Abstract
Background:
Methylmalonic acidemia (MMA) and propionic acidemia (PA) are related disorders of mitochondrial propionate metabolism, caused by defects in methylmalonyl-CoA mutase (MUT) and propionyl-CoA carboxylase (PCC), respectively. These biochemical defects lead to a complex cascade of downstream metabolic abnormalities, and identification of these abnormal pathways has important implications for understanding disease pathophysiology. Using a multi-omics approach in cellular models of MMA and PA, we identified serine and thiol metabolism as important areas of metabolic dysregulation.
Methods:
We performed global proteomic analysis of fibroblasts and untargeted metabolomics analysis of plasma from individuals with MMA to identify novel pathways of dysfunction. We probed these novel pathways in CRISPR-edited, MUT and PCCA null HEK293 cell lines via targeted metabolomics, gene expression analysis, and flux metabolomics tracing utilization of 13C-glucose.
Results:
Proteomic analysis of fibroblasts identified upregulation of multiple proteins involved in serine synthesis and thiol metabolism including: phosphoserine amino transferase, cystathionine beta synthase and mercaptopyruvate sulfurtransferase. Metabolomics analysis of plasma revealed significantly increased levels of cystathionine and glutathione, central metabolites in thiol metabolism. CRISPR edited MUT and PCCA HEK293 cells recapitulate primary defects of MMA and PA and have upregulation of transcripts associated with serine and thiol metabolism including PSAT1 13C- Glucose flux metabolomics in MUT and PCCA null HEK293 cells identified increases in serine de novo biosynthesis, serine transport, and abnormal downstream TCA cycle utilization.
Conclusion:
We identified abnormal serine metabolism as a novel area of cellular dysfunction in MMA and PA, thus introducing a potential new target for therapeutic investigation.
Keywords: Methylmalonic acidemia, propionic acidemia, serine metabolism
1. INTRODUCTION
Serine is a non-essential amino acid that can be synthesized de novo from glucose via 3-phosphoglycerate, a glycolytic and gluconeogenic intermediate. Serine has multiple critical roles in the synthesis of fundamental cellular metabolites including phosphatidylcholine, creatine, one-carbon metabolites, and S-adenosylmethionine, the key methyl donor in cells [1].
Recently, alterations in serine metabolism have been described in several models of mitochondrial disease including the Deletor mouse model of mtDNA deletions, as well as in affected humans with POLG-associated disease, Myoclonic epilepsy with ragged red fibers (MERRF) and Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) [2–4]. Evidence points towards redirected flux towards one carbon metabolism and glutathione as a protective mechanism for managing mitochondrial bioenergetic stress. Interestingly, similar changes have also been reported in multiple studies of various forms of malignancy [5–8], suggesting that serine metabolism represents a central response to diverse forms of mitochondrial metabolic challenges. In this study, we explored the role of serine metabolism in propionic acidemia (PA) and methylmalonic acidemia (MMA).
PA and MMA are closely related autosomal recessive disorders of isoleucine and valine metabolism and result in defective entry of propiogenic metabolites into the TCA cycle at the level of succinyl CoA [9, 10]. PA is caused by genetic defects in propionyl CoA carboxylase (encoded by PCC and PCCB). MMA is caused by genetic defects in methylmalonyl-CoA mutase (encoded by MUT), or in vitamin B12 metabolism, among other rarer causes [10–13]. There is a strong clinical overlap between these disorders, but also important phenotypic differences [9, 14–16]. In both disorders, there are multiple layers of evidence for mitochondrial dysfunction and oxidative stress[17–19].
We used global proteomic analysis to identify novel pathways of cellular dysfunction in fibroblasts from individuals with PA and MMA. In both disorders, we uncovered enrichment of proteins involved in serine biosynthesis and thiol metabolism. We next employed untargeted metabolomics to evaluate the plasma of individuals with PA and MMA and identified increased excretion of key metabolites associated with serine metabolism, cystathionine and glutathione.
In order to obtain further mechanistic insight into disturbances of serine metabolism in PA and MMA, we developed CRISPR/Cas9 edited PCCA-null and MUT-null HEK293 cell lines, which showed increased expression of genes associated with serine metabolism, notably phosphoserine amino transferase (PSAT). Finally, we employed flux metabolomics and showed significant increases in de novo serine metabolism in these cells.
2. METHODS
2.1. Fibroblast cell culture.
Primary fibroblast lines were derived from unrelated individuals with mut (0 (null)) MMA (n=2), PA (n=2), and healthy controls (n=2). Patient fibroblast experiments and the collection of clinical history had institutional IRB approval via Johns Hopkins University protocols NA_00026986 and NA_00036487. Individuals were diagnosed with MMA via lack of MUT enzyme activity in fibroblasts, the molecular confirmation of two pathological variants in MUT, and a clinical history of diagnostic urine organic acid analysis that includes massive elevations of methylmalonic acid. Individuals were diagnosed with PA via lack of functional PCC enzyme activity in fibroblasts, the molecular confirmation of two pathological variants in PCCA, and a clinical history of diagnostic urine organic acid analysis that includes massive elevations of propionylglycine, methylcitrate, and tiglyglycine. Additionally, all 4 individuals, with MMA or PA had a clinical history consistent with the infantile onset, severe form of the respective disease. The primary fibroblast cell lines were grown in RPMI 1640 medium (Gibco, Thermo Fisher Scientific) with 10% FBS (Gemini Bio-Products) at 37°C with 5% CO, unless otherwise noted. Control fibroblasts were obtained from healthy individuals without a known metabolic disorder.
Prior to harvesting, each primary fibroblast cell line (n=2 for each condition) was grown to 70% confluence and then maintained in serum-free medium for 12 hours. Cells were lysed and the protein concentration was estimated using BCA (Pierce) assay. Sample processing for proteomics was performed based on modified filter aided sample preparation (FaSp) protocol. 100 |jg of protein lysate from each cell line was subjected to filtration using 30 kDa filters and 8M urea to reduce the amount of SDS in the lysate. The reduced protein lysate was alkylated using 20 mM iodoacetamide (IAA) and incubated for 10 minutes in the dark at room temperature (RT). Prior to digestion, the concentration of urea was reduced to < 1M using 50mM Triethylammonium bicarbonate buffer (TEABC). Trypsin digestion was performed using 6-(1-tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK) treated trypsin (Promega) at an enzyme: substrate ratio of 1:20 for 12–16 hours at 37°C.
2.2. Global Proteomics
2.2.1. Tandem mass tag (TMT) labeling and Reversed phase liquid chromatography (RPLC) fractionation of proteins.
Digested peptide samples (n=2 for each condition) were labeled using TMT 6-plex reagents as per manufacturer’s instruction (Thermo Fischer Scientific). Briefly, peptide digests from each condition were reconstituted in 50 mM TEABC buffer and mixed with the TMT reagent and incubated at RT for 1 h. After incubation, the reaction was quenched by adding 5% hydroxylamine. The TMT-labeled samples were pooled at 1:1:1:1:1:1 ratio, evaporated to dryness and subjected to desalting using Sep-Pak C18 cartridges.
Peptides were fractionated by bRPLC [20]. Briefly, TMT-labeled and lyophilized peptide mixtures were resuspended in buffer A (10 mM TEABC) and fractionated by bRPLC chromatography on an Agilent 1100 LC system using a linear gradient of 8–60% buffer B (10 mM TEABC in 90% acetonitrile (ACN)) for 60 min at a flow rate of 1 mL/min. A total of 96 fractions were collected, concatenated to 12 fractions, vacuum dried and stored at −80 C until LC-MS/MS analysis.
2.2.2. LC-MS/MS analysis.
TMT-labeled samples were analyzed on LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) interfaced with Proxeon Easy nLC system (Thermo Fisher Scientific). Each fraction was loaded and concentrated on a on a trap column (75μm × 2cm) packed in-house using C18 material (Magic C18AQ, 5pm, 100Å, Michrom Biosciences Inc.) with a flow rate of 3μL/min and resolved on an analytical column (75 μm × 30 cm, Magic C18AQ, 5um, 100A, Michrom Biosciences Inc.) at a flow rate of 200 nl/min using a linear gradient of 5–30% ACN over 90 min. Precursor MS scan (m/z 350–1,700) and MS/MS was acquired with a mass resolution of 60,000 and 30,000 at 400 m/z in orbitrap mass analyzer respectively. The top ten intense peaks were selected for MS/MS fragmentation in each scan and fragmented using higher-energy collision dissociation (HCD) mode with 45 % normalized collision energy. The isolation width was set to 1.9 m/z. Acquired ions selected in a given scan were selectively excluded for 30 sec. The maximum ion accumulation time was set to 200 msec for MS and 300 msec for MS/MS scans. The lock mass option was enabled using polysiloxane ion (m/z, 445.120025) from ambient air for internal calibration.
2.2.3. Proteomic data analysis.
Mass spectrometry-derived data were processed and quantified using Proteome Discoverer (Version 1.4) software suite (Thermo Fisher Scientific). The data were searched against NCBI Human RefSeq protein database (Release 59) containing common contaminants using Mascot (version 2.2.0) and Sequest search algorithms. The precursor mass tolerance was set to 10 ppm and the fragment mass tolerance was set to 0.05 Da. The search parameters included oxidation of methionine set as a dynamic modification, carbamidomethylation of cysteine (+57.0215 Da) and TMT tag (+229.1629 Da) at N-termini of peptides and lysine residues. A maximum of 2 missed cleavage were allowed for tryptic peptides. Identified peptides were validated using a Percolator algorithm with a q-value threshold of 0.01. The ratios were calculated by the quantitation node in proteome discoverer.
Protein signals (as a proxy for quantity) were analyzed as log base 10 ratios of relative abundance between cell lines. Using the statistical program “R” for analysis, we confirmed that each result set showed a normal Gaussian distribution, (R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/). In order to increase the specificity and avoid false positives, only proteins that were within two standard deviations of the mean within genotypes (MMA1 vs. MMA2, PA1 vs. PA2, WT1 vs. WT2) were considered for further analysis. Therefore, proteins which were highly discordant in abundance within each genotype were discarded. Subsequently, in order to determine differentially expressed proteins in MMA and PA fibroblast cell lines, the ratios of protein abundance were analyzed as log base 10 ratios of relative abundance between disease groups (MMA vs. control, PA vs. control, MMA vs. PA). Proteins that were at least 2 standard deviations from the mean were assessed for functional annotation.
2.2.4. Functional annotation of differentially expressed proteins in fibroblasts.
To appreciate the functional characteristics of the differentially expressed proteins, we employed the DAVID Bioinformatics Resource v6.7 to determine the KEGG pathways and GO TERMS that are enriched proteins differentially expression in PA and MMA as compared to WT [21, 22]. Pathways and GO terms were tested for significant enrichment via Fisher Exact test.
2.3. Untargeted metabolomics analysis
2.3.1. Metabolomics subjects
Fifty-two plasma samples were selected to identify discriminating biomarkers that correlate with PA and MMA Samples included: 17 samples from individual with phenotypes consistent with the classical, neonatal onset form of PA (average age 10y 9.6m; age range 4y 2m to 25y 9.9m), 15 samples from individuals with the classical mut(0) type of MMA (average age 12y 3m; age range 8m to 31y0m, and 20 sex-matched control samples (average age 9y 8.8m; age range 7m to 25y 6m). Control samples were obtained from individuals who underwent routine laboratory testing and were not known to have a genetic or metabolic condition. All samples were acquired during a “well” outpatient visit to mitigate effects of intercurrent illness or metabolic decompensation. Patient plasma experiments had institutional IRB approval via Johns Hopkins University protocol NA_00069372.
2.3.2. Metabolomics sample preparation and analysis
Sample preparation, data acquisition and analysis were performed as previously described [23–25]. Briefly, each of the fifty-two plasma samples were prepared by adding 400 μl cold methanol with tryptophan-d5 internal standard to 50 μl of plasma for protein precipitation. After centrifugation at 16,000 rcf, 350 μl of supernatant was lyophilized overnight and re-constituted in 100 μl of 95:5 water:methanol. In addition, phenotypic pooled samples were prepared by combining 30 ul from each of the study samples belonging to the same phenotype (MMA, PA, Control). In addition, total pools were created by taking a 250 ul aliquot from each of the phenotypic pools. Pooled plasma samples (phenotypic and total) were prepared identically to the individual study samples.
Samples were analyzed on a SYNAPT G2-Si QTOF mass spectrometer coupled to an Acquity UPLC (Waters Corporation, MA). Prior to analyzing the study samples, the column and the system were equilibrated with five injections of quality control samples. The compounds were separated on a Waters Acquity UPLC HSS T3 column (2.1 × 100 mm, 1.8 μm particle size) operating at 50°C using a reversed-phase chromatographic method. A gradient mobile phase consisting of water with 0.1% formic acid (A) and methanol with 0.1% formic acid (B) were used. All MS data were collected over 50–1000 m/z in electrospray ionization (ESI) positive ion mode. Leucine enkephalin was used as the lock mass and a lock mass scan was collected every 45 sec and averaged over 3 scans to perform correction for any mass drift over the course of the analytical run. Source and desolvation temperatures were set at 110 °C and 400 °C, respectively.
2.3.3. Metabolomics Data Processing.
The raw mass spectrometric data was processed (alignment and peak picking) using Progenesis QI (Waters Corporation). The normalized data were mean centered and scaled by dividing by the standard deviation and multivariate analysis methods (principal component analysis (PCA), orthogonal partial least squares discriminant analysis (OPLS-DA)) were used to reduce the dimensionality and to visualize the study groups (SIMCA 14.1, MKS Umetrics, Umea, Sweden) [26, 27]. The PCA scores plot was used to ensure that the quality control samples clustered in the center of the study samples from which they were derived, which has become a standard method for assessing the quality of untargeted metabolomics data. OPLS-DA, which is a supervised analysis method, was used to identify peaks important to differentiating the study groups. Loadings plots and variable influence on projection (VIP) plots were inspected. All models used a 7-fold crossvalidation to assess the predictive variation of the model (Q2). Descriptive statistics and t-tests using the Satterthwaite approximation for unequal variances were conducted using SAS 9.4 (SAS Institute Inc, Cary, NC). In this exploratory study, p-values were not adjusted for multiple testing. Peaks that had a VIP ≥ 2.0 with a jack-knife confidence interval that did not include 0 or had a p-value < 0.001 were determined to be important for differentiating the study groups.
Putative identification of the signals differentiating the study groups was made using a database search against the ERCMRC’s in-house exact-mass-retentiontime library of standards and the Human Metabolome Database (HMDB). Peaks that could not be library-matched were classified as unknown.
2.4. MUT-null and PCCA-null CRISPR/Cas9 edited cells
2.4.1. Development of MUT-null and PCCA-null CRISPR/Cas9 edited HEK293 cells.
Cas9-nickase with two adjacent guide RNAs (gRNAs) was used to make the null HEK293 cell lines as previously described [28]. MUT exon 5 and PCCA exon 12 were targeted for editing, based on the location of previously reported human disease-causing mutations [12, 29]. gRNAs were designed using http://tools.genome-engineering.org(Supplementary Table 1) [30]. Synthesized gRNAs were inserted into a CR-BluntII-TOPO plasmid (Invitrogen) and transformed into E.coli. A plasmid co-expressing Cas9 and GFP (Addgene) was co-transfected with the gRNA-plasmids into WT HEK293 cells via lipofectamine. GFP positive single cells were FACS sorted into 96-well plates. Positive clones were identified by PCR and Sanger sequencing (Supplementary Table 2). For whole cell lysate extraction, confluent culture dishes were lysed with RIPA lysis buffer (1% (v/v) Triton X-100, 20 mm HEPES-KOH, pH 7.4, 50 mm NaCl, 1 mm EDTA, 2.5 mm MgCl2, 0.1% (w/v) SDS) spiked with 1 mm PMSF). Insoluble material was removed by centrifugation, the supernatant collected, and protein quantified using a BCA (Pierce). Absence of expressed PCCA (Novus Biologicals) or MUT (Abcam) protein was then confirmed in whole cell lysates via immunoblot of 3 individual cell passages for each genotype.
HEK293 cells were obtained from Thermo Fisher Scientific (Waltham, MA). Cells were cultured with DMEM (Corning Cellgro, ThermoFisher Scientific) supplemented with 10% FBS (Gemini Bio-Products), and 1% L-glutamine (Gibco, Thermo Fisher Scientific), and grown at 37°C with 5% CO, unless otherwise noted.
2.4.2. Metabolite measurements in HEK293 cells.
Acylcarnitines were analyzed in spent media from confluent cell cultures at 4 days, 11 days and 18 days after cell plating via tandem mass spectrometry (MS/MS) without chromatographic separation in the positive-ion mode using multiple-reaction monitoring (MRM) as previously published [31, 32]. Methylmalonate was quantified in spent media from confluent cell cultures at 4 days, 11 days and 18 days via stable-isotope dilution and gas chromatography/mass spectrometry (GC/MS) as previously published [33]. Metabolite measurements were performed with n=4 (4 individual cell culture wells assayed on every genotype for each metabolite at all time points).
2.4.3. Gene expression in HEK293 cells.
150,000 cells from each genotype (MUT-null, PCCA-null, and WT HEK293 cells) were plated in 6 well culture dishes. Following plating, gene expression was measured at day 1, 2, 4 and 8. SYBR green (Bio-Rad) labeled reverse transcription-quantitative PCR (RT-qPCR) was carried out on cDNA derived from cells as previously described [34] for expression of the following genes: PSAT1, CBS, ASNS, SLC7A5, SLC3A2 and 18S rRNA (as a loading control) (Supplementary Table 3). Gene expression from each genotype at each time point was tested in triplicate. Each of the three measurements were made on cells derived from individual wells.
2.5. Glucose-13C6 flux metabolomics.
2.5.1. Cell preparation for flux metabolomics
225,000 cells from each genotype (MUT-null, PA-null, WT) were plated into five 100mm culture dishes (n=5 per genotype) in DMEM (Gibco, Thermo Fisher Scientific 11965092) with 10% FBS (Gemini Bio-Products) and 1% L-glutamine (Gibco, Thermo Fisher Scientific). When cells reached 70% confluency, log phase of growth, growth media was replaced with DMEM-no glucose (Gibco, Thermo Fisher Scientific 11966025), with 10% FBS (Gemini Bio-Products), 1% L-glutamine (Gibco, Thermo Fisher Scientific), and D-Glucose-13C6 to a final concentration of 4500 mg/L (Cambridge Isotope Laboratories, Inc.). After 8 hrs, cells were washed, and metabolites were extracted from each dish (n=5 for each genotype). An internal control (Human Metabolome Technologies, Inc.) was added to each sample, followed by ultrafiltration of cell extracts and sample evaporation. Samples were stored at −80 degrees Celsius.
2.5.2. Metabolite measurements.
The metabolite samples were further processed and analyzed by Human Metabolome Technologies (HMT). Briefly, the compounds were measured in the Cation and Anion modes of Capillary Electrophoresis Time-of-Flight Mass Spectrometry (CE-TOFMS) followed by metabolome analysis on a Agilent CE-TOFMS system (Agilent Technologies Inc.) Cationic samples were run in ESI positive mode and anionic samples were run in ESI negative mode [35, 36].
2.5.3. Data processing and quantification.
Peaks detected by CE-TOFMS analysis were extracted using automatic integration software (MasterHands ver. 2.17.1.11 developed at Keio University) in order to obtain peak information including m/z, migration time (MT), and peak area. Putative metabolites were then assigned from HMT’s target library and their isotopic ions on the basis of m/z and MT. Absolute quantification was performed for total amount of each detected metabolite. All the metabolite concentrations were calculated by normalizing the peak area of each metabolite with respect to the area of the internal standard and by using standard curves, which were obtained by single-point (100 μM) calibrations.
3. RESULTS
3.1. Differentially expressed proteins in fibroblasts from propionic acidemia (PA) and methylmalonic acidemia (MMA) patients.
Discovery proteomics analysis was performed on fibroblasts from individuals with PA (n=2), MMA (n=2), and WT (n=2) in order to generate hypotheses about novel adaptive pathways in MMA and PA, with the goal of performing targeted studies downstream to validate pathways of interest.
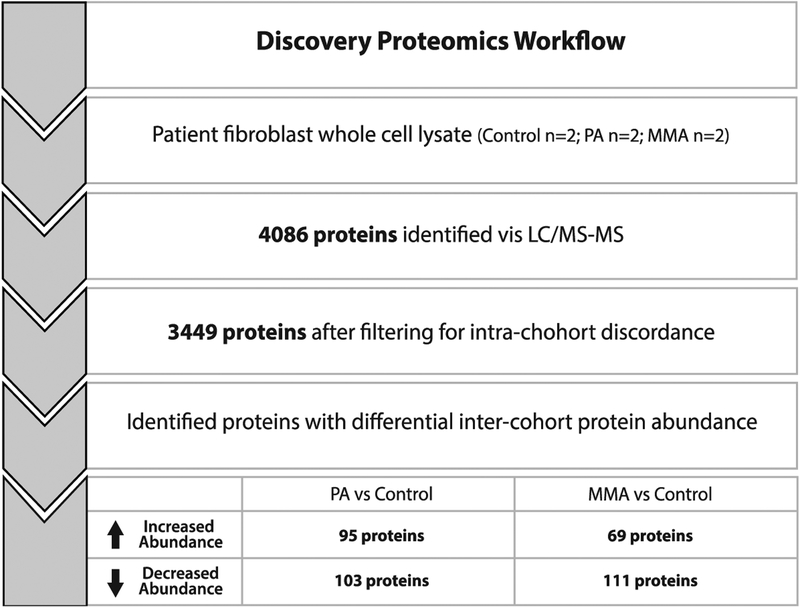
A total of 4,086 proteins were identified across the MMA, PA and WT fibroblast cohorts. To eliminate intra-cohort discordance, we removed all proteins that fell beyond 2 standard deviations of the mean protein abundance for each matched pair (i.e. MMA1/MMA2, PA1/PA2, and WT1/WT2). After this exclusion, 3,449 proteins were considered for inter-cohort analysis (Figure 1, Supplementary Figure 1).
Figure 1.
Workflow for proteomics analysis. A total of 4,086 proteins were identified across the MMA (n=2), PA (n=2) and WT (n=2) fibroblast cohorts. We removed all proteins that fell beyond 2 standard deviations of the mean protein abundance for each matched pair from further analysis. After this exclusion, 3,449 proteins were considered for further analysis to determine differentially expressed proteins between cases and controls. Proteins with abundances that that fell above +2 or below −2 standard deviations when comparing PA/Control and MMA/Control were then considered for functional analysis. MMA, methylmalonic acidemia. PA, propionic acidemia. CON, control.
We then identified proteins with significant differential expression between disease groups (intercohort) by selecting proteins that fell beyond 2 standard deviations from the mean protein abundance across all proteins measured (for each disease pair i.e. PA/WT and MMA/WT). In the control vs PA fibroblast cohort comparison, a total of 103 proteins fell at least 2 standard deviation below the mean, and 95 proteins fell at least 2 standard deviations above the mean. In the control vs MMA fibroblast cohort comparison, a total of 111 proteins fell at least 2 standard deviations below the mean, and 69 proteins fell at least 2 standard deviations above the mean (Figure 1). The 378 differentially expressed proteins were then assessed via in silico functional analysis using DAVID to determine KEGG pathways and GO TERMS enriched for the proteins of interest (Supplementary Table 4). Pathways were tested for significant enrichment via Fisher Exact test.
Of the 69 proteins with significantly increased abundance in the MMA fibroblast group, KEGG pathway analysis showed significant enrichment for proteins involved in the biosynthesis of amino acids (p=2.3e−3), and cysteine and methionine metabolism (p=8.2e−3). Significantly enriched GO TERMS included: cellular amino acid biosynthetic processes (p=1.1e−4), neutral amino acid transport (p=4.6e−2), hydrogen sulfide biosynthetic processes (p=1.4e−2), and transsulfuration (p=1.8e−2). Of the 95 proteins with significantly increased abundance in the PA fibroblast group, KEGG pathway analysis showed increased enrichment of proteins involved in biosynthesis of amino acids (approaching statistical significance, p=7.1e−2). Significantly enriched GO TERMS included: cellular amino acid biosynthetic processes (p=7.1e−3), hydrogen sulfide biosynthetic processes (p=1.9e−2), and transsulfuration (p=2.4e−2) and neutral amino acid transport (p=1.3e−2).
Of the 111 proteins with significantly decreased abundance in MMA fibroblast group, KEGG pathway analysis showed significant enrichment of proteins in pyruvate metabolism (p=3.8e−3), valine, leucine and isoleucine degradation (p=6.00e−3), among other pathways. Significantly enriched GO TERMS included: oxidation-reduction process (p=2.7e−4), autophagy (p=9.1e−3), and response to hypoxia (p=2.1e−2). Of the 103 proteins with significantly decreased abundance in PA fibroblast group, KEGG pathway analysis showed significant enrichment of proteins in the following pathways: valine, leucine and isoleucine degradation (p=4.60e−4), protein digestion and absorption (p=3.1e−2), and pyruvate metabolism (p=3.9e−2), and other pathways. Significantly enriched GO TERMS included: oxidation-reduction process (p=2.4e−3), response to hypoxia (p=5.1e−4) and branched-chain amino acid catabolic process (5.40e−3).
3.2. Plasma Metabolomics shows altered levels of metabolites in MMA and PA.
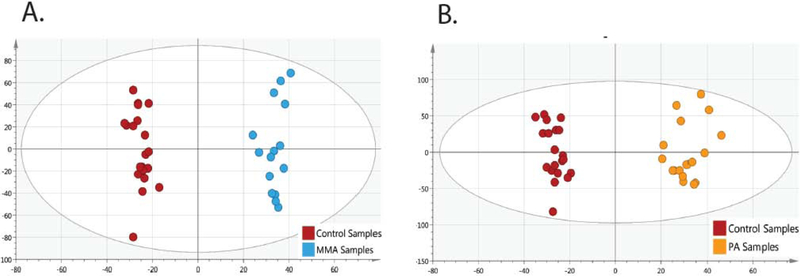
Supervised multivariate data analysis (OPLS-DA) showed clear differentiation of control subjects with MMA (Figure 2A, R2X=0.45, R2Y=0.98, Q2=0.75) and PA (Figure 2B, R2X=0.37, R2Y=0.97, Q2=0.78).
Figure 2.
OPLS-DA score plots show clustering of cohorts. Plots showing the separation of (A) MMA (blue filled circles) to control (red filled circles) and (B) and (orange filled circles) to control (red filled circles) (B) show that disease cohorts are tightly clustered, and there are no significant outliers. MMA, methylmalonic acidemia. PA, propionic acidemia.
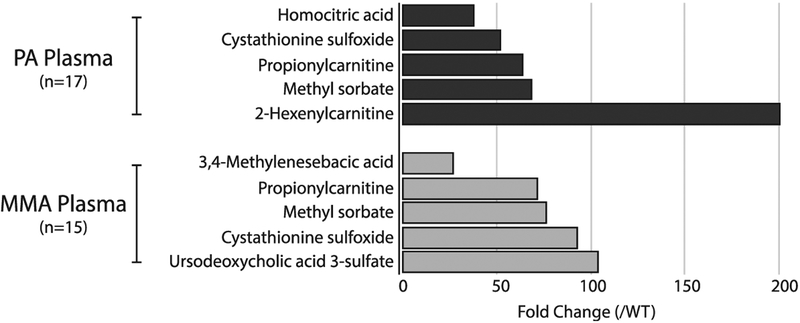
Seven hundred ninety five (795) peaks were identified as being important to differentiating individuals with PA compared to controls (VIP ≥ 2.0 with a jack-knife confidence interval that did not include 0 or a p-value < 0.001) and 60 peaks were library-matched to metabolites. Thirty four (34) individual metabolites were reduced compared with controls (range of −43.3 fold to −1.3 fold), and 25 metabolites were significantly increased compared with controls (range of 1.5 fold to 199.6 fold). Of the 5 metabolites with the most highly increased abundance, one is a known metabolite of propionic acid (propionylcarnitine, 64 fold increase). Cystathionine sulfoxide was increased 52-fold compared with controls, which is consistent with the upregulation of sulfur metabolism found in our proteomics studies. The other three most highly increased metabolites were homocitric acid, which was increased at 38-fold, methyl sorbate, which was increased at 67.9 fold, and 2-hexenoylcarnitine, which was increased 199.6 fold (Figure 3).
Figure 3.
Metabolites with increased abundance in propionic acidemia (PA) and methylmalonic acidemia (MMA) plasma samples. Fold change of top 5 increased plasma metabolites (p < 0.001) in PA and MMA comDared to controls.
Eight hundred fifty seven (857) peaks were identified as being important to differentiating individuals with MMA compared to controls (VIP ≥ 2.0 with a jack-knife confidence interval that did not include 0 or a p-value < 0.001) and 56 peaks were library-matched to metabolites. Twenty (20) metabolites had increased abundance compared with controls (range of 2.5 fold to 103.6 fold), and 36 metabolites had decreased abundance compared with controls (range of 1.3 fold to 21.3 fold). Of the 5 metabolites with the most highly increased abundance in samples from MMA patients, one is a well-known metabolite of propionic acid (propionylcarnitine at a 71.5 fold increase). Cystathionine sulfoxide was increased 92.9 fold compared with controls, which is consistent with the upregulation of sulfur metabolism found in functional protein annotation. Similar to the PA individuals, methyl sorbate had a 76.5 fold increase, which is flavoring agent and may be a formula artifact but is also a biological product that has precursor structures similar to MMA. The remaining two metabolites of the top 5 include ursodeoxycholic acid 3-sulfate (103.6 fold increase) and 3,4 methylenesebacic acid (27.3 fold increase), whose significance are not known at this time. Additionally, outside of the top 5 metabolites, glutathione (15.2 fold) and S-formylmethylglutathione (16.3 fold) were also significantly increased in MMA individuals.
3.3. Characterization of CRIPSR edited MUT-null and PCCA-null HEK293 cells.
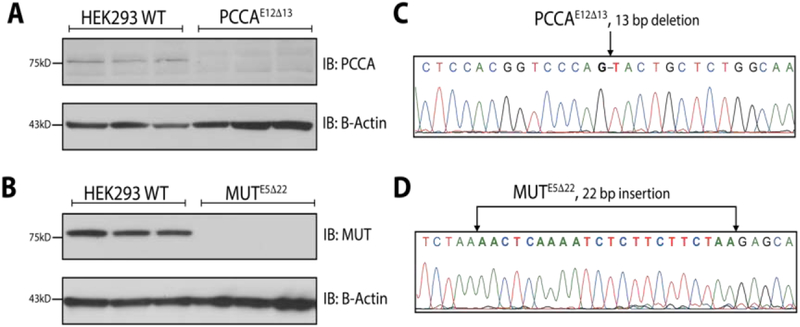
CRISPR/Cas9 editing of HEK293 cells targeting exon 12 of PCCA resulted in a homozygous 13bp deletion (PCCAE5Δ13). Immunoblotting against PCCA revealed no detectable expression of PCCA. CRISPR/Cas9 editing of HEK293 cells targeting exon 5 of MUT resulted in a homozygous 22bp insertion (MU7E5Δ22). Immunoblotting against MUT revealed no detectable MUT protein (Figure 4).
Figure 4. Generation of MUT-null and PCCA-null HEK293 cells.
(A) Immunoblot shows absent expression of PCCA in PCCAE5Δ13 cells (n=3, serial passages) compared to WT cells (n=3, serial passages). β-actin was used as a loading control. (B) Immunoblot shows absent expression of MUT in MUTE5Δ22 cells (n=3, serial passages) compared to WT cells (n=3, serial passages). β-actin was used as a loading control. (C) Sanger sequencing trace of PCCAE5Δ13 highlights a 13 bp deletion in exon 12 of PCCA. (D) Sanger sequencing trace of MUTE5Δ22 cells highlights a 22 bp insertion within exon 5 of MUT.
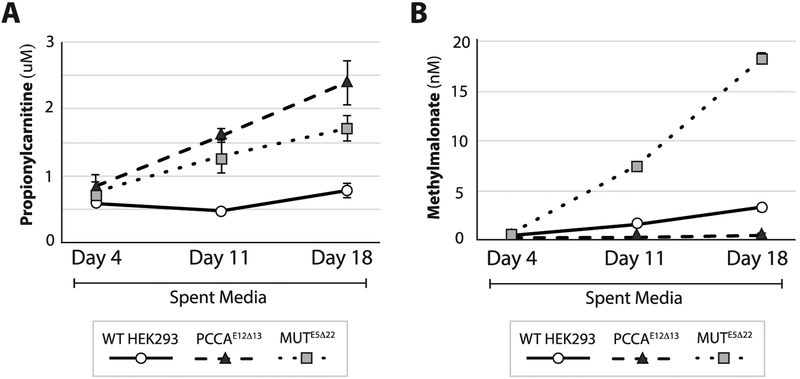
Propionylcarnitine was measured in spent media from confluent PCCAE5Δ13 (n=3 individual culture wells), MU7E5Δ22 (n=3 individual cell culture wells), and WT (n=3 individual cell culture wells) HEK293 cells at days 4, 11 and 18. Both PCCAE5Δ13 and MUTE5Δ22 cells showed a significant increase in the excretion of propionylcarnitine compared to controls at days 11 and 18. Methylmalonylcarnitine (C5DC) was below the limit of detection in all 3 cell lines (Figure 5).
Figure 5.
Metabolites in spent media from in PCCAE12Δ13, MUTE5Δ22 and WT cells. A) Methylmalonate levels are significantly higher in spent media at 11 and 18 days of cell growth in mUtE5Δ22 (dotted line) compared to PCCAE12Δ13 (dashed line) and WT (solid lines) cells, and in WT compared to PCCAE12Δ13 cells. B) Propionylcarnitine (C3) levels are significantly higher in spent media at 11 and 18 days of cell growth in PCCAE12Δ13 (dashed line) compared MUTE5Δ22 (dotted line) and WT (solid lines) cells, and in MUTE5Δ22 compared WT cells. (n=4 culture wells per cohort)
Methylmalonic acid was measured in spent media confluent PCCAE5Δ13(n=3 individual cell culture wells), MUTE5Δ22 (n=3 individual cell culture wells), and WT (n=3 individual cell culture wells) HEK293 cells at days 4, 11 and 18. At days 11 and 18, MUTE5Δ22 cells had significantly increased excretion of methylmalonic acid compared to both control and PCCAE5Δ13 cells (Figure 5). Additionally, the methylmalonic acid in spent media was decreased in PCCAE5Δ13 cells compared to controls, which is expected because the metabolic block in PA inhibits the production of methylmalonic acid.
3.4. Altered gene expression in PCCAE5Δ13, MUTE5Δ22’ and WT cells.
Based on our proteomic and metabolomic findings we selected 5 genes to assess for RNA expression in the PCCAE12Δ13 and MUTE5Δ22 cells: CBS, PSAT, SLC7A5, SLC3A2, and ASNS. CBS and PSAT have critical roles in serine and thiol metabolism and have significantly increased protein expression in MMA and PA patient-derived fibroblasts. SLC7A5 and SLC3A2 are involved in the transport of serine across the cellular membrane and have increased expression in MMA and PA patient derived fibroblasts. ASNS is a canonical target of the ATF4 transcription factor, which helps to regulate the integrated cellular stress response [37].
We measured RNA expression in PCCAE5Δ13 (n=3), MUTE5Δ22 (n=3) and WT cells (n=3) at 1, 2, 4 and 8 days after plating to capture gene expression through the log phase of growth and at confluence (Supplementary Figure 2). Each experiment was carried with n=3 individual culture wells analyzed per genotype per time point.
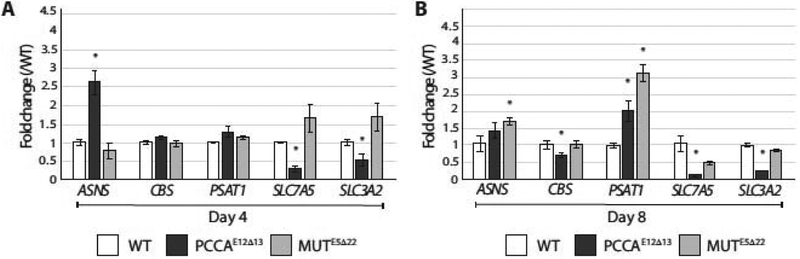
At day 4 (~70% confluence, log phase of growth), MUTE5Δ22 had increased expression of serine transporter gene SLC7A5 (1.6-fold) and SLC3A2 (1.7 fold) compared to WT. PCCAE12Δ13 had increased expression of ASNS compared to WT (2.7-fold). Additionally, PCCAE12Δ13 had decreased expression of both serine transporters gene SLC7A5 and SLC3A2 (0.3 fold and 0.5-fold, respectively) compared to WT. CBS and PSAT1 expression was comparable to WT for both MUTE5Δ22 and PCCAE12Δ13. (Figure 6).
Fig. 6.
mRNA expression in PCCAE12Δ13 and MUTE5Δ22 cells at 4 and 8 days of growth. (A) At day 4 (log phase of growth), MUTE5Δ22 had increased expression of serine transporter gene SLCJA5 (1.6–fold) and SLC3A2 (1.7–fold) compared to WT, though this did not reach statistical significance. Additionally, PCCAE12Δ13 had significantly decreased expression of both serine transporters gene SLCJA5 andSLC3A2 (0.3 fold and 0.5–fold, respectively) compared to WT. (B) At day 8 (cells at confluence), PCCAE12Δ13 and MUTE5Δ22 had increased expression of ASNS (1.4–fold and 1.7–fold, respectively) and PSAT1 (2.0–fold and 3.1–fold, respectively) compared to WT. Additionally, PCCAE12Δ13 had significantly decreased expression of SLC7A5 (0.1–fold) and SLC3A2 (0.2–fold) compared to WT. (n=3, 3 individual 6–well culture wells per genotype per time point) (n=3, 3 individual 6–well culture wells analyzed per genotype per time point). * indicates a statisticially significant change (p < 0.05) compared to WT.
At day 8 (cells at confluence), PCCAE12Δ13 and MUTE5Δ22 had increased expression of ASNS (1.4-fold and 1.7-fold, respectively) and PSAT1 (2.0-fold and 3.1-fold, respectively) compared to WT. Additionally, both PCCAE12Δ13 and MUTE5Δ22 had decreased expression of SLC7A5 (0.1-fold and 0.3-fold, respectively) and SLC3A2 (0.2-fold and 0.9-fold, respectively) compared to WT (Figure 6).
3.5. Flux metabolomics reveals alteration in serine synthesis and transport
From CE-TOFMS measurement, 231 peaks (112 in Cation and 119 in Anion Mode, respectively) of isotopic ions and their total amount for each compound were identified.
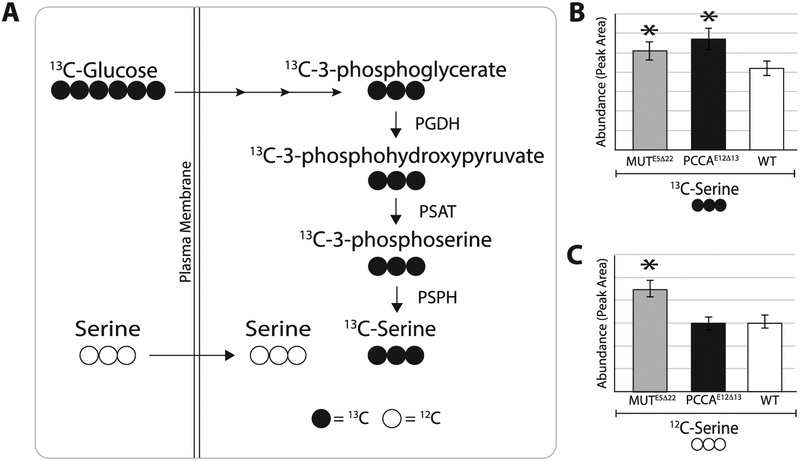
De novo serine synthesis from glucose was increased in both PCCAE12Δ13 and MUTE5Δ22 cells compared to WT, as shown by the amount of serine with all 3 carbons labeled with 13C. Additionally, glucose-6-phosphate, with all 6 carbons labeled with 13C, was significantly reduced in both PCCAE12Δ13 and MUTE5Δ22, indicating a rapid flux towards serine biosynthesis as glucose-6-phosphate is a pool of ‘trapped” intracellular glucose for cellular metabolite biosynthesis. Lastly, glycine with both carbons labeled with 13C, which is the product of SHMT from serine, was significantly increased in PCCAE12Δ1Z but not MUTE5Δ22 (Figure 7). Interestingly, serine transport (as measured by serine with no 13C labeled carbons) was only increased in MUTE5Δ22.
Figure 7. Serine biosynthesis and transport in MUTE5Δ22 and PCCAE12Δ13 cells.
(A) 13C labeled glucose (13C-Glucose) is metabolized to 13C labeled serine (13C-Serine). (B) MUTE5Δ22 (n=5) and PCCAE12Δ13 (n=5) cells have significantly increased de novo synthesis of serine from extracellular glucose compared to WT cells (n=5) (1.4-fold increase (p=0.001) and 1.2-fold increase (p=0.007), respectively). (C) Additionally, the intracellular pool of unlabeled serine (12C-Serine) is significantly higher in MUTE5Δ22 compared to both WT and PCCAE12Δ13 (1.7-fold increase (p=5.6E−05) and 1.8 fold increase (p=4.6E−05), respectively) indicating increased uptake from the extracellular milieu. Values (y-axis) are represented as average MS peak heights +/− standard deviation for each genotype. Raw values were corrected for protein content.
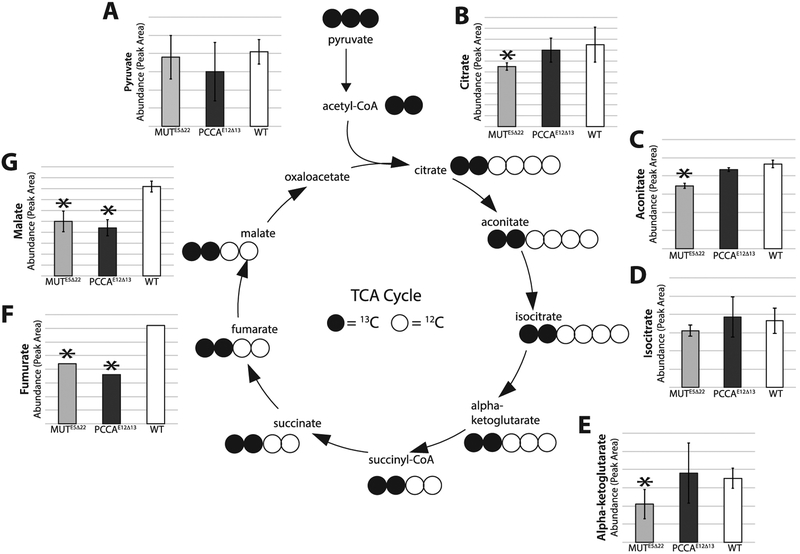
In addition to evaluating serine biosynthetic intermediates, we also evaluated the entrance of pyruvate with all three carbons labeled with 13C, into the TCA cycle (Figure 8). The TCA cycle intermediates we assessed are from the first “turn” of the cycle and therefore have two carbons labeled with 13C. In MUTE5Δ22 cells, proximal TCA cycle intermediates, with the exception of isocitrate, are significantly decreased compared to WT (Figure 8). In PCCAE12Δ13 cells, we found that the proximal TCA cycle intermediates are not significantly different compared to WT. Proximal intermediates include: citrate, aconitate, isocitrate, and alpha-ketoglutarate. However, the distal TCA cycle intermediates malate and fumarate are significantly decreased compared to WT (Figure 8). The differences noted are not due to the availability of labeled pyruvate, since the amount of pyruvate at the start of the cycle is not significantly different between cell types (Figure 8).
Figure 8. 13C-labeled TCA intermediates MUTE5Δ22and PCCAE12Δ13 cells.
(A) 13C labeled pyruvate is not significantly different in and MUTE5Δ22 (n=5, separate culture dishes) and PCCAE12Δ13 cells (n=5, separate culture dishes) compared to WT (n=5, separate culture dishes) (B) Citrate is significantly lower in MUTE5Δ22 compared to WT (0.7-fold, p=0.045). (C) Aconitate is significantly lower in MUTE5Δ22 compared to WT (0.7-fold, p=0.035). (D) Isocitrate is not significantly different between cell types. (E) Alpha-ketoglutarate is significantly lower in MUTE5Δ22 compared to WT (0.6-fold, p=0.025). (F) Fumarate is significantly lower in MUTE5Δ22 and PCCAE12Δ13 compared to WT (0.6-fold (p=0.002) and 0.5 fold (p=1.9E−04) respectively). (G) Malate is significantly lower in MUTE5Δ22 and PCCAE12Δ13 compared to wT (0.6-fold (p=7.8E−04) and 0.6 fold (p=3.6E−05) respectively). Values (y-axis) are represented as average Ms peak heights +/− standard deviation for each genotype. Raw values were corrected for protein content.13C labeled succinate, succinyl CoA, oxaloacetate and acetyl CoA were not measured in this assay due to technical limitations.
4. DISCUSSION
Disturbances in serine metabolism are increasingly implicated as a mechanism of response to mitochondrial stress in primary mitochondrial disease [2–4, 38]. Evidence points towards shifting of serine flux towards synthesis of one-carbon metabolite intermediates and glutathione [2, 3]. In this study, we provide multiple lines of evidence to demonstrate similar secondary metabolic disturbances in methylmalonic acidemia (MMA) and propionic acidemia (PA), suggesting that adaptations in serine metabolism represent a central theme of the cellular response to diverse mitochondrial stressors. This has significant implications for identifying both novel targets for therapeutic intervention, and targets for clinical monitoring in both primary and secondary disorders of mitochondrial function.
We employed a discovery proteomics approach in fibroblasts from individuals with PA (n=2), MMA (n=2), and WT (n=2) in order to generate hypotheses about novel adaptive pathways in MMA and PA, Here we uncovered potential adaptations in serine and thiol metabolism. We then undertook a series of targeted studies downstream to validate these adaptations, and to understand their potential role in the pathogenesis of MMA and PA.
In PA and MMA patient derived fibroblasts, we uncovered an enrichment of proteins involved in serine and thiol metabolism, including phosphoserine aminotransferase (PSAT), cystathionine beta synthase (CBS), and mercaptopyruvate sulfurtransferase (MPST). Additionally, we also identified increased expression of multiple canonical proteins whose corresponding gene expression is regulated by the transcription factor ATF4. This is particularly relevant because ATF4 activation is known to activate serine biosynthesis and has also been reported in various models of primary mitochondrial disease [2]. In the MMA fibroblasts, proteins with significantly increased expression correspond with classic ATF4 response genes BCAT, ASNS, PYCS, SLC7A5, and PSAT. These same genes had increased protein expression in the PA fibroblasts, as well as one other classic ATF4 response gene, SLC38A2 (SNAT2) [39, 40]. Activation of ATF4 in response to mitochondrial stress results in the upregulation of genes involved in the integrated response to adapt and alter cellular metabolism [37].
We chose to model MMA and PA in HEK293 cells due to their well-characterized mitochondrial function, retention of relevant biochemical pathways, amenability to metabolomic tracer studies, and prior evidence for in vivo relevance in other disorders of mitochondrial intermediary metabolism [2, 41]. Once our PCCA-null and MUT-null CRISPR edited HEK293 cells were shown to faithfully replicate the canonical biochemical defects in MMA and PA, we showed that these cells had alterations in expression of genes associated with serine biosynthesis and ATF4 regulation. Serial measurements of gene expression during the lag, exponential (log) and stationary phases of cell growth of the PCCA-null and MUT-null cells show differential expression compared to WT. Genes encoding for neutral amino acid transport including serine (SLC3A2 and SLC7A5) had increased expression during early phases of growth, while genes associated with amino acid metabolism had increased expression during later phases of growth (ASNS, PSAT).
In order to determine the biochemical etiology and downstream intracellular fate of serine, we performed 13C-labeled glucose flux experiments in PCCA and MUT null HEK293 cells. We found that both cell types have an increased de novo synthesis from extracellular glucose. In addition to elevations of 13C-serine in MUT null cells, we also observed increased unlabeled serine compared to WT and PCCA null cells. This could indicate either increased transport of serine from the periphery in the MUT null cells, or increased serine biosynthesis from other intracellular sources. At this time, it is not clear why the MUT-and PCCA-null cells are different in this regard.
Interestingly, 13C-flux experiments also showed increased de novo synthesis of glycine from 13C-serine in PCCA-null cells. Massive elevations in plasma glycine are commonly seen in individuals with PA. In fact, before the canonical organic acid was identified, PA disorder was originally named “non-ketotic hyperglycinemia” [42]. While increased glycine levels may also be seen in MMA compared to unaffected individuals, typically it is at a significantly lower level than the plasma glycine levels observed in PA [43]. It has been previously hypothesized that the increased plasma glycine could be due to inhibition of the glycine cleavage system [44]. However, our results would suggest that increased synthesis from serine may contribute to elevated plasma levels of glycine found in PA compared to MMA and unaffected individuals [43]. Perhaps introduction of mitochondrial stress in the MUT-null cellular model could induce elevated glycine synthesis.
In addition to alterations in biosynthesis of serine, we also examined the flux of 13C-labeled glucose into the TCA cycle. Anaplerotic deficiencies of the TCA cycle have been previously described in MMA and PA, and anaplerotic therapy has been suggested as a mechanism of treatment in these disorders [45, 46]. These deficiencies are intuitive considering that the metabolic flux of propionyl-CoA metabolism is towards succinyl-CoA. However, we found that MUT-null cells have proximal defects in the TCA cycle, which may represent defective conversion of pyruvate to acetyl CoA at the step of pyruvate dehydrogenase. Thus, the TCA cycle defects in MMA cannot be solely due to limited flux of propionyl-CoA into succinyl-CoA.
Furthermore, in PCCA-null cells, we found that flux of 13C labeled alpha-ketoglutarate to the distal segment of the TCA cycle is limited. This could be at the step of either alpha-ketoglutarate dehydrogenase or succinate dehydrogenase. Within the current experiment, we were unable to measure 13C labeled succinate, and therefore we are unable to resolve the precise location of this relative metabolic inhibition. However, as seen in MUT-null cells, the anaplerotic abnormalities in PA are not solely due to limited flux of propionyl-CoA into succinyl-CoA.
From a clinical standpoint, abnormally low plasma serine has been reported in a number of individuals with various primary mitochondrial disorders, and has also been reported in an individual with MMA after liver/kidney transplant [4, 47]. Importantly, re-disturbed flux of serine has been shown to paradoxically deprive some biologically important synthetic pathways of serine (i.e. sphingolipid synthesis), thus resulting in biochemical and clinical pathogenesis [4]. Therefore, correction of serine deficits is potentially highly clinically relevant.
Further studies are necessary to determine if serine levels vary with metabolic stress, i.e. depletion of bioenergetic precursors. Our evidence would suggest that provision of glucose is particularly important for the de novo biosynthesis of serine in MMA and PA. It follows, that serine supplementation could represent an avenue for therapy, though our evidence would suggest that this could be more beneficial in MMA, where transport of extracellular serine (as opposed to de novo synthesis) may be the dominant mechanism.
There are several limitations to this work. The measurements of unlabeled TCA cycle intermediates in our experiment are unable to differentiate between cytoplasmic and mitochondrial metabolites. Therefore, we are unable to estimate the total amounts of mitochondrial TCA intermediates. Additionally, while HEK293 cells are derived from human kidney cells, they do not necessarily represent the affected tissues in MMA and PA. Therefore, we cannot be sure our HEK293 cellular models are capturing the full extent of the adaption and alteration of cellular metabolism in response to mitochondrial stress in MMA and PA. However, the confluence of evidence from the three biological experimental systems (patient-derived fibroblasts, patient-derived plasma, and cellular modeling) provides a strong argument for the importance of serine metabolism in the cellular pathogenesis of these disorders.
In future work, we plan to expand upon these findings, and explore how substrate utilization differs in the setting of bioenergetic stress, such as starvation or increased reactive oxygen stress. This may provide insight into the biochemical adaptations that occur in the setting of physiologic stressors and metabolic crises in individuals with MMA and PA.
Supplementary Material
Highlights.
We evaluated downstream biochemical abnormalities in methylmalonic and propionic acidemia
Serine biosynthesis and utilization are altered in both disorders
This has important implications for disease therapeutic targeting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Kalhan SC, Hanson RW: Resurgence of serine: an often neglected but indispensable amino Acid. JBiol Chem 2012, 287(24):19786–19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F et al. : Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikkanen J, Forsstrom S, Euro L, Paetau I, Kohnz RA, Wang L, Chilov D, Viinamaki J, Roivainen A, Marjamaki P et al. : Mitochondrial DNA Replication Defects Disturb Cellular dNTP Pools and Remodel One-Carbon Metabolism. CellMetab 2016, 23(4):635–648. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira CR, Goorden SMI, Soldatos A, Byers HM, Ghauharali-van der Vlugt JMM, Beers-Stet FS, Groden C, van Karnebeek CD, Gahl WA, Vaz FM et al. : Deoxysphingolipid precursors indicate abnormal sphingolipid metabolism in individuals with primary and secondary disturbances of serine availability. Mol Genet Metab 2018, 124(3):204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samanta D, Semenza GL: Serine Synthesis Helps Hypoxic Cancer Stem Cells Regulate Redox. Cancer Res 2016, 76(22):6458–6462. [DOI] [PubMed] [Google Scholar]
- 6.Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X, Ma J, Yin X, Mao A, Shang M: Overexpression of Phosphoserine Aminotransferase 1 (PSAT1) Predicts Poor Prognosis and Associates with Tumor Progression in Human Esophageal Squamous Cell Carcinoma. Cell Physiol Biochem 2016, 39(1):395–406. [DOI] [PubMed] [Google Scholar]
- 7.Qian C, Xia Y, Ren Y, Yin Y, Deng A: Identification and validation of PSAT1 as a potential prognostic factor for predicting clinical outcomes in patients with colorectal carcinoma.Oncol Lett 2017, 14(6):8014–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Wu J, Cai J, He Z, Yuan J, Zhu X, Li Y, Li M, Guan H: PSAT1 regulates cyclin D1 degradation and sustains proliferation of non-small cell lung cancer cells. IntJ Cancer 2015,136(4):E39–50. [DOI] [PubMed] [Google Scholar]
- 9.Shchelochkov OA, Carrillo N, Venditti C: Propionic Acidemia In: GeneReviews((R)). Edited by Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. Seattle (WA); 1993. [Google Scholar]
- 10.Manoli I, Sloan JL, Venditti CP: Isolated Methylmalonic Acidemia In: GeneReviews((R)). Edited by Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. Seattle (WA); 1993. [Google Scholar]
- 11.Kraus JP, Spector E, Venezia S, Estes P, Chiang PW, Creadon-Swindell G, Mullerleile S, de Silva L, Barth M, Walter M et al. : Mutation analysis in 54 propionic acidemia patients. J Inherit Metab Dis 2012, 35(1):51–63. [DOI] [PubMed] [Google Scholar]
- 12.Forny P, Schnellmann AS, Buerer C, Lutz S, Fowler B, Froese DS, Baumgartner MR: Molecular Genetic Characterization of 151 Mut-Type Methylmalonic Aciduria Patients and Identification of 41 Novel Mutations in MUT. Hum Mutat 2016, 37(8):745–754. [DOI] [PubMed] [Google Scholar]
- 13.Fowler B, Leonard JV, Baumgartner MR: Causes of and diagnostic approach to methylmalonic acidurias. JInherit Metab Dis 2008, 31(3):350–360. [DOI] [PubMed] [Google Scholar]
- 14.Fraser JL, Venditti CP: Methylmalonic and propionic acidemias: clinical management update. Curr Opin Pediatr 2016, 28(6):682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgartner MR, Horster F, Dionisi-Vici C, Haliloglu G, Karall D, Chapman KA, Huemer M, Hochuli M, Assoun M, Ballhausen D et al. : Proposed guidelines for the diagnosis and management of methylmalonic and propionic acidemia. Orphanet J Rare Dis 2014, 9:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunert SC, Mullerleile S, De Silva L, Barth M, Walter M, Walter K, Meissner T, Lindner M, Ensenauer R, Santer R et al. : Propionic acidemia: clinical course and outcome in 55 pediatric and adolescent patients. Orphanet J Rare Dis 2013, 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandler RJ, Zerfas PM, Shanske S, Sloan J, Hoffmann V, DiMauro S, Venditti CP: Mitochondrial dysfunction in mut methylmalonic acidemia. FASEB J 2009, 23(4):1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zsengeller ZK, Aljinovic N, Teot LA, Korson M, Rodig N, Sloan JL, Venditti CP, Berry GT, Rosen S: Methylmalonic acidemia: a megamitochondrial disorder affecting the kidney. Pediatr Nephrol 2014, 29(11):2139–2146. [DOI] [PubMed] [Google Scholar]
- 19.Chapman KA, Ostrovsky J, Rao M, Dingley SD, Polyak E, Yudkoff M, Xiao R, Bennett MJ, Falk MJ: Propionyl-CoA carboxylase pcca-1 and pccb-1 gene deletions in Caenorhabditis elegans globally impair mitochondrial energy metabolism. J Inherit Metab Dis 2018, 41(2):157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinto SM, Nirujogi RS, Rojas PL, Patil AH, Manda SS, Subbannayya Y, Roa JC, Chatterjee A, Prasad TS, Pandey A: Quantitative phosphoproteomic analysis of IL-33-mediated signaling.Proteomics 2015, 15(2–3):532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009, 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA: Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009, 37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-Mclntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN et al. : Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 2011, 6(7):1060–1083. [DOI] [PubMed] [Google Scholar]
- 24.Dhungana S, Carlson JE, Pathmasiri W, McRitchie S, Davis M, Sumner S, Appt SE: Impact of a western diet on the ovarian and serum metabolome. Maturitas 2016, 92:134–142. [DOI] [PubMed] [Google Scholar]
- 25.Williamson BK, Hawkey NM, Blake DA, Frenkel JW, McDaniel KP, Davis JK, Satija C, Beazer A, Dhungana S, Carlson J et al. : The Effects of Glaucoma Drainage Devices on Oxygen Tension, Glycolytic Metabolites, and Metabolomics Profile of Aqueous Humor in the Rabbit. Transl Vis Sci Technol 2018, 7(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trygg J, Holmes E, Lundstedt T: Chemometrics in metabonomics. J Proteome Res 2007, 6(2):469–479. [DOI] [PubMed] [Google Scholar]
- 27.Eriksson LBT, Johansson E, Trygg J, Vikstrom C.: Multi- and Megavariate Data Analysis: Basic Principles and Applications. 3rd ed. Umeå, Sweden: Umetric Academy; 2013. [Google Scholar]
- 28.Yang L, Yang JL, Byrne S, Pan J, Church GM: CRISPR/Cas9-Directed Genome Editing of Cultured Cells. Curr Protoc Mol Biol 2014, 107:31 31 31–17. [DOI] [PubMed] [Google Scholar]
- 29.Desviat LR, Perez B, Perez-Cerda C, Rodriguez-Pombo P, Clavero S, Ugarte M: Propionic acidemia: mutation update and functional and structural effects of the variant alleles. Mol Genet Metab 2004, 83(1–2):28–37. [DOI] [PubMed] [Google Scholar]
- 30.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F: Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013, 8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinaldo P, Cowan TM, Matern D: Acylcarnitine profile analysis. Genet Med 2008, 10(2):151–156. [DOI] [PubMed] [Google Scholar]
- 32.Smith EH, Matern D: Acylcarnitine analysis by tandem mass spectrometry. Curr Protoc Hum Genet 2010, Chapter 17:Unit 17 18 11–20. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen K: Solid-phase sample extraction for rapid determination of methylmalonic acid in serum and urine by a stable-isotope-dilution method. Clin Chem 1989, 35(2):260–264. [PubMed] [Google Scholar]
- 34.Green MR, Sambrook J: Quantification of RNA by Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR). Cold Spring Harb Protoc 2018, 2018(10):pdb prot095042. [DOI] [PubMed] [Google Scholar]
- 35.Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T: Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 2002, 74(10):2233–2239. [DOI] [PubMed] [Google Scholar]
- 36.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T: Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. JProteome Res 2003, 2(5):488–494. [DOI] [PubMed] [Google Scholar]
- 37.Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, Auwerx J: Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 2017, 216(7):2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buzkova J, Nikkanen J, Ahola S, Hakonen AH, Sevastianova K, Hovinen T, Yki-Jarvinen H, Pietilainen KH, Lonnqvist T, Velagapudi V et al. : Metabolomes of mitochondrial diseases and inclusion body myositis patients: treatment targets and biomarkers. EMBO Mol Med 2018, 10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M et al. : ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013, 15(5):481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broer S, Broer A: Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem J 2017, 474(12):1935–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiff M, Haberberger B, Xia C, Mohsen AW, Goetzman ES, Wang Y, Uppala R, Zhang Y, Karunanidhi A, Prabhu D et al. : Complex I assembly function and fatty acid oxidation enzyme activity of ACAD9 both contribute to disease severity in ACAD9 deficiency. Hum Mol Genet 2015, 24(11):3238–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Childs B, Nyhan WL, Borden M, Bard L, Cooke RE: Idiopathic hyperglycinemia and hyperglycinuria: a new disorder of amino acid metabolism. I. Pediatrics 1961, 27:522–538. [PubMed] [Google Scholar]
- 43.Nizon M, Ottolenghi C, Valayannopoulos V, Arnoux JB, Barbier V, Habarou F, Desguerre I, Boddaert N, Bonnefont JP, Acquaviva C et al. : Long-term neurological outcome of a cohort of 80 patients with classical organic acidurias. Orphanet JRare Dis 2013, 8:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayasaka K, Narisawa K, Satoh T, Tateda H, Metoki K, Tada K, Hiraga K, Aoki T, Kawakami T, Akamatsu H et al. : Glycine cleavage system in ketotic hyperglycinemia: a reduction of H-protein activity. Pediatr Res 1982, 16(1):5–7. [DOI] [PubMed] [Google Scholar]
- 45.Knerr I, Weinhold N, Vockley J, Gibson KM: Advances and challenges in the treatment of branched-chain amino/keto acid metabolic defects. J Inherit Metab Dis 2012, 35(1):29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longo N, Price LB, Gappmaier E, Cantor NL, Ernst SL, Bailey C, Pasquali M: Anaplerotic therapy in propionic acidemia. Mol Genet Metab 2017, 122(1–2):51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vernon HJ, Sperati CJ, King JD, Poretti A, Miller NR, Sloan JL, Cameron AM, Myers D, Venditti CP, Valle D: A detailed analysis of methylmalonic acid kinetics during hemodialysis and after combined liver/kidney transplantation in a patient with mut (0) methylmalonic acidemia. J Inherit Metab Dis 2014, 37(6):899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.