Abstract
Restriction endonuclease analysis (REA) and PCR ribotyping are two typing systems that have been frequently utilized for molecular epidemiologic characterization of Clostridioides (Clostridium) difficile. To correlate typing data obtained from each method, we performed both REA and PCR ribotyping on a large and diverse set of historical and contemporary C. difficile infection clinical isolates. Eighty isolates were selected from each reference laboratory in the United States (Microbiology Reference Laboratory, Hines VA Medical Center) and United Kingdom (Clostridium difficile Network for England and Northern Ireland laboratory, University of Leeds). The 160 isolates were assigned to 82 unique ribotypes and 51 unique REA groups (116 unique REA types). In general, concordance between typing methods was good. Dendrogram analysis of PCR ribotype band patterns demonstrated close genetic relationships among strain types with discordant REA and ribotype assignments. While REA typing was more discriminatory, several REA types in this study were further discriminated by PCR ribotyping, indicating that discriminatory value of these typing methods may be strain dependent. These data will assist with molecular epidemiologic surveillance of strains identified by these two commonly used C. difficile typing systems.
Keywords: Clostridium difficile, Molecular epidemiology, Typing, Restriction endonuclease analysis, PCR ribotyping
1. Introduction
Clostridioides (Clostridium) difficile has emerged as an important public health threat that is associated with considerable morbidity, mortality, and increased healthcare expenditures [1]. The emergence and global dissemination of C. difficile has been associated with the spread of antibiotic-resistant and potentially hypervirulent epidemic strains. The most notable strain, which caused outbreaks of severe C. difficile infection (CDI) first in North America [2,3] and later in the UK [4], is the strain identified as group BI by restriction endonuclease analysis (REA), ribotype 027 by polymerase chain reaction (PCR) ribotyping, and NAP1 by pulsed-field gel electrophoresis (PFGE). However, the molecular epidemiology of C. difficile is dynamic and oftentimes highly variable among different regions of the world; BI/NAP1/027 is declining in both the US [5] and UK [6], and new strain types are emerging. Thus, rigorous surveillance and investigation of the molecular epidemiology of CDI is an important public health responsibility. Identification of epidemic strains has guided research to better understand C. difficile pathogenesis [1], human transmission [7], global dissemination [8], and identification of novel potential reservoirs for C. difficile, such as animals and food [4].
However, molecular epidemiologic investigation of CDI has presented several challenges, in particular the lack of a single portable typing system that is shared amongst the public health and academic communities. Several typing methods are in use, each with unique benefits and limitations [9]. PCR ribotyping and REA are two typing systems that have been frequently utilized for molecular epidemiologic characterization of C. difficile. However, particularly for REA, very few laboratories perform these analyses, and generated data are not portable, presenting challenges for cross-typing of strains analyzed by these methods. Several previous studies have compared REA and PCR ribotyping data (in addition to other typing methods), but these prior studies are limited by small sample size and restricted strain diversity [10], omission of several epidemiologically important ribotypes [11], or lack of inclusion of strains collected outside of North America [12]. The objective of this study was to correlate REA and PCR ribotyping performed by two highly experienced reference laboratories for identification of a diverse collection of clinical C. difficile strains obtained from a multinational group of patients predominantly in the US and UK.
2. Materials and methods
2.1. Study isolates
This study included isolates collected from two reference laboratories: the Microbiology Reference Laboratory (MRL) at Hines VA Medical Center, Hines, Illinois, USA, where REA [13] is performed; and the Clostridium difficile Network for England and Northern Ireland (CDRN) laboratory at University of Leeds, Leeds, UK, where PCR ribotyping [14] is performed. Clinical isolates were predominantly from patients in the US and UK, although the CDRN laboratory also occasionally receives strains from other European countries. Saved clinical C. difficile isolates from each reference laboratory were selected to undergo both REA and PCR ribotyping. From each site, a diverse collection of historical and contemporary strain types known to commonly cause CDI in humans was selected. These isolates were originally collected between 1982 and 2009. From the Hines MRL, 80 unique isolates were selected. Eighty isolates were also selected from the Leeds CDRN laboratory, and this subset included a reference panel of 70 well-characterized unique isolates and 5 pairs of duplicate isolates [14,15]. The isolates selected for inclusion in this study were de-identified, and a waiver of informed consent was granted by the institutional review board.
2.2. Restriction endonuclease analysis
As previously described [13], REA was performed by analyzing unique electrophoretic DNA band patterns from extracted whole genomic DNA after restriction digestion with the HindIII. Isolates were typed based on manual comparison of electrophoretic band patterns of the isolate to the band patterns of a large collection of reference isolate band patterns. Band patterns with a 90% similarity index are assigned to a REA group (letter designation) and unique patterns are given a specific REA type (number designation). Thus, an REA type is assigned to an isolate band pattern that is identical to an existing REA type in the reference isolate library. REA groups and types are categorized in chronological order as new groups/types are identified. Currently, >120 REA groups (i.e., REA groups A through DS) and >600 REA types have been identified.
Potentially new REA types (i.e., those isolates with as few as a single band difference from all isolates in the reference library) [16] identified in this study were confirmed only for REA groups of known epidemiologic and/or clinical significance. For those types with subtle band differences, isolates were further assessed to determine whether the band difference represents a new REA type or an isolate of an existing REA type that also contains a plasmid. Plasmid preparations of the strain of interest and closely related reference strains were prepared as previously described [17]. On the same gel, electrophoresis was performed as described above on whole genomic DNA and plasmid preparations of the strain of interest and closely related reference strains.
2.3. PCR ribotyping
As previously described [14], PCR ribotyping was performed by analyzing capillary electrophoresis banding patterns from PCR products of the 16S–23S rRNA intergenic spacer region. Unique PCR ribotypes were identified based on the patterns of major peaks in fluorescent signal obtained from PCR product analysis of each isolate. BioNumerics v5.1 (Applied Maths, Sint-Martens-Latum, Belgium) was used to discriminate PCR ribotypes based on inter-comparison of isolate peak profiles. PCR ribotype identities are assigned to isolates following basic maximum similarity scoring against a validated reference library. New PCR ribotypes (i.e., those isolates with as few as a single peak difference [>5 base-pairs in length], when compared with all profiles in the reference library) were further tested for pattern stability and reproducibility before new library assignments were opened. New PCR-ribotypes are categorized in chronological order as new ribotypes are identified. Currently, >900 distinct PCR ribotypes are present in the library.
2.4. Comparison of REA and PCR ribotyping
To correlate REA and PCR ribotyping data, each site provided the other site with 80 previously typed C. difficile isolates. Each site subsequently analyzed those 80 isolates by the typing method performed at their reference laboratory. Thus, 155 unique isolates (plus 1 duplicate of each of 5 strains) underwent both REA and PCR ribotyping. Both sites were initially blinded to the previous typing results obtained by the other laboratory. To assess REA precision, the 5 pairs of duplicate isolates provided by the Leeds CDRN laboratory underwent REA. The Hines MRL was blinded to the identification of the duplicate isolates. After REA and PCR ribotyping were completed, the investigators were unblinded to the identification of the duplicate isolates. Discordance between anticipated REA groups/types and ribotypes in three of the duplicate pairs prompted reassessment to confirm the preliminary typing data. REA groups/types reported here are the final typing data assigned after unblinding.
3. Results
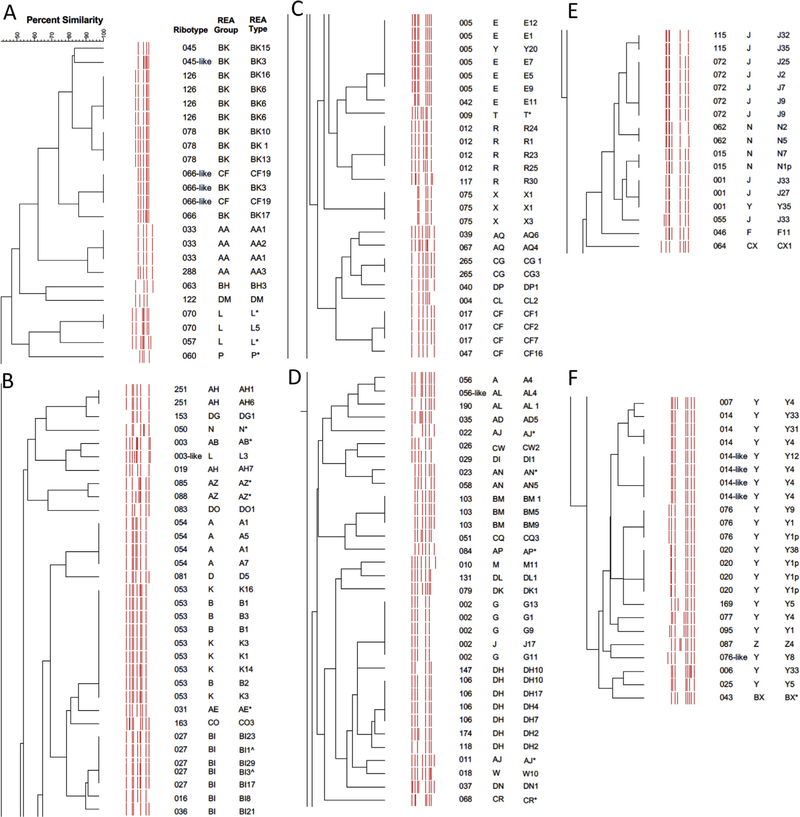
In total, 160 isolates underwent both REA and PCR ribotyping (155 unique isolates, and 5 additional duplicates among those 75 unique isolates provided by the Leeds CDRN laboratory). Among these, strains were assigned to 82 unique ribotypes and 51 unique REA groups (further characterized into 116 unique REA types). There were an additional 15 potential newly identified REA types, but because of the unclear epidemiologic and/or clinical significance, they were only identified to the level of REA group and a specific REA type was not assigned. PCR ribotypes and the corresponding REA identifications, and vice versa, are provided in Tables 1 and 2, respectively. In general, REA typing provided greater discrimination of strain types. The genetic relationships between PCR ribotypes and REA groups/types are illustrated in the PCR ribotype dendrogram aligned by ribotype pattern similarity (Fig. 1- dendrogram separated into sections for print; Figure S1- complete dendrogram online).
Table 1.
C. difficile PCR ribotypes and corresponding REA groups and types.
| Ribotype (n) | REA Groups (n) | REA Types (n) |
|---|---|---|
| 001 (7) | J (6), Y (1) | J2 (1), J7 (1), J9 (2), J27 (1), J33 (1), Y35 (1) |
| 002 (5) | G (4), J (1) | G1 (1), G9 (1), G11 (1), G13 (1), J17 (1) |
| 003 (2) | L (1), AB (1) | L3 (1), AB* (1) |
| 004 (1) | CL (1) | CL2 (1) |
| 005 (6) | E (5), Y (1) | E1 (1), E5 (1), E7 (1), E9 (1), E12 (1), Y20 (1) |
| 006 (1) | Y (1) | Y33 (1) |
| 007 (1) | Y (1) | Y4 (1) |
| 009 (1) | T (1) | T* (1) |
| 010 (1) | M (1) | M11 (1) |
| 011 (1) | AJ (1) | AJ* (1) |
| 012 (4) | R (4) | R1 (1), R23 (1), R24 (1), R25 (1) |
| 014 (7) | Y (7) | Y4 (4), Y12 (1), Y31 (1), Y33 (1) |
| 015 (2) | N (2) | N1p (1), N7 (1) |
| 016 (1) | BI (1) | BI8 (1) |
| 017 (3) | CF (3) | CF1 (1), CF2 (1), CF7 (1) |
| 018 (1) | W (1) | W10 (1) |
| 019 (2-dup) | AH (2-dup) | AH7 (2-dup) |
| 020 (4) | Y (4) | Y1p (3), Y38 (1) |
| 022 (1) | AJ (1) | AJ* (1) |
| 023 (1) | AN (1) | AN* (1) |
| 025 (1) | Y (1) | Y5 (1) |
| 026 (1) | CW (1) | CW2 (1) |
| 027 (5) | BI (5) | BI1ˆ (1), BI3ˆ (1), BI17 (1), BI23 (1), BI29 (1) |
| 029 (1) | DI (1) | DI1 (1) |
| 031 (1) | AE (1) | AE* (1) |
| 033 (3) | AA (3) | AA1 (2), AA2 (1) |
| 035 (1) | AD (1) | AD5 (1) |
| 036 (1) | BI (1) | BI21 (1) |
| 037 (1) | DN (1) | DN1 (1) |
| 039 (1) | AQ (1) | AQ6 (1) |
| 040 (1) | DP (1) | DP1 (1) |
| 042 (1) | E (1) | E11 (1) |
| 043 (1) | BX (1) | BX* (1) |
| 045 (2) | BK (2) | BK3, BK15 |
| 046 (1) | F (1) | F11 (1) |
| 047 (1) | CF (1) | CF16 (1) |
| 050 (1) | N (1) | N* (1) |
| 051 (1) | CQ (1) | CQ3 (1) |
| 053 (9) | K (5), B (4) | K1 (1), K3 (2), K14 (1), K16 (1), B1 (2), B2 (1), B3 (1) |
| 054 (4) | A (4) | A1 (2), A5 (1), A7 (1) |
| 055 (2-dup) | J (2-dup) | J33 (2-dup) |
| 056 (2) | A (1), AL (1) | A4 (1), AL4 (1) |
| 057 (1) | L (1) | L* (1) |
| 058 (1) | AN (1) | AN5 (1) |
| 060 (1) | P (1) | P* (1) |
| 062 (2) | N (2) | N2 (1), N5 (1) |
| 063 (1) | BH (1) | BH3 (1) |
| 064 (1) | CX (1) | CX1 (1) |
| 066 (4) | BK (2), CF (2) | BK3 (1), BK17 (1), CF19 (2) |
| 067 (2-dup) | AQ (2-dup) | AQ4 (2-dup) |
| 068 (1) | CR (1) | CR* (1) |
| 070 (2) | L (2) | L5 (1), L* (1) |
| 072 (1) | J (1) | J25 (1) |
| 075 (3) | X (3) | X1 (2), X3 (1) |
| 076 (4) | Y (4) | Y1 (1), Y1p (1), Y8 (1), Y9 (1) |
| 077 (1) | Y (1) | Y4 (1) |
| 078 (4 [2-dup]) | BK (4 [2-dup]) | BK1 (1), BK10 (2-dup), BK13 (1) |
| 079 (1) | DK (1) | DK1 (1) |
| 081 (2-dup) | D (2-dup) | D5 (2-dup) |
| 083 (1) | DO (1) | DO1 (1) |
| 084 (1) | AP (1) | AP* (1) |
| 085 (1) | AZ (1) | AZ* (1) |
| 087 (1) | Z (1) | Z4 (1) |
| 088 (1) | AZ (1) | AZ* (1) |
| 095 (1) | Y (1) | Y1 (1) |
| 103 (3) | BM (3) | BM1 (1), BM5 (1), BM9 (1) |
| 106 (4) | DH (4) | DH4 (1), DH7 (1), DH10 (1), DH17 (1) |
| 115 (2) | J (2) | J32 (1), J35 (1) |
| 117 (1) | R (1) | R30 (1) |
| 118 (1) | DH (1) | DH2 (1) |
| 122 (1) | DM (1) | DM1 (1) |
| 126 (4) | BK (4) | BK6 (3), BK16 (1) |
| 131 (1) | DL (1) | DL1 (1) |
| 147 (1) | DH (1) | DH10 (1) |
| 153 (1) | DG (1) | DG1 (1) |
| 163 (1) | CO (1) | CO3 (1) |
| 169 (1) | Y (1) | Y5 (1) |
| 174 (1) | DH (1) | DH2 (1) |
| 190 (1) | AL (1) | AL1 (1) |
| 251 (2) | AH (2) | AH1 (1), AH6 (1) |
| 265 (2) | CG (2) | CG1 (1), CG3 (1) |
| 288 (1) | AA (1) | AA3 (1) |
Isolates identified by REA group, but specific REA type not assigned (see methods).
Historic BI strains isolated in the late 1980s/early 1990s, well before the BI/NAP1/027 epidemic, which started in the early 2000s.
Dup-two of the isolates analyzed were blinded duplicates of the same isolate.
p-Plasmid accounts for a distinguishing band in the REA pattern of the whole genome HindIII digest.
Table 2.
C. difficile REA groups and corresponding ribotypes.
| REA Group (n) |
Ribotype (n) [REA type(s) within each ribotype] |
|---|---|
| A (5) | 054 (4) [A1, A5, A7], 056(1) [A4] |
| B (4) | 053 (4) [B1. B2. B3] |
| D (2) | 081 (2) [D5] |
| E (6) | 005 (5) [E1, E5, E7, E9, E12], 042 (1) [E11] |
| F (1) | 046 (1) [F11] |
| G (4) | 002 (4) [G1, G9, G11, G13] |
| J (12) | 001 (6) [J2, J7, J9, J27, J33], 002 (1) [J17], 055 (2) [J33], 072 (1) [J25], 115 (2) [J32] |
| K (5) | 053 (5) [K1, K3, K14, K16] |
| L (4) | 003 (1) [L3], 057 (1) [L*], 070 (2) [L5, L*] |
| M (1) | 010 (1) [M11] |
| N (5) | 015 (2) [N1p, N7], 050 (1) [N*], 062 (2) [N2, N5] |
| P (1) | 060 (1) [P*] |
| R (5) | 012 (4) [R1, R23, R24, R25], 117 (1) [R30] |
| T (1) | 009 (1) [T*] |
| W (1) | 018 (1) [W10] |
| X (3) | 075 (3) [X1, X3] |
| Y (23) | 001 (1) [Y35], 005 (1) [Y20], 006(1) [Y33], 007 (1) [Y4], 014 (7) [Y4, Y12, Y31, Y33], 020 (4) [Y1p,Y38], 025 (1) [Y5], 076(4) [Y1, Y1p, Y8, Y9], 077 (1) [Y4], 095 (1) [Y1], 169 (1) [Y5] |
| Z (1) | 087 (1) [Z4] |
| AA (4) | 033 (3) [AA1, AA2], 288 (1) [AA3] |
| AB (1) | 003 (1) [AB*] |
| AD (1) | 035 (1) [AD5] |
| AE (1) | 031 (1) [AE*] |
| AH (4) | 019 (2) [AH7], 251 (2) [AH1, AH6] |
| AJ (2) | 011 (1) [AJ*], 022 (1) [AJ*] |
| AL (2) | 056 (1) [AL4], 190 (1) [AL1] |
| AN (2) | 023 (1) [AN*], 058 (1) [AN5] |
| AP (1) | 084 (1) [AP*] |
| AQ (3) | 039 (1) [AQ6], 067 (2) [AQ4] |
| AZ (2) | 085 (1) [AZ*], 088 (1) [AZ*] |
| BH (1) | 063 (1) [BH3] |
| BI (7) | 016 (1) [BI8], 027 (5) [BI1ˆ, BI3ˆ, BI17, BI23, BI29], 036 (1) [BI21] |
| BK (12) | 045 (2) [BK3, BK15], 066 (2) [BK3, BK17], 078 (4) [BK10], 126 (4) [BK6, BK16] |
| BM (3) | 103 (3) [BM1, BM5, BM9] |
| BX (1) | 043 (1) [BX*] |
| CF (6) | 017 (3) [CF1, CF2, CF7], 047 (1) [CF16], 066 (2) [CF19] |
| CG (2) | 265 (2) [CG1, CG3] |
| CL (1) | 004 (1) [CL2] |
| CO (1) | 163 (1) [CO3] |
| CQ (1) | 051 (1) [CQ3] |
| CR (1) | 068 (1) [CR*] |
| CW (1) | 026 (1) [CW2] |
| CX (1) | 064 (1) [CX1] |
| DG (1) | 153 (1) [DG1] |
| DH (7) | 106 (4) [DH4, DH7, DH10, DH17], 118 (1) [DH2], 147 (1) [DH10], 174 (1) [DH2] |
| DI (1) | 029 (1) [DI1] |
| DK (1) | 079 (1) [DK1] |
| DL (1) | 131 (1) [DL1] |
| DM (1) | 122 (1) [DM1] |
| DN (1) | 037 (1) [DN1] |
| DO (1) | 083 (1) [DO1] |
| DP (1) | 040 (1) [DP1] |
Isolates identified by REA group, but specific REA type not assigned (see methods).
Historic BI strains isolated in the late 1980s/early 1990s, well before the BI/NAP1/027 epidemic, which started in the early 2000s.
p-Plasmid accounts for a distinguishing band in the REA pattern of the whole genome HindIII digest
Fig. 1.
PCR ribotyping dendrogram of C. difficile isolates and corresponding REA groups and types. The complete dendrogram has been separated into 6 sections for print (A-F). *Isolates identified by REA group, but specific REA type not assigned (see methods). ˆHistoric BI strains isolated in the late 1980s/early 1990s, well before the BI/NAP1/027 epidemic, which started in the early 2000s. dup-two of the isolates analyzed were blinded duplicates of the same isolate. p- Plasmid accounts for a distinguishing band in the REA pattern of the whole genome HindIII digest.
Of the 27 ribotypes represented by multiple unique isolates per ribotype, all 27 of these ribotypes were further distinguished into distinct REA groups and/or types (Table 1). However, the discriminatory value of PCR ribotyping was also identified. Of the 17 REA types represented by multiple unique isolates per REA type, 9 (53%) of these REA types were further distinguished into distinct ribotypes (Table 3). In these cases, the PCR ribotyping dendrogram confirmed the close relationships of the different ribotypes represented by distinct REA types (Fig. 1 and S1). For example, among the 23 isolates that were identified as REA group Y, 11 different ribotypes were represented (Table 3). However, 21 of the group Y isolates were closely related in the ribotype dendrogram (Fig. 1F and S1). Multilocus sequence typing (MLST) of 11 of these isolates (4, RT020; 7, RT014 or 014-like; 3, RT076) indicated that they were all ST-2 (data not shown).
Table 3.
REA types with multiple unique isolates per type and corresponding ribotypes.
| REA Type (n) | Ribotypes (n) |
|---|---|
| A1 (2) | 054 (2) |
| AA1 (2) | 033 (2) |
| B1 (2) | 053 (2) |
| BK3 (2) | 045 (1), 066 (1) |
| BK6 (3) | 126 (3) |
| CF19 (2) | 066 (2) |
| DH10 (2) | 106 (1), 147 (1) |
| DH2 (2) | 118 (1), 174 (1) |
| J33 (3) | 001 (1), 055 (2) |
| J9 (2) | 001 (2) |
| K3 (2) | 053 (2) |
| X1 (2) | 075 (2) |
| Y1 (2) | 076 (1), 095 (1) |
| Y1p (4) | 020 (3), 076 (1) |
| Y33 (2) | 006 (1), 014 (1) |
| Y4 (6) | 007 (1), 014 (4), 077 (1) |
| Y5 (2) | 025 (1), 169 (1) |
p-Plasmid accounts for a distinguishing band in the REA pattern of the whole genome HindIII digest.
Of the 5 pairs of duplicate strains, the same REA group was assigned to 4 of the pairs in the blinded analysis (AH, J, BK, D). The ribotype 067 duplicates were initially identified as REA groups AQ and CI. After unblinding and reviewing the REA patterns for AQ and CI in the reference library, it was determined that AQ and CI have similar band patterns and were in fact the same REA group (REA group CI is now removed from the master REA reference library). It was also confirmed that the REA band pattern of these duplicate isolates were identical, now correctly identified as AQ4. There were two additional duplicate pairs (i.e., ribotypes 019 and 078) with preliminarily assigned REA types that differed. Repeat REA was performed on these 4 isolates confirming identical REA types for each duplicate pair (AH7 for ribotype 019 and BK10 for ribotype 078).
4. Discussion
These data serve as an important reference to permit cross-identification of strains identified by PCR ribotyping and REA, two typing systems that have been frequently utilized for molecular epidemiologic characterization of C. difficile. In general, concordance was good between typing systems. As anticipated, REA provided better discrimination of strains of the same ribotype. This is consistent with previous studies of REA and PCR ribotyping that have demonstrated better strain discrimination with REA [10,11,18]. However, there were several REA types in this study that were further discriminated by PCR ribotyping, indicating that discriminatory value of these typing methods may be strain dependent.
The relationship between REA types and PCR ribotypes was more complex in this study compared to the previous formal comparison of these 2 typing systems [11]. Previously, a particular REA group typically correlated with a specific PCR ribotype (for example REA group J and RT001). As experience with PCR ribotyping has grown, new types continue to be recognized, and the correlation with REA grouping is not as tight as previously considered. Newly recognized strains in each of these systems are added numerically or alphabetically, and the relative similarity of these strains is not always apparent by the strain designations. The relatedness of the diverse REA types and ribotypes within REA group Y, however, was clarified with the PCR ribotype dendrogram and by MLST analysis.
Despite the better discriminatory value, a significant limitation of REA was highlighted by this study. REA types are assigned by humans in the laboratory through comparison of electrophoretic band patterns to reference library strains. The electrophoretic band patterns of REA are quite complex. For these reasons, particularly for relatively infrequently encountered and closely related REA groups, the reliability of REA assignments may be impacted. The impact of potentially decreased inter-and intra-individual reliability was highlighted after unblinding of the identification of the duplicate isolates. Because data obtained by PCR ribotyping are computer analyzed, the data are likely to be reported more reliably. Furthermore, genetic relationships can be visualized by a dendrogram created from ribotype data, which is an additional benefit of PCR ribotyping.
Unlike previous studies comparing typing methods [10,11,18], the present study included several emerging and recent epidemic strain types in the US [5] and UK [6], including Bl/027, BK/078, DH/ 106/174, N/15, G/002, and Y/014/020. However, because this study only included a single isolate from several ribotypes and REA groups, the relative discriminatory value of the typing methods could not be evaluated for those strain types. This study was predominantly limited to strains collected in the US and by the CDRN (predominantly UK); characterization of strains from outside the US and Europe may require further investigation.
5. Conclusions
This international study of C. difficile molecular epidemiology provides important data to permit cross-typing of epidemiologically and clinically significant strains commonly countered in the US and UK, as well as several strains of historical significance. Until a portable typing system is widely adopted, these data will assist with molecular epidemiologic surveillance of strains identified by these two commonly used C. difficile typing systems.
Supplementary Material
Acknowledgements
The authors acknowledge Adam Cheknis, Laurica Petrella-Zitko, and Susan Sambol for their assistance with performance of REA. D.N.G. and S.J. are both supported by the US Department of Veterans Affairs Research Service. L.K.K. is supported by a grant from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health [K23 A1123525].
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.anaerobe.2018.07.004.
Conflicts of interest
None relevant to this manuscript.
References
- [1].Kelly CP, Lamont JT, Clostridium difficile- More difficult than ever, N. Engl. J. Med. 359 (2008) 1932–1940. [DOI] [PubMed] [Google Scholar]
- [2].Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, et al. , A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality, N. Engl. J. Med. 353 (2005) 2442–2449. [DOI] [PubMed] [Google Scholar]
- [3].McDonald LC, Killgore GE, Thompson A, Owens RC, Kazakova SV, Sambol SP, et al. , An epidemic, toxin-gene variant strain of Clostridium difficile, N. Engl. J. Med. 353 (2005) 2433–2441. [DOI] [PubMed] [Google Scholar]
- [4].Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. , The changing epidemiology of Clostridium difficile infections, Clin. Microbiol. Rev. 23 (2010) 529–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Karlsson M, Paulick A, Albreght V, Granade M, Guh A, Rasheed JK, et al. , ElP CDl Pathogen Group. 2016. Abstr 13th Biennial Congress of the Anaerobe Society of the Americas, Abstr Plll-4. Molecular Epidemiology of Clostridium difficile Isolated in the United States, 2014. http://www.anaerobe.org/2016/2016abBook.pdf. [Google Scholar]
- [6].Public Health England, Clostridium difficile Ribotyping Network (CDRN) for England and Northern Ireland, Biennial report, 2013–2015, https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/491253/CDRN_2013-15_Report.pdf.
- [7].Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O’Connor L, et al. , Diverse sources of C. difficile infection identified on whole-genome sequencing, N. Engl. J. Med. 369 (2013) 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, et al. , Emergence and global spread of epidemic healthcare-associated Clostridium difficile, Nat. Genet. 45 (2013) 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Huber CA, Foster NF, Riley TV, Paterson DL, Challenges for standardization of Clostridium difficile typing methods, J. Clin. Microbiol. 51 (2013) 2810–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J, et al. , Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing, J. Clin. Microbiol. 46 (2008) 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Manzo CE, Merrigan MM, Johnson S, Gerding DN, Riley TV, Silva J Jr., et al. , International typing study of Clostridium difficile, Anaerobe 28C (2014) 4–7. [DOI] [PubMed] [Google Scholar]
- [12].Tenover FC, Akerlund T, Gerding DN, Goering RV, Bostrom T, Jonsson AM, et al. , Comparison of strain typing results for Clostridium difficile isolates from North America, J. Clin. Microbiol. 49 (2011) 1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Clabots CR, Johnson S, Bettin KM, Mathie PA, Mulligan ME, Schaberg DR, et al. , Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems, J. Clin. Microbiol. 31 (1993) 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fawley WN, Knetsch CW, MacCannell DR, Harmanus C, Du T, Mulvey MR, et al. , Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile, PLoS One 10 (2015), e0118150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Knetsch CW, Terveer EM, Lauber C, Gorbalenya AE, Harmanus C, Kuijper EJ, et al. , Comparative analysis of an expanded Clostridium difficile reference strain collection reveals genetic diversity and evolution through six lineages, Infect. Genet. Evol. 12 (2012) 1577–1585. [DOI] [PubMed] [Google Scholar]
- [16].Sambol SP, Johnson S, Gerding DN, Restriction endonuclease analysis typing, in: Mullany P, Roberts AP (Eds.), Clostridium difficile, Methods and Protocols (Methods in Molecular Biology), second ed, Humana Press, New Yprk, NY, 2016, pp. 1–13. [DOI] [PubMed] [Google Scholar]
- [17].Preparation of plasmid DNA by alkaline lysis with SDS: minipreparation, in: Sambrook J, Russell DW (Eds.), Molecular Cloning: a Laboratory Protocol, third ed, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2001, pp. 1.32–1.34. [Google Scholar]
- [18].Kristjansson M, Samore MH, Gerding DN, DeGirolami PC, Bettin KM, Karchmer AW, et al. , Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains, J. Clin. Microbiol. 32 (1994) 1963–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.