Abstract
Triple-negative breast cancer (TNBC) is characterized by a deficiency in the estrogen receptor (ER), progesterone receptor (PR) and HER2/neu genes. Patients with TNBC have an increased likelihood of distant recurrence and mortality, compared with patients with other subtypes of breast cancer. The current study aimed to identify novel biomarkers for TNBC. Weighted gene co-expression network analysis (WGCNA) was applied to construct gene co-expression networks; these were used to explore the correlation between mRNA profiles and clinical data, thus identifying the most significant co-expression network associated with the American Joint Committee on Cancer-TNM stage of TNBC. Using RNAseq datasets from The Cancer Genome Atlas, downloaded from the University of California, Santa Cruz, WGCNA identified 23 modules via K-means clustering. The most significant module consisted of 248 genes, on which gene ontology analysis was subsequently performed. Differently Expressed Gene (DEG) analysis was then applied to determine the DEGs between normal and tumor tissues. A total of 42 genes were positioned in the overlap between DEGs and the most significant module. Following survival analysis, 5 genes [GIPC PDZ domain containing family member 1 (GIPC1), hes family bHLH transcription factor 6 (HES6), calmodulin-regulated spectrin-associated protein family member 3 (KIAA1543), myosin light chain kinase 2 (MYLK2) and peter pan homolog (PPAN)] were selected and their association with the American Joint Committee on Cancer-TNM diagnostic stage was investigated. The expression level of these genes in different pathological stages varied, but tended to increase in more advanced pathological stages. The expression of these 5 genes exhibited accurate capacity for the identification of tumor and normal tissues via receiver operating characteristic curve analysis. High expression of GIPC1, HES6, KIAA1543, MYLK2 and PPAN resulted in poor overall survival (OS) in patients with TNBC. In conclusion, via unsupervised clustering methods, a co-expressed gene network with high inter-connectivity was constructed, and 5 genes were identified as biomarkers for TNBC.
Keywords: triple-negative breast cancer, weighted gene co-expression network analysis, progression
Introduction
Triple-negative breast cancers (TNBCs) are characterized by the lack of HER2/neu, progesterone receptor (PR) and estrogen receptor (1) (ER) gene expression. The TNBC subtype constitutes 10–15% of total breast tumors and 80% of basal-like breast cancers (1). Triple-negative and basal-like tumors commonly have a high histological grade (1). TNBCs also have a poor prognosis and tend to result in earlier relapse compared with other subtypes of breast cancer (1). Additionally, TBNCs exhibit increased chemosensitivity compared with other genotypes of breast cancer (2), thus, chemotherapeutic methods are currently the most prevalently used medical treatments (3). These include epidermal growth factor receptor (EGFR)-targeted therapies, multi-tyrosine kinase inhibitors, poly-ADP ribose polymerase (PARP) inhibitors and anti-angiogenic agents (4). However, patients with TNBCs usually exhibit poorer outcomes following chemotherapy, compared with patients with breast cancers of other subtypes (3). Hence, it would be beneficial to identify novel biomarkers associated with the progression of TNBCs, and identify new targets to improve their precise diagnosis and treatment.
Weighted-correlation network analysis (WGCNA) has previously been performed to construct non-scale co-expressed gene networks (5–8). In the present study, WGCNA was performed to identify the hub-module that contained genes showing a strong correlation with the pathological stage of TNBC. Subsequently, differentially expressed gene (DEG) analysis was performed on RNAsequencing (RNAseq) data of TNBC. The hub-genes were defined as all genes contained in the overlap between the DEGs and the hub-module. Gene ontology (GO) and gene enrichment analyses were then employed, and identified several important terms in biological process, molecular function and cellular components. Survival analysis was also conducted, and 5 genes were selected from the hub-genes. Receiver operating characteristic (ROC) and Kaplan-Meier (KM) curves were plotted to indicate the capacity of these genes to differentiate tumor and para-tumor tissues, and to confirm the influence of these genes on the overall survival (OS) of patients with TNBC.
Materials and methods
Data processing and co-expression gene network construction using RNAseq data
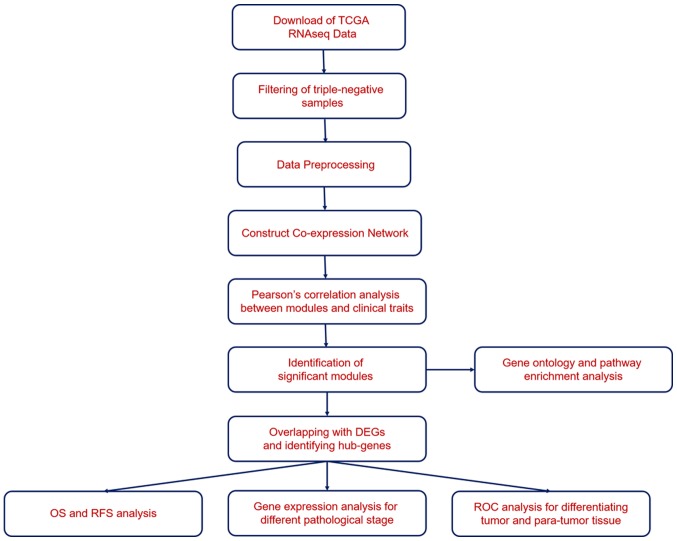
A total of 1,240 RNAseq datasets from The Cancer Genome Atlas (TCGA) and clinical data for patients with breast cancer were downloaded from the University of California, Santa Cruz (UCSC) database using the Xena browser (https://xenabrowser.net/). The workflow is displayed in Fig. 1. The TNBC samples were filtered using the criteria of ‘not expressing genes for ER, PR and HER2/neu’. R software (version 3.5.2; http://www.r-project.org) was applied to perform WGCNA analysis. WGCNA was used to construct the gene co-expression network; co-expression similarity (Si,j) was defined as the absolute value of the correlation coefficient between the mRNA expression profile of nodes i and j:
Figure 1.
Workflow of the present study.
Where Xi and Xj are mRNA expression values for genes i and j, Si,j was calculated using the Pearson's correlation coefficient between genes i and j. Weighted-network adjacency was defined by raising the co-expression similarity to a power:
β≥1. The power of β=4 and scale free R2=0.95 were selected as the soft-thresholding parameters to ensure a signed scale-free gene network.
By evaluating the correlation between the pathological stage of TNBC and the module membership with the ‘p. weighted’, a high-correlated module was identified. The tan modules which had the most significant adjusted P-values were selected. Genes involved in the tan modules were presented using Cytoscape v3.4.0 (https://cytoscape.org). The genes in the tan module were selected as the input for GO and KEGG analysis, which was performed using Metascape (http://metascape.org/gp/index.html).
Statistical analysis
Statistical analysis was performed using R (R Foundation for Statistical Computing; http://www.R-project.org/). The fold-change and Q-value (adjusted P-value) for para-tumor and tumor samples were calculated using the Limma package (9). A Q-value <0.05 was considered to be statistically significant. The overall survival analysis was conducted using the Survminer package (10), and the P-values in the KM curve were obtained using the log-rank test. The false discovery rate was set as 0.05 for analysis.
Results
WGCNA on RNAseq dataset of TNBC
In order to determine the co-expression network most highly associated with the progress and prognosis of TNBC, TNBCTCGA RNAseq datadownloaded from UCSC, was analyzed using WGCNA. The analysis showed TNBC clustering using the average linkage and Pearson's correlation methods. The scale-free network was constructed by raising the power of β to 4 and by ensuring that the scale-free R2 reached 0.95 (Fig. 2A and B). The clustering dendrogram of TNBC tissues is shown in Fig. 2C.
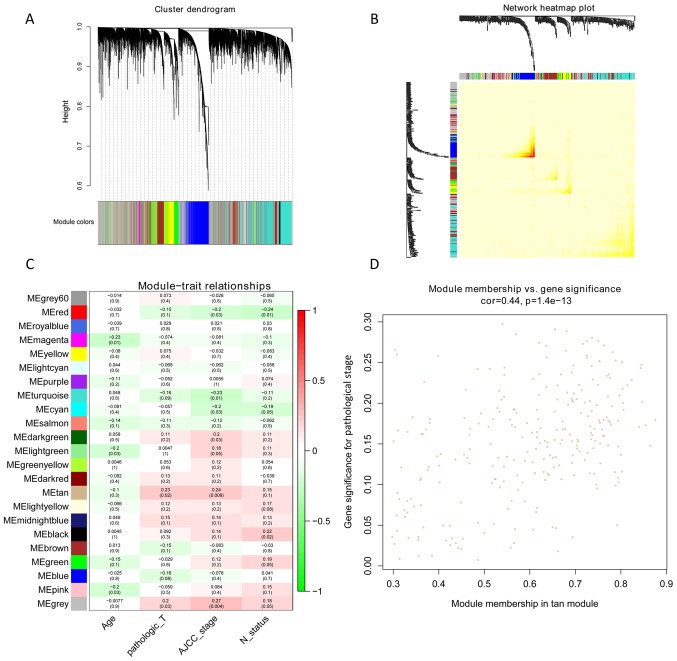
Figure 2.
Soft-threshold power in WGCNA and K-means clustering of TNBC samples. (A) Relationship between scale-free topology model fit and soft-thresholds (powers). (B) Relationship between the mean connectivity and various soft-thresholds (powers). (C) Clustering dendrogram of TNBC tissues. WGNCA, weighted correlation network analysis; TNBC, triple-negative breast cancer.
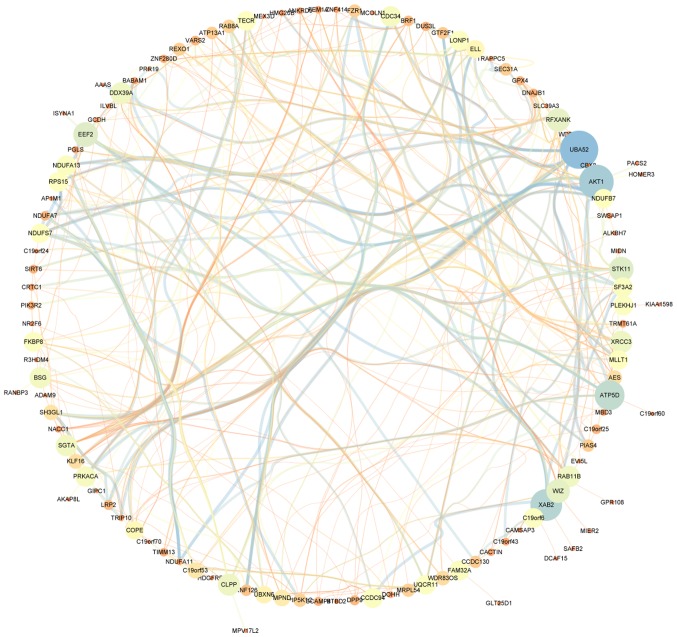
A total of 23 modules were found to be clustered, and this gene clustering is displayed as a dendrogram in Fig. 3A. The weighted network of all genesis exhibited in a heat map, depicting the topological overlap matrix amongst the mRNA expression profiles (Fig. 3B). The tan module was determined using a trait-heat map to be the module with the strongest correlation with the pathological stage of TNBC (Fig. 3C). Fig. 3D illustrates the correlation of genes with pathological stage, as well as module membership (the correlation of genes with clusters) in the tan module. The results revealed that genes, which had high a correlation with tan modules were also strongly associated with the pathological stage of TNBC. Based on the cut-off criteria (|GS|>0.4), 129 genes with high connectivity were selected for the construction of the co-expression network. The inner connectivity in the tan module with the threshold (|GS|>0.4) was plotted. This showed strong co-expression relationships in the tan module (Fig. 4).
Figure 3.
Identification of modules associated with the clinical traits of triple-negative breast cancer. (A) Dendrogram of modules identified by WGCNA. (B) Topological overlap matrix among detected genes from RNAseq. Genes with high intramodular connectivity are located at the tip of the module branches. (C) Heatmap of Module-clinical trait associations. (D) Scatterplot of gene significance for pathological stage vs. module membership in the tan module. WGNCA, weighted correlation network analysis.
Figure 4.
Innerconnectivity in the tan module. Node size indicates the betweenness centrality, which reflects the amount of control that this node exerts over the interactions of other nodes in the network. Edge sizes depict the weight between connected genes.
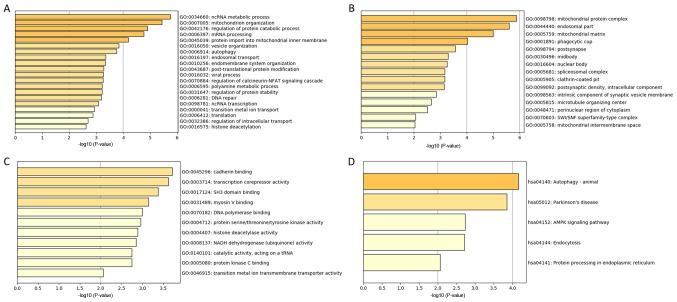
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
The genes in the tan module were divided into three groups (biological process, cellular component, and molecular function), which were then analyzed using GO and KEGG analysis. In the biological process group, the most enriched genes concerned epigenetic regulation, cell metabolism, DNA repair and mRNA processing (Fig. 5A). The genes in the cellular component group that were most enriched comprised ‘mitochondrial protein complex’, ‘endosomal part’ and ‘phagocytic cup’ (Fig. 5B). The most enriched genes in the molecular function group were in ‘cadherin binding’, ‘transcription corepressor activity’ and ‘SH3 domain binding’ (Fig. 5C). KEGG pathway analysis indicated that ‘autophagy’ and ‘AMPK signaling pathway’ were involved in the regulation of genes in the tan module (Fig. 5D).
Figure 5.
Gene ontology and pathway enrichment analysis of genes in the tan module. (A) Biological process analysis. (B) Cellular component analysis. (C) Molecular function analysis. (D) KEGG pathway analysis. KEGG, Kyoto Encyclopedia of Genes and Genomes.
DEG analysis of TNBC tissues
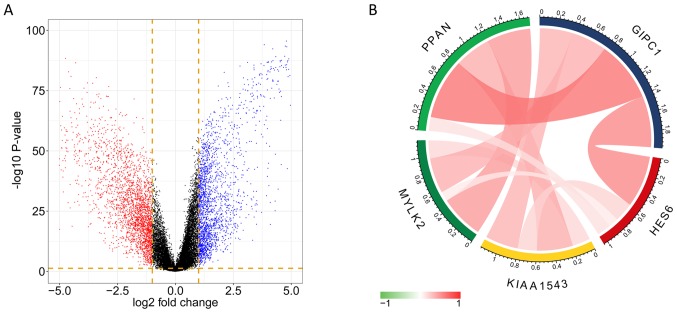
The DEGs were obtained from the analysis of RNAseq datasets, and a total of 2,169 significant genes were identified (Q-value<0.05; fold change >2) from the comparison of TNBT tissues and para-tumor tissues. The volcano plot indicates the fold-change and P-value of DEGs (Fig. 6A). The overlap between the genes discovered in DEG analysis and the genes in the tan module comprised 42 genes, which were selected as hub-genes. Among the 42 genes, 5 of which showed high correlation between expression level and survival probability, and were used for further analysis.
Figure 6.
Differentially expressed gene analysis and correlation of the 5 selected genes. (A) The volcano plot for differentially expressed genes in the tumor group and the para-tumor group. (B) Correlation between the 5 selected genes.
Survival and expression of the 5 selected genes
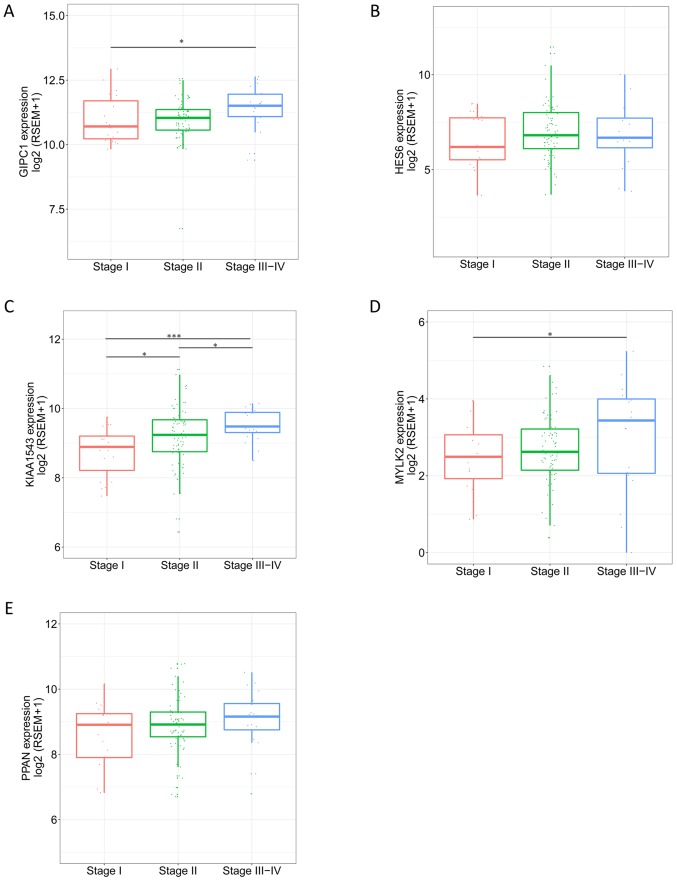
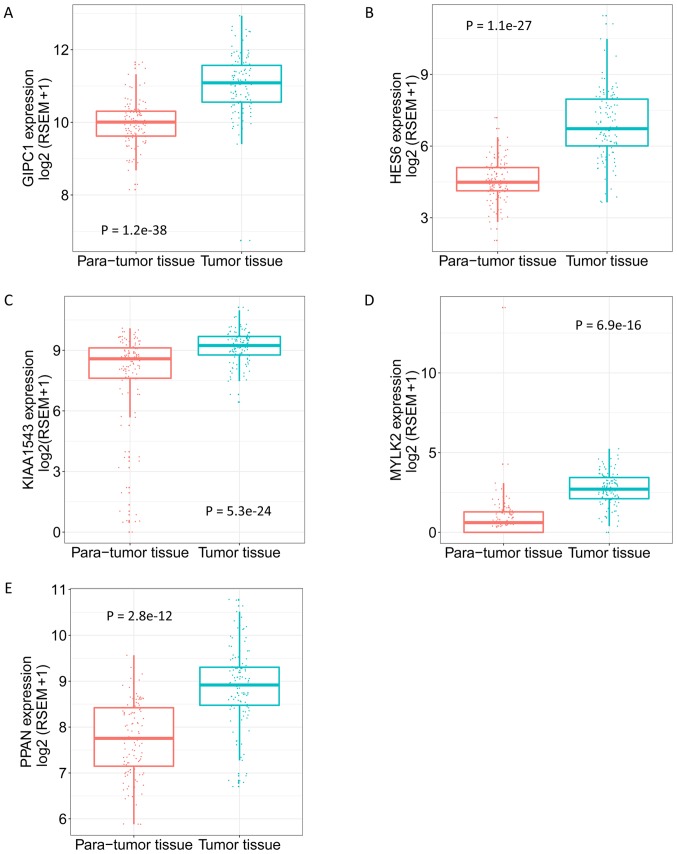
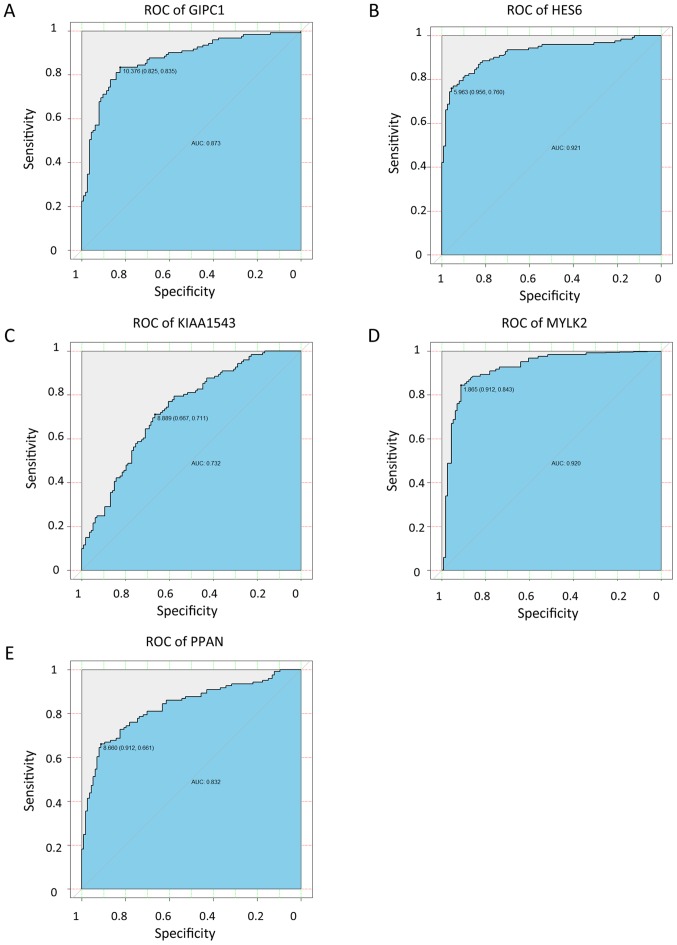
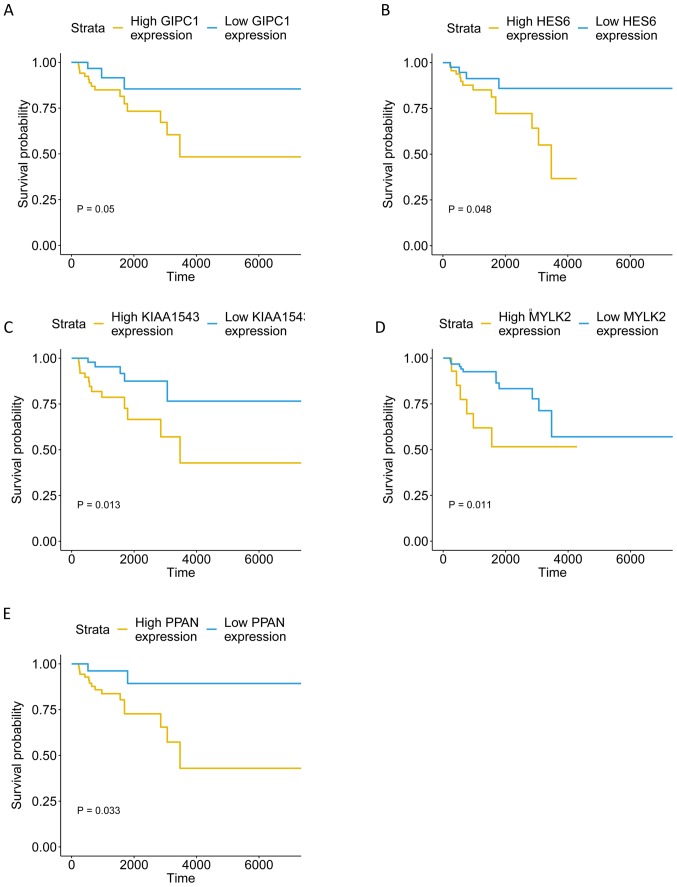
From the aforementioned hub-genes, 5 genes [GIPC PDZ domain containing family member 1 (GIPC1), hes family bHLH transcription factor 6 (HES6), calmodulin-regulated spectrin-associated protein family member 3 (KIAA1543), myosin light chain kinase 2 (MYLK2) and peter pan homolog (PPAN)] were selected to explore their association with the pathological progress. The unsupervised clustering results illustrate the co-expression of the 5 genes in the tan module and the extent of correlation between the 5 genes in TNBC was determined. Fig. 6B indicates high correlation between HES6 and GIPC1, and also between PPAN and GIPC1. The expression level of the genes in different pathological stages varied, and tended to be higher in more progressed pathological stages (Fig. 7). The expression levels of the 5 genes in tumor and para-tumor tissues were also plotted in Fig. 8, and showed significantly higher expression in tumor tissues (P<0.01). The ROC curves indicate that GIPC1, HES6, KIAA1543, MYLK2 and PPAN all exhibited high diagnostic accuracy for the identification of para-tumor and tumor tissues (Fig. 9). The KM curves illustrate that the expression levels of the 5 genes were associated with the OS of patients with TNBC (Fig. 10). Furthermore, high-expression groups of KIAA1543, MYLK2 and PPAN were significantly associated with lower OS times when compared with low-expression of the same proteins (P<0.05).
Figure 7.
Expression of GIPC1, HES6, KIAA1543, MYLK2, and PPAN in different pathological stages of triple-negative breast cancer. (A) GIPC1. (B) HES6. (C) KIAA1543. (D) MYLK2. (E) PPAN. *P<0.05 and ***P<0.005 indicated a significant difference between groups. Wilcoxon signed-rank test was used to evaluate the statistical significance of differences. GIPC1, GIPC PDZ domain containing family member 1; HES6, hes family bHLH transcription factor 6; KIAA1543, calmodulin-regulated spectrin-associated protein family member 3; MYLK2, myosin light chain kinase 2; PPAN, peter pan homolog.
Figure 8.
Expression of GIPC1, HES6, KIAA1543, MYLK2 and PPAN in para-tumor and tumor tissues. (A) GIPC1. (B) HES6. (C) KIAA1543. (D) MYLK2. (E) PPAN. Adjusted Q value, determined using the R Limma package, was used to evaluate the statistical significance of differences. GIPC1, GIPC PDZ domain containing family member 1; HES6, hes family bHLH transcription factor 6; KIAA1543, calmodulin-regulated spectrin-associated protein family member 3; MYLK2, myosin light chain kinase 2; PPAN, peter pan homolog.
Figure 9.
ROC curve of GIPC1, HES6, KIAA1543, MYLK2 and PPAN expression levels to differentiate tumor and para-tumor tissues. (A) GIPC1. (B) HES6. (C) KIAA1543. (D) MYLK2. (E) PPAN. ROC, receiver operating characteristic; GIPC1, GIPC PDZ domain containing family member 1; HES6, hes family bHLH transcription factor 6; KIAA1543, calmodulin-regulated spectrin-associated protein family member 3; MYLK2, myosin light chain kinase 2; PPAN, peter pan homolog.
Figure 10.
Overall survival of the 5 hub-genes in triple-negative breast cancer. (A) GIPC1. (B) HES6. (C) KIAA1543. (D) MYLK2. (E) PPAN. GIPC1, GIPC PDZ domain containing family member 1; HES6, hes family bHLH transcription factor 6; KIAA1543, calmodulin-regulated spectrin-associated protein family member 3; MYLK2, myosin light chain kinase 2; PPAN, peter pan homolog.
Discussion
Breast cancer is an umbrella term summarizing carcinomas originating from the breast, and is one of the most prevalent cancer types worldwide. TNBCs are described as breast cancers that simultaneously lack expression of the ER, PR and HER2 genes. Triple-negative tumors represent 80% of basal-like molecular breast cancers (1). They have less favorable outcomes compared with other molecular subtypes of breast cancer, as they are commonly associated with a higher risk of early relapse and poor survival (11). Currently, no drug is available that specifically targets TNBC, and the results of chemotherapy are unsatisfactory. The molecular characteristics of TNBC include disruption of BRCA1 DNA repair associated (BRCA)-1 function (12,13). The low expression level of BRCA-1 and inhibitor of DNA binding 4, HLH protein (ID4) lead to the change from homologous repair (HR) to non-homologous end joining (NHEJ) or single-strand annealing (SSA) pathways (14,15). HR is the most important DNA repair mechanism in healthy tissues and maintains genomic stability. However, the change to NHEJ (the alternative, more error-prone DNA-repair mechanism) can lead to genetic instability in tumor tissues. The inhibitor olaparib, which targets PARP-1, can block the NHEJ of breast cancer types and ultimately inhibit DNA repair (16). Moreover, epigenetic dysregulation is well established as having a crucial role in cancer pathology and progression (17,18), and the current study identified enrichment of epigenetic regulation in TNBC.
However, the molecular characteristics of TNBC are not well understood. Exploring the mechanisms involved in the progress and prognosis of TNBC would be helpful for improving diagnosis and treatment, and new targets or biomarkers are required. The present study determined a number of candidates using TGCA RNAseq datasets downloaded from UCSC database to identify promising biomarkers related to TNBC progression. WGCNA is a commonly used bioinformatics analysis tool used to identify the key modules and genes, which are associated with specific clinical traits. A study applied WGCNA analysis in breast cancer, identifying the association between cyclin B2 (CCNB2), F-box protein 5 (FBXO5), kinesin family member 4A (KIF4A), minichromosomal maintenance 10 replication initiation factor (MCM10) and TPX2 microtubule nucleation factor (TPX2) expression, and the survival of breast cancer patients (19). Another study used WGCNA to reveal the association between ATRX chromatin remodeler (ATRX) and TNBC (20). In the present study, WGCNA was performed to determine the association between American Joint Committee on Cancer-TNM stage and the gene co-expression module. By overlapping the genes from the tan module and DEG analysis, 42 hub genes were identified. Of these 42 genes, GIPC1, HES6, KIAA1543, MYLK2 and PPAN were selected to validate the strength of association with TNBC progression.
GIPC1 (also known as C19orf3) is a cytoplasmic protein that acts as an adaptor protein, linking receptor interactions to intracellular signaling pathways, including cell cycle regulation (21). GIPC1 protein is highly expressed both in cultured human breast cancer cells and in primary and metastatic tumor tissues (22). It is a cancer-associated immunogenic antigen in breast cancer which is also associated with bone metastasis development in breast cancer (23). The functions of GIPC1 include the regulation of apoptotic cell death, G2 cell-cycle arrest, modified cell adhesion and the migration of breast cancer cells (24). In the present analysis, GIPC1 was significantly upregulated in both triple-negative breast tumor tissue and a progressed pathological stage.
HES6 encodes a subfamily of basic helix-loop-helix transcription repressors with homology to the Drosophila enhancer of split genes (25). The overexpression of HES6 is found in metastatic carcinomas of different origins. Hes6 ectopic expression stimulates cell proliferation, not only in breast cancer T47D cells, but also in breast tumor growth in xenografts. The overexpression of HES6 also led to the induction of E2F transcription factor 1, a crucial target gene for the transcriptional repressor HES1 (26). Furthermore, the ER α-negative breast cancer cell lines MDA-MB-231 and SKBR3express 4 to 10 times higher levels of Hes-6 mRNA, compared with ERα-positive T47D and MCF-7 cells. The aforementioned studies complement the present findings of an association between HES6 expression and estrogen in TNBC.
KIAA1543, also known as Nezha, was shown to bridge the minus ends of non-centrosome anchored microtubules with p120, which plays a crucial role in stabilizing cadherin-catenin mediated cell-cell adhesion complexes (27,28). KIAA1543 is also involved in epithelial-mesenchymal transition, and mediates the interaction between cadherin and microtubules. It is overexpressed in invasive lobular carcinomas but its role is poorly characterized (29,30). The underlying mechanisms of KIAA1543 on TNBCs need to be further established. The current study found KIAA1543 to be significantly upregulated in TNBC tissues and at advanced pathological stages. Moreover, high KIAA1543 expression was associated with poor OS in TNBC patients.
MYLK2 encodes a calcium/calmodulin-dependent serine/threonine kinase (31). A somatic mutation in, or amplification of, MYLK2 has been detected in several cancer tissues, compared with normal tissues (31). Moreover, proteomic analyses conducted on serum protein profiling results revealed the upregulation of MYLK2 in pancreatic cancer patients (32). The present study showed that in TNBC tissues, MYLK2 was overexpressed and was also associated with the poor patient OS.
PPAN encodes an evolutionarily conserved protein similar to the Drosophila gene peter pan. A signature inferred from Drosophila mitotic genes revealed an association between PPAN expression and the survival of breast cancer patients (33). Tumor patients with high expression levels of PPAN have been shown to respond more favorably to treatment with anti-EGFR antibodies such as cetuximab (34). The data presented reveal a significant association between high PPAN expression and a poor OS in patients with TNBC.
Taken together, WGCNA and DEG analysis on RNAseq datasets from TNBC tissues revealed that the tan module was the most significantly associated with AJCC-TNM stage. The genes in the tan module showed high inter-connectivity with the co-expression network. Furthermore, GO and KEGG analysis revealed the enrichment of the genes in the related terms from GO. The overlap between the module and DEG analysis identified 42 genes, 5 of which were negatively associated with the OS of patients with TNBC. Therefore, the current study provides 5 novel biomarkers for TNBC, which exhibit potential as targets in the diagnosis and treatment of the disease.
Acknowledgements
Not applicable.
Funding
This study was supported by the Jinhua technology division fund (grant no. 2018-4-120).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available in the University of California, Santa Cruz repository, (https://xenabrowser.net/datapages/).
Authors' contributions
MB, JW, LB and KZ conceived the experiment design. LB, JW, KZ and TG performed the data analysis. JW and KZ revised the manuscript. LB, TG, JW, KZ and MB wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Dawood S. Triple-negative breast cancer: Epidemiology and management options. Drugs. 2010;70:2247–2258. doi: 10.2165/11538150-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 3.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 4.Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 5.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Zhang Q, Gao Z, Xin S, Zhao Y, Zhang K, Shi R, Bao X. A novel 4-gene signature for overall survival prediction in lung adenocarcinoma patients with lymph node metastasis. Cancer Cell Int. 2019;19:100. doi: 10.1186/s12935-019-0822-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang K, Maruyama T, Fan G. The naive state of human pluripotent stem cells: A synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell. 2014;15:410–415. doi: 10.1016/j.stem.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Xin S, Zhang K, Shi R, Bao X. Low GAS5 levels as a predictor of poor survival in patients with lower-grade gliomas. J Oncol. 2019;2019:1785042. doi: 10.1155/2019/1785042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smyth GK. Limma: linear models for microarray data. Bioinformatics and computational biology solutions using R and Bioconductor. In: Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Statistics for Biology and Health. Springer; New York, NY: 2005. pp. 397–420. [DOI] [Google Scholar]
- 10.Kassambara A, Kosinski M, Biecek P. Survminer: Drawing Survival Curves using ‘ggplot2’. R package version 03. 2017 Jan [Google Scholar]
- 11.Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, Harris L, Hait W, Toppmeyer D. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–5657. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 12.Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, Hortobagyi GN, Arun BK. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26:4282–4288. doi: 10.1200/JCO.2008.16.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: Therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 14.Powell SN, Kachnic LA. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene. 2003;22:5784–5791. doi: 10.1038/sj.onc.1206678. [DOI] [PubMed] [Google Scholar]
- 15.Evers B, Helleday T, Jonkers J. Targeting homologous recombination repair defects in cancer. Trends Pharmacol Sci. 2010;31:372–380. doi: 10.1016/j.tips.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Deng H, Xin S, Zhang K, Shi R, Bao X. Prognostic and predictive value of three DNA methylation signatures in lung adenocarcinoma. Front Genet. 2019;10:349. doi: 10.3389/fgene.2019.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang J, Kong D, Cui Q, Wang K, Zhang D, Gong Y, Wu G. Prognostic genes of breast cancer identified by gene co-expression network analysis. Front Oncol. 2018;8:374. doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan L, Xu B, Chen Q, Jiang J, Shen Y. Weighted correlation network analysis of triple-negative breast cancer progression: Identifying specific modules and hub genes based on the GEO and TCGA database. Oncol Lett. 2019;18:1207–1217. doi: 10.3892/ol.2019.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valdembri D, Caswell PT, Anderson KI, Schwarz JP, König I, Astanina E, Caccavari F, Norman JC, Humphries MJ, Bussolino F, Serini G. Neuropilin-1/GIPC1 signaling regulates alpha5beta1 integrin traffic and function in endothelial cells. PLoS Biol. 2009;7:e25. doi: 10.1371/journal.pbio.1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudchenko S, Scanlan M, Kalantarov G, Yavelsky V, Levy C, Estabrook A, Old L, Chan GL, Lobel L, Trakht I. A human monoclonal autoantibody to breast cancer identifies the PDZ domain containing protein GIPC1 as a novel breast cancer-associated antigen. BMC Cancer. 2008;8:248. doi: 10.1186/1471-2407-8-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westbrook JA, Cairns DA, Peng J, Speirs V, Hanby AM, Holen I, Wood SL, Ottewell PD, Marshall H, Banks RE, et al. CAPG and GIPC1: Breast cancer biomarkers for bone metastasis development and treatment. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chittenden TW, Pak J, Rubio R, Cheng H, Holton K, Prendergast N, Glinskii V, Cai Y, Culhane A, Bentink S, et al. Therapeutic implications of GIPC1 silencing in cancer. PLoS One. 2010;5:e15581. doi: 10.1371/journal.pone.0015581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000;127:2933–2943. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- 26.Hartman J, Lam EW, Gustafsson JA, Ström A. Hes-6, an inhibitor of Hes-1, is regulated by 17beta-estradiol and promotes breast cancer cell proliferation. Breast Cancer Res. 2009;11:R79. doi: 10.1186/bcr2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akhmanova A, Hoogenraad CC. Microtubule minus-end-targeting proteins. Curr Biol. 2015;25:R162–R171. doi: 10.1016/j.cub.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 28.Fu Y, Zheng S, An N, Athanasopoulos T, Popplewell L, Liang A, Li K, Hu C, Zhu Y. β-catenin as a potential key target for tumor suppression. Int J Cancer. 2011;129:1541–1551. doi: 10.1002/ijc.26102. [DOI] [PubMed] [Google Scholar]
- 29.Castellana B, Escuin D, Pérez-Olabarria M, Vázquez T, Muñoz J, Peiró G, Barnadas A, Lerma E. Genetic up-regulation and overexpression of PLEKHA7 differentiates invasive lobular carcinomas from invasive ductal carcinomas. Hum Pathol. 2012;43:1902–1909. doi: 10.1016/j.humpath.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka N, Meng W, Nagae S, Takeichi M. Nezha/CAMSAP3 and CAMSAP2 cooperate in epithelial-specific organization of noncentrosomal microtubules. Proc Natl Acad Sci USA. 2012;109:20029–20034. doi: 10.1073/pnas.1218017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soung YH, Lee JW, Kim SY, Nam SW, Park WS, Lee JY, Yoo NJ, Lee SH. Mutational analysis of the kinase domain of MYLK2 gene in common human cancers. Pathol Res Pract. 2006;202:137–140. doi: 10.1016/j.prp.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Rong Y, Jin D, Hou C, Hu J, Wu W, Ni X, Wang D, Lou W. Proteomics analysis of serum protein profiling in pancreatic cancer patients by DIGE: Up-regulation of mannose-binding lectin 2 and myosin light chain kinase 2. BMC Gastroenterol. 2010;10:68. doi: 10.1186/1471-230X-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damasco C, Lembo A, Somma MP, Gatti M, Di Cunto F, Provero P. A signature inferred from drosophila mitotic genes predicts survival of breast cancer patients. PLoS One. 2011;6:e14737. doi: 10.1371/journal.pone.0014737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabernero J, Cervantes A, Rivera F, Martinelli E, Rojo F, von Heydebreck A, Macarulla T, Rodriguez-Braun E, Eugenia Vega-Villegas M, Senger S, et al. Pharmacogenomic and pharmacoproteomic studies of cetuximab in metastatic colorectal cancer: Biomarker analysis of a phase I dose-escalation study. J Clin Oncol. 2010;28:1181–1189. doi: 10.1200/JCO.2009.22.6043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the present study are available in the University of California, Santa Cruz repository, (https://xenabrowser.net/datapages/).