Abstract
Background
Alzheimer’s disease (AD) is a fatal neurodegenerative disease. APOE4 is the greatest genetic risk factor for AD, increasing risk up to 15-fold compared to the common APOE3. Importantly, female (♀) APOE4 carriers have a greater risk for developing AD and an increased rate of cognitive decline compared to male (♂) APOE4 carriers. While recent evidence demonstrates that AD, APOE genotype, and sex affect the gut microbiome (GM), how APOE genotype and sex interact to affect the GM in AD remains unknown.
Methods
This study analyzes the GM of 4-month (4 M) ♂ and ♀ E3FAD and E4FAD mice, transgenic mice that overproduce amyloid-β 42 (Aβ42) and express human APOE3+/+ or APOE4+/+. Fecal microbiotas were analyzed using high-throughput sequencing of 16S ribosomal RNA gene amplicons and clustered into operational taxonomic units (OTU). Microbial diversity of the EFAD GM was compared across APOE, sex and stratified by APOE + sex, resulting in 4-cohorts (♂E3FAD, ♀E3FAD, ♂E4FAD and ♀E4FAD). Permutational multivariate analysis of variance (PERMANOVA) evaluated differences in bacterial communities between cohorts and the effects of APOE + sex. Mann-Whitney tests and machine-learning algorithms identified differentially abundant taxa associated with APOE + sex.
Results
Significant differences in the EFAD GM were associated with APOE genotype and sex. Stratification by APOE + sex revealed that APOE-associated differences were exhibited in ♂EFAD and ♀EFAD mice, and sex-associated differences were exhibited in E3FAD and E4FAD mice. Specifically, the relative abundance of bacteria from the genera Prevotella and Ruminococcus was significantly higher in ♀E4FAD compared to ♀E3FAD, while the relative abundance of Sutterella was significantly higher in ♂E4FAD compared to ♂E3FAD. Based on 29 OTUs identified by the machine-learning algorithms, heatmap analysis revealed significant clustering of ♀E4FAD separate from other cohorts.
Conclusions
The results demonstrate that the 4 M EFAD GM is modulated by APOE + sex. Importantly, the effect of APOE4 on the EFAD GM is modulated by sex, a pattern similar to the greater AD pathology associated with ♀E4FAD. While this study demonstrates the importance of interactive effects of APOE + sex on the GM in young AD transgenic mice, changes associated with the development of pathology remain to be defined.
Keywords: Alzheimer’s disease, Gut microbiome, APOE genotype, Sex
Background
The gut microbiome (GM), the collective genome of gastrointestinal bacteria, is an integral component of human physiology [1–5]. Recent studies link dysbiotic GM profiles with neurological disorders, with multiple sclerosis the first identified [6–12]. While subsequent studies have linked dysbiosis with Alzheimer’s disease (AD) pathology [13–22], the effects of AD risk factors, specifically APOE genotype, sex and their interaction, on the GM remain unclear.
The APOE4 genotype is the greatest genetic risk factor for AD, increasing risk up to 15-fold compared to the more common APOE3 genotype [23, 24]. Apolipoprotein E (apoE) is a member of the apolipoprotein family, the protein components of lipoproteins. Both humans and AD transgenic (−Tg) mice with APOE4 exhibit an increase in amyloid-β (Aβ) peptide accumulation, both as amyloid plaques, a hallmark of the disease, and small soluble aggregates. Thus, one explanation for the APOE4-associated AD risk is a loss of function in Aβ clearance. Tran and colleagues demonstrated significant differences between the GM of human APOE3 and APOE4 carriers, as well as differences between the GM of APOE3 and APOE4 targeted replacement (−TR) mice [25]. These differences were attributed to a loss of apoE4 function in lipid homeostasis, as APOE4 is associated with higher levels of cholesterol, triglycerides and low-density lipoproteins compared to APOE3 [26–29], changes that significantly affect the GM [30–37]. Sex is another risk factor for AD as females (♀) exhibit almost two-fold greater lifetime AD risk compared to males (♂) [38]. Additionally, sex plays an important role in the GM as the bacterial composition and metabolic function differ significantly between ♂ and ♀ [37, 39–46]. Importantly, ♀APOE4 carriers have a greater lifetime risk for developing AD, an increased rate of cognitive decline and an accelerated accumulation of Aβ compared to ♂APOE4 carriers [47–61]. While the underlying mechanism is unclear, evidence suggests this interaction modulates the GM.
EFAD-Tg mice [62] overexpress Aβ42 via five familial AD (FAD) mutations [63] and express h-APOE3 or APOE4, allowing for the study of the interaction among AD risk factors [64–66]. EFAD mice expressing the APOE4+/+ genotype (E4FAD), compared to E3FAD mice, exhibit increased behavioral deficits, Aβ deposition and neuroinflammation. Importantly, these differences are reproduced in ♀ vs ♂EFAD mice, resulting in 4 pathologically-distinct cohorts when the EFAD mice are stratified by APOE + sex (♀E4FAD > ♂E4FAD = ♀E3FAD > ♂E3FAD), a phenotype that develops with age [65, 66]. For this study, we focused on 4 M EFAD mice to evaluate the interactive effects of APOE + sex on the GM at an age prior to, or early in, the development of pathology. Microbial analysis of fecal samples demonstrated that APOE + sex have a significant effect on the GM at various taxonomic levels.
Methods
Mouse model
As previously described, the EFAD (5xFAD+/−/APOE+/+) mice are homozygous for APOE2, APOE3, or APOE4 and heterozygous for the 5x familial AD (5xFAD) mutations [62, 63]. Although APOE2 is considered neuroprotective, 100% of APOE2+/+ mice have type III hyperlipoproteinemia, compared to only 15% of human ε2/2 carriers [67–69]; thus, E2FAD mice were excluded from the current study. At 4 M, fecal samples were obtained from the 4 cohorts (9 ♂E3FAD, 8 ♂E4FAD, 19 ♀E3FAD, 12 ♀E4FAD) by individually placing mice in clean disposable Styrofoam cups. Feces were flash frozen and stored at − 80 °C until DNA isolation.
Bacteria identification
Fecal DNA was isolated using a PowerSoil DNA isolation kit (Mo Bio Laboratories) and DNA concentrations determined by UV absorbance (Nanodrop, ThermoFisher). The V4 variable region of 16S ribosomal RNA gene was PCR-amplified using target-specific primers containing bar codes and linker sequences [70]. PCR reaction conditions included an initial denaturation step of 30 s (s) at 98 °C, followed by 28 cycles of 10s at 98 °C, 15 s at 60 °C, 30s at 72 °C, and a final elongation step of 7 min at 72 °C. The PCR master mix (20 μl volume) contained 100 ng of DNA template, 0.5 μM forward and reverse primers, Phusion Hot Start DNA polymerase and high-fidelity buffer (New England Biolabs), dNTPs and sterile water. Results were checked by polyacrylamide gel electrophoresis and samples pooled in equimolar ratio. The samples were sequenced on an Illumina MiSeq sequencer at the University of Kentucky Advanced Genetic Technologies Center, with sequence merging, trimming, chimera removal, clustering and annotation performed using the software package QIIME [71]. The Greengenes database was implemented for Operational Taxonomic Unit (OTU) annotation at a threshold of 97% sequence similarity [72]. To avoid effects of uneven sequencing depth [73], datasets were rarified to 3000 sequences/sample prior to analysis. For statistical analyses, OTUs with a frequency below 0.1% across the dataset were removed [71].
Data analysis
The Shannon H α-diversity index was used to assess bacterial richness and evenness. The interaction between APOE + sex in α-diversity measures was evaluated using a mixed effects model, similar to a two-way analysis of variance (ANOVA), that analyzes repeated measures with missing values. This analysis was performed in the software package GraphPad Prism (version 8.2.0). For β-diversity, permutational ANOVA (PERMANOVA) was used to compare microbial community structure within and among the EFAD cohorts based on Bray-Curtis dissimilarity [74, 75]. Pair-wise PERMANOVA was used to assess the effect of the interaction among universal biological variables on the microbiome composition [76]. Principal coordinate analysis plots (PCoA; Bray-Curtis distances) with 95% confidence ellipses were used to visualize microbial communities [75, 77, 78]. The Mann-Whitney U (MWU) test under the Monte Carlo simulation, corrected with Benjamini-Hochberg False Discovery Rate (p < 0.05), was used to identify differentially abundant taxa associated with APOE + sex at the taxonomic level of genus. The Random Forest based Boruta algorithm was used to determine OTUs significant in distinguishing samples by APOE + sex compared to randomly generated probes or “shadow scores” [79]. Heatmaps were generated using the R package, “pheatmaps”, calculating the Euclidean distance among cohorts.
Results and discussion
Mouse fecal microbial community structure was analyzed using high-throughput sequencing of 16S rRNA gene amplicons, followed by sequence clustering (97% similarity) into a total of 2063 OTUs. No significant difference in α-diversity (Shannon H index) was observed between E3FAD and E4FAD mice (p = 0.975; Additional file 1: Figure S1A) or between ♂EFAD and ♀EFAD (p = 0.949; Additional file 1: Figure S1B). In comparing across cohorts stratified by APOE + sex, Shannon H indices were significantly higher in ♂E4FAD and ♀E3FAD, compared to ♂E3FAD and ♀E4FAD (p < 0.05; Additional file 1: Figure S1C). Additionally, the interaction of APOE + sex significantly modulated α-diversity measures (p < 0.05; Additional file 1: Figure S1C), suggesting that analyses by APOE genotype or sex alone will mask effects on microbial community structure.
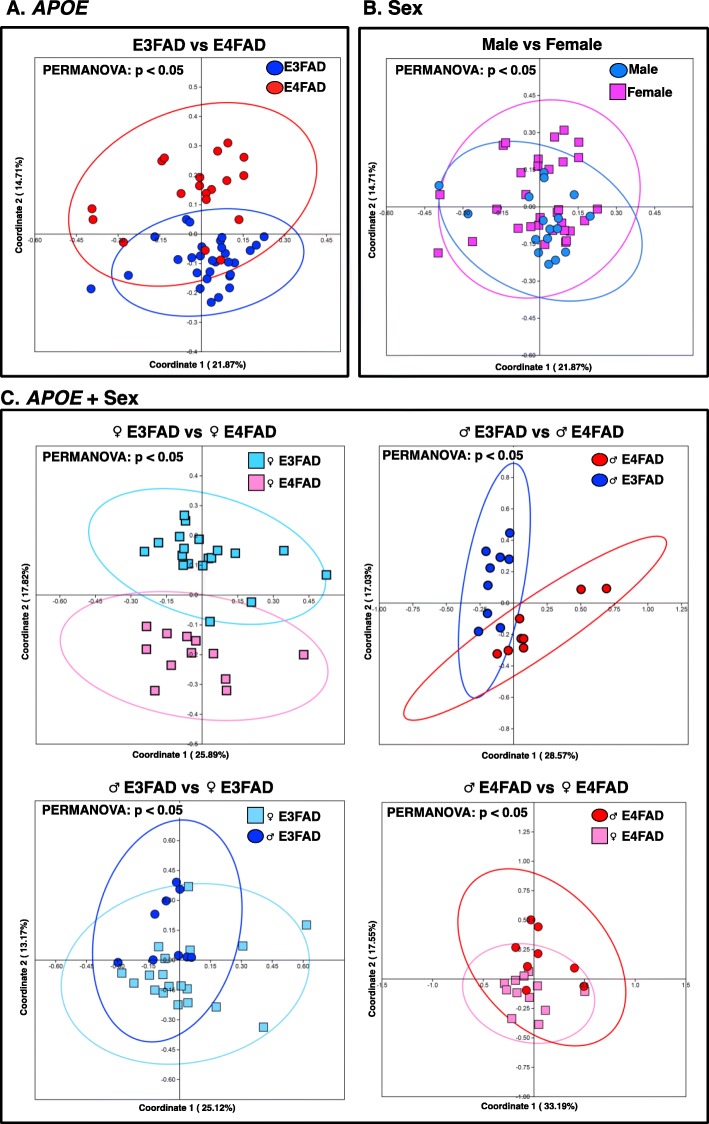
Differences in microbial community structure between EFAD cohorts (β-diversity) were examined with PERMANOVA (Additional file 3: Table S1) and visualized with PCoA plots (Fig. 1). At the taxonomic level of OTU, significant differences in microbial communities were observed between E3FAD and E4FAD mice (p < 0.05; Fig. 1a) and between ♂EFAD and ♀EFAD mice (p < 0.05; Fig. 1b). Differences associated with APOE genotype were also exhibited in the taxonomic levels of Family and Genus (Additional file 3: Table S1A), suggesting that APOE genotype is an important modulator of the GM, consistent with findings in APOE-TR mice [25]. Importantly, the interaction between APOE + sex significantly modulated the GM across taxonomic levels of Family, Genus and OTU (p < 0.05; Additional file 3: Table S1A). Comparisons at the OTU level among samples stratified by APOE + sex demonstrated significant differences between ♂E4FAD and ♂E3FAD mice (p < 0.05; Fig. 1c), and between ♀E4FAD and ♀E3FAD mice (p < 0.05; Fig. 1c), indicating that the effect of APOE genotype is consistent across sex. Furthermore, significant differences associated with sex were observed between ♂E4FAD and ♀E4FAD and between ♂E3FAD and ♀E3FAD (p < 0.05; Fig. 1c). These data demonstrate that the APOE genotype interacts with sex, leading to sex differentiation in E3FAD and E4FAD mice. While a recent paper by Dodiya and colleagues demonstrated no sex effect on α- or β-diversity in FAD-Tg mice that express mouse APOE [80], the current findings may suggest that the sex effect is specific to carriers of human APOE. This mirrors the synergistic effects of ♀sex and APOE4 genotype on AD risk in humans, greatest in ♀APOE4 > ♂APOE4 [47–50].
Fig. 1.
Differences in microbial community between EFAD mice stratified by APOE, sex and APOE + sex. Analysis of β-diversity associated with (a) APOE, (b) sex and (c) APOE + sex in the GM of 4 M EFAD mice. PCoA plots with 95% confidence ellipses were generated based on the Bray-Curtis dissimilarity. Significant differences between cohorts were determined by PERMANOVA, with significance (bold) defined by p < 0.05. Additional file 1: Table S1 contains the complete PERMANOVA dataset
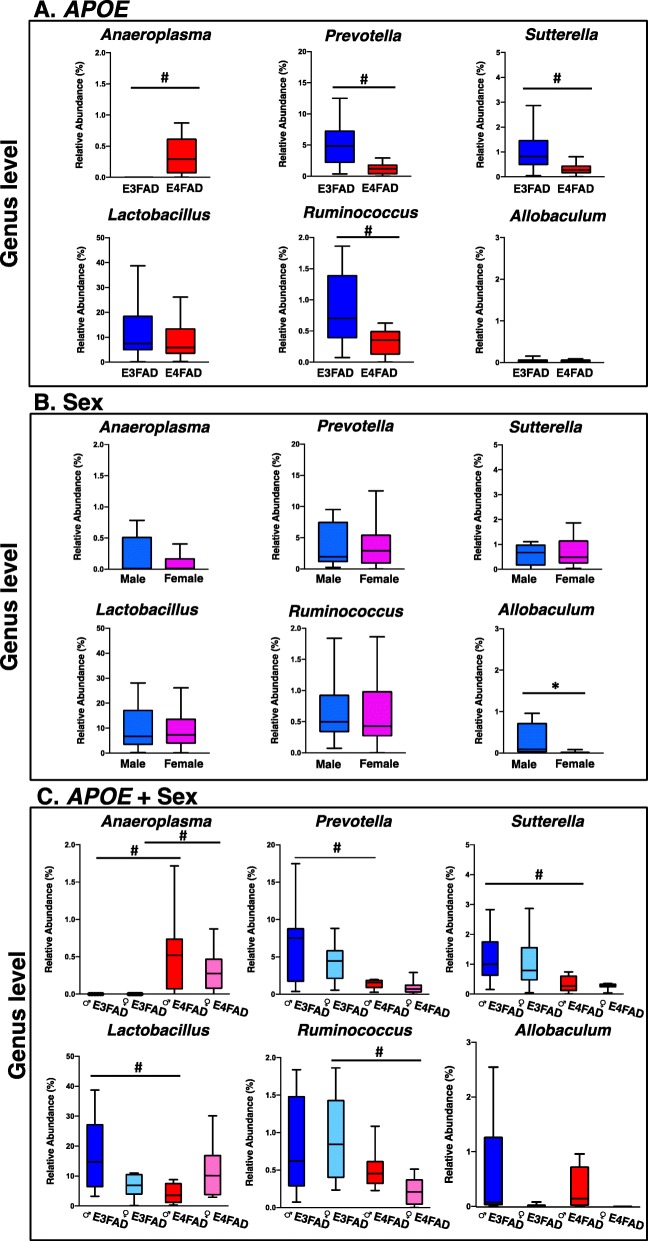
A taxon-by-taxon analysis at the genus level was performed to identify microbial genera significantly different between cohorts. The relative abundance of the genera Prevotella, Ruminonoccous and Sutterella were significantly higher in E3FAD mice compared to E4FAD mice, while the relative abundance of Anaeroplasma was significantly lower (Fig. 2a). Interestingly, FAD-Tg mice also exhibited significantly higher relative abundance of Anaeroplasma compared to wild-type mice [81, 82], suggesting that Anaeroplasma may have a role in AD pathology. Tran and colleagues demonstrated that APOE4-TR mice exhibit greater relative abundance of bacteria from the genera Mucispirillum, Desulfovibrio, Butyricicoccus and lower relative abundance of Bacteroides, Alistipes, Johnsonella compared to APOE3-TR mice [25]. Thus, our results together suggest that the effects of APOE genotype on the GM is modulated by AD pathology. Additionally, Org and colleagues determined that Allobaculum, Anaeroplasma and Erwinia are the most abundant genera in ♂mice relative to ♀mice [83]. Similarly, ♂EFAD exhibited a significantly greater relative abundance of Allobaculum compared to ♀EFAD (Fig. 2b). Comparing the stratified cohorts, the fecal microbiota of ♂E4FAD mice had lower relative abundance of Sutterella and Lactobacillus compared to ♂E3FAD. ♀E4FAD mice had lower relative abundance of Prevotella and Ruminococcus compared to ♀E3FAD (Fig. 2c). Similarly, these differences are significant at the OTU level (Additional file 4: Table S2). Therefore, the results suggest that the effect of APOE genotype on differentially abundant bacteria is modulated by sex, as specific genera and OTUs are significantly different in males or females.
Fig. 2.
Relative abundance of bacterial genera in EFAD mice stratified by APOE, sex, APOE + sex. Significantly different relative abundance of genus-level bacterial taxa associated with (a) APOE, (b) sex and (c) APOE + sex, identified by Mann-Whitney U test with a Monte Carlo Simulation corrected for false discovery rate (*p < 0.05 vs sex; #p < 0.05 vs genotype). Tukey plots show the median and interquartile range, with outliers removed from the graph. Significantly different relative abundance of unclassified genera and taxa from other taxonomic levels are found in the Additional file 4: Table S2.
Compared to ♀E3FAD mice, ♀E4FAD mice exhibited a lower relative abundance of bacterial genera associated with short chain fatty acid (SCFA) production, including Prevotella and Ruminococcus [84–89]. The GM is crucial for the production of SCFAs that, while the underlying mechanism is not completely understood, serve as energy sources for intestinal epithelial cells, regulators of plasma lipid levels, and modulators of immune cells [90–95]. The current results suggest a metabolic dysfunction in the ♀E4FAD GM. However, metabolomic and metagenomic analyses will be required to interpret accurately the interactive effects of APOE + sex on the metabolic function of the EFAD GM.
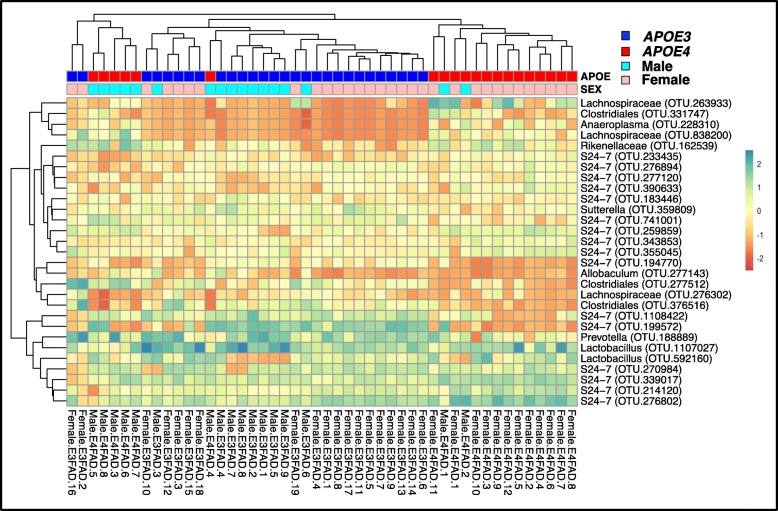
The Boruta algorithm identified 29 OTUs significant in distinguishing EFAD samples by APOE + sex (Additional file 2: Figure S2). These 29 bacterial OTUs were annotated at varying taxonomic levels, including the genera Prevotella, Lactobacillus, Allobaculum, Anaeroplasma, and Sutterella, consistent with the results of differentially abundant bacteria (Fig. 2). Based on the abundance of these 29 OTUs, a hierarchical heatmap demonstrates that EFAD samples clustered by APOE + sex (Fig. 3). Clustering of ♀E4FAD samples is further demonstration that the murine GM is affected by a specific interaction between APOE4 genotype and ♀sex, consistent with human ♀APOE4 carriers exhibiting greater AD risk compared to ♂APOE4 carriers [47–50].
Fig. 3.
Two-way clustered heatmap of microbial OTUs from EFAD mice stratified by APOE + sex. Heatmap generated with hierarchical clustering (Euclidean distance, complete linkage) based on bacterial OTUs identified by Boruta, a Random Forest based machine learning algorithm (Additional file 2: Fig. S2)
Conclusions
This short report demonstrates: 1) the EFAD GM is modulated by APOE + sex, 2) the synergistic effects of ♀sex and APOE4 genotype yield a specific GM profile in ♀E4FAD mice, and 3) clustering samples by only APOE genotype or sex masks the interactive effects of APOE + sex on the EFAD GM. Notably, these findings are consistent with AD readouts from EFAD mice varying in severity of pathology by APOE + sex, including behavioral deficits, Aβ deposition and neuroinflammation greatest in ♀E4FAD > ♂E4FAD = ♀E3FAD > ♂E3FAD [65, 66]. Therefore, the GM would potentially serve as an AD readout, reflecting the interaction between APOE + sex. Although the use of 16S rRNA sequencing has more limited taxonomic resolution than shotgun metagenome sequencing [96], 16S rRNA sequencing is sufficiently robust to identify significant effects on the GM. This study demonstrates the importance of stratifying the EFAD population by APOE + sex to better understand the relationship between AD and the GM. Future studies will examine the composition and metabolic function of the GM throughout the development of EFAD pathology through the use of metagenomic and metabolomic analyses. In conclusion, the interactive effects of APOE + sex on AD play an important role in modulating the GM composition, and the current report is the first step in identifying and understanding these effects.
Supplementary information
Additional file 1: Figure S1. Analysis of α-diversity of EFAD mice stratified by APOE, sex, APOE + sex. Based on bacterial evenness and richness, Shannon H index scores were generated and compared across EFAD mice stratified by (A) APOE, (B) sex and (C) APOE + sex with Mann-Whitney U test (*p < 0.05 vs sex; #p < 0.05 vs genotype). A mixed-model analysis was used to evaluate the interactive effects of APOE + sex on richness, evenness and α-diversity (¶ p < 0.05 vs APOE + sex).
Additional file 2: Figure S2. Boruta-identified bacterial OTUs from EFAD mice stratified by APOE + sex. Implementing the R package “randomForest”, Boruta is a feature-selection algorithm that determined the OTUs that were significant in distinguish samples by APOE + sex compared to randomly generated probes (“shadow scores” in blue). Significance is defined by a z-score > max shadow z-score (green; listed in the table). OTUs with a z-score that trends towards significance are labeled in yellow.
Additional file 3: Table S1. Permutational multivariate analysis of variance (PERMANOVA) of EFAD mice stratified by APOE, sex, APOE + sex. (A) PERMANOVA was used to assess the effect of the interaction between universal biological variables on the microbiome composition at various taxonomic levels. P-values were obtained using 9999 permutations under a reduced model. Pseudo-F ratio is defined by the difference between cohorts over the difference within each cohort and the degrees of freedom. Each term is contributing a fixed component to the overall model. Estimated sizes of components of variation are multivariate analogs to the classical ANOVA unbiased estimators. Significance (bold) is defined by a p < 0.05. (B) As the interaction between APOE + sex is significant, pair-wise PERMANOVAs at the OTU level evaluated the effects of APOE on β-diversity within ♂EFAD and ♀EFAD mice, and the effects of sex in E3FAD and E4FAD. Significance (bold) is defined by a p < 0.05.
Additional file 4: Table S2. Results of Mann-Whitney U tests at specific taxonomic levels in EFAD mice. Significantly different relative abundance of bacterial genera associated with APOE, sex, and APOE + sex, identified by Mann-Whitney U under the Monte Carlo Simulation corrected for false discovery rate (p < 0.05) at the levels of Phylum, Class, Order, Family, Genus and OTU.
Acknowledgements
Not applicable.
Abbreviations
- AD
Alzheimer’s disease
- apoE
Apolipoprotein E
- Aβ
Amyloid-β
- FAD
Familial AD
- GM
Gut microbiome
- MWU
Mann-Whitney U
- OTU
Operational taxonomic units
- Perm
Permutation
- PERMANOVA
Permutational multivariate analysis of variance
- SCFA
Short chain fatty acid
- Tg
Transgenic
Authors’ contributions
ML and JMW analyzed the data and wrote the manuscript, IP and SE purified DNA and performed PCR, JMW and SJG performed statistical analyses, JY bred the mice, SJG and SE contributed to data analysis and manuscript editing. All authors read and approved the final manuscript.
Funding
The authors acknowledge funding from the NIH (R56-AG057589 and P01-AG030128) and the BrightFocus Foundation (A2014210S) to the Estus lab. The LaDu lab funding includes funds from NIH/NIA (R01 AG058068, R01 AG057008), institutional funds from the College of Medicine at the University of Illinois, Chicago, and philanthropic contributions. JMW was supported by an administrative supplement to R01 AG057008 and by the NSF Bridge to the Doctorate Fellowship.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Raw sequence data files were submitted in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI). The BioProject identifier of the samples is PRJNA556445.
Ethics approval
Animal studies were performed in compliance with IACUC (Institutional Animal Care and Use Committee) at University of Illinois-Chicago.
Consent for publication
All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Juan Maldonado Weng, Email: jmaldo27@uic.edu.
Ishita Parikh, Email: ishita.parikh@uky.edu.
Ankur Naqib, Email: ankur_naqib@rush.edu.
Jason York, Email: jmyork@uic.edu.
Stefan J. Green, Email: greendna@uic.edu
Steven Estus, Email: steve.estus@uky.edu.
Mary Jo LaDu, Email: mladu@uic.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13024-019-0352-2.
References
- 1.Laukens D, Brinkman BM, Raes J, De Vos M, Vandenabeele P. Heterogeneity of the gut microbiome in mice: guidelines for optimizing experimental design. FEMS Microbiol Rev. 2016;40(1):117–132. doi: 10.1093/femsre/fuv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012;70(Suppl 1):S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 4.Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28(8):1221–1238. doi: 10.1210/me.2014-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyte M, Cryan JF. Dealing with ability of the microbiota to influence the brain, and ultimately cognition and behavioral. Adv Exp Med Biol. 2014;817:ix–xi. [PubMed] [Google Scholar]
- 6.DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8(9):523–531. doi: 10.1038/nrgastro.2011.133. [DOI] [PubMed] [Google Scholar]
- 7.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tremlett H, Fadrosh DW, Faruqi AA, Zhu F, Hart J, Roalstad S, et al. Gut microbiota in early pediatric multiple sclerosis: a case-control study. Eur J Neurol. 2016;23(8):1308–1321. doi: 10.1111/ene.13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhargava P, Mowry EM. Gut microbiome and multiple sclerosis. Curr Neurol Neurosci Rep. 2014;14(10):492. doi: 10.1007/s11910-014-0492-2. [DOI] [PubMed] [Google Scholar]
- 11.Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochoa-Reparaz J, Magori K, Kasper LH. The chicken or the egg dilemma: intestinal dysbiosis in multiple sclerosis. Ann Transl Med. 2017;5(6):145. doi: 10.21037/atm.2017.01.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, et al. Gut microbiome alterations in Alzheimer's disease. Sci Rep. 2017;7(1):13537. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging. 2017;49:60–68. doi: 10.1016/j.neurobiolaging.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 15.MahmoudianDehkordi S, Arnold M, Nho K, Ahmad S, Jia W, Xie G, et al. Altered bile acid profile associates with cognitive impairment in Alzheimer's disease-an emerging role for gut microbiome. Alzheimers Dement. 2019;15(1):76–92. doi: 10.1016/j.jalz.2018.07.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox M, Knorr DA, Haptonstall KM. Alzheimer's disease and symbiotic microbiota: an evolutionary medicine perspective. Ann N Y Acad Sci. 2019;1449(1):3–24. [DOI] [PMC free article] [PubMed]
- 17.Jiang C, Li G, Huang P, Liu Z, Zhao B. The gut microbiota and Alzheimer's disease. J Alzheimers Dis. 2017;58(1):1–15. doi: 10.3233/JAD-161141. [DOI] [PubMed] [Google Scholar]
- 18.Angelucci F, Cechova K, Amlerova J, Hort J. Antibiotics, gut microbiota, and Alzheimer's disease. J Neuroinflammation. 2019;16(1):108. doi: 10.1186/s12974-019-1494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcez Michelle L., Jacobs Kelly R., Guillemin Gilles J. Microbiota Alterations in Alzheimer’s Disease: Involvement of the Kynurenine Pathway and Inflammation. Neurotoxicity Research. 2019;36(2):424–436. doi: 10.1007/s12640-019-00057-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang ZQ, Shen LL, Li WW, Fu X, Zeng F, Gui L, et al. Gut microbiota is altered in patients with Alzheimer's disease. J Alzheimers Dis. 2018;63(4):1337–1346. doi: 10.3233/JAD-180176. [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Wu L, Peng G, Han Y, Tang R, Ge J, et al. Altered microbiomes distinguish Alzheimer's disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav Immun. 2019;80:633–643. doi: 10.1016/j.bbi.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Li Binyin, He Yixi, Ma Jianfang, Huang Pei, Du Juanjuan, Cao Li, Wang Yan, Xiao Qin, Tang Huidong, Chen Shengdi. Mild cognitive impairment has similar alterations as Alzheimer's disease in gut microbiota. Alzheimer's & Dementia. 2019;15(10):1357–1366. doi: 10.1016/j.jalz.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Reitz C, Mayeux R. Use of genetic variation as biomarkers for mild cognitive impairment and progression of mild cognitive impairment to dementia. J Alzheimers Dis. 2010;19(1):229–251. doi: 10.3233/JAD-2010-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leoni V. The effect of apolipoprotein E (ApoE) genotype on biomarkers of amyloidogenesis, tau pathology and neurodegeneration in Alzheimer's disease. Clin Chem Lab Med. 2011;49(3):375–383. doi: 10.1515/CCLM.2011.088. [DOI] [PubMed] [Google Scholar]
- 25.Tran Tam T. T., Corsini Simone, Kellingray Lee, Hegarty Claire, Le Gall Gwénaëlle, Narbad Arjan, Müller Michael, Tejera Noemi, O’Toole Paul W., Minihane Anne-Marie, Vauzour David. APOE genotype influences the gut microbiome structure and function in humans and mice: relevance for Alzheimer’s disease pathophysiology. The FASEB Journal. 2019;33(7):8221–8231. doi: 10.1096/fj.201900071R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/S0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 27.Davignon J, Gregg RE, Sing CF. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8(1):1–21. doi: 10.1161/01.ATV.8.1.1. [DOI] [PubMed] [Google Scholar]
- 28.de Knijff P, van den Maagdenberg AM, Frants RR, Havekes LM. Genetic heterogeneity of apolipoprotein E and its influence on plasma lipid and lipoprotein levels. Hum Mutat. 1994;4(3):178–194. doi: 10.1002/humu.1380040303. [DOI] [PubMed] [Google Scholar]
- 29.Dallongeville J, Lussier-Cacan S, Davignon J. Modulation of plasma triglyceride levels by apoE phenotype: a meta-analysis. J Lipid Res. 1992;33(4):447–454. [PubMed] [Google Scholar]
- 30.Caparros-Martin JA, Lareu RR, Ramsay JP, Peplies J, Reen FJ, Headlam HA, et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome. 2017;5(1):95. doi: 10.1186/s40168-017-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natividad JM, Lamas B, Pham HP, Michel ML, Rainteau D, Bridonneau C, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun. 2018;9(1):2802. doi: 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, et al. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 2015;22(3):516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des. 2009;15(13):1546–1558. doi: 10.2174/138161209788168164. [DOI] [PubMed] [Google Scholar]
- 35.Rebolledo C, Cuevas A, Zambrano T, Acuna JJ, Jorquera MA, Saavedra K, et al. Bacterial community profile of the gut microbiota differs between Hypercholesterolemic subjects and controls. Biomed Res Int. 2017;2017:8127814. doi: 10.1155/2017/8127814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caesar R, Nygren H, Oresic M, Backhed F. Interaction between dietary lipids and gut microbiota regulates hepatic cholesterol metabolism. J Lipid Res. 2016;57(3):474–481. doi: 10.1194/jlr.M065847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haro C, Rangel-Zuniga OA, Alcala-Diaz JF, Gomez-Delgado F, Perez-Martinez P, Delgado-Lista J, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016;11(5):e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chene G, Beiser A, Au R, Preis SR, Wolf PA, Dufouil C, et al. Gender and incidence of dementia in the Framingham heart study from mid-adult life. Alzheimers Dement. 2015;11(3):310–320. doi: 10.1016/j.jalz.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominianni C, Sinha R, Goedert JJ, Pei Z, Yang L, Hayes RB, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015;10(4):e0124599. doi: 10.1371/journal.pone.0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Yong Sung, Unno Tatsuya, Kim Byung-Yong, Park Mi-Sung. Sex Differences in Gut Microbiota. The World Journal of Men's Health. 2020;38(1):48. doi: 10.5534/wjmh.190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10:253. doi: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jasarevic E, Morrison KE, Bale TL. Sex differences in the gut microbiome-brain axis across the lifespan. Philos Trans R Soc Lond Ser B Biol Sci. 2016;371(1688):20150122. doi: 10.1098/rstb.2015.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39(2):400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker JM, Al-Nakkash L, Herbst-Kralovetz MM. Estrogen-gut microbiome axis: physiological and clinical implications. Maturitas. 2017;103:45–53. doi: 10.1016/j.maturitas.2017.06.025. [DOI] [PubMed] [Google Scholar]
- 45.Chen KL, Madak-Erdogan Z. Estrogen and microbiota crosstalk: should we pay attention? Trends Endocrinol Metab. 2016;27(11):752–755. doi: 10.1016/j.tem.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72(2):1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farrer LA, Cupples LA, Haines JL. Hyman B, Kukull WA, Mayeux R, et al. effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease Meta analysis consortium. JAMA. 1997;278(16):1349–1356. doi: 10.1001/jama.1997.03550160069041. [DOI] [PubMed] [Google Scholar]
- 48.Altmann A, Tian L, Henderson VW, Greicius MD. Alzheimer's disease neuroimaging initiative I. sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol. 2014;75(4):563–573. doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bretsky PM, Buckwalter JG, Seeman TE, Miller CA, Poirier J, Schellenberg GD, et al. Evidence for an interaction between apolipoprotein E genotype, gender, and Alzheimer disease. Alzheimer Dis Assoc Disord. 1999;13(4):216–221. doi: 10.1097/00002093-199910000-00007. [DOI] [PubMed] [Google Scholar]
- 50.Payami H, Zareparsi S, Montee KR, Sexton GJ, Kaye JA, Bird TD, et al. Gender difference in apolipoprotein E-associated risk for familial Alzheimer disease: a possible clue to the higher incidence of Alzheimer disease in women. Am J Hum Genet. 1996;58(4):803–811. [PMC free article] [PubMed] [Google Scholar]
- 51.Breitner JC, Wyse BW, Anthony JC, Welsh-Bohmer KA, Steffens DC, Norton MC, et al. APOE-epsilon4 count predicts age when prevalence of AD increases, then declines: the Cache County study. Neurology. 1999;53(2):321–331. doi: 10.1212/WNL.53.2.321. [DOI] [PubMed] [Google Scholar]
- 52.Martinez M, Campion D, Brice A, Hannequin D, Dubois B, Didierjean O, et al. Apolipoprotein E epsilon4 allele and familial aggregation of Alzheimer disease. Arch Neurol. 1998;55(6):810–816. doi: 10.1001/archneur.55.6.810. [DOI] [PubMed] [Google Scholar]
- 53.Andersen K, Launer LJ, Dewey ME, Letenneur L, Ott A, Copeland JR, et al. Gender differences in the incidence of AD and vascular dementia: the EURODEM studies. EURODEM Incidence Research Group. Neurology. 1999;53(9):1992–1997. doi: 10.1212/WNL.53.9.1992. [DOI] [PubMed] [Google Scholar]
- 54.Molero AE, Pino-Ramirez G, Maestre GE. Modulation by age and gender of risk for Alzheimer's disease and vascular dementia associated with the apolipoprotein E-epsilon4 allele in Latin Americans: findings from the Maracaibo aging study. Neurosci Lett. 2001;307(1):5–8. doi: 10.1016/S0304-3940(01)01911-5. [DOI] [PubMed] [Google Scholar]
- 55.Corder EH, Ghebremedhin E, Taylor MG, Thal DR, Ohm TG, Braak H. The biphasic relationship between regional brain senile plaque and neurofibrillary tangle distributions: modification by age, sex, and APOE polymorphism. Ann N Y Acad Sci. 2004;1019:24–28. doi: 10.1196/annals.1297.005. [DOI] [PubMed] [Google Scholar]
- 56.Altmann A, Tian L, Henderson VW, Greicius MD. Sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol. 2014;75(4):563–573. doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyman BT, Gomez-Isla T, Briggs M, Chung H, Nichols S, Kohout F, et al. Apolipoprotein E and cognitive change in an elderly population. Ann Neurol. 1996;40(1):55–66. doi: 10.1002/ana.410400111. [DOI] [PubMed] [Google Scholar]
- 58.Mortensen EL, Hogh P. A gender difference in the association between APOE genotype and age-related cognitive decline. Neurology. 2001;57(1):89–95. doi: 10.1212/WNL.57.1.89. [DOI] [PubMed] [Google Scholar]
- 59.Holland D, Desikan RS, Dale AM, McEvoy LK. Alzheimer's disease neuroimaging I. higher rates of decline for women and apolipoprotein E epsilon4 carriers. AJNR Am J Neuroradiol. 2013;34(12):2287–2293. doi: 10.3174/ajnr.A3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fleisher A, Grundman M, Jack CR, Jr, Petersen RC, Taylor C, Kim HT, et al. Sex, apolipoprotein E epsilon 4 status, and hippocampal volume in mild cognitive impairment. Arch Neurol. 2005;62(6):953–957. doi: 10.1001/archneur.62.6.953. [DOI] [PubMed] [Google Scholar]
- 61.Jack CR, Jr, Wiste HJ, Weigand SD, Knopman DS, Vemuri P, Mielke MM, et al. Age, sex, and APOE epsilon4 effects on memory, brain structure, and beta-amyloid across the adult life span. JAMA Neurol. 2015;72(5):511–519. doi: 10.1001/jamaneurol.2014.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Youmans KL, Tai LM, Nwabuisi-Heath E, Jungbauer L, Kanekiyo T, Gan M, et al. APOE4-specific changes in Abeta accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem. 2012;287(50):41774–41786. doi: 10.1074/jbc.M112.407957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J Neurosci. 2006;26(40):10129–40. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tai LM, Ghura S, Koster KP, Liakaite V, Maienschein-Cline M, Kanabar P, et al. APOE-modulated Abeta-induced neuroinflammation in Alzheimer's disease: current landscape, novel data and future perspective. J Neurochem. 2015;133(4):465–88. [DOI] [PMC free article] [PubMed]
- 65.Tai Leon M., Balu Deebika, Avila-Munoz Evangelina, Abdullah Laila, Thomas Riya, Collins Nicole, Valencia-Olvera Ana Carolina, LaDu Mary Jo. EFAD transgenic mice as a humanAPOErelevant preclinical model of Alzheimer’s disease. Journal of Lipid Research. 2017;58(9):1733–1755. doi: 10.1194/jlr.R076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balu Deebika, Karstens Aimee James, Loukenas Efstathia, Maldonado Weng Juan, York Jason M., Valencia-Olvera Ana Carolina, LaDu Mary Jo. The role of APOE in transgenic mouse models of AD. Neuroscience Letters. 2019;707:134285. doi: 10.1016/j.neulet.2019.134285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sullivan PM, Mezdour H, Quarfordt SH, Maeda N. Type III hyperlipoproteinemia and spontaneous atherosclerosis in mice resulting from gene replacement of mouse Apoe with human Apoe*2. J Clin Invest. 1998;102(1):130–135. doi: 10.1172/JCI2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corsetti JP, Sparks CE, Bakker SJL, Gruppen EG, Dullaart RPF. Roles of high apolipoprotein E blood levels and HDL in development of familial dysbetalipoproteinemia in epsilon2epsilon2 subjects. Clin Biochem. 2018;52:67–72. doi: 10.1016/j.clinbiochem.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Koopal C, Marais AD, Westerink J, Visseren FL. Autosomal dominant familial dysbetalipoproteinemia: a pathophysiological framework and practical approach to diagnosis and therapy. J Clin Lipidol. 2017;11(1):12–23. doi: 10.1016/j.jacl.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth's multiscale microbial diversity. Nature. 2017;551(7681):457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gihring TM, Green SJ, Schadt CW. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ Microbiol. 2012;14(2):285–290. doi: 10.1111/j.1462-2920.2011.02550.x. [DOI] [PubMed] [Google Scholar]
- 74.Clarke KR, Warwick RM. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. Plymouth: Primer-E Ltd; 2001. [Google Scholar]
- 75.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 76.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18(1):117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 77.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, et al. Conducting a microbiome study. Cell. 2014;158(2):250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammer Ø, Harper DAT, Ryan PD. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electron. 2001;4(1):9. [Google Scholar]
- 79.Kursa MB, Rudnicki WR. Feature selection with the Boruta package. J Stat Softw. 2010;36(11):1–13. doi: 10.18637/jss.v036.i11. [DOI] [Google Scholar]
- 80.Dodiya Hemraj B., Kuntz Thomas, Shaik Shabana M., Baufeld Caroline, Leibowitz Jeffrey, Zhang Xulun, Gottel Neil, Zhang Xiaoqiong, Butovsky Oleg, Gilbert Jack A., Sisodia Sangram S. Sex-specific effects of microbiome perturbations on cerebral Aβ amyloidosis and microglia phenotypes. The Journal of Experimental Medicine. 2019;216(7):1542–1560. doi: 10.1084/jem.20182386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harach T, Marungruang N, Duthilleul N, Cheatham V, Mc Coy KD, Frisoni G, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep. 2017;7:41802. doi: 10.1038/srep41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanguinetti E, Collado MC, Marrachelli VG, Monleon D, Selma-Royo M, Pardo-Tendero MM, et al. Microbiome-metabolome signatures in mice genetically prone to develop dementia, fed a normal or fatty diet. Sci Rep. 2018;8(1):4907. doi: 10.1038/s41598-018-23261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7(4):313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen T, Long W, Zhang C, Liu S, Zhao L, Hamaker BR. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci Rep. 2017;7(1):2594. doi: 10.1038/s41598-017-02995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hiippala K, Kainulainen V, Kalliomaki M, Arkkila P, Satokari R. Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp. Front Microbiol. 2016;7:1706. doi: 10.3389/fmicb.2016.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.La Reau AJ, Suen G. The Ruminococci: key symbionts of the gut ecosystem. J Microbiol. 2018;56(3):199–208. doi: 10.1007/s12275-018-8024-4. [DOI] [PubMed] [Google Scholar]
- 87.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65(11):1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 88.Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int. 2012;95(1):50–60. doi: 10.5740/jaoacint.SGE_Macfarlane. [DOI] [PubMed] [Google Scholar]
- 89.Kolmeder CA, Salojarvi J, Ritari J, de Been M, Raes J, Falony G, et al. Faecal Metaproteomic analysis reveals a personalized and stable functional microbiome and limited effects of a probiotic intervention in adults. PLoS One. 2016;11(4):e0153294. doi: 10.1371/journal.pone.0153294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Natarajan N, Pluznick JL. From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. Am J Phys Cell Phys. 2014;307(11):C979–C985. doi: 10.1152/ajpcell.00228.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holzer P, Farzi A, Hassan AM, Zenz G, Jacan A, Reichmann F. Visceral inflammation and immune activation stress the brain. Front Immunol. 2017;8:1613. doi: 10.3389/fimmu.2017.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caballero-Villarraso J, Galvan A, Escribano BM, Tunez I. Interrelationships among gut microbiota and host: paradigms, role in neurodegenerative diseases and future prospects. CNS Neurol Disord Drug Targets. 2017;16(8):945–964. doi: 10.2174/1871527316666170714120118. [DOI] [PubMed] [Google Scholar]
- 94.Yamada T, Takahashi D, Hase K. The diet-microbiota-metabolite axis regulates the host physiology. J Biochem. 2016;160(1):1–10. doi: 10.1093/jb/mvw022. [DOI] [PubMed] [Google Scholar]
- 95.Dopkins N, Nagarkatti PS, Nagarkatti M. The role of gut microbiome and associated metabolome in the regulation of neuroinflammation in multiple sclerosis and its implications in attenuating chronic inflammation in other inflammatory and autoimmune disorders. Immunology. 2018;154(2):178–185. doi: 10.1111/imm.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469(4):967–977. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Analysis of α-diversity of EFAD mice stratified by APOE, sex, APOE + sex. Based on bacterial evenness and richness, Shannon H index scores were generated and compared across EFAD mice stratified by (A) APOE, (B) sex and (C) APOE + sex with Mann-Whitney U test (*p < 0.05 vs sex; #p < 0.05 vs genotype). A mixed-model analysis was used to evaluate the interactive effects of APOE + sex on richness, evenness and α-diversity (¶ p < 0.05 vs APOE + sex).
Additional file 2: Figure S2. Boruta-identified bacterial OTUs from EFAD mice stratified by APOE + sex. Implementing the R package “randomForest”, Boruta is a feature-selection algorithm that determined the OTUs that were significant in distinguish samples by APOE + sex compared to randomly generated probes (“shadow scores” in blue). Significance is defined by a z-score > max shadow z-score (green; listed in the table). OTUs with a z-score that trends towards significance are labeled in yellow.
Additional file 3: Table S1. Permutational multivariate analysis of variance (PERMANOVA) of EFAD mice stratified by APOE, sex, APOE + sex. (A) PERMANOVA was used to assess the effect of the interaction between universal biological variables on the microbiome composition at various taxonomic levels. P-values were obtained using 9999 permutations under a reduced model. Pseudo-F ratio is defined by the difference between cohorts over the difference within each cohort and the degrees of freedom. Each term is contributing a fixed component to the overall model. Estimated sizes of components of variation are multivariate analogs to the classical ANOVA unbiased estimators. Significance (bold) is defined by a p < 0.05. (B) As the interaction between APOE + sex is significant, pair-wise PERMANOVAs at the OTU level evaluated the effects of APOE on β-diversity within ♂EFAD and ♀EFAD mice, and the effects of sex in E3FAD and E4FAD. Significance (bold) is defined by a p < 0.05.
Additional file 4: Table S2. Results of Mann-Whitney U tests at specific taxonomic levels in EFAD mice. Significantly different relative abundance of bacterial genera associated with APOE, sex, and APOE + sex, identified by Mann-Whitney U under the Monte Carlo Simulation corrected for false discovery rate (p < 0.05) at the levels of Phylum, Class, Order, Family, Genus and OTU.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. Raw sequence data files were submitted in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI). The BioProject identifier of the samples is PRJNA556445.