Abstract
Fibronectin 1 (FN1) is involved in the occurrence and development of various tumors and is upregulated in multiple cancer types. FN1 has been demonstrated to promote cell proliferation and migration in gastric cancer cell lines. However, the relationship between the expression of FN1 and clinicopathological factors and prognosis is not clear in gastric cancer (GC). The aim of the present study was to investigate the association between FN1 expression and clinicopathology and prognosis of gastric cancer. In this study, 17 publicly available GC cohorts (n=2,376) with gene expression data from the Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) and Oncomine databases were tested. In addition, FN1 protein expression was validated by immunohistochemistry in a separate cohort (n=190). The meta-analysis results demonstrated an increase in FN1 expression at the protein and mRNA level in GC tissues, and the FN1 gene was highly expressed at the mRNA level in the advanced T stage (T2 + T3 + T4) group compared with that in the early T stage (T1) group. In addition, the expression of epithelial FN1 at the protein level was positively correlated with tumor size. FN1 expression at the protein and mRNA level was a predictor of poor prognosis following radical resection of GC. In conclusion, the expression of FN1 in GC tissues is upregulated compared with adjacent normal tissues, and it is a potential biomarker of poor prognosis in patients with GC.
Keywords: fibronectin 1, gastric cancer, prognosis, The Cancer Genome Atlas cohort, Gene Expression Omnibus cohort
Introduction
Gastric cancer is the fifth most commonly diagnosed cancer (5.7% of total cases) and the third leading cause of cancer mortality (8.2% of total cancer mortality) worldwide (1). Gastric cancer is also the third leading cause of cancer-related mortality in China (2). Although surgery combined with radiotherapy, chemotherapy and targeted therapy prolongs survival, the 5-year overall survival rate of patients with advanced gastric cancer remains poor. The 5-year overall survival rates of patients with pathological T stage 2, 3 and 4 disease were 68.3, 33.0 and 24.0% respectively (3,4). Therefore, new biomarkers of gastric cancer to determine prognosis are necessary.
Fibronectin 1 (FN1) mediates the interaction between cells and the extracellular matrix and serves an important role in cell adhesion, migration, growth and differentiation (5). FN1 is a ligand for numerous members of the integrin receptor family (6). FN1 is involved in the occurrence and development of various tumors. FN1 activates the PI3K/Akt pathway by binding to its integrin receptor α5β1 in breast cancer (7). In addition, FN1 has been demonstrated to promote cell proliferation and migration in esophageal squamous cell carcinoma, oral squamous cell carcinoma (OSCC), nasopharyngeal carcinoma, colorectal, ovarian, renal and thyroid cancer (8–14). However, little is known about the expression of FN1 in gastric cancer. FN1 is upregulated in GC tissues compared with normal gastric tissues (15). FN1 knockdown inhibits cell migration and invasion in vitro, and FOXF1 adjacent non-coding developmental regulatory RNA and microRNA-200c promote the proliferation, migration and invasion of GC cells by negatively targeting FN1 (15–17). Overall, FN1 is a potential biomarker candidate for GC prognosis, but the relationship between FN1 expression and clinical factors and prognosis has not been reported, and thus it is necessary to verify and clarify the role of FN1 in GC.
The aim of the present study was to investigate FN1 gene expression in GC and its association with clinicopathological factors and prognosis by examining 17 publicly available GC cohorts. Furthermore, FN1 protein expression was validated by immunohistochemistry in a separate cohort. The results demonstrated that FN1 may serve as a new prognostic marker for GC.
Materials and methods
Data collection
Microarray data were downloaded from the following datasets in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), The Cancer Genome Atlas (https://xenabrowser.net/datapages/?cohort=TCGA) and Oncomine (https://www.oncomine.org/resource/login.html): GSE13861, GSE13911, GSE14208, GSE15456, GSE15459, GSE19826, GSE26253, GSE26899, GSE26901, GSE29272, GSE34942, GSE35809, GSE54129, GSE66229, GSE79973, Chen Gastric and TCGA STAD. Several of these datasets have been previously published (18–32). The 17 datasets comprised 2,376 cancer tissues and 294 adjacent normal tissues. Datasets with no clinical data (GSE13861, GSE13911, GSE19826, GSE54129, GSE79973 and Chen Gastric), GSE29272 and TCGA STAD were used to analyze the differences between tumor and adjacent tissues. The remaining datasets were used to analyze the relationship between FN1 expression and clinicopathological factors. Clinical information for the cohorts with respective clinical data included in this study is presented in Table I.
Table I.
Clinicopathological characteristics of patients in different datasets.
| Characteristic | IHC cohort, n (%) | GSE13861, n (%) | GSE15456, n (%) | GSE15459, n (%) | GSE26253, n (%) | GSE26899, n (%) | GSE26901, n (%) | GSE29272, n (%) | GSE34942, n (%) | GSE35809, n (%) | GSE66229, n (%) | TCGA, n (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | ||||||||||||
| Total | 190 (100) | 65 (100) | 30 (100) | 192 (100) | 432 (100) | 92 (100) | 109 (100) | 134 (100) | 56 (100) | 70 (100) | 300(100) | 415(100) |
| Male | 144 (75.8) | 46 (70.8) | 17 (56.7) | 125 (65.1) | 280 (64.8) | 73 (79.3) | 69 (63.3) | 103 (76.9) | 36 (64.3) | 48 (68.6) | 199 (66.3) | 268 (64.6) |
| Female | 46 (24.2) | 19 (29.2) | 13 (43.3) | 67 (34.9) | 152 (35.2) | 19 (20.7) | 40 (36.7) | 31 (23.1) | 20 (35.7) | 22 (31.4) | 101 (33.7) | 147 (35.4) |
| Median age, years (min, max) | 59 (25, 85) | 63 (32, 83) | 73 (53, 83) | 66 (23, 92) | 53 (23, 74) | 59 (36, 83) | 58 (28, 74) | 59 (23, 73) | 69 (43, 84) | 67 (32, 85) | 64 (24, 86) | 67 (30, 90) |
| T stage | ||||||||||||
| 1 | 15 (7.9) | 2 (3.1) | – | 8 (4.2) | – | – | – | – | – | – | 2 (0.7) | 22 (5.3) |
| 2 | 32 (16.8) | 23 (35.4) | – | 45 (23.4) | – | – | – | – | – | – | 186 (62) | 88 (21.2) |
| 3 | 24 (12.6) | 34 (52.3) | – | 107 (55.7) | – | – | – | – | – | – | 91 (30.3) | 181 (43.6) |
| 4 | 119 (62.6) | 1 (1.5) | – | 1 (0.5) | – | – | – | – | – | – | 21 (7) | 115 (27.7) |
| Unknown | 0 (0) | 5 (7.7) | – | 31 (16.1) | – | – | – | – | – | – | 0 (0) | 9 (2.2) |
| TNM stage | ||||||||||||
| I | 29 (15.3) | 12 (18.5) | 6 (20) | 31 (16.1) | 68 (15.7) | 11 (12.0) | 38 (29.5) | 5 (3.7) | 11 (19.6) | 13 (18.6) | 31 (10.3) | 57 (13.7) |
| II | 60 (31.6) | 2 (3.1) | 4 (13.3) | 29 (15.1) | 167 (38.7) | 18 (19.6) | 40 (31) | 5 (3.7) | 11 (19.6) | 16 (22.9) | 97 (32.3) | 123 (29.6) |
| III | 91 (47.9) | 35 (53.8) | 15 (50) | 72 (37.5) | 130 (30.1) | 27 (29.3) | 36 (27.9) | 115 (85.8) | 19 (33.9) | 33 (47.1) | 95 (31.7) | 169 (40.7) |
| IV | 10 (5.3) | 16 (24.6) | 5 (16.7) | 60 (31.3) | 67 (15.5) | 36 (39.1) | 15 (11.6) | 9 (6.7) | 13 (23.2) | 7 (10) | 77 (25.7) | 41 (9.9) |
| Unknown | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (3.6) | 1 (1.4) | 0 (0) | 25 (6.0) |
| Tumor grade | ||||||||||||
| Low | 48 (24) | 20 (30.8) | 2 (6.7) | 6 (3.1) | – | – | – | 1 (0.7) | – | 2 (2.9) | – | 12 (2.9) |
| Intermediate | 66 (33) | 15 (23.1) | 13 (43.3) | 53 (27.6) | – | – | – | 48 (35.8) | – | 22 (31.4) | – | 148 (35.7) |
| High | 76 (38) | 6 (9.2) | 15 (50) | 86 (44.8) | – | – | – | 85 (63.4) | – | 26 (37.1) | – | 246 (59.3) |
| Undifferentiated | 10 (5) | 24 (36.9) | 0 (0) | 47 (24.5) | – | – | – | 0 (0) | – | 20 (28.6) | – | 9 (2.2) |
| Follow-up endpoint (death) | ||||||||||||
| Occurred | 101 (50.5) | – | – | 95 (49.5) | – | – | – | 31 (23.1) | 27 (48.2) | – | 159 (53) | 144 (34.7) |
| Not occurred | 99 (49.5) | – | – | 97 (50.5) | – | – | – | 95 (70.9) | 29 (51.8) | – | 141 (47) | 214 (51.6) |
| No data | 0 (0) | – | – | 0 (0) | – | – | – | 8 (6) | 0 (0) | – | 0 (0) | 57 (13.7) |
-, data not available; TNM, Tumor-Node-Metastasis.
Validation dataset
Immunohistochemistry (IHC) was used for validation. Gastric cancer tissues and adjacent normal gastric tissues were obtained during surgery from 190 randomly selected patients between June 2011 and June 2012 at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). The study was approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from all patients. The tissues were fixed with formalin and embedded in paraffin for subsequent experiments. All patients were followed up for ≥5 years, and 102 succumbed to any cause during the follow-up period.
IHC
Formalin-fixed paraffin-embedded tissue samples from the IHC cohort were sliced into 4-µm sections. A mouse monoclonal antibody against FN1 (cat. no. 66042-1-Ig; ProteinTech Group, Inc.) was used at a 1:600 dilution at pH 9.0. The immunohistochemical staining of the specimens was performed as previously described (16). The results of FN1 expression were separately scored in epithelial cancer cells and intertumoral stroma. The scoring method described by Sung et al (33) was used. For epithelial FN1 (E-FN1) expression, staining intensity and the proportion of stained tumor cells were considered. Staining intensity was classified as follows: 1, weak; 2, moderate; and 3, strong. Positive cells were quantified as a percentage of the total number of tumor cells and assigned to one of the following categories: 0, <5%; 1, 5–24%; 2, 25–49%; 3, 50–74%; and 4, ≥75%. The percentage of positive tumor cells and staining intensity were multiplied to generate an immunoreactivity score (IS) for each case. IS values ranged from 0 to 12; IS≥3 was considered positive, whereas IS<3 was considered negative. Stromal FN1 (S-FN1) expression was graded into three categories: No or weak staining, no staining or a low number of FN1-positive strands; moderate staining, fine FN1-positive strands; and strong staining, coarse FN1-positive strands (34).
Statistical analysis
When >1 FN1 probe was present in a group, the probe with the highest variance was selected for statistical analysis (35). All FN1 gene expression data normalization and probe summarization were performed by Robust Multichip Analysis and transformed by log2. SPSS 22.0 (IBM Corp.) and RevMan 5.3 (Cochrane Community) were used to perform all statistical analyses.
Independent sample t-tests were used in SPSS for continuous data analysis and Pearson's χ2 tests were used for categorical data analysis. The gene expression value was equal to three, ≥1/3 were defined as high expression and the <1/3 as low expression. Overall survival (OS) rate was analyzed using Kaplan-Meier plots and the log-rank test or Gehan-Breslow-Wilcoxon test. When the two survival functions were parallel, the log-rank test was used, whereas the Gehan-Breslow-Wilcoxon test was used if the data crossed over. A Cox regression model was used to assess the hazard ratio (HR) and perform multivariate analysis. All tests were two-sided, and P<0.05 was considered to indicate a statistically significant difference.
Meta-analyses were performed using RevMan 5.3. First, the heterogeneity between the results of each study was analyzed by the χ2 test. The threshold was set to α=0.100, and the extent of heterogeneity was assessed by combining I2. If P>0.10 and I2≤50%, the homogeneity between the results was considered high, and the fixed effect model was used; if P≤0.10 or I2>50%, the random effects model was used.
Results
Patient cohorts
Data from 17 independent GC cohorts were downloaded from the Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) and Oncomine, including 2,670 samples, which comprised 2,376 cancer tissues and 294 adjacent normal tissues. Eight of the 17 cohorts included tumor and normal samples. The IHC cohort comprised 190 GC samples and 20 adjacent tissue samples. The clinicopathological characteristics of the patients are presented in Table I.
FN1 expression in gastric cancer
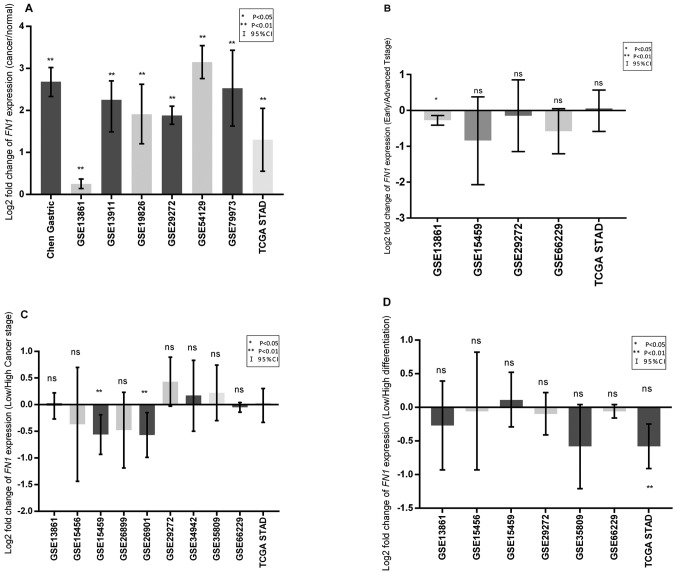
A total of eight independent cohorts that included expression data from cancer and normal samples were analyzed; the results revealed upregulated FN1 mRNA levels in tumor tissues compared with normal tissues (Fig. 1A). Meta-analysis of all the cohorts revealed a significant combined mean difference of 1.99 (P<0.001; Fig. 2A). These results indicated that FN1 expression was significantly higher in GC tissues compared with that in adjacent normal tissues.
Figure 1.
Log2 fold change in FN1 gene expression in GC tissues. (A) FN1 expression in tumor tissues compared with that in normal tissues. (B) FN1 expression in the advanced T stage (T2 + T3 + T4) group compared with that in the early T stage (T1) group. (C) FN1 expression in the low differentiation group (high tumor grade) compared with that in the high differentiation group (intermediate and low tumor grade). (D) FN1 expression in the high clinical TNM stage (III+IV) group compared with that in the low clinical TNM stage (I+II) group. Error bars represent 95% confidence interval. *P<0.05 and **P<0.01 vs. normal tissue. FN1, fibronectin 1; TNM, Tumor-Node-Metastasis; ns, non-significant.
Figure 2.
Forest plot and meta-analysis of FN1 expression in gastric cancer. (A) Forest plot of the log2 fold change in FN1 expression in tumor tissues compared with that in normal tissues. (B) Forest plot of the log2 fold change in FN1 expression in the advanced T stage group (T2 + T3 + T4) compared with that in the early T stage group (T1). (C) Forest plot of the log2 fold change in FN1 expression in the low differentiation group (high tumor grade) compared with that in the high differentiation group (intermediate and low tumor grade). (D) Forest plot of the log2 fold change in FN1 expression in the high clinical TNM stage (III+IV) group compared with that in the low clinical TNM stage (I+II) group. (E) Forest plot of the comparison of overall survival in patients with gastric cancer with high and low FN1 expression (The gene expression value was equal to three, the first two-thirds were defined as high expression and the last one-third as low expression). FN1, fibronectin 1; TNM, Tumor-Node-Metastasis; CI, confidence interval.
Association between FN1 expression and clinicopathological factors
Compared with that in the early T stage (T1) group, the expression of FN1 was significantly increased in the advanced T stage (T2+T3+T4) group (P=0.002; Fig. 1B) in one cohort, which was further confirmed by meta-analysis in all examined cohorts (P<0.001; Fig. 2B). The expression of FN1 was not associated with differentiation in any cohort (Figs. 1C and 2C). Only two cohorts exhibited increased FN1 expression in patients with high clinical Tumor-Node-Metastasis (TNM) stage (36) (III + IV) compared with that in patients with low clinical TNM stage (I + II) (Fig. 1D). No significant differences between patients with high and low TNM stage were observed in the meta-analysis of all cohorts (Fig. 2D).
High FN1 expression level indicates poor clinical outcomes
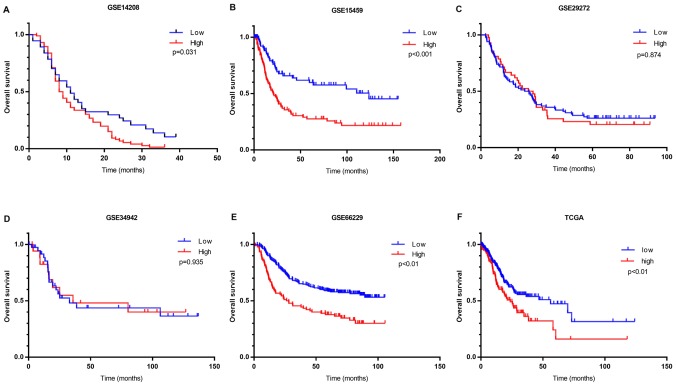
Kaplan-Meier survival analysis was performed using clinical data. OS analysis demonstrated that high FN1 expression was associated with unfavorable prognosis compared with low FN1 expression in four of the six cohorts containing prognostic information (Fig. 3). A meta-analysis of all cohorts validated this result, as it exhibited a significant combined FN1 hazard ratio (HR) of 1.67 (P<0.001; Fig. 2E). This indicated that the expression of FN1 is a potential indicator of clinical outcome in patients with GC.
Figure 3.
Kaplan-Meier survival curves of the high- and low-FN1 expression groups in six cohorts. Survival curves for the following datasets: (A) GSE14208; (B) GSE15459; (C) GSE29272; (D) GSE34942; (E) GSE66229 and (F) TCGA. FN1, fibronectin 1; TCGA, The Cancer Genome Atlas.
FN1 immunohistochemistry
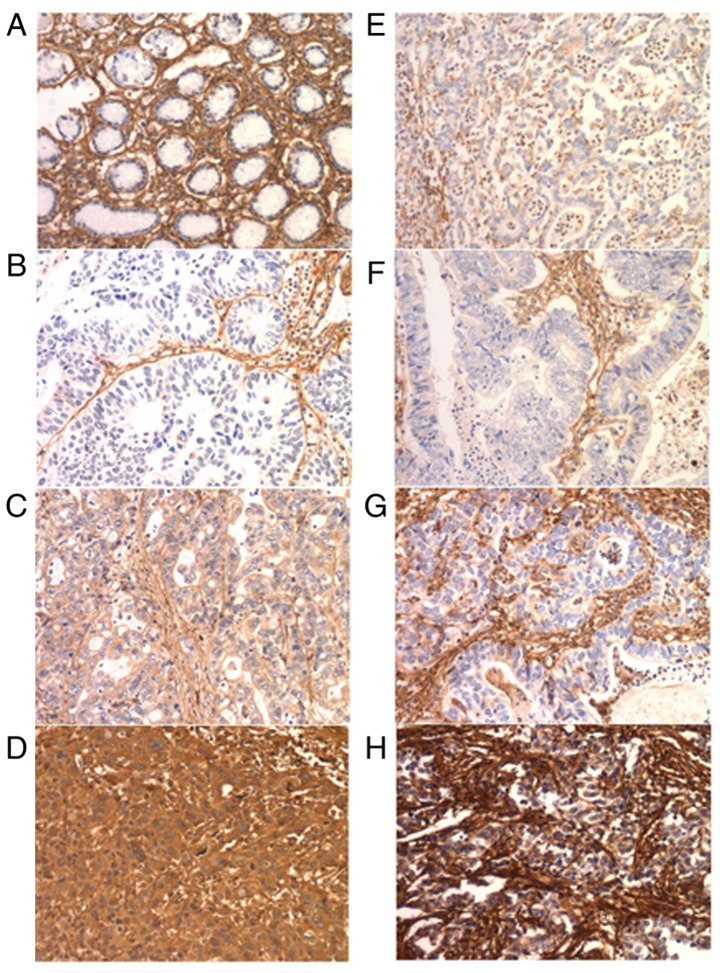
FN1 is expressed in cancer cells and the intratumoral matrix in GC (Fig. 4). In the IHC cohort, normal epithelial cells exhibited no FN1 expression. E-FN1 expression was positive in 85 of the 190 cases (44.7%). S-FN expression was graded as no/weak in 11 (5.8%), moderate in 71 (37.4%) and strong in 108 (56.8%) cases (Table II). No association was identified between E-FN1 and S-FN1 expression (P=0.112; Table III). E-FN1 expression in GC exhibited a significant association with tumor size (P=0.037), whereas S-FN1 expression was associated with sex (P=0.027) (Table II).
Figure 4.
Immunohistochemical analysis of FN1 in GC. (A) Normal gastric mucosa exhibited negative staining for FN1. (B-D) E-FN1 expression in GS tissues. (B) Expression score, 0; (C) expression score, 6; and (D) expression score, 12. (E-H) S-FN1 expression in in GS tissues. (E) No expression; (F) low expression; (G) medium expression; and (H) high expression. Magnification, ×200. GC, gastric cancer; FN1, fibronectin 1; S-FN1, stromal FN1; E-FN1, epithelial FN1.
Table II.
Patient characteristics based on the immunohistochemistry results of FN1 expression in gastric cancer.
| Expression of E-FN1 (%) | Expression of S-FN1 (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | No. of patients (n=190) | Negative (n=85) | Positive (n=105) | P-value | No/weak (n=11) | Moderate (n=71) | Strong (n=108) | P-value |
| Sex | 0.380 | 0.027a | ||||||
| Female | 46 | 18 (39.14) | 28 (60.9) | 0 (0.0) | 13 (28.3) | 33 (71.7) | ||
| Male | 144 | 67 (46.5) | 77 (53.5) | 11 (7.6) | 58 (40.3) | 75 (52.1) | ||
| Age (years) | 0.508 | 0.361 | ||||||
| <60 | 100 | 47 (47.0) | 53 (53.0) | 4 (4.0) | 41 (41.0) | 55 (55.0) | ||
| ≥60 | 90 | 38 (42.2) | 52 (57.8) | 7 (7.8) | 30 (33.3) | 53 (58.9) | ||
| Tumor diameter (cm) | 0.037a | 0.639 | ||||||
| <5 | 114 | 58 (50.9) | 56 (49.1) | 8 (7.0) | 41 (36.0) | 65 (57.0) | ||
| ≥5 | 76 | 27 (35.5) | 49 (64.5) | 3 (3.9) | 30 (39.5) | 43 (56.6) | ||
| T stage | 0.742 | 0.962 | ||||||
| T1 + T2 | 47 | 22 (46.8) | 25 (53.2) | 3 (6.4) | 18 (38.3) | 26 (55.3) | ||
| T3 + T4 | 143 | 63 (44.1) | 80 (55.9) | 8 (5.6) | 53 (37.1) | 82 (57.3) | ||
| N stage | 0.080 | 0.616 | ||||||
| N0 + N1 | 112 | 56 (50.0) | 56 (50.0) | 8 (7.1) | 42 (37.5) | 62 (55.4) | ||
| N2 + N3 | 78 | 29 (37.2) | 49 (62.8) | 3 (3.8) | 29 (37.2) | 46 (59.0) | ||
| TNM stage | 0.352 | 0.510 | ||||||
| I + II | 89 | 43 (48.3) | 46 (51.7) | 7 (7.9) | 32 (36.0) | 50 (56.2) | ||
| III + IV | 101 | 42 (41.6) | 59 (58.4) | 4 (4.0) | 39 (38.6) | 58 (57.4) | ||
P<0.05, FN1, fibronectin 1; S-FN1, stromal FN1; E-FN1, epithelial FN1; TNM, Tumor-Node-Metastasis.
Table III.
Association between epithelial and stromal expression of FN1 in gastric cancer.
| Expression of E-FN1 | ||||
|---|---|---|---|---|
| Expression of S-FN1 | Negative (%) | Positive (%) | Total (%) | P-value |
| No/Weak | 7 (63.6) | 4 (36.4) | 11 (5.8) | |
| Moderate | 34 (47.9) | 37 (52.1) | 71 (37.4) | |
| Strong | 44 (40.7) | 64 (59.3) | 108 (56.8) | |
| Total (%) | 85 (100.0) | 105 (100.0) | 190 (100.0) | 0.112 |
FN1, fibronectin 1; S-FN1, stromal FN1; E-FN1, epithelial FN1.
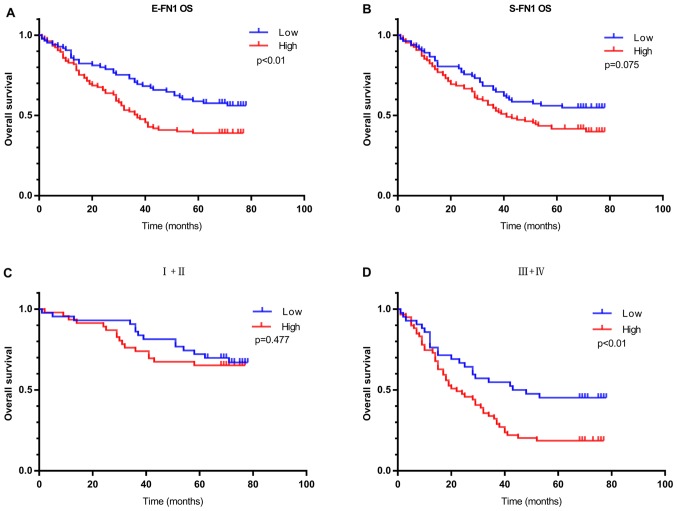
E-FN1-positive patients with GC in the IHC cohort exhibited worse OS compared with E-FN1-negative patients (P=0.009; Fig. 5A). S-FN1 expression exhibited no significant effect on OS (P=0.075, Fig. 5B). In addition, in patients with high clinical TNM stage (III + IV), E-FN1 positivity was strongly associated with OS; however, in patients with low clinical TNM stage (I + II), no difference was observed in overall survival between patients with low and high E-FN1 expression (Fig. 5C and D). E-FN1 was also confirmed as an independent predictor of overall survival in GC by multivariate analysis (HR, 2.115; 95% CI, 1.343–3.333; P=0.001; Table IV).
Figure 5.
The prognostic significance of FN1 in patients with gastric cancer based on immunohistochemistry. (A) Kaplan-Meier analysis of OS based on E-FN1 expression in 190 patients. (B) Kaplan-Meier analysis of OS based on S-FN1 expression in 190 patients. (C and D) Kaplan-Meier analysis of OS based on S-FN1 expression in patients with (C) TNM stage I+II (D) and III+IV gastric cancer. FN1, fibronectin 1; S-FN1, stromal FN1; E-FN1, epithelial FN1; TNM, Tumor-Node-Metastasis; OS, overall survival.
Table IV.
Multivariate analysis of overall survival in 190 patients with gastric cancer.
| 95.0% CI for HR | ||||
|---|---|---|---|---|
| Characteristic | HR | Lower | Upper | P-value |
| E-FN1 expression | 0.001b | |||
| Negative vs. positive | 2.115 | 1.343 | 3.333 | |
| Tumor size | 0.083 | |||
| <5 cm vs. ≥5 cm | 1.442 | 0.954 | 2.181 | |
| Tumor grade | 0.024a | |||
| Low + intermediate vs. high | 1.286 | 1.034 | 1.601 | |
| Depth of invasion | 0.028a | |||
| T1+T2 vs. T3+T4 | 2.352 | 1.097 | 5.043 | |
| TNM stage | 0.005b | |||
| I + II vs. III + IV | 2.124 | 1.258 | 3.585 | |
P<0.05
P<0.01, HR, hazard ratio; E-FN1, epithelial fibronectin 1; TNM, Tumor-Node-Metastasis.
Discussion
In this study, FN1 gene expression was analyzed in 17 independent GC cohorts. The results demonstrated an increase in FN1 expression in GC compared with normal tissues and a possible increase in the advanced T stage (T2+T3+T4) group compared with that in the early T stage (T1) group in one cohort; however, no association between FN1 expression levels and differentiation or clinical TNM stage was identified. In addition, upregulation of the FN1 gene may be a predictor of poor prognosis following radical gastrectomy for GC. In summary, the results of the present study support FN1 as a biomarker of poor prognosis in GC.
FN1, which is an extracellular matrix glycoprotein, is involved in cell proliferation, embryogenesis, wound healing, host defense, epithelial-mesenchymal transition (EMT) and metastasis, as well as oncogenic transformation (5). FN1 is involved in the occurrence and development of various tumors and is upregulated in multiple cancer types, such as esophageal squamous cell carcinoma, colorectal cancer, OSCC, and thyroid cancer (8–10,14). For instance, FN1 is upregulated in OSCC with lymph node metastasis (LNM); FN1 increases the expression of vascular endothelial growth factor C, lymphangiogenesis and LNM through FAK activation and promotes EMT in SAS human OSCC cells (37). FN1 is a key mediator of glioma progression, as its inhibition delays tumor progression and immunosuppression through a mechanism that involves the maintenance of integrin β1 FN receptors (38). In GC, FN1 is highly expressed in tumor tissues compared with that in non-tumor tissues, and knockdown of FN1 represses GC cell proliferation, adhesion and metastasis in vitro (15). The present study aimed to analyze the relationship between FN1 expression in GC and clinicopathological factors and prognoses.
The results of the present study demonstrated that the FN1 gene was upregulated in gastric cancer tissues compared with that in normal tissues in eight cohorts, and these data were confirmed by meta-analysis of combinations of all datasets. This result was consistent with the results of Xu et al (15) and Zhang et al (16), who used immunohistochemical methods to analyze tumor and normal tissue specimens from 40 and 52 patients with gastric cancer, respectively. In summary, previous studies have reported that the expression of the FN1 gene was increased in GC tissues compared with that in normal gastric tissues, but the studies were all small-scale. The present study used multiple cohorts to provide substantial validation of increased FN1 expression in GC.
To the best of our knowledge, the association between FN1 expression and clinicopathological features or patient prognosis, have not been reported previously. In the present study, compared with that in the early T stage group, the expression of FN1 was significantly increased in the advanced T stage group, which was further confirmed by meta-analysis in all the examined groups. OS analysis revealed that high FN1 expression was associated with unfavorable prognosis in four of the six cohorts containing prognostic information. A meta-analysis of all cohorts further validated this finding. These results indicated that the expression of FN1 may be a potential indicator of clinical outcomes in patients with GC.
FN1 is expressed in cancer cells and the intratumoral matrix in GC. Hanamura et al (39) reported that the expression of S-FN1 mRNA was positively correlated with deep invasion and LNM of colon cancer. Bae et al (34) reported that E-FN1-positive patients exhibited lower OS and disease-free survival compared with FN1-negative breast cancer patients. E-FN1 was an independent predictor for survival in breast cancer in multivariate analysis, but the expression of S-FN1 had no significant effect on patient survival (34). In the present study, E-FN1-positive patients with GC exhibited worse OS compared with E-FN1-negative patients, whereas S-FN1 expression had no significant effect on OS. In addition, in patients with high clinical TNM stage (III + IV), E-FN1 positivity was strongly associated with OS. FN1 was also confirmed as an independent predictor of overall survival in patients with GC by multivariate analysis.
Xu et al (15) and Zhang et al (16) demonstrated no FN1 expression in the stroma of gastric cancer. In the IHC cohort of the present study, FN1 was expressed in tumor cells and stromal cells, but not in normal epithelial cells. No association was observed between E-FN1 and S-FN1. E-FN1 expression in GC was significantly associated with tumor size. Soikkeli et al (40) reported that FN1 is required for tumor and stromal cell growth. It may be speculated in large tumors, the central region is likely to be necrotic, and the expression of FN1 may promote the migration of tumor cells and reduce necrosis.
In the present study, increased expression of the FN1 gene at the protein and mRNA level in GC tissues was observed; FN1 was highly expressed at the mRNA level in the advanced T stage group compared with that in the early T stage group, and the expression of FN1 at the protein level was positively associated with tumor size. In addition, FN1 expression at the protein and mRNA level was a predictor of poor prognosis following radical resection of GC. In conclusion, the expression of FN1 in GC tissues may be upregulated, and FN1 may be a biomarker of poor prognosis in patients with GC.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
YS wrote the manuscript and performed the majority of the experiments. CZ and YS participated in the study design, data acquisition and revision of the manuscript. YL and YH performed immunohistochemistry scoring, followed up the patients and collected clinical information. YY, HM and ZW analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients, and the study was approved by the Biomedical Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Patient consent for publication
Patients provided their consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Katai H, Ishikawa T, Akazawa K, Isobe Y, Miyashiro I, Oda I, Tsujitani S, Ono H, Tanabe S, Fukagawa T, et al. Five-year survival analysis of surgically resected gastric cancer cases in Japan: A retrospective analysis of more than 100,000 patients from the nationwide registry of the Japanese gastric cancer association (2001–2007) Gastric Cancer. 2018;21:144–154. doi: 10.1007/s10120-017-0716-7. [DOI] [PubMed] [Google Scholar]
- 4.Peng PL, Zhou XY, Yi GD, Chen PF, Wang F, Dong WG. Identification of a novel gene pairs signature in the prognosis of gastric cancer. Cancer Med. 2018;7:344–350. doi: 10.1002/cam4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 6.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275:21785–21788. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 7.Korah R, Boots M, Wieder R. Integrin alpha5beta1 promotes survival of growth-arrested breast cancer cells: An in vitro paradigm for breast cancer dormancy in bone marrow. Cancer Res. 2004;64:4514–4522. doi: 10.1158/0008-5472.CAN-03-3853. [DOI] [PubMed] [Google Scholar]
- 8.Xiao J, Yang W, Xu B, Zhu H, Zou J, Su C, Rong J, Wang T, Chen Z. Expression of fibronectin in esophageal squamous cell carcinoma and its role in migration. BMC Cancer. 2018;18:976. doi: 10.1186/s12885-018-4850-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X, Liu C, Zhang TN, Zhu YW, Dong X, Xue P. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J Cell Biochem. 2018;119:4717–4728. doi: 10.1002/jcb.26651. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa Y, Nakayama H, Nagata M, Yoshida R, Kawahara K, Hirosue A, Tanaka T, Yuno A, Matsuoka Y, Kojima T, et al. Overexpression of fibronectin confers cell adhesion-mediated drug resistance (CAM-DR) against 5-FU in oral squamous cell carcinoma cells. Int J Oncol. 2014;44:1376–1384. doi: 10.3892/ijo.2014.2265. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Deng L, Huang J, Cai R, Zhu X, Liu F, Wang Q, Zhang J, Zheng Y. High expression of fibronectin 1 suppresses apoptosis through the NF-κB pathway and is associated with migration in nasopharyngeal carcinoma. Am J Transl Res. 2017;9:4502–4511. [PMC free article] [PubMed] [Google Scholar]
- 12.Lou X, Han X, Jin C, Tian W, Yu W, Ding D, Cheng L, Huang B, Jiang H, Lin B. SOX2 targets fibronectin 1 to promote cell migration and invasion in ovarian cancer: New molecular leads for therapeutic intervention. OMICS. 2013;17:510–518. doi: 10.1089/omi.2013.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waalkes S, Atschekzei F, Kramer MW, Hennenlotter J, Vetter G, Becker JU, Stenzl A, Merseburger AS, Schrader AJ, Kuczyk MA, Serth J. Fibronectin 1 mRNA expression correlates with advanced disease in renal cancer. BMC Cancer. 2010;10:503. doi: 10.1186/1471-2407-10-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sponziello M, Rosignolo F, Celano M, Maggisano V, Pecce V, De Rose RF, Lombardo GE, Durante C, Filetti S, Damante G, et al. Fibronectin-1 expression is increased in aggressive thyroid cancer and favors the migration and invasion of cancer cells. Mol Cell Endocrinol. 2016;431:123–132. doi: 10.1016/j.mce.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Sun Z, Li Y, Fan D, Jiang H. MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomed Pharmacother. 2017;88:285–292. doi: 10.1016/j.biopha.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Chin-Sheng H, Kuo LJ, Wei PL, Lien YC, Lin FY, Liu HH, Ho YS, Wu CH, Chang YJ. NNK enhances cell migration through alpha7-nicotinic acetylcholine receptor accompanied by increased of fibronectin expression in gastric cancer. Ann Surg Oncol. 2012;19(Suppl):S580–S588. doi: 10.1245/s10434-011-2064-x. [DOI] [PubMed] [Google Scholar]
- 18.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 20.Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Michalowski A, Green JE. A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS One. 2011;6:e16694. doi: 10.1371/journal.pone.0016694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi CH, Ivanova T, Wu J, Lee M, Tan IB, Tao J, Ward L, Koo JH, Gopalakrishnan V, Zhu Y, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5:e1000676. doi: 10.1371/journal.pgen.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chia NY, Deng N, Das K, Huang D, Hu L, Zhu Y, Lim KH, Lee MH, Wu J, Sam XX, et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64:707–719. doi: 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- 23.Lei Z, Tan IB, Das K, Deng N, Zouridis H, Pattison S, Chua C, Feng Z, Guan YK, Ooi CH, et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–565. doi: 10.1053/j.gastro.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Muratani M, Deng N, Ooi WF, Lin SJ, Xing M, Xu C, Qamra A, Tay ST, Malik S, Wu J, et al. Nanoscale chromatin profiling of gastric adenocarcinoma reveals cancer-associated cryptic promoters and somatically acquired regulatory elements. Nat Commun. 2014;5:4361. doi: 10.1038/ncomms5361. [DOI] [PubMed] [Google Scholar]
- 25.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 26.He J, Jin Y, Chen Y, Yao HB, Xia YJ, Ma YY, Wang W, Shao QS. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther. 2016;9:6099–6109. doi: 10.2147/OTT.S110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao J, Deng NT, Ramnarayanan K, Huang B, Oh HK, Leong SH, Lim SS, Tan IB, Ooi CH, Wu J, et al. CD44-SLC1A2 gene fusions in gastric cancer. Sci Transl Med. 2011;3:77ra30. doi: 10.1126/scitranslmed.3001423. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Sohn I, Do IG, Kim KM, Park SH, Park JO, Park YS, Lim HY, Sohn TS, Bae JM, et al. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One. 2014;9:e90133. doi: 10.1371/journal.pone.0090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li WQ, Hu N, Burton VH, Yang HH, Su H, Conway CM, Wang L, Wang C, Ding T, Xu Y, et al. PLCE1 mRNA and protein expression and survival of patients with esophageal squamous cell carcinoma and gastric adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2014;23:1579–1588. doi: 10.1158/1055-9965.EPI-13-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 31.Wang G, Hu N, Yang HH, Wang L, Su H, Wang C, Clifford R, Dawsey EM, Li JM, Ding T, et al. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in china. PLoS One. 2013;8:e63826. doi: 10.1371/journal.pone.0063826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, Grabsch H, Ivanova T, Tan IB, Murray J, Ooi CH, Wright AI, West NP, Hutchins GG, Wu J, et al. Comprehensive genomic meta-analysis identifies intra-tumoural stroma as a predictor of survival in patients with gastric cancer. Gut. 2013;62:1100–1111. doi: 10.1136/gutjnl-2011-301373. [DOI] [PubMed] [Google Scholar]
- 33.Sung CO, Park CK, Kim SH. Classification of epithelial-mesenchymal transition phenotypes in esophageal squamous cell carcinoma is strongly associated with patient prognosis. Mod Pathol. 2011;24:1060–1068. doi: 10.1038/modpathol.2011.59. [DOI] [PubMed] [Google Scholar]
- 34.Bae YK, Kim A, Kim MK, Choi JE, Kang SH, Lee SJ. Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum Pathol. 2013;44:2028–2037. doi: 10.1016/j.humpath.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Sandsmark E, Andersen MK, Bofin AM, Bertilsson H, Drablos F, Bathen TF, Rye MB, Tessem MB. SFRP4 gene expression is increased in aggressive prostate cancer. Sci Rep. 2017;7:14276. doi: 10.1038/s41598-017-14622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 37.Morita Y, Hata K, Nakanishi M, Omata T, Morita N, Yura Y, Nishimura R, Yoneda T. Cellular fibronectin 1 promotes VEGF-C expression, lymphangiogenesis and lymph node metastasis associated with human oral squamous cell carcinoma. Clin Exp Metastasis. 2015;32:739–753. doi: 10.1007/s10585-015-9741-2. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta S, Nandi S, Hindi ES, Wainwright DA, Han Y, Lesniak MS. Short hairpin RNA-mediated fibronectin knockdown delays tumor growth in a mouse glioma model. Neoplasia. 2010;12:837–847. doi: 10.1593/neo.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanamura N, Yoshida T, Matsumoto E, Kawarada Y, Sakakura T. Expression of fibronectin and tenascin-C mRNA by myofibroblasts, vascular cells and epithelial cells in human colon adenomas and carcinomas. Int J Cancer. 1997;73:10–15. doi: 10.1002/(SICI)1097-0215(19970926)73:1<10::AID-IJC2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Soikkeli J, Podlasz P, Yin M, Nummela P, Jahkola T, Virolainen S, Krogerus L, Heikkila P, von Smitten K, Saksela O, Hölttä E. Metastatic outgrowth encompasses COL-I, FN1, and POSTN up-regulation and assembly to fibrillar networks regulating cell adhesion, migration, and growth. Am J Pathol. 2010;177:387–403. doi: 10.2353/ajpath.2010.090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.