Abstract
Retinoblastoma (RB) is the most prevalent childhood intraocular cancer type. Previous studies have demonstrated that c-myc (a proto-oncogene) is associated with tumorigenesis. However, at present, the influence of the expression profile and bioactivity of c-Myc on RB occurrence and progression is yet to be characterised. Notably, the present study demonstrated that c-myc is downregulated in the RB cell line WERI-Rb1. However, treatment with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) was revealed to significantly upregulate the expression of c-Myc mRNA and protein in WERI-Rb1 cells. Moreover, TSA increased the activity of the c-myc promoter in WERI-Rb1 cells, and the expression of c-Myc was also regulated by other HDAC inhibitors, including vorinostat (SAHA), valproic acid sodium salt (VPA) and entinostat. Notably, although c-myc was silenced in the Y79 cell line, the HDAC inhibitor TSA did not induce upregulation of mRNA and protein in Y79 cells. By contrast, certain HDAC inhibitors (TSA, VPA and SAHA) were discovered to significantly decrease the activity of the c-myc promoter in Y79 cells. Furthermore, the current data indicated that exogenous c-myc expression has a mild inhibitory effect on WERI-Rb1 and Y79 cell viability. Therefore, the present study revealed novel insights into the expression mechanism and bioactivity of c-Myc in RB cells.
Keywords: c-Myc, histone deacetylase inhibitor, retinoblastoma, WERI-Rb1 cells, Y79 cells
Introduction
Retinoblastoma (RB) is the most prevalent intraocular cancer that affects children (1). It occurs worldwide, with an incidence of one case per 15,000-20,000 live births (2) and an estimated mortality rate of 50% (3). Therefore, elucidation of the characteristics of RB is necessary in order to identify more effective therapeutic strategies. Although the most significant event contributing to the oncogenesis of RB is the inactivation of the two RB alleles on chromosome 13 (4), other oncogenes may also contribute to RB tumorigenesis.
The c-myc proto-oncogene belongs to the MYC family (5). Expression of c-myc or its protein product c-Myc is upregulated in the majority of malignant tumour types, including lymphoma, neuroblastoma, melanoma, breast, ovarian, prostate and liver cancer (6–9). c-Myc upregulation in tumours may result from gene amplification, increased c-myc transcription, or an increase in c-Myc protein stability and activity via post-translational regulation (10). Thus, it has been hypothesized that the oncogenicity of c-myc is dependent on elevated expression levels. However, the expression level of c-Myc in human cancer types ranges from lower than average to greatly elevated (11), and it is differentially expressed depending on the cell type. The expression level of c-Myc in RB is yet to be identified, to the best of our knowledge.
Additionally, it has been determined that c-Myc is regulated via different pathways in different cell lines. Histone acylation and DNA methylation are involved in the transcriptional regulation of c-myc. For example, c-myc is downregulated by the demethylating reagent 5-azacytidine in human prostate cancer cells (12,13), whereas 5-aza-deoxycytidine induces the upregulation of c-myc in lung cancer cells (14). Moreover, c-myc expression is regulated via histone deacetylation in human cervical cancer cells (15). Nonetheless, whether c-myc is regulated via histone acylation or DNA methylation in RB cells has not yet been elucidated.
Furthermore, c-Myc is a pleiotropic transcription factor that binds to the promoters, and regulates the expression, of a large number of genes regulating metabolic processes, macromolecular synthesis, the cell cycle and apoptosis (16). In a similar manner to the majority of oncoproteins, c-Myc enhances cell proliferation and regulates cell cycle (17). In both healthy and tumorous cells, MYC-dependent signalling is an important regulator of cell cycle progression from the G1 to S phases (18), and inactivation of c-Myc expression results in tumour regression accompanied by apoptosis, differentiation or tumour dormancy (19). However, unlike most oncoproteins, c-Myc also significantly enhances certain mechanisms of programmed cell death (PCD), including senescence and apoptosis (20). Therefore, under conditions of limited energy sources, downregulation of c-Myc may represent a survival strategy enabling cancer cell proliferation (21). The conflicting roles discovered indicate a complex role served by c-Myc, which varies depending on cancer cell type. Thus, investigation of the bioactivity of c-Myc may greatly improve the present understanding of RB pathophysiology.
Based on the aforementioned findings, the present study sought to determine the expression profile and bioactivity of c-Myc in RB cells. It was discovered that c-Myc was downregulated in the RB cell lines WERI-Rb1 and Y79. Moreover, the expression of c-Myc was significantly upregulated following cell treatment with HDAC inhibitors, such as trichostatin A (TSA), vorinostat (SAHA) and entinostat (MS-275). The activity of the c-myc promoter was significantly increased following TSA treatment in WERI-Rb1 cells. However, the low level of c-Myc expression in Y79 cells was not upregulated by the HDAC inhibitors. Furthermore, exogenous c-myc significantly reduced the viability of both WERI-Rb1 and Y79 cells. Therefore, the present data provide new insights into the c-Myc expression mechanism and its bioactivity in RB cells.
Materials and methods
Cell culture and transfection
Human retinoblastoma cell lines WERI-Rb1 and Y79 [both American Type Culture Collection (ATCC)], and the human colon cancer cell line RKO (ATCC), were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) and supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (both Gibco; Thermo Fisher Scientific, Inc.), in a humidified 5% CO2 incubator at 37°C. The cells selected for the assays were collected during the exponential growth phase. TSA was obtained from Sigma-Aldrich; Merck KGaA, and SAHA, MS-275 and VPA were obtained from Selleck Chemicals. WERI-Rb1 cells and Y79 cells were seeded at a density of 1×106 cells per well in a 6 well plate and were stably transfected with a plasmid expressing c-Myc or an empty vector control (pMXs-c-Myc or vector; Addgene, Inc.), using Lipofectamine® (Invitrogen; Thermo Fisher Scientific Inc.) in Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The plasmid or vector was used at a concentration of 0.8 µg/well. Medium was changed to complete growth medium (10% FBS) after 4 h.
Reverse-Transcription (RT) PCR
Total RNA was extracted from WERI-Rb1 and Y79 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). One microgram of total RNA was subjected to reverse transcription with the PrimeScriptTM RT-PCR kit (Takara Bio, Inc.) according to the manufacture's protocol. Reverse transcription was performed at 37°C for 15 min and 85°C for 5 sec. The PCR reaction was carried out using a My Cycler thermal cycler (Bio-Rad Laboratories, Inc.) with TaKaRa Premix Taq® Version 2.0 (Takara Bio, Inc.). For c-Myc, PCR reaction was performed for 35 cycles each at 98°C for 10 sec, 60°C for 30 sec, 72°C for 30 sec and a final extension at 72°C for 10 min. PCR reaction for β-actin was performed for 20 cycles, with the same temperature and time parameters as for c-Myc. The primer sequences are presented in Table SI.
Reverse transcription-quantitative (RT-q)PCR
The mRNA expression levels of histone deacetylases (HDACs) in WERI-Rb1 cells were identified using a Roche 480 system (Roche Diagnostics) and assays-on-demand primers (Invitrogen; Thermo Fisher Scientific, Inc.) for HDACs. Total RNA was extracted from WERI-Rb1 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). One microgram of total RNA was subjected to reverse transcription with the PrimeScriptTM RT-PCR kit (Takara Bio, Inc.) according to the manufacture's protocol. Reverse transcription was performed at 37°C for 15 min and 85°C for 5 sec. qPCR was then performed using the SYBR® Prime Script TM RT-PCR kit (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. qPCR was conducted to determine the expression levels of HDACs using the Roche 480 system (Roche Diagnostics). PCR was performed as follows: 94°C for 5 min, followed by 40 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec. Relative target gene expression levels were quantified using the 2−ΔΔCq method and normalized to the internal reference gene, β-actin (22). The data are presented as the inverse of the normalized Cq value (InvCq), or as the relative fold change compared with the untreated control. The primer sequences are presented in Table SI.
Western blot analysis
Total proteins were extracted following cell lysis using RIPA buffer (Beyotime Institute of Biotechnology) at 4°C. Protein concentration was determined using bicinchoninic acid method (Beyotime Institute of Biotechnology). Western blotting was performed according to standard protocols. Proteins (30 µg/well) were separated by 8% SDS-PAGE and transferred onto polyvinylidene difluoride membrane. The membranes were blocked with 5% non-fat milk dissolved in TBS containing 0.1% Tween-20 (TBST) for 1 h at room temperature, and incubated with primary antibodies diluted in milk dissolved in TBST at 4°C overnight. The following primary antibodies were used: Rabbit anti-c-Myc (1:500; cat. no. ab32072; Abcam) and rabbit anti-GAPDH (1:20,000; cat. no. 10494-1-AP; ProteinTech Group, Inc.). Proteins were visualized using horseradish peroxidase-conjugated anti-rabbit secondary antibodies (1:10,000; cat. no. 7074; Cell Signaling Technology, Inc.) incubated for 1 h at room temperature followed by use of the ECL system (Merck KGaA). Densitometric analysis was performed on the western blotting data using computerized image analysis and software (Gel-Pro Analyzer software v. 6.0; Media Cybernetics, Inc.).
Immunofluorescence assay
WERI-Rb1 or RKO cells were fixed in ice-cold 95% methanol at −20°C for 20 min. Then the fixed cells were incubated with 0.1% Triton X-100 for 10 min, and blocked with 10% normal goat serum (Beyotime Institute of Biotechnology) at room temperature for 30 min. Cells were stained with rabbit anti-c-Myc (1:100; cat. no. ab32072; Abcam) at 4°C overnight. Secondary anti-rabbit antibody (1:500; cat. no. 4413S; Cell Signaling Technology, Inc.) was added at room temperature for 1 h, and the nuclei were stained using DAPI at room temperature for 10 min. Images were acquired by fluorescence microscopy (magnification, ×100; Leica Microsystems GmbH).
Cell Counting Kit (CCK)-8 assay
The viability of WERI-Rb1 and Y79 cells was assessed using a CCK-8 assay (Dojindo Molecular Technologies, Inc.) according to the manufacture's protocol. The CCK-8 reagent was added to each well and cells were incubated for 2 h at 37°C. The absorbance (optical density) at 450 nm was measured.
Luciferase assay
A plasmid encoding the human c-Myc promoter upstream of firefly luciferase (pDEL1) was obtained from Addgene, Inc., and a pGL2-Control was purchased from Promega Corporation. WERI-Rb1 cells (1×106 cells in 60 mm dishes) were transfected using Lipofectamine® (Invitrogen; Thermo Fisher Scientific Inc.) according to the manufacturer's instructions. The transfected plasmids contained 2 µg expression plasmid or pGL2-Control, and 100 ng Renilla luciferase reporter plasmid, pCMV-RL (Promega Corporation). The pCMV-RL plasmid encoding Renilla luciferase was included in all samples to monitor transfection efficiency. At 24 h following initial transfection, WERI-Rb1 and Y79 cells were further treated with TSA, SAHA, MS-275 or VPA for another 24 h. Subsequently, the cells were harvested and a Dual-Luciferase reporter assay system (Promega Corporation) was performed to identify the sequential measurements of the firefly and Renilla luciferase activities. The luciferase activities and calculation of the relative ratios were quantified using a Turner Designs Luminometer Model TD-20/20 (TD-20/20; Turner Designs, Inc.). The levels of firefly luciferase activity were normalized against Renilla luciferase activity.
Statistical analysis
The data shown are representative of three independent experiments with each experiment performed in triplicate. The data are expressed as the mean ± SD. Statistical analyses were performed using the SPSS for Windows software package, version 10.5 (SPSS, Inc.). The differences between mean values were evaluated using either the Student's two-tailed t-test (for comparisons between 2 groups) or ANOVA followed by Tamhane T2 post hoc test (for comparisons of mRNA expression levels between different HDACs in WERI-Rb1 cells). P<0.05 was considered to indicate a statistically significant difference.
Results
c-Myc is downregulated in WERI-Rb1 cells
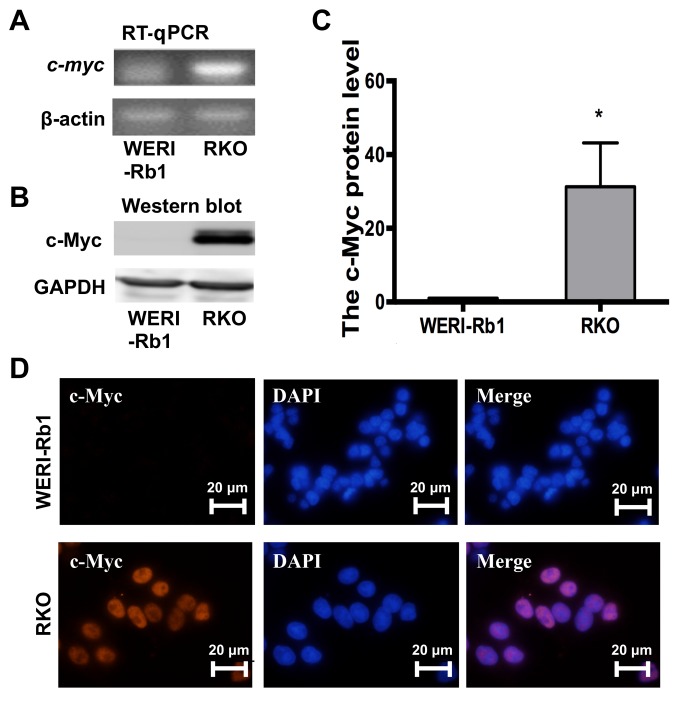
c-myc is typically reported as an oncogene with high expression levels in a variety of cancer types, including glioma, colon and gastric cancer (6–9), though its expression level in RB is yet to be reported. To investigate the influence of c-Myc activity on RB progression, a quantitative examination of c-myc expression in WERI-Rb1 cells was performed; the RT-PCR results indicated that c-myc was almost undetectable in WERI-Rb1 cells but significantly upregulated in RKO colon cancer cells (Fig. 1A). Western blotting further supported this result (Fig. 1B and C; c-Myc protein expression; WERI-Rb1 cells, 1; RKO cells, 31.26±11.92; P<0.05). Moreover, as exhibited in Fig. 1D, nuclear staining of c-Myc (red) was observed in RKO cells but not WERI-Rb1 cells. Thus, the current results indicated that c-myc was not expressed in WERI-Rb1 cells.
Figure 1.
c-Myc is downregulated in WERI-Rb1 cells. c-Myc was not detectable in WERI-Rb1 compared with RKO cells according to (A) RT-PCR and (B) western blot assays. (C) Relative expression levels of the c-Myc protein in WERI-Rb1 and RKO cells were quantified using densitometry and the data are presented as histograms. (D) Cells were stained for c-Myc (red). Immunofluorescence showed that compared with the positive control (RKO cells), c-Myc (red) was not expressed in WERI-Rb1 cells. Magnification, ×100. All experiments were performed in triplicate. *P<0.05 vs. WERI-Rb1. RT-PCR, reverse transcription PCR.
c-Myc is upregulated in WERI-Rb1 cells following treatment with the HDAC inhibitor TSA
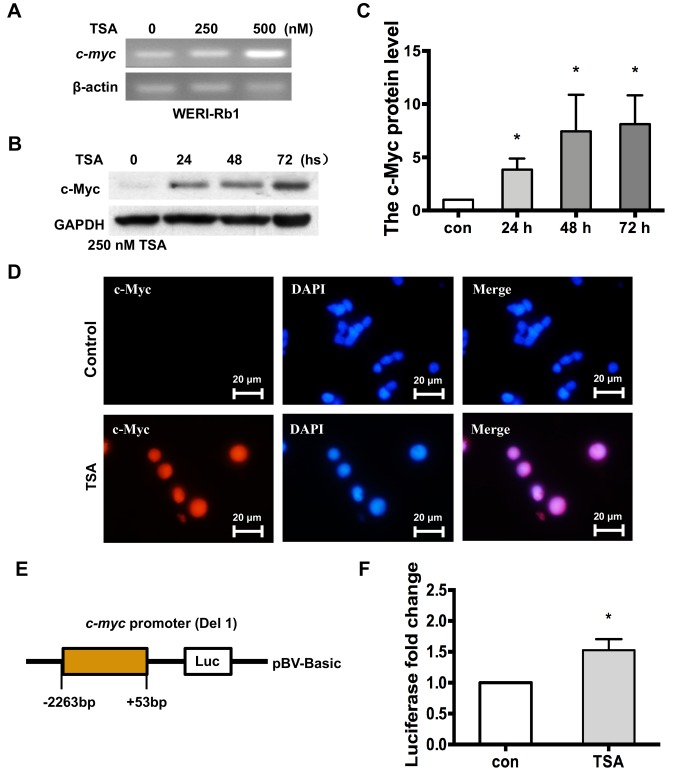
To determine the influence of epigenetic modifications on the silencing of the c-myc gene, WERI-Rb1 cells were treated with the HDAC inhibitor TSA. Fig. 2A indicates that c-myc expression was increased in WERI-Rb1 cells treated with 250 or 500 nM TSA, according to the results of the RT-PCR analysis. Western blotting was performed to further validate the aforementioned results, and it was revealed that the level of c-Myc protein expression was significantly increased in WERI-Rb1 cells treated with 250 nM TSA for 24, 48 and 72 h, compared with the control group (Fig. 2B and C; c-Myc relative protein expression: Control, 1; 24 h, 3.84±1.06; 48 h, 7.44±3.43; 72 h, 8.12±2.69; P<0.05). Furthermore, nuclear staining of c-Myc (red) was observed in WERI-Rb1 cells treated with TSA, but not in the untreated cells (Fig. 2D).
Figure 2.
c-myc is transcriptionally upregulated in WERI-Rb1 cells following treatment with the histone deacetylase inhibitor TSA. (A) RT-PCR analysis indicated that c-myc was upregulated by TSA in a dose-dependent manner in WERI-Rb1 cells. (B) Western blot analysis of c-Myc in WERI-Rb1 cells following TSA treatment. GAPDH is shown as an internal control. (C) Relative expression of the c-Myc protein in WERI-Rb1 following TSA treatment at different time points. Data are presented as histograms. (D) Expression of c-Myc (red) was visualized in WERI-Rb1 cells after TSA treatment, and compared with the control. Magnification, ×100. (E) Luciferase plasmid structure, which contains a c-myc promoter sequence from −2,263 to +53 bp. (F) WERI-Rb1 cells were transfected with the reporter containing the c-myc promoter and subsequently treated with 250 nM TSA for 24 h. Levels of luciferase activity were normalized to those of Renilla luciferase (control, 1; TSA, 1.53±0.18; n=3 for each group). All results were confirmed in triplicate. *P<0.05 vs. respective control. RT-PCR, reverse transcription PCR; BP, base pairs; TSA, trichostatin A; con, control.
Moreover, to further investigate whether histone deacetylation is specific to the expression of the endogenous c-myc gene, a construct containing the human c-myc promoter sequence between positions −2,263 and +53 upstream of the luciferase reporter gene (Fig. 2E) was utilised. WERI-Rb1 cells (transfected with a plasmid expressing the aforementioned construct) were treated with 250 nM TSA and subjected to luciferase assays. As exhibited in Fig. 2F, the basal activity of the c-myc promoter was significantly higher in WERI-Rb1 cells treated with TSA, compared to the control (control, 1; TSA, 1.53±0.18; P<0.05). Therefore, the current results indicated that the expression of c-myc was significantly increased by treatment with TSA in WERI-Rb1 cells.
c-Myc expression is differentially regulated by three HDAC inhibitors in WERI-Rb1 cells
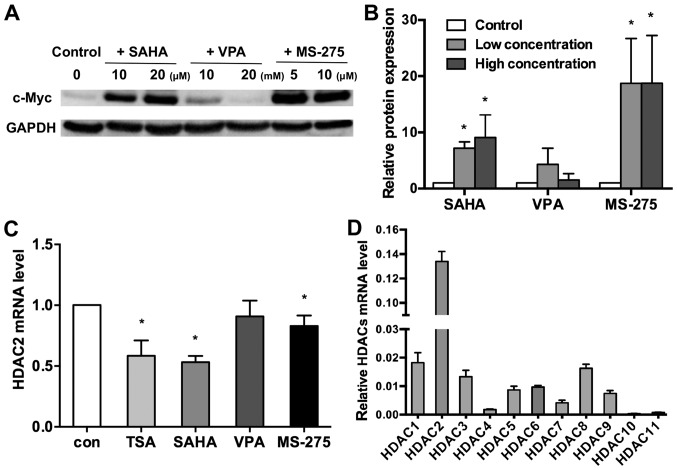
To further determine whether histone deacetylation is implicated in c-myc silencing, three different HDAC inhibitors (SAHA, VPA and MS-275) were used to investigate the mechanism of c-myc silencing in WERI-Rb1 cells, via treatment of the cells with various concentrations of the HDAC inhibitors SAHA, VPA and MS-275. As indicated in Fig. 3A and B, c-Myc expression was significantly upregulated following treatment with 10 and 20 µM SAHA, and 5 and 10 µM MS-275. However, 10 and 20 mM VPA did not influence the expression of c-Myc (control, 1; 10 µM SAHA, 7.159±1.157; 20 µM SAHA, 9.027±4.066; 10 mM VPA, 4.301±2.856; 20 mM VPA, 1.477±1.156; 5 µM MS-275, 18.750±7.971; 10 µM MS-275, 18.742±8.480; P<0.05).
Figure 3.
c-Myc expression is differentially regulated by three HDAC inhibitors in WERI-Rb1 cells. (A) WERI-Rb1 cells were treated with different concentrations of SAHA, VPA and MS-275, and c-Myc expression was detected via western blotting. GAPDH was used as an internal control. (B) Relative expression of c-Myc in WERI-Rb1 cells was quantified using densitometry; data are presented as histograms. (C) Relative HDAC2 mRNA expression in WERI-Rb1 cells treated with HDAC inhibitors. (D) HDAC mRNA expression in healthy WERI-Rb1 cells was determined via RT-qPCR. Results were normalized to β-actin expression. All results were confirmed in triplicate. *P<0.05 vs. respective control. RT-qPCR, reverse transcription-quantitative PCR; HDAC, histone deacetylase; TSA, trichostatin A; SAHA, vorinostat; VPA, valproic acid sodium salt; MS-275, entinostat; con, control.
To investigate the hypothesis that VPA does not increase c-Myc expression in WERI-Rb1 cells, HDAC mRNA was assessed in WERI-Rb1 cells treated with 500 nM TSA, 20 µM SAHA, 20 mM VPA and 10 µM MS-275. The present results indicated that different HDAC inhibitors differentially affected the mRNA expression of specific HDAC family members in WERI-Rb1 cells, 6 h after treatment (Fig. S1 and Table SII). All four HDAC inhibitors downregulated HDAC7 and 8 in WERI-Rb1 cells. Notably, VPA did not decrease HDAC2 mRNA expression like the other HDAC inhibitors did (Fig. 3C; control, 1; TSA, 0.584±0.127; SAHA, 0.532±0.051; VPA, 0.909±0.130; MS-275, 0.829±0.087; P<0.05). The current data implied that HDAC2 may influence HDAC inhibitor-mediated regulation of c-Myc.
Thus, the mRNA expression level of HDAC1-11 in WERI-Rb1 cells was quantified using RT-qPCR. It was revealed that HDAC2 expression was significantly upregulated in comparison with the other members of the HDAC family (relative HDAC mRNA expression levels: HDAC1, 0.0183±0.0035; HDAC2, 0.1339±0.0082; HDAC3, 0.0133±0.0023; HDAC4, 0.0018±0.0002; HDAC5, 0.0087±0.0013; HDAC6, 0.0097±0.0005; HDAC7, 0.0041±0.0009; HDAC8, 0.0163±0.0014; HDAC9, 0.0075±0.0010; HDAC10, 0.0003±0.0001; and HDAC11, 0.0007±0.0002; Fig. 3D). Therefore, it is speculated that HDAC2 may serve a key role in the regulation of c-Myc in WERI-Rb1 cells.
Exogenous c-Myc influences the viability of WERI-Rb1 cells
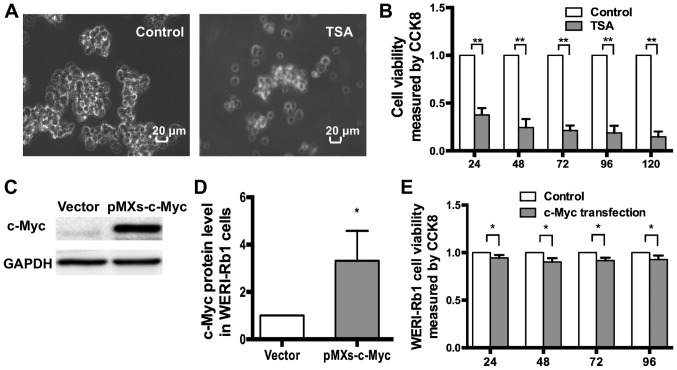
TSA treatment resulted in an increase in c-Myc expression, but also affected the morphology of WERI-Rb1 cells. Normally, WERI-Rb1 cells appear as small, round cells in large clusters. However, following treatment with 250 nM TSA for 72 h, the clusters reduced in size, and single cells were observed (Fig. 4A). The CCK-8 assay indicated that the viability of WERI-Rb1 cells was markedly reduced (in a continuous manner), following treatment with 250 nM TSA (Fig. 4B; TSA, 24 h, 0.377±0.072; 48 h, 0.244±0.089; 72 h, 0.213±0.053; 96 h, 0.188±0.075; 120 h, 0.147±0.056) compared to the respective controls (control, 1; P<0.01). Therefore, TSA significantly inhibited the viability of WERI-Rb1 cells.
Figure 4.
Exogenous c-myc affects the viability of WERI-Rb1 cells. (A) Morphological changes in WERI-Rb1 cells treated with TSA (250 nM) for 72 h. (B) CCK-8 assays indicated that TSA significantly decreased the viability of WERI-Rb1 cells compared with the negative controls. **P<0.01. (C) Western blotting showing the expression of c-Myc in WERI-Rb1 cells after transfection. (D) Relative expression of c-Myc in WERI-Rb1 cells was quantified using densitometry; the data are presented as histograms. *P<0.05 vs. vector. (E) CCK-8 assays indicated that the viability of WERI-Rb1 cells was reduced following c-myc transfection, compared with the controls. All results were confirmed in triplicate. *P<0.05. TSA, trichostatin A; CCK-8, Cell Counting Kit-8.
The c-Myc protein possesses versatile functions in human cancer (16,20). To investigate whether the TSA-induced inhibition of viability in WERI-Rb1 cells was associated with c-Myc, the cells were transfected with either pMXs-c-Myc or an empty control vector. The CCK-8 assay was then performed to evaluate the effects of exogenous c-Myc on WERI-Rb1 proliferation. As indicated in Fig. 4C and D, western blot analysis revealed a higher level of c-Myc expression in WERI-Rb1 cells transfected with pMXs-c-Myc compared with the empty vector (c-Myc protein expression: Vector, 1; pMXs-c-Myc, 3.32±1.27; P<0.05). Notably, the viability of WERI-Rb1 cells was significantly reduced following pMXs-c-Myc transfection (exogenous c-Myc; 24 h, 0.929±0.014; 48 h, 0.886±0.028; 72 h, 0.901±0.012; and 96 h, 0.913±0.030) compared to vector transfection (control, 1) (Fig. 4E; P<0.05). Therefore, the present results demonstrated that exogenous c-myc may significantly decrease the viability of WERI-Rb1 cells.
c-Myc downregulation results from histone deacetylation in WERI-Rb1 cells, but not in Y79 cells
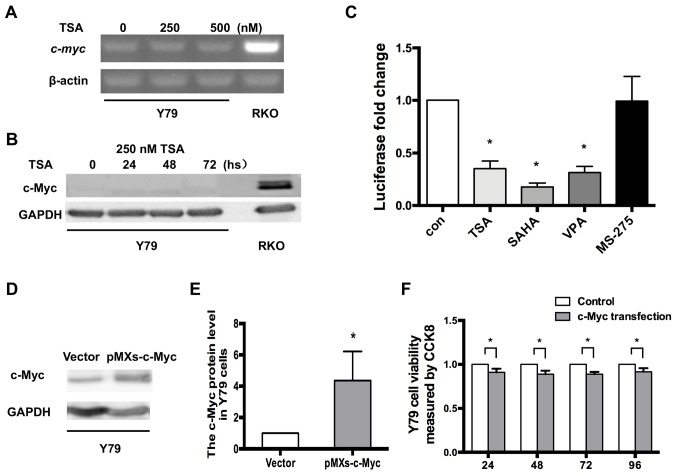
To examine whether the silencing mechanism was conserved across other RB cell lines, Y79 cells were chosen for comparison with WERI-Rb1 cells. Compared with RKO cells, c-myc was also nearly silenced in Y79 cells, similar to WERI-Rb1 cells (Fig. 5A). However, the expression level of c-myc was not upregulated in Y79 cells after treatment with 250 nM and 500 nM TSA. Moreover, western blot analysis demonstrated that the protein level of c-Myc was not induced by 250 nM TSA at different time points (Fig. 5B).
Figure 5.
c-Myc is not upregulated by HDAC inhibitors in RB Y79 cells. (A) RT-PCR analysis indicated that c-myc expression was not induced by treatment with 250 or 500 nM TSA, in Y79 cells. (B) Western blot analysis of c-Myc in Y79 cells following TSA treatment. GAPDH was used as a reference gene. (C) Y79 cells were transfected with the c-myc promoter reporter and then treated with 250 nM TSA, 10 µM SAHA, 10 mM VPA or 5 µM MS-275 for 24 h. Levels of luciferase activity were normalized to those of Renilla luciferase (control, 1; TSA, 0.35±0.07; SAHA, 0.18±0.04; VPA, 0.31±0.59; MS-275, 0.99±0.24; n=3 for each group) *P<0.05 vs. control. (D) Expression level of c-Myc in Y79 cells after c-Myc transfection. (E) Relative expression of c-Myc in Y79 cells was quantified using densitometry, and the data are presented as histograms. *P<0.05 vs. vector. (F) CCK-8 assays indicated that the viability of Y79 cells may also be reduced by exogenous c-Myc, similar to WERI-Rb1 cells. All the results were confirmed in triplicate. *P<0.05. RT-PCR, reverse transcription PCR; TSA, trichostatin A; SAHA, vorinostat; VPA, valproic acid sodium salt; MS-275, entinostat; con, control.
Y79 cells were also transfected with the plasmid carrying the human c-myc promoter with the firefly luciferase gene (Fig. 2E) and treated with 250 nM TSA, 10 µM SAHA, 10 mM VPA or 5 µM MS-275. After 24 h, the cells were harvested and subjected to luciferase assays. As detailed in Fig. 5C, unlike in WERI-Rb1 cells, TSA, SAHA, VPA and MS-275 did not upregulate the basal activity of the c-myc promoter in Y79 cells. By contrast, c-myc promoter activity was decreased in Y79 cells treated with TSA, SAHA and VPA. In addition, c-myc promoter activity was not influenced by MS-275 compared to the controls (control, 1; TSA, 0.35±0.07; SAHA, 0.18±0.04; VPA, 0.31±0.59; MS-275, 0.99±0.24; P<0.05). Therefore, the present results indicated that the mechanism of c-myc silencing is specific to WERI-Rb1 cells compared with Y79 cells.
Moreover, to determine whether exogenous c-myc is able to inhibit the viability of Y79 cells, the cells were transfected with either pMXs-c-Myc or a control vector, and analysed using a CCK-8 assay. As revealed in Fig. 5D and E, western blot analysis confirmed the change in the c-Myc protein expression level in Y79 cells when transfected with pMXs-c-Myc, compared with the vector (c-Myc protein expression: Vector, 1; pMXs-c-Myc: 4.36±1.85; P<0.05). Consistent with WERI-Rb1 cells, Fig. 5F shows that the viability of Y79 cells was also significantly reduced by exogenous c-myc at 24 h (0.923±0.033), 48 h (0.898±0.037), 72 h (0.903±0.004) and 96 h (0.92±0.040) (all P<0.05). Thus, according to the current results, exogenous c-myc also significantly inhibits the proliferation of Y79 cells.
Discussion
It has been revealed that c-Myc is highly expressed in a variety of tumour types (6–9), though the expression level of c-Myc in RB cells is yet to be reported, to the best of our knowledge. In the present study, it was revealed that the expression level of c-Myc was very low in the RB cell line WERI-Rb1. It was confirmed that c-Myc expression in WERI-Rb1 cells was increased at the transcriptional level by the HDAC inhibitor TSA, and was also differentially regulated by three other HDAC inhibitors. However, none of the HDAC inhibitors induced the upregulation of c-Myc expression in Y79 cells. Moreover, exogenous c-Myc significantly inhibited the proliferation of WERI-Rb1 and Y79 cells. Thus, the results of the current study revealed the expression mechanism of c-Myc and the role it serves in RB cells.
c-myc has been reported to be regulated by histone acylation or DNA methylation in numerous types of cancer cell (12–15). The present data revealed that expression levels of c-Myc were significantly upregulated in WERI-Rb1 cells following treatment with TSA, SAHA and MS-275. This result is consistent with a previous study, which revealed that TSA significantly increased the expression of c-myc mRNA resulting from nerve growth factor (NGF), and blocked both oncogenic ras- and NGF-induced neurite outgrowth from PC12 cells (23). By contrast, previous studies have indicated that c-myc may also be regulated by certain DNA demethylating reagents, such as 5-azacytidine (12–14). Certain previous studies on different cancer cells have reported that c-myc expression was downregulated or unaffected by treatment with HDAC inhibitors. For example, Kretzner et al (24) demonstrated that both TSA and SAHA decreased c-Myc mRNA and protein expression, as well as c-Myc-regulated microRNA expression. Nebbioso et al (25) reported that treatment with SAHA and MS-275 resulted in both c-Myc acetylation at lysine residue 323, and c-Myc downregulation in acute myeloid leukaemia cell lines. Furthermore, silencing of c-Myc was not influenced by TSA in prostate cancer PC3 cells (26). Thus, the present data indicated that histone deacetylation was implicated in the silencing mechanism of c-Myc in WERI-Rb1 cells.
Moreover, in the present study, the expression of c-Myc in WERI-Rb1 cells was discovered not to be increased by VPA, by contrast to TSA, SAHA and MS-275. In addition, HDAC2 was reduced by all the other HDAC inhibitors except VPA. HDAC2 might be involved in HDAC inhibitor-medicated regulation of c-Myc. HDAC2 has a pivotal role in the modulation chromatin architecture leading to transcriptional changes (27). Previous studies reported that HDAC2 is upregulated in numerous cancer types, including breast and prostate cancer (28–30). Additionally, upregulation of HDAC2 has been revealed to significantly enhance cancer cell proliferation, migration and invasion (28,30–33). These findings support the results of the present study which indicated that the HDAC2 mRNA level was the higher than that of any other HDACs in WERI-Rb1 cells. Additionally, several studies have demonstrated that VPA treatment may decrease or have no effect on HDAC2 expression (34–37). This may be partially attributable to the fact that VPA is tumour-selective, similar to the other HDAC inhibitors (38). However, HDAC2 has been reported to enhance c-Myc expression in numerous cell types (39,40), by contrast to the present study which discovered that c-Myc expression increased when HDAC2 was downregulated. Further research should investigate the association between HDAC2 and c-Myc expression in RB cells. Taken together, the current data suggest that HDAC2 may serve an important role in the regulation of c-Myc expression in WERI-Rb1 cells treated with TSA, SAHA and MS-275.
Notably, none of the HDAC inhibitors significantly altered c-Myc expression in Y79 cells. This result is partially consistent with the different effects on gene expression caused by SAHA in human breast carcinoma cell line MDA 468, compared with MDA 435 (41). Moreover, the RB mutation in WERI-Rb1 cells is a complete deletion of the gene, whereas a partial deletion is present in Y79 cells (42). The two RB cell lines possess differing growth characteristics (43), which may contribute to the variation in their gene expression patterns (44) and result in contrasting c-myc-silencing mechanisms. Overall, the current results demonstrated that histone acetylation of the c-myc gene was specific to WERI-Rb1 cells.
Furthermore, the present data revealed that exogenous c-Myc mildly reduced the viability of WERI-Rb1 and Y79 cells. However, as suggested in previous reports, c-Myc may be highly expressed in a variety of tumour types, and may also promote proliferation and metastasis (6–9,17). By contrast, c-Myc was revealed to induce apoptosis in myeloid progenitor cells and fibroblasts (45–47). Notably, c-Myc induces massive programmed cell death (PCD) in the majority of transgenic mouse models, with greater efficiency than other oncogenes (20). Downregulation of c-Myc protein levels contributes to cancer cell survival under certain conditions (21). The aforementioned studies are consistent with the current results, which revealed that exogenous c-Myc reduced the viability of RB cells, to a certain extent. However, the results of c-Myc on PCD were thought to promote cancer cells to adapt to living conditions and environmental changes (20,21). Thus, further studies exploring the inhibition of exogenous c-Myc on retinoblastoma may help elucidate its role.
In conclusion, the present study indicated that c-myc was downregulated in WERI-Rb1 and Y79 cells. Moreover, HDAC inhibitors TSA, SAHA and MS-275 were able to transcriptionally induce the expression of c-Myc in WERI-Rb1 cells, but not in Y79 cells. Exogenous c-myc also decreased WERI-Rb1 and Y79 cell viability. Therefore, the current data support the hypothesis that c-Myc regulates tumorigenesis at an epigenetic level in the WERE-Rb1 human RB cell line.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science Foundation of China (grant no. 81670848 and 81470626).
Availability of data and materials
All data generated or analyzed during the present study are included in the published article.
Authors' contributions
KMY, JZ, JG and NY made substantial contributions to the design of the study. NY, ML, JQ, PZ, MY, PY, YW and XH performed the experiments. PC and QW analyzed the data. All authors contributed to writing the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dimaras H, Kimani K, Dimba EA, Gronsdahl P, White A, Chan HS, Gallie BL. Retinoblastoma. Lancet. 2012;379:1436–1446. doi: 10.1016/S0140-6736(11)61137-9. [DOI] [PubMed] [Google Scholar]
- 2.Fabian ID, Onadim Z, Karaa E, Duncan C, Chowdhury T, Scheimberg I, Ohnuma SI, Reddy MA, Sagoo MS. The management of retinoblastoma. Oncogene. 2018;37:1551–1560. doi: 10.1038/s41388-017-0050-x. [DOI] [PubMed] [Google Scholar]
- 3.Kivela T. The epidemiological challenge of the most frequent eye cancer: Retinoblastoma, an issue of birth and death. Br J Ophthalmol. 2009;93:1129–1131. doi: 10.1136/bjo.2008.150292. [DOI] [PubMed] [Google Scholar]
- 4.Knudson AG., Jr Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmerman K, Alt FW. Expression and function of myc family genes. Crit Rev Oncog. 1990;2:75–95. [PubMed] [Google Scholar]
- 6.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 7.Brodeur GM. Neuroblastoma: Biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 8.Bociek RG. Adult Burkitt's lymphoma. Clin Lymphoma. 2005;6:11–20. doi: 10.3816/CLM.2005.n.021. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi D, Anderton B, Goga A. Taking on challenging targets: Making MYC druggable. Am Soc Clin Oncol Educ Book. 2014:e497–e502. doi: 10.14694/EdBook_AM.2014.34.e497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung HJ, Levens D. c-myc expression: Keep the noise down! Mol Cells. 2005;20:157–166. [PubMed] [Google Scholar]
- 12.Kitagawa Y, Kyo S, Takakura M, Kanaya T, Koshida K, Namiki M, Inoue M. Demethylating reagent 5-azacytidine inhibits telomerase activity in human prostate cancer cells through transcriptional repression of hTERT. Clin Cancer Res. 2000;6:2868–2875. [PubMed] [Google Scholar]
- 13.Grandjenette C, Schnekenburger M, Karius T, Ghelfi J, Gaigneaux A, Henry E, Dicato M, Diederich M. 5-aza-2′- deoxycytidine-mediated c-myc Down-regulation triggers telomere-dependent senescence by regulating human telomerase reverse transcriptase in chronic myeloid leukemia. Neoplasia. 2004;16:511–528. doi: 10.1016/j.neo.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long Y, Tsai WB, Chang JT, Estecio M, Wangpaichitr M, Savaraj N, Feun LG, Chen HH, Kuo MT. Cisplatin-induced synthetic lethality to arginine-starvation therapy by transcriptional suppression of ASS1 is regulated by DEC1, HIF-1α, and c-Myc transcription network and is independent of ASS1 promoter DNA methylation. Oncotarget. 2016;7:82658–82670. doi: 10.18632/oncotarget.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Wu X. Histone deacetylase inhibitor, Trichostatin A, activates p21WAF1/CIP1 expression through downregulation of c-myc and release of the repression of c-myc from the promoter in human cervical cancer cells. Biochem Biophys Res Commun. 2004;324:860–867. doi: 10.1016/j.bbrc.2004.09.130. [DOI] [PubMed] [Google Scholar]
- 16.Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 17.Guo P, Nie Q, Lan J, Ge J, Qiu Y, Mao Q. C-Myc negatively controls the tumor suppressor PTEN by upregulating miR-26a in glioblastoma multiforme cells. Biochem Biophys Res Commun. 2013;441:186–190. doi: 10.1016/j.bbrc.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 18.Karn J, Watson JV, Lowe AD, Green SM, Vedeckis W. Regulation of cell cycle duration by c-myc levels. Oncogene. 1989;4:773–787. [PubMed] [Google Scholar]
- 19.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, Sundberg CD, Bishop JM, Felsher DW. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Tai Y, Lisanti MP, Liao DJ. c-Myc induction of programmed cell death may contribute to carcinogenesis: A perspective inspired by several concepts of chemical carcinogenesis. Cancer Biol Ther. 2011;11:615–626. doi: 10.4161/cbt.11.7.14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuyama H, Endo H, Akashika T, Kato K, Inoue M. Downregulation of c-MYC protein levels contributes to cancer cell survival under dual deficiency of oxygen and glucose. Cancer Res. 2010;70:10213–10223. doi: 10.1158/0008-5472.CAN-10-2720. [DOI] [PubMed] [Google Scholar]
- 22.Liva KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Futamura M, Monden Y, Okabe T, Fujita-Yoshigaki J, Yokoyama S, Nishimura S. Trichostatin A inhibits both ras-induced neurite outgrowth of PC12 cells and morphological transformation of NIH3T3 cells. Oncogene. 1995;10:1119–1123. [PubMed] [Google Scholar]
- 24.Kretzner L, Scuto A, Dino PM, Kowolik CM, Wu J, Ventura P, Jove R, Forman SJ, Yen Y, Kirschbaum MH. Combining histone deacetylase inhibitor vorinostat with aurora kinase inhibitors enhances lymphoma cell killing with repression of c-Myc, hTERT, and microRNA levels. Cancer Res. 2011;71:3912–3920. doi: 10.1158/0008-5472.CAN-10-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nebbioso A, Carafa V, Conte M, Tambaro FP, Abbondanza C, Martens J, Nees M, Benedetti R, Pallavicini I, Minucci S, et al. c-Myc modulation & acetylation is a key HDAC inhibitor target in cancer. Clin Cancer Res. 2017;23:2542–2555. doi: 10.1158/1078-0432.CCR-15-2388. [DOI] [PubMed] [Google Scholar]
- 26.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conte M, Dell'Aversana C, Sgueglia G, Carissimo A, Altucci L. HDAC2-dependent miRNA signature in acute myeloid leukemia. FEBS Lett. 2019;593:2574–2584. doi: 10.1002/1873-3468.13521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan W, Jiang Y, Yu H, Huang Q, Liu L, Guo X, Li L, Mi Q, Zhang K, Yang Z. HDAC2 overexpression correlates with aggressive clinicopathological features and DNA-damage response pathway of breast cancer. Am J Cancer Res. 2017;7:1213–1226. [PMC free article] [PubMed] [Google Scholar]
- 29.Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, Kristiansen G. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JK, Noh JH, Eun JW, Jung KH, Bae HJ, Shen Q, Kim MG, Chang YG, Kim SJ, Park WS, et al. Targeted inactivation of HDAC2 restores p16INK4a activity and exerts antitumor effects on human gastric cancer. Mol Cancer Res. 2013;11:62–73. doi: 10.1158/1541-7786.MCR-12-0332. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Wang F, Qu Y, Chen X, Gao M, Yang J, Zhang D, Zhang N, Li W, Liu H. HDAC2 regulates cell proliferation, cell cycle progression and cell apoptosis in esophageal squamous cell carcinoma EC9706 cells. Oncol Lett. 2017;13:403–409. doi: 10.3892/ol.2016.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Mei DT, Zeng Y. HDAC2 promotes the migration and invasion of non-small cell lung cancer cells via upregulation of fibronectin. Biomed Pharmacother. 2016;84:284–290. doi: 10.1016/j.biopha.2016.09.030. [DOI] [PubMed] [Google Scholar]
- 33.Zhao H, Yu Z, Zhao L, He M, Ren J, Wu H, Chen Q, Yao W, Wei M. HDAC2 overexpression is a poor prognostic factor of breast cancer patients with increased multidrug resistance-associated protein expression who received anthracyclines therapy. Jpn J Clin Oncol. 2016;46:893–902. doi: 10.1093/jjco/hyw096. [DOI] [PubMed] [Google Scholar]
- 34.Krämer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, Göttlicher M. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwiecińska P, Wróbel A, Taubøll E, Gregoraszczuk EŁ. Valproic acid, but not levetiracetam, selectively decreases HDAC7 and HDAC2 expression in human ovarian cancer cells. Toxicol Lett. 2014;224:225–232. doi: 10.1016/j.toxlet.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 36.Jones J, Juengel E, Mickuckyte A, Hudak L, Wedel S, Jonas D, Blaheta RA. The histone deacetylase inhibitor valproic acid alters growth properties of renal cell carcinoma in vitro and in vivo. J Cell Mol Med. 2009;13:2376–2385. doi: 10.1111/j.1582-4934.2008.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travaglini L, Vian L, Billi M, Grignani F, Nervi C. Epigenetic reprogramming of breast cancer cells by valproic acid occurs regardless of estrogen receptor status. Int J Biochem Cell Biol. 2009;41:225–234. doi: 10.1016/j.biocel.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Nebbioso A, Clarke N, Voltz E, Germain E, Ambrosino C, Bontempo P, Alvarez R, Schiavone EM, Ferrara F, Bresciani F, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 39.Noh JH, Jung KH, Kim JK, Eun JW, Bae HJ, Xie HJ, Chang YG, Kim MG, Park WS, Lee JY, Nam SW. Aberrant regulation of HDAC2 mediates proliferation of hepatocellular carcinoma cells by deregulating expression of G1/S cell cycle proteins. PLoS One. 2011;6:e28103. doi: 10.1371/journal.pone.0028103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ecker J, Oehme I, Mazitschek R, Korshunov A, Kool M, Hielscher T, Kiss J, Selt F, Konrad C, Lodrini M, et al. Targeting class I histone deacetylase 2 in MYC amplified group 3 medulloblastoma. Acta Neuropathol Commun. 2015;3:22. doi: 10.1186/s40478-015-0201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: Defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther. 2003;2:151–163. [PubMed] [Google Scholar]
- 42.Bookstein R, Lee EY, To H, Young LJ, Sery TW, Hayes RC, Friedmann T, Lee WH. Human retinoblastoma susceptibility gene: Genomic organization and analysis of heterozygous intragenic deletion mutants. Proc Natl Acad Sci USA. 1988;85:2210–2214. doi: 10.1073/pnas.85.7.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Busch M, Philippeit C, Weise A, Dünker N. Re-characterization of established human retinoblastoma cell lines. Histochem Cell Biol. 2015;143:325–338. doi: 10.1007/s00418-014-1285-z. [DOI] [PubMed] [Google Scholar]
- 44.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 45.Askew DS, Ashmun RA, Simmons BC, Cleveland JL. Constitutive c-Myc expression in an IL-3 dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 46.Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-Myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-T. [DOI] [PubMed] [Google Scholar]
- 47.Harrington EA, Bennett MR, Fanidi A, Evan GI. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in the published article.