Abstract
Dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein (DC-SIGNR) is a transmembrane receptor primarily involved in pathogen recognition by the innate immune system, with particular importance for viral recognition. DC-SIGNR may also be associated with tumorigenesis. The aim of the present study was to investigate the association between DC-SIGNR expression, development of hepatocellular carcinoma (HCC), and clinicopathological features. Immunohistochemistry was used to assess DC-SIGNR protein expression in HCC and paired non-cancerous tissue samples. DC-SIGNR expression was lower in HCC tissues compared with adjacent non-tumor tissue samples. The expression of DC-SIGNR was associated with small tumor size, low Edmondson grade and high patient long term survival rates. Bioinformatics analyses were performed on several datasets to assess the potential function of DC-SIGNR and related genes; the data revealed that DC-SIGNR mRNA expression was lower in HCC tissues compared with non-cancerous controls, and analyses of ten-year survival rates indicated patients with low DC-SIGNR expression exhibited shorter average survival times. In conclusion, decreased DC-SIGNR expression in HCC tissues may be a relevant predictive biomarker of clinical prognosis, in addition to being a viable therapeutic target for HCC treatment.
Keywords: dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein, hepatocellular carcinoma, immunohistochemistry, bioinformatics
Introduction
Hepatocellular carcinoma (HCC) is a severe form of cancer with extensive global morbidity and mortality. It is the fifth most common tumor type and the third leading source of cancer-related mortality, causing 500,000 deaths annually (1,2). Notably, more than 50% of global HCC morbidity arises in China (3,4). As it imposes a serious social and economic burden on society, it is vital that doctors identify liver cancer at an early stage, when treatment is less complicated and more likely to succeed. Identification of meaningful tumor biomarkers is a valuable approach to improving early diagnostic efforts.
Dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein (DC-SIGNR) is a type II transmembrane protein receptor that has been linked to the innate immune response against pathogens and tumors, which makes it a relevant target for cancer biomarker-related studies (5,6). DC-SIGNR is mainly expressed in liver sinuses, lymphoid tissues and placental capillaries (7,8). DC-SIGNR may therefore serve a specialized role in these tissues.
Previous studies on DC-SIGNR in the context of oncology have yielded new insights. For example, DC-SIGNR expression levels are altered in certain tumor types or blood samples from cancer patients; Jiang et al reported that secreted DC-SIGNR levels in patient's serum samples with colon cancer were significantly higher compared with healthy controls; however no detectable upregulation was observed in DC-SIGNR expression in tumor cells relative to normal tissue (9). By contrast, Liu et al reported that serum DC-SIGNR levels in patients with lung cancer were lower compared with healthy controls, whereas in a subset of patients with brain metastases, serum DC-SIGNR levels were higher compared with patients without metastases (10). Another study identified significant increases in serum DC-SIGNR in patients with gastric cancer relative to healthy controls (11).
Considering the limited number of studies assessing DC-SIGNR expression in HCC tissues, the aim of the present study was to compare the levels of this protein in tumor tissue samples and in adjacent non-cancerous tissues from patients with HCC. A combination of immunohistochemical and bioinformatics analyses was used to evaluate DC-SIGNR expression in HCC and characterize its potential functions.
Materials and methods
Patients and samples
A total of 267 HCC samples and 166 adjacent non-tumor liver tissue samples were collected from patients who underwent surgery at Zhejiang Provincial People's Hospital (Hangzhou, China) between January 2010 and December 2017. The tissues were confirmed to be cancerous or non-cancerous by hospital pathologists, fixed with 4% formalin for 24 h at room temperature and embedded in paraffin. Information on patient sex, age, tumor size, number, location, Edmondson grade and site of tumor metastasis was collected during patient hospitalization and treatment. Because some clinical data were missing or unavailable, the total number of some clinical indicators was <267. Overall survival (OS) was determined using either the date of patients' death or the last follow-up time point. This study was approved by the Review Board of the Hospital Ethics Committee, and written informed consent was obtained from each participant prior to data collection.
Immunohistochemical staining
Paraffin-embedded specimens were used to design three microarrays with the help of the Shanghai BioChip Co., Ltd. (Shanghai, China). Immunohistochemistry (IHC) was performed using the following protocol: Three tissue section microarrays (5 µm) were heated at 70°C for 2 h, washed three times in a xylene solution to remove paraffin, rehydrated in decreasing concentrations of ethanol (100, 95, 85 and 75%; each for 5 min), and boiled in Tris-EDTA (TE) buffer (Tris, 1.21 g/l; EDTA, 0.37 g/l; Tween-20, 0.5 ml/l) under high pressure (103 kPa) for 3 min to facilitate antigen retrieval. The samples were incubated with 3% hydrogen peroxide for 15 min to prevent endogenous peroxidase activity, blocked with 10% goat non-immune serum (reagent A; Histostain®-plus Bulk kit; Thermo Fisher Scientific, Inc., Waltham, MA, USA) for 20 min to reduce non-specific binding and incubated with anti-DC-SIGNR antibody (1:400, cat. no. ab169783; Human CD299 Antibody; Abcam, Cambridge, UK) overnight at 4°C. Sections were washed and incubated with a biotinylated secondary antibody (reagent B; Histostain®-plus Bulk kit; Thermo Fisher Scientific, Inc.) for 15 min. Samples were exposed to streptavidin-peroxidase (reagent C; Histostain®-plus Bulk kit; Thermo Fisher Scientific, Inc.) for an additional 15 min and a chromogenic reaction was performed using the 3,3′-diaminobenzidine color substrate solution (OriGene Technologies, Inc.; Beijing, China) according to the manufacturer's protocol. Color development was terminated when a brown signal indicative of staining was evident in the sample. Hematoxylin (cat. no. C0107; Beyotime Institute of Biotechnology, Haimen, China) staining was performed for 3–5 min. To finalize the staining process, samples were dehydrated in increasing concentrations of ethyl alcohol (75, 85, 95, and 100%) for 5 min each, soaked three times in xylene for 10 min and mounted with a gelatin resin. All procedures were performed at room temperature unless otherwise specified.
Immunohistochemical staining evaluation
Immunohistochemically stained samples were independently evaluated by two pathologists from the Department of Pathology of the Zhejiang Provincial People's Hospital based on the intensity and proportion of stained cells. The intensity of stained cells was scored from 0–3 as follows: 0, no staining; 1, weak staining; 2, medium staining; 3, strong staining. The proportion of stained cells were scored from 0–4: 0, no cells stained; 1, 1–25% cells stained; 2, 26–50% cells stained; 3, 51–75% cells stained; 4, >75% cells stained. The intensity and frequency scores were multiplied to produce an overall score used to confirm the level of DC-SIGNR expression in HCC tissues; tissues scoring <6 were defined as having low expression, whereas those with scores ≥6 were defined as having high expression.
Bioinformatics analysis of online databases
Using the Oncomine databases (www.oncomine.org), DC-SIGNR mRNA levels in HCC and normal tissues were compared. The analysis was performed under the following filtering conditions: Gene, DC-SIGNR; analysis type, cancer versus normal analysis; cancer type, hepatocellular carcinoma; data type, mRNA. Co-expression was also analyzed in the top four datasets using the screening conditions: Gene, DC-SIGNR; analysis type, co-expression analysis; cancer type, liver cancer. Ten-year survival analyses of patients with specific DC-SIGNR levels were performed using the Kaplan-Meier Plotter (http://kmplot.com). Biological process and signaling pathway analysis and annotation for DC-SIGNR were performed using the STRING data platform (https://string-db.org).
Statistical analysis
SPSS 16.0 software (SPSS, Inc.) was used for all statistical analyses. χ2 test was performed to assess the association between DC-SIGNR expression and different clinicopathological parameters. OS curves were generated using the Kaplan-Meier method, with the P-value derived from the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics of patients
HCC samples from 267 patients, including 219 male and 48 female patients between 25 and 90 years old (mean age, 58±11.5 years) were analyzed. At the time of the last follow-up visit, 103 patients remained alive and 65 were deceased; the status of the remaining patients was unavailable (Table I).
Table I.
Expression of DC-SIGNR in hepatocellular carcinoma tissue samples.
| DC-SIGNR expression | ||||
|---|---|---|---|---|
| Clinical parameters | Number | Low | High | P-value |
| Sex | 0.454 | |||
| Male | 219 | 164 | 55 | |
| Female | 48 | 37 | 11 | |
| Age (years) | 0.338 | |||
| <55 | 105 | 81 | 24 | |
| ≥55 | 162 | 120 | 42 | |
| Location | 0.255 | |||
| Left | 51 | 34 | 17 | |
| Right | 153 | 119 | 34 | |
| Left + right | 15 | 12 | 3 | |
| Tumor size (cm) | 0.012 | |||
| <5 | 144 | 100 | 44 | |
| ≥5 | 118 | 97 | 21 | |
| Tumor number | 0.384 | |||
| Single | 217 | 162 | 55 | |
| Multiple | 50 | 39 | 11 | |
| Edmondson grade | 0.001 | |||
| I+II | 157 | 106 | 51 | |
| III | 109 | 94 | 15 | |
| Metastasis | 0.074 | |||
| M0 | 241 | 179 | 62 | |
| M1 | 21 | 19 | 2 | |
| Microvascular invasion | 0.523 | |||
| Absent | 99 | 77 | 22 | |
| Present | 102 | 80 | 22 | |
| Cirrhosis | 0.246 | |||
| Negative | 84 | 66 | 18 | |
| Positive | 183 | 135 | 48 | |
| Hepatitis B antigen | 0.479 | |||
| Negative | 51 | 39 | 12 | |
| Positive | 210 | 157 | 53 | |
| AFP | 0.280 | |||
| <50 | 115 | 82 | 33 | |
| ≥50 | 103 | 78 | 25 | |
| Status | 0.031 | |||
| Deceased | 65 | 56 | 9 | |
| Alive | 103 | 75 | 28 | |
AFP, α-fetoprotein; DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein.
DC-SIGNR expression in HCC tissues
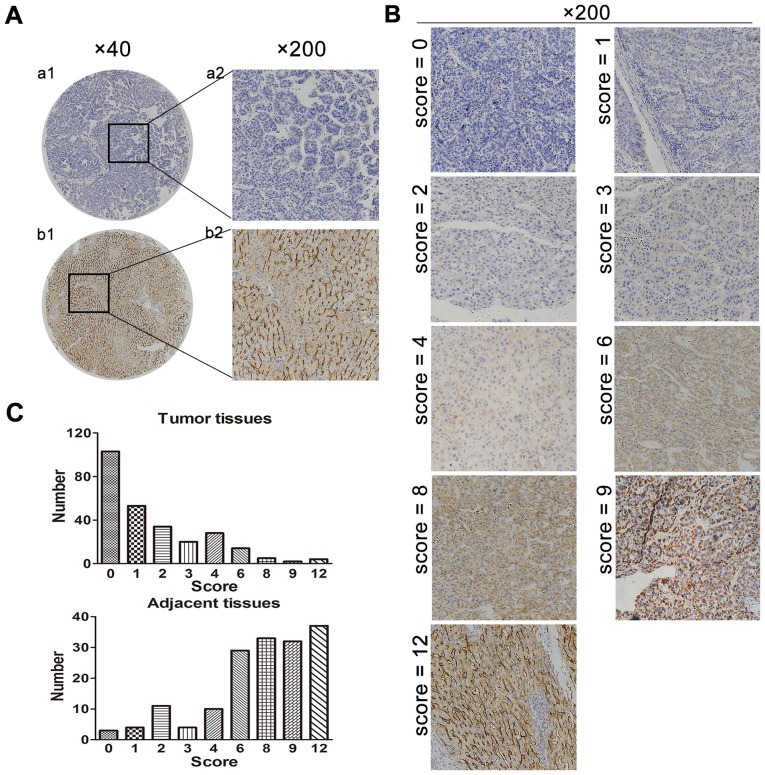
DC-SIGNR expression was analyzed by IHC in 267 HCC specimens and 166 adjacent non-cancerous tissues. There were 201/267 (75.3%) HCC tissue samples and 31/166 (18.7%) adjacent non-cancerous samples with low expression of DC-SIGNR. DC-SIGNR expression in HCC tissues was significantly lower compared with expression in adjacent non-cancerous tissue (P<0.001; Table II; Fig. 1A and B). The distribution of DC-SIGNR staining scores in the samples demonstrated a higher incidence of low scores in tumor tissues compared with adjacent non-cancerous tissues (Fig. 1C).
Table II.
Low-expression of DC-SIGNR in HCC.
| DC-SIGNR expression | ||||
|---|---|---|---|---|
| Tissue type | Number | Low | High | P-value |
| HCC | 267 | 201 | 66 | <0.001 |
| Adjacent tissue | 166 | 31 | 135 | |
| Average survival months | 26.79 | 36.70 | ||
DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein; HCC, hepatocellular carcinoma.
Figure 1.
DC-SIGNR expression in tumor and adjacent non-cancerous tissues. (A) The expression of DC-SIGNR is low in a tumor specimen (a1 and a2) and high in paired adjacent tissue (b1 and b2). (B) Immunohistochemical staining of DC-SIGNR expression in HCC samples and adjacent normal tissues ranked by staining scores. (C) DC-SIGNR expression score values in HCC tissues and adjacent healthy tissues. DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein; HCC, hepatocellular carcinoma.
Association between DC-SIGNR expression and clinicopathological features
Tumor size was identified as significantly associated with low levels of DC-SIGNR expression (P=0.012; Table I); low DC-SIGNR expression was also associated with Edmondson grade (P=0.001; Table I). Sex, age, tumor location, metastasis, microvascular invasion, cirrhosis, α-fetoprotein (AFP) concentration, and hepatitis B infection were not associated with DC-SIGNR expression (Table I).
Association between DC-SIGNR expression and clinical outcome in patients
To evaluate the association between DC-SIGNR expression and patient survival, a Kaplan-Meier curve of OS was generated. The five-year survival rate differed significantly between patients with high and low DC-SIGNR expression (P=0.026; Fig. 2). Patients with low DC-SIGNR expression survived for an average of 26.79 months, whereas those with high expression survived for an average of 36.70 months (Table II). Therefore, the data suggested that patients with HCC who exhibited low levels of DC-SIGNR expression were likely to survive for a shorter period of time.
Figure 2.
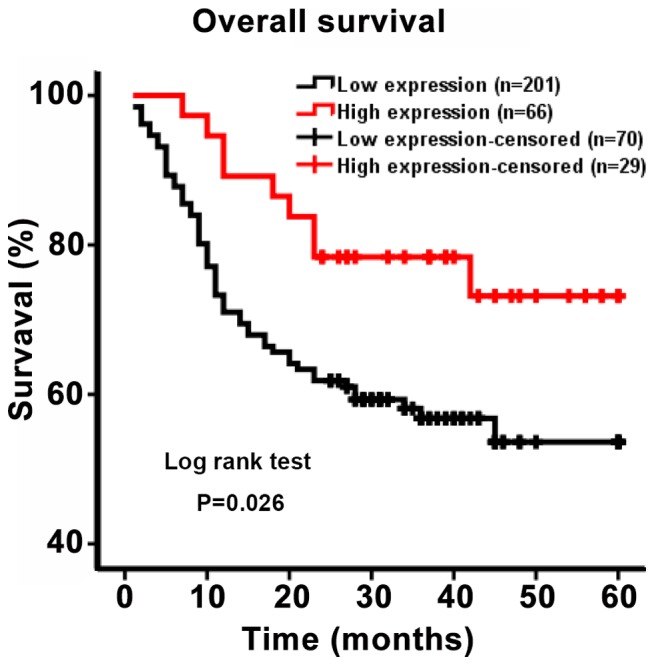
Kaplan-Meier survival curves of patients with HCC based on DC-SIGNR expression levels. Patients with low DC-SIGNR expression levels had shorter survival time compared with patients with high DC-SIGNR expression. DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein; HCC, hepatocellular carcinoma.
Bioinformatics analysis for DC-SIGNR expression in patients with HCC
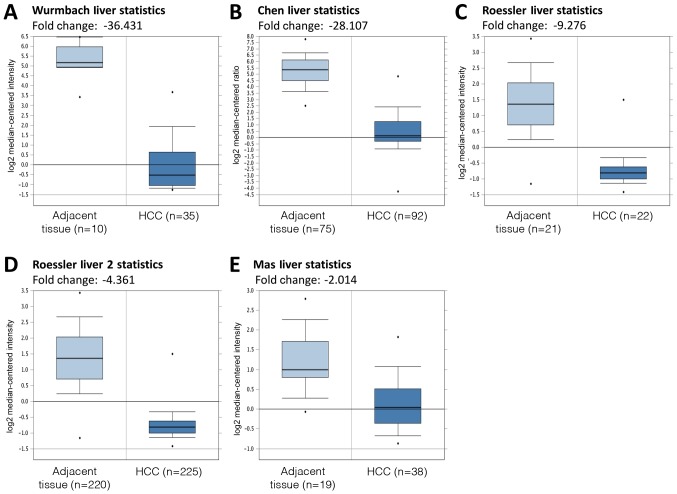
DC-SIGNR mRNA expression levels in HCC and adjacent non-cancerous tissues were compared using the Oncomine databases. Five datasets were selected for analysis: Wurmbach Liver Statistics (12), Chen Liver Statistics (13), Roessler Liver Statistics, Roessler Liver 2 Statistics (14) and Mas Liver Statistics (15). The results of this analysis revealed that DC-SIGNR mRNA expression was significantly lower in HCC tissues compared with adjacent tissues (P<0.05; Fig. 3).
Figure 3.
Analysis of DC-SIGNR gene expression in HCC and normal tissues using online data. (A-E) The expression level of DC-SIGNR mRNA was significantly lower in HCC in (A) Wurmbach Liver Statistics, (B) Chen Liver Statistics, (C) Roessler Liver Statistics, (D) Roessler Liver 2 Statistics and (E) Mas Liver Statistics datasets. The data were obtained from https://www.oncomine.org. DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein; HCC, hepatocellular carcinoma.
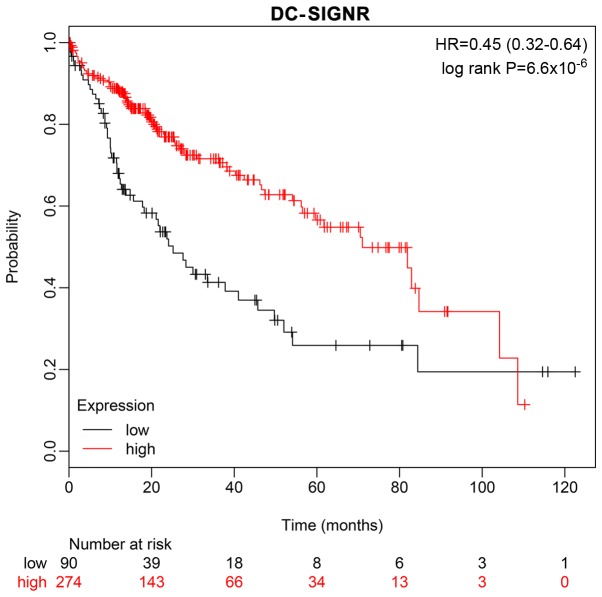
A ten-year survival rate prediction analysis was performed using the Kaplan-Meier Plotter database, which confirmed that low expression of DC-SIGNR was associated with shorter survival (Fig. 4).
Figure 4.
The ten-year survival rate of patients with HCC is associated with DC-SIGNR expression. Patients with low DC-SIGNR expression exhibited a lower ten-year survival rate compared with patients with high DC-SIGNR expression. DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein.
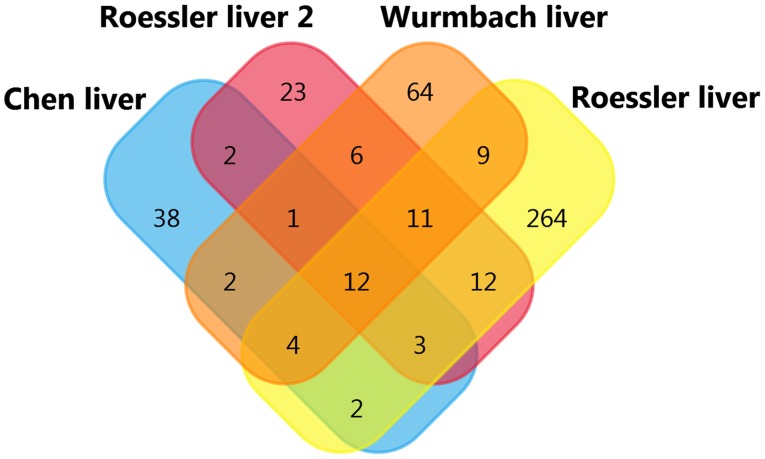
DC-SIGNR co-expression analysis was conducted using the Oncomine databases. The top four datasets were selected for analysis: Wurmbach Liver statistics (12), Roessler Liver Statistics (14), Chen Liver Statistics (13), and Roessler Liver 2 Statistics (14), with gene correlation values >0.50. A total of twelve co-expressed genes were found (Table III; Fig. 5). This indicates that the function or product of DC-SIGNR may be associated with these genes.
Table III.
DC-SIGNR co-expression genes with the cut-off for selection defined as an appearance in four datasets.
| Gene | Gene name | Number of appearances |
|---|---|---|
| CETP | Cholesteryl ester transfer protein | 4 |
| CLEC1B | C-type lectin domain family 1 member B | 4 |
| CRHBP | Corticotropin-releasing hormone-binding protein | 4 |
| DNASE1L3 | Deoxyribonuclease 1 like 3 | 4 |
| ECM1 | Extracellular matrix protein 1 | 4 |
| GPM6A | Glycoprotein M6A | 4 |
| MARCO | Macrophage receptor with collagenous structure | 4 |
| MT1E | Metallothionein 1E | 4 |
| MT1F | Metallothionein 1F | 4 |
| MT1H | Metallothionein 1H | 4 |
| MT1X | Metallothionein 1X | 4 |
| VIPR1 | Vasoactive intestinal peptide receptor 1 | 4 |
DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein.
Figure 5.
Gene co-expression analysis of DC-SIGNR in four databases. Four datasets were collected for gene co-expression analysis on Oncomine, and twelve genes were found. DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein.
Biological process and signaling pathway enrichment analyses related to DC-SIGNR were conducted using STRING. The results of these analyses indicated that DC-SIGNR expression was strongly associated with the immune response, as indicated by pathway ID, GO.0002376, GO.0006955, GO.0050776, GO.0002682, GO.0002684 GO.0045087, 4660, 4650 and 4662 (Tables IV and V).
Table IV.
Top 10 statistically meaningful biological processes associated with DC-SIGNR.
| Rank | Pathway ID | Pathway description | Matching proteins |
|---|---|---|---|
| 1 | GO.0002223 | Stimulatory C-type lectin receptor signaling pathway | DC-SIGN, HRAS, ICAM2, ICAM3, KRAS, NRAS, RAF1 |
| 2 | GO.0031347 | Regulation of defense response | DC-SIGN, HRAS, ICAM2, ICAM3, IL10, IL4, KRAS, NRAS, RAF1 |
| 3 | GO.0006952 | Defense response | DC-SIGN, CD83, HRAS, ICAM2, ICAM3, IL10, ITIH4, KRAS, NRAS, RAF1 |
| 4 | GO.0002376 | Immune system process | DC-SIGN, CD83, HRAS, ICAM2, ICAM3, IL10, IL4, KRAS, NRAS, RAF1 |
| 5 | GO.0006955 | Immune response | DC-SIGN, CD83, HRAS, ICAM2, ICAM3, IL10, KRAS, NRAS, RAF1 |
| 6 | GO.0050776 | Regulation of immune response | DC-SIGN, HRAS, ICAM2, ICAM3, IL10, KRAS, NRAS, RAF1 |
| 7 | GO.0002682 | Regulation of immune system processes | DC-SIGN, CD83, HRAS, ICAM2, ICAM3, IL10, KRAS, NRAS, RAF1 |
| 8 | GO.0002684 | Positive regulation of immune system processes | DC-SIGN, CD83, HRAS, ICAM2, ICAM3, KRAS, NRAS, RAF1 |
| 9 | GO.0045087 | Innate immune response | DC-SIGN, HRAS, ICAM2, ICAM3, IL4, KRAS, NRAS, RAF1 |
| 10 | GO.0000186 | Activation of MAPKK activity | HRAS, KRAS, NRAS, RAF1 |
DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein; MAPKK, mitogen-activated protein kinase kinase.
Table V.
Top 10 statistically meaningful signaling pathways associated with DC-SIGNR.
| Rank | Pathway ID | Pathway description | Matching proteins |
|---|---|---|---|
| 1 | 4660 | T cell receptor signaling pathway | HRAS, IL10, IL4, NRAS, RAF1 |
| 2 | 4664 | Fc ε receptor antibody signaling pathway | HRAS, IL4, NRAS, RAF1 |
| 3 | 4068 | FoxO signaling pathway | HRAS, IL10, NRAS, RAF1 |
| 4 | 4650 | Natural killer cell mediated cytotoxicity | HRAS, ICAM2, NRAS, RAF1 |
| 5 | 5219 | Bladder cancer | HRAS, NRAS, RAF1 |
| 6 | 4370 | Vascular endothelial growth factor signaling pathway | HRAS, NRAS, RAF1 |
| 7 | 4662 | B cell receptor signaling pathway | HRAS, NRAS, RAF1 |
| 8 | 4720 | Long-term potentiation | HRAS, NRAS, RAF1 |
| 9 | 4730 | Long-term depression | HRAS, NRAS, RAF1 |
| 10 | 4917 | Prolactin signaling pathway | HRAS, NRAS, RAF1 |
DC-SIGNR, dendritic cell-specific intercellular adhesion molecule-grabbing non-integrin-related protein.
Discussion
Liver tumor development is a complex process, with many distinct factors participating in tumorigenesis and progression, often through key changes in gene expression (16,17). To the best of our knowledge, DC-SIGNR expression in this context has not been extensively studied, and whether it is expressed in HCC tissues has not been clarified to date. To resolve this question, 267 HCC specimens were collected and the association between DC-SIGNR expression and clinical outcomes was evaluated.
Patients with low DC-SIGNR expression in tumor tissue were more likely to have larger tumors and a higher Edmondson grade. These results were similar to those reported by Liu et al, which revealed low DC-SIGNR expression in serum samples from patients with lung cancer (10). Similarly, Jiang et al reported that DC-SIGNR expression was not detectable in colon cancer foci or in matched normal tissues (9). Lymphoid tissues from patients with non-Hodgkin's lymphoma have also been demonstrated to be DC-SIGNR-negative by IHC (18). In contrast to these findings, other studies have demonstrated that DC-SIGNR is upregulated in gastric cancer and colon cancer liver metastases (11,19). These distinct findings indicated that DC-SIGNR expression may vary between tumor types, tissues samples, or at different stages of the disease.
DC-SIGN and DC-SIGNR are homologous genes involved in immune response (20,21). DC-SIGN is expressed on dendritic cells; it binds to the T cell receptor to mediate T cell activation (22). DC-SIGN is also expressed on macrophages and is involved in phagocytosis (23). Therefore, it may suggest that normal expression of DC-SIGNR on the surface of liver cells may be a required gene product for the removal of abnormal cells by signaling the mutation status of hepatocytes to immune cells including T cells, NK cells or macrophages; downregulation of this protein may facilitate tumor formation.
Based on the IHC and bioinformatics analyses of DC-SIGNR, DC-SIGNR downregulation in liver cells may not be essential for tumor physiology, as tumor location, number, metastasis, microvascular invasion and AFP were not associated with DC-SIGNR expression. Instead, DC-SIGNR downregulation may act by suppressing the local immune response to the tumor.
In conclusion, DC-SIGNR is an important marker of immunity in HCC tissues, and its downregulation is associated with reduced patient survival. However, multiple conclusions have been drawn on the role and significance of DC-SIGNR. Therefore, further study is required to examine its function in HCC, to guide HCC treatment and to facilitate predictions for long-term survival in patients with HCC.
Acknowledgements
Not applicable.
Funding
This work was supported by the grants from The National Science Foundation of China (grant. no. 81672474 to DSH), Funds of Science Technology Department of Zhejiang Province (grant no. 2015C0303 to DSH; grant no. LGF18H160024 to ZMH; grant no. LGF18H160025 to XLH) and Zhejiang Province Bureau of Health (grant nos. WKJ-ZJ-1812 and 2018ZZ002 to ZMH).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
HBX and HJW designed and conducted the experiments. SSS and JGZ were in charge of sample processing. XLH, ZMH and CWZ performed data analysis. DSH and XZM revised the manuscript critically for important intellectual content, participated in online data analysis and gave final approval of the version to be published. HBX reviewed and wrote the manuscript. All authors have read and approved of the final version of the manuscript.
Ethics approval and consent to participate
The research was approved by the Review Board of Hospital Ethics Committee, Zhejiang Provincial People's Hospital (Hangzhou, China), and written informed consent from was obtained each participant before data collection.
Patient consent for publication
Patients agreed that the researchers would use their clinical specimens and that the results would be published.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 3.Zuo T, Zheng R, Zeng H, Zhang S, Chen W. Analysis of liver cancer incidence and trends in China. Zhonghua Zhong Liu Za Zhi. 2015;37:691–696. (In Chinese) [PubMed] [Google Scholar]
- 4.Zhu Q, Qiao GL, Zeng XC, Li Y, Yan JJ, Duan R, Du ZY. Elevated expression of eukaryotic translation initiation factor 3H is associated with proliferation, invasion and tumorigenicity in human hepatocellular carcinoma. Oncotarget. 2016;7:49888–49901. doi: 10.18632/oncotarget.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary O, Kumar S, Bala M, Singh J, Hazarika A, Luthra K. Association of DC-SIGNR expression in peripheral blood mononuclear cells with DC-SIGNR genotypes in HIV-1 infection. Viral Immunol. 2015;28:472–475. doi: 10.1089/vim.2014.0148. [DOI] [PubMed] [Google Scholar]
- 7.Lalor PF, Lai WK, Curbishley SM, Shetty S, Adams DH. Human hepatic sinusoidal endothelial cells can be distinguished by expression of phenotypic markers related to their specialised functions in vivo. World J Gastroenterol. 2006;12:5429–5439. doi: 10.3748/wjg.v12.i34.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engering A, van Vliet SJ, Hebeda K, Jackson DG, Prevo R, Singh SK, Geijtenbeek TB, van Krieken H, van Kooyk Y. Dynamic populations of dendritic cell-specific ICAM-3 grabbing nonintegrin-positive immature dendritic cells and liver/lymph node-specific ICAM-3 grabbing nonintegrin-positive endothelial cells in the outer zones of the paracortex of human lymph nodes. Am J Pathol. 2004;164:1587–1595. doi: 10.1016/S0002-9440(10)63717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y, Zhang C, Chen K, Chen Z, Sun Z, Zhang Z, Ding D, Ren S, Zuo Y. The clinical significance of DC-SIGN and DC-SIGNR, which are novel markers expressed in human colon cancer. PLoS One. 2014;9:e114748. doi: 10.1371/journal.pone.0114748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Zhang H, Su L, Yang P, Xin Z, Zou J, Ren S, Zuo Y. Low expression of dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin-related protein in lung cancer and significant correlations with brain metastasis and natural killer cells. Mol Cell Biochem. 2015;407:151–160. doi: 10.1007/s11010-015-2465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang Q, Zhang M, Yuan M, Wang Z, Zhang J, Zhou X, Zhang Y, Lin F, Na H, et al. DC-SIGNR by influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4 in gastric cancer liver metastasis. Mol Cancer. 2017;16:78. doi: 10.1186/s12943-017-0639-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P, Fisher R. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu AK. DNA damage, mutagenesis and cancer. Int J Mol Sci. 2018;19:970. doi: 10.3390/ijms19061767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falco M, Palma G, Rea D, De Biase D, Scala S, D'Aiuto M, Facchini G, Perdonà S, Barbieri A, Arra C. Tumour biomarkers: Homeostasis as a novel prognostic indicator. Open Biol. 2016;6:160254. doi: 10.1098/rsob.160254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Chen K, Yan L, Yang Z, Zhu Z, Chen C, Zeng J, Wei W, Qi X, Ren S, Zuo Y. Low expression of dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin-related protein in non-Hodgkin lymphoma and significant correlations with lactic acid dehydrogenase and β2-microglobulin. Biochem Cell Biol. 2013;91:214–220. doi: 10.1139/bcb-2012-0110. [DOI] [PubMed] [Google Scholar]
- 19.Na H, Liu X, Li X, Zhang X, Wang Y, Wang Z, Yuan M, Zhang Y, Ren S, Zuo Y. Novel roles of DC-SIGNR in colon cancer cell adhesion, migration, invasion, and liver metastasis. J Hematol Oncol. 2017;10:28. doi: 10.1186/s13045-016-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox N, Pilling D, Gomer RH. DC-SIGN activation mediates the differential effects of SAP and CRP on the innate immune system and inhibits fibrosis in mice. Proc Natl Acad Sci USA. 2015;112:8385–8390. doi: 10.1073/pnas.1500956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soilleux EJ. DC-SIGN (dendritic cell-specific ICAM-grabbing non-integrin) and DC-SIGN-related (DC-SIGNR): Friend or foe? Clin Sci (Lond) 2003;104:437–446. doi: 10.1042/CS20020092. [DOI] [PubMed] [Google Scholar]
- 22.Gringhuis SI, Kaptein TM, Wevers BA, Mesman AW, Geijtenbeek TB. Fucose-specific DC-SIGN signalling directs T helper cell type-2 responses via IKKe- and CYLD-dependent Bcl3 activation. Nat Commun. 2014;5:3898. doi: 10.1038/ncomms4898. [DOI] [PubMed] [Google Scholar]
- 23.Yang K, Liu X, Liu Y, Wang X, Cao L, Zhang X, Xu C, Shen W, Zhou T. DC-SIGN and Toll-like receptor 4 mediate oxidized low-density lipoprotein-induced inflammatory responses in macrophages. Sci Rep. 2017;7:3296. doi: 10.1038/s41598-017-03740-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.