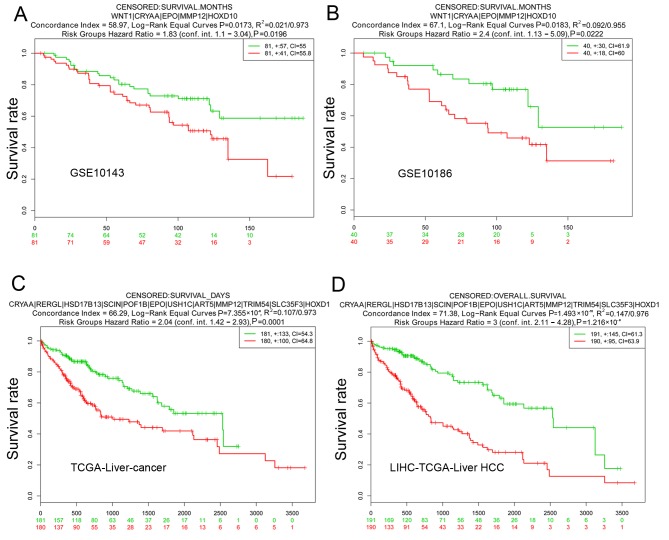
For validation of SurvExpress, five datasets, including GSE10143, GSE17856, GSE10186, TCGA-Liver-Cancer, and LIHC-TCGA-Liver HCC associated with HCC were included for validation in SurvExpress. The 51 screened candidate genes were inputted and the results revealed that the OS times were all significantly different between the high-risk group and the low-risk group in GSE10143, GSE10186, TCGA-Liver-cancer and LIHC-TCGA-Liver HCC (Fig. 6). This result supported the reliability of the gene signature.
Figure 6.
Validation analysis of the gene signature by SurvExpress. (A) GSE10143, (B) GSE10186, (C) LIHC-TCGA-Liver hepatocellular carcinoma and (D) TCGA-liver cancer datasets. CI, confidence interval.
Stratified analysis
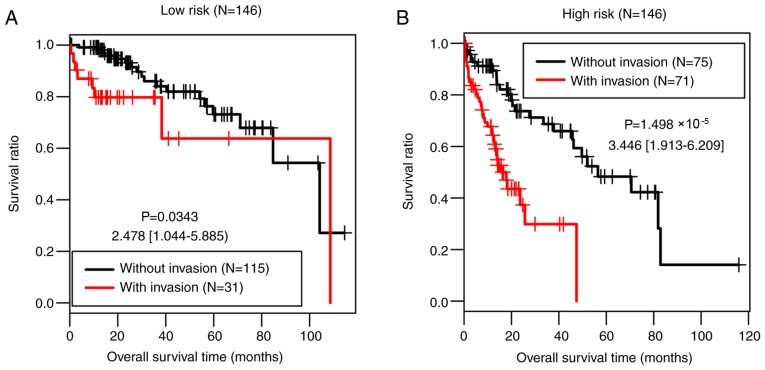
The present study further investigated the associations between the clinical factors and OS in the low-risk group and the high-risk group of the training set by performing Cox regression analyses. Vascular invasion was significantly associated with OS time in both risk groups (P=0.034 and P=1.50×10−05, respectively; Table V; Fig. 7).
Table V.
Results of Cox regression analysis for the high- and low-risk groups of The Cancer Genome Atlas set.
| Low risk group | High risk group | |||
|---|---|---|---|---|
| Clinical characteristics | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age, years, mean ± SD | 1.018 (0.986–1.052) | 0.273 | 1.012 (0.992–1.032) | 0.257 |
| Sex (male/female) | 0.568 (0.246–1.308) | 0.178 | 1.129 (0.676–1.886) | 0.643 |
| Pathological M (M0/M1/-) (28) | 8.721 (1.090–69.77) | 0.138 | 3.227 (0.770–13.520) | 0.090 |
| Pathological N (N0/N1/-) | 3.01 (1.052–33.22) | 0.763 | 1.429 (0.195–10.490) | 0.724 |
| Pathological T (T1/T2/T3/T4/-) | 1.527 (0.955–2.443) | 0.072 | 1.240 (0.914–1.681) | 0.165 |
| Pathological stage (I/II/III/IV/-) | 1.346 (0.805–2.252) | 0.254 | 1.215 (0.885–1.668) | 0.228 |
| Histological grade (G1/G2/G3/G4/-) | 1.115 (0.630–1.971) | 0.709 | 0.916 (0.642–1.307) | 0.629 |
| Virus infection (HBV/HCV/Mixed/-) | 2.333 (1.962–5.655) | 0.038 | 0.932 (0.613–1.416) | 0.741 |
| Vascular invasion (yes/no) | 2.478 (1.044–5.885) | 0.034 | 3.446 (1.913–6.209) | <0.001 |
| Recurrence (yes/no/-) | 1.569 (0.670–3.672) | 0.296 | 0.924 (0.526–1.623) | 0.783 |
M, metastasis; T, tumor; N, node; HBV, hepatitis B virus; HCV, hepatitis C virus; HR, hazard ratio; CI, confidence interval.
Figure 7.
Kaplan-Meier and receiver operating characteristic curves for patients with and without vascular invasion in (A) the low-risk group and (B) the high-risk group of the training set.
Identification and pathway analysis of DEGs between the two risk groups in the training set
In the training set, 599 upregulated genes and 163 downregulated genes were identified in the high-risk group compared with the low-risk group. These genes were significantly involved in pathways of ‘retinol metabolism’, ‘drug metabolism other enzymes’, ‘drug metabolism cytochrome P450’, ‘peroxisome proliferator-activated receptor (PPAR) signaling pathway’, ‘primary bile acid biosynthesis’, ‘steroid hormone biosynthesis’ and ‘histidine metabolism pathways’ (Table VI).
Table VI.
Significant signaling pathways.
| Pathway | ES | NES | Normal P-value | FDR | Count | Gene |
|---|---|---|---|---|---|---|
| Retinol metabolism | −0.7987 | −2.3043 | 0 | 0 | 6 | CYP4A22, CYP26A1, CYP3A43, CYP2A7, CYP2A6, CYP2A13 |
| Drug metabolism other enzymes | −0.9022 | −2.2834 | 0 | 0 | 4 | CYP3A43, CYP2A7, CYP2A6, CYP2A13 |
| Drug metabolism cytochrome P450 | −0.9011 | −2.0480 | 0 | 0.0047 | 4 | CYP3A43, CYP2A7, CYP2A6, CYP2A13 |
| PPAR signaling pathway | −0.7106 | −1.9354 | 0.0026 | 0.0121 | 3 | CYP4A22, CYP8B1, ACADL |
| Primary bile acid biosynthesis | −0.9631 | −1.9038 | 0 | 0.0124 | 3 | CYP8B1, AKR1D1, CYP7A1 |
| Steroid hormone biosynthesis | −0.5989 | −1.8162 | 0.0084 | 0.0188 | 6 | AKR1D1, CYP7A1, HSD3B2, HSD3B1, CYP11A1, CYP3A43 |
| Histidine metabolism | −0.8709 | −1.6875 | 0.01 | 0.0468 | 3 | HDC, CNDP1, UROC1 |
ES, enrichment score; NES, normalized enrichment score; count of genes, the number of genes enriched in a pathway; FDR, false discovery rate.
Discussion
HCC is an aggressive malignancy characterized by high incidence rates of recurrence and metastasis (29). Vascular invasion is an unfavorable prognostic factor for patients with HCC (30). Therefore, unraveling the underlying molecular landscape of vascular invasion is of significance for the prognosis of HCC. In the present study, a total of 175 DEGs were identified between patients with the presence and absence of vascular invasion. An SVM classifier was built that consisted of 42 feature genes by implementing an RFE-SVM algorithm. In both the training and validation sets, the classifier had high C-index values, low Brier scores and significant log-rank P-values, indicating good performances in separating patients with vascular invasion from patients without vascular invasion. Furthermore, through using a LASSO Cox-PH model, a 14-gene prognostic signature was obtained and consequently, a prognostic scoring model was established. The 14-gene signature was able to predict those patients with HCC that would have a shorter survival time, as evidenced by the result that OS time was significantly different between the predicted high-risk patients and the predicted low-risk patients. T prognostic performance of the 14-gene signature was successfully confirmed in the validation set.
The 14-gene prognostic combination included WNT1, CRYAA, RERGL, HSD17B13, SCIN, POF1B, EPO, USH1C, ART5, MMP12, TRIM54, SLC35F3, HOXD10 and PPP2R2C. Proto-oncogene protein Wnt-1 encoded by the WNT1 gene has been demonstrated as upregulated in HCC, acting as a direct target of miR-122 (31). RERGL is a member of the RAS superfamily of GTPases that participates in regulating several biological processes, such as cell proliferation, differentiation and apoptosis (32). There was one HSD17B13 protein, namely 17β-HSD type 13, that was downregulated in HCC (33). There is evidence to suggest that HSD17B13 suppresses HCC progression by delaying the G1/S phase transition of HCC cells (34). Furthermore, HSD17B13 is a novel liver-specific protein associated with lipid droplet, and may be a promising biomarker of liver cancer (35). SCIN encodes scinderin, which is an actin-severing protein of the gelsolin superfamily. It acts as a regulator of HCC cell apoptosis and growth, and has been identified as a transcriptional target of tumor suppressor factor breast cancer metastasis-suppressor 1 (36). It has long been established that the EPO/EPO-receptor plays an important role in angiogenesis and progression of HCC (37). EPO protein expression is positively correlated with vasculogenic mimicry in HCC, and has been identified as an independent predictor of prognosis in patients with HCC (38). Furthermore, EPO is upregulated in HCC and could promote HCC cell proliferation through translocation of its specific receptor induced by hypoxia (39). MMP12 belongs to the MMP family implicated in the degradation of the extracellular matrix. It is upregulated in HCC and is an independent predictive factor for OS in patients with HCC (40,41). TRIM54 is a member of the TRIM protein family. Several members in the TRIM family have been reported to be involved in biological processes, such as cell proliferation, differentiation and apoptosis, and may play a role in cancer initiation and progression (42). However, to the best of our knowledge TRIM54 has not been reported previously. HOXD10, a member of the Abd-B homeobox family, exhibits decreased expression levels in HCC and serves as a tumor-suppressor gene through prohibiting extracellular signal-regulated kinase signaling (43). PPP2R2C encodes serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B γ isoform, and has been identified as upregulated in HCC (44). To the best of our knowledge, there are little studies that focus on the function of CRYAA, RERGL, POF1B, POF1B, USH1C, TRIM54 and SLC35F3 in HCC. The results of the present study indicate that the 14 vascular invasion-associated genes may be prognostic biomarkers of HCC.
Another aim of the present study was identifying the potential roles of DEGs between the high- and low-risk groups of the training set. There were 762 DEGs between the two risk groups, which were significantly involved in a number of signaling pathways, such as ‘retinol metabolism’, ‘drug metabolism cytochrome P450’, and ‘PPAR signaling pathway’. The association between retinol metabolism and HCC has been demonstrated previously and a synthetic retinoid has been indicated to prevent HCC recurrence (45). Drug-metabolizing cytochrome P450 enzyme activities are severely disrupted in HCC (46). The PPAR signaling pathway plays a part in tumorigenesis and tumor progression via different metabolic pathways: Glycolysis/gluconeogenesis, lipid, glycerolipid and glycerophospholipid metabolism, protein synthesis and degradation and purine metabolism (47). These findings reveal the critical roles of these pathways in HCC.
There are some limitations in the present study; though the 14-gene prognostic signature has been validated by an independent dataset, the expression levels of these 14 genes have not been confirmed by individual gene expression experiments.
In summary, using TCGA data, the present study defined a classifier of 42 feature genes for classification of patients with HCC with and without vascular invasion, and identified a vascular invasion-associated 14-gene prognostic signature for HCC. Several genes and pathways have been revealed to be critical for HCC. These results further the current knowledge on the molecular mechanisms underlying HCC and may aid in the development of personalized treatment for patients with HCC. Large-scale studies are required in order to further validate the results of the present study.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BY performed the data analyses and wrote the manuscript. CT and YT contributed significantly towards the data analyses and revised the manuscript. ZZ conceived and designed the study. All authors read and approved the final published version of the manuscript.
Ethics approval and consent to participate
In the original article of the datasets, the trials were approved by the local institutional review boards of all participating centers, and informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Balogh J, Victor D, III, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP., Jr Hepatocellular carcinoma: A review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, Negri E. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302–309. doi: 10.1016/j.jhep.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Lo YC, Hsu FC, Hung SK, Tseng KC, Hsieh YH, Lee MS, Tseng CW, Lin HY, Chen LC, Chiou WY. Prognosticators of hepatocellular carcinoma with intrahepatic vascular invasion. Ci Ji Yi Xue Za Zhi. 2019;31:40–46. doi: 10.4103/tcmj.tcmj_14_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawlik TM, Poon RT, Abdalla EK, Zorzi D, Ikai I, Curley SA, Nagorney DM, Belghiti J, Ng IO, Yamaoka Y, et al. Critical appraisal of the clinical and pathologic predictors of survival after resection of large hepatocellular carcinoma. Arch Surg. 2005;140:450–458. doi: 10.1001/archsurg.140.5.450. [DOI] [PubMed] [Google Scholar]
- 5.Sakata J, Shirai Y, Wakai T, Kaneko K, Nagahashi M, Hatakeyama K. Preoperative predictors of vascular invasion in hepatocellular carcinoma. Eur J Surg Oncol. 2008;34:900–905. doi: 10.1016/j.ejso.2008.01.031. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh CH, Wei CK, Yin WY, Chang CM, Tsai SJ, Wang LY, Chiou WY, Lee MS, Lin HY, Hung SK. Vascular invasion affects survival in early hepatocellular carcinoma. Mol Clin Oncol. 2015;3:252–256. doi: 10.3892/mco.2014.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hui Z, Chen C, Xu F, Yan X, Jia W, Mao L, Jin H, Qiu Y. Prognostic value of a novel risk classification of microvascular invasion in patients with hepatocellular carcinoma after resection. Oncotarget. 2016;8:5474–5486. doi: 10.18632/oncotarget.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho MC, Lin JJ, Chen CN, Chen CC, Lee H, Yang CY, Ni YH, Chang KJ, Hsu HC, Hsieh FJ, Lee PH. A gene expression profile for vascular invasion can predict the recurrence after resection of hepatocellular carcinoma: A microarray approach. Ann Surg Oncol. 2006;13:1474–1484. doi: 10.1245/s10434-006-9057-1. [DOI] [PubMed] [Google Scholar]
- 9.Mínguez B, Hoshida Y, Villanueva A, Toffanin S, Cabellos L, Thung S, Mandeli J, Sia D, April C, Fan JB, et al. Gene-expression signature of vascular invasion in hepatocellular carcinoma. J Hepatol. 2011;55:1325–1331. doi: 10.1016/j.jhep.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Z, Cai YJ, Chen RC, Chen BC, Zhao L, Xu SH, Wang XD, Song M, Wu JM, Wang YQ, et al. A microRNA expression profile for vascular invasion can predict overall survival in hepatocellular carcinoma. Clin Chim Acta. 2017;469:171–179. doi: 10.1016/j.cca.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Ravishankar H, Madhavan R, Mullick R, Shetty T, Marinelli L, Joel SE. Recursive feature elimination for biomarker discovery in resting-state functional connectivity. Conf Proc IEEE Eng Med Biol Soc. 2016;2016:4071–4074. doi: 10.1109/EMBC.2016.7591621. [DOI] [PubMed] [Google Scholar]
- 12.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P, Wang Y, Hang B, Zou X, Mao JH. A novel gene expression-based prognostic scoring system to predict survival in gastric cancer. Oncotarget. 2016;7:55343–55351. doi: 10.18632/oncotarget.10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng Z, Zhu G, Qi J, Ma H, Nian H, Wang Y. RNA-seq analyses of multiple meristems of soybean: Novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol. 2014;14:169. doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Yang Y, Wu F, Gao M, Xu Y, Zhang Y, Yao Y, Du X, Li C, Wu L, et al. Discriminative analysis of schizophrenia using support vector machine and recursive feature elimination on structural MRI images. Medicine (Baltimore) 2016;95:e3973. doi: 10.1097/MD.0000000000003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deist TM, Dankers FJMM, Valdes G, Wijsman R, Hsu IC, Oberije C, Lustberg T, van Soest J, Hoebers F, Jochems A, et al. Machine learning algorithms for outcome prediction in (chemo)radiotherapy: An empirical comparison of classifiers. Med Phys. 2018;45:3449–3459. doi: 10.1002/mp.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Liu X. Screening of feature genes in distinguishing different types of breast cancer using support vector machine. Onco Targets Ther. 2015;8:2311–2317. doi: 10.2147/OTT.S85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayr A, Schmid M. Boosting the concordance index for survival data-a unified framework to derive and evaluate biomarker combinations. PLoS One. 2014;9:e84483. doi: 10.1371/journal.pone.0084483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Li Y, Akinyemiju T, Ojesina AI, Buckhaults P, Liu N, Xu B, Yi N. Pathway-structured predictive model for cancer survival prediction: A two-stage approach. Genetics. 2017;205:89–100. doi: 10.1534/genetics.116.189191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schroder MS, Culhane AC, Quackenbush J, Haibe-Kains B. Survcomp: An R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–395. doi: 10.1002/(SICI)1097-0258(19970228)16:4<385::AID-SIM380>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez- Barrientos A, Tamez-Peña JG, Treviño V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varotti G, Ramacciato G, Ercolani G, Grazi GL, Vetrone G, Cescon M, Del Gaudio M, Ravaioli M, Ziparo V, Lauro A, Pinna A. Comparison between the fifth and sixth editions of the AJCC/UICC TNM staging systems for hepatocellular carcinoma: Multicentric study on 393 cirrhotic resected patients. Eur J Surg Oncol. 2005;31:760–767. doi: 10.1016/j.ejso.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: Translating knowledge into practice. Clin Gastroenterol Hepatol. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasai Y, Hatano E, Seo S, Taura K, Yasuchika K, Uemoto S. Hepatocellular carcinoma with bile duct tumor thrombus: Surgical outcomes and the prognostic impact of concomitant major vascular invasion. World J Surg. 2015;39:1485–1493. doi: 10.1007/s00268-015-2985-9. [DOI] [PubMed] [Google Scholar]
- 31.Ahsani Z, Mohammadi-Yeganeh S, Kia V, Karimkhanloo H, Zarghami N, Paryan M. WNT1 Gene from WNT signaling pathway is a direct target of miR-122 in hepatocellular carcinoma. Appl Biochem Biotechnol. 2017;181:884–897. doi: 10.1007/s12010-016-2256-8. [DOI] [PubMed] [Google Scholar]
- 32.Goitre L, Trapani E, Trabalzini L, Retta SF. The Ras superfamily of small GTPases: The unlocked secrets. Methods Mol Biol. 2014;1120:1–18. doi: 10.1007/978-1-62703-791-4_1. [DOI] [PubMed] [Google Scholar]
- 33.Xing X, Huang Y, Wang S, Chi M, Zeng Y, Chen L, Li L, Zeng J, Lin M, Han X, et al. Dataset for the quantitative proteomics analysis of the primary hepatocellular carcinoma with single and multiple lesions. Data Brief. 2015;5:226–240. doi: 10.1016/j.dib.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Zhuo JY, Yang F, Liu ZK, Zhou L, Xie HY, Xu X, Zheng SS. 17-beta-hydroxysteroid dehydrogenase 13 inhibits the progression and recurrence of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2018;17:220–226. doi: 10.1016/j.hbpd.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Su W, Mao Z, Liu Y, Zhang X, Zhang W, Gustafsson JA, Guan Y. Role of HSD17B13 in the liver physiology and pathophysiology. Mol Cell Endocrinol. 2019;489:119–125. doi: 10.1016/j.mce.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Qiao X, Zhou Y, Xie W, Wang Y, Zhang Y, Tian T, Dou J, Yang X, Shen S, Hu J, et al. Scinderin is a novel transcriptional target of BRMS1 involved in regulation of hepatocellular carcinoma cell apoptosis. Am J Cancer Res. 2018;8:1008–1018. [PMC free article] [PubMed] [Google Scholar]
- 37.Ribatti D, Marzullo A, Gentile A, Longo V, Nico B, Vacca A, Dammacco F. Erythropoietin/erythropoietin-receptor system is involved in angiogenesis in human hepatocellular carcinoma. Histopathology. 2007;50:591–596. doi: 10.1111/j.1365-2559.2007.02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Sun B, Zhao X, Shao B, An J, Gu Q, Wang Y, Dong X, Zhang Y, Qiu Z. Erythropoietin and erythropoietin receptor in hepatocellular carcinoma: Correlation with vasculogenic mimicry and poor prognosis. Int J Clin Exp Pathol. 2015;8:4033–4043. [PMC free article] [PubMed] [Google Scholar]
- 39.Miao S, Wang SM, Cheng X, Li YF, Zhang QS, Li G, He SQ, Chen XP, Wu P. Erythropoietin promoted the proliferation of hepatocellular carcinoma through hypoxia induced translocation of its specific receptor. Cancer Cell Int. 2017;17:119. doi: 10.1186/s12935-017-0494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng KT, Qi X, Kong KL, Cheung BY, Lo CM, Poon RT, Fan ST, Man K. Overexpression of matrix metalloproteinase-12 (MMP-12) correlates with poor prognosis of hepatocellular carcinoma. Eur J Cancer. 2011;47:2299–2305. doi: 10.1016/j.ejca.2011.05.032. [DOI] [PubMed] [Google Scholar]
- 41.He MK, Le Y, Zhang YF, Ouyang HY, Jian PE, Yu ZS, Wang LJ, Shi M. Matrix metalloproteinase 12 expression is associated with tumor FOXP3+ regulatory T cell infiltration and poor prognosis in hepatocellular carcinoma. Oncol Lett. 2018;16:475–482. doi: 10.3892/ol.2018.8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cambiaghi V, Giuliani V, Lombardi S, Marinelli C, Toffalorio F, Pelicci PG. TRIM proteins in cancer. Adv Exp Med Biol. 2012;770:77–91. doi: 10.1007/978-1-4614-5398-7_6. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Peng Y, Gao D, Zhang M, Yang W, Linghu E, Herman JG, Fuks F, Dong G, Guo M. Silencing HOXD10 by promoter region hypermethylation activates ERK signaling in hepatocellular carcinoma. Clin Epigenetics. 2017;9:116. doi: 10.1186/s13148-017-0412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho DW, Kai AK, Ng IO. TCGA whole-transcriptome sequencing data reveals significantly dysregulated genes and signaling pathways in hepatocellular carcinoma. Front Med. 2015;9:322–330. doi: 10.1007/s11684-015-0408-9. [DOI] [PubMed] [Google Scholar]
- 45.Shirakami Y, Sakai H, Shimizu M. Retinoid roles in blocking hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2015;4:222–228. doi: 10.3978/j.issn.2304-3881.2015.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan T, Lu L, Xie C, Chen J, Peng X, Zhu L, Wang Y, Li Q, Shi J, Zhou F, et al. Severely impaired and dysregulated cytochrome P450 expression and activities in hepatocellular carcinoma: Implications for personalized treatment in patients. Mol Cancer Ther. 2015;14:2874–2886. doi: 10.1158/1535-7163.MCT-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fanale D, Amodeo V, Caruso S. The interplay between metabolism, PPAR signaling pathway, and cancer. PPAR Res. 2017;2017:1830626. doi: 10.1155/2017/1830626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.