Abstract
Background
Meningeal carcinomatosis (MC) is the most severe form of brain metastasis and causes significant morbidity and mortality. Currently, the diagnosis of MC is routinely confirmed on the basis of clinical manifestation, positive cerebrospinal fluid (CSF) cytology, and/or neuroimaging features. However, negative rate of CSF cytology and neuroimaging findings often result in a failure to diagnose MC from the patients who actually have the disease. Here we evaluate the CSF circulating tumor DNA (ctDNA) in the diagnosis of MC.
Methods
A total of 35 CSF samples were collected from 35 patients with MC for CSF cytology examination, CSF ctDNA extraction and cancer-associated gene mutations detection by next-generation sequencing (NGS) at the same time.
Results
The most frequent primary tumor in this study was lung cancer (26/35, 74%), followed by gastric cancer (2/35, 6%), breast cancer (2/35, 6%), prostatic cancer (1/35, 3%), parotid gland carcinoma (1/35, 3%) and lymphoma (1/35, 3%) while no primary tumor could be found in the remaining 2 patients in spite of using various inspection methods. Twenty-five CSF samples (25/35; 71%) were found neoplastic cells in CSF cytology examination while all of the 35 CSF samples (35/35; 100%) were revealed having detectable ctDNA in which cancer-associated gene mutations were detected. All of 35 patients with MC in the study underwent contrast-enhanced brain MRI and/or CT and 22 neuroimaging features (22/35; 63%) were consistent with MC. The sensitivity of the neuroimaging was 88% (95% confidence intervals [95% CI], 75 to 100) (p = 22/25) and 63% (95% CI, 47 to 79) (p = 22/35) compared to those of CSF cytology and CSF ctDNA, respectively. The sensitivity of the CSF cytology was 71% (95% CI, 56 to 86) (n = 25/35) compared to that of CSF ctDNA.
Conclusions
This study suggests a higher sensitivity of CSF ctDNA than those of CSF cytology and neuroimaging findings. We find cancer-associated gene mutations in ctDNA from CSF of patients with MC at 100% of our cohort, and utilizing CSF ctDNA as liquid biopsy technology based on the detection of cancer-associated gene mutations may give additional information to diagnose MC with negative CSF cytology and/or negative neuroimaging findings.
Keywords: Meningeal Carcinomatosis, Cerebrospinal fluid ctDNA, Next-generation sequencing, Cancer-associated gene mutations
Background
Meningeal carcinomatosis (MC), also called neoplastic meningitis (NM) is the most severe form of brain metastasis and causes significant morbidity and mortality [1]. The early diagnosis and timely treatment are likely to have the greatest impact on improving outcome. However, MC is easily to be missed in diagnosis and easily misdiagnosed because of diverse clinical manifestations and the lack of sensitive and specific diagnostic tools, which has presented difficulties for early treatment of patients with MC [2].
Currently, the diagnosis of MC is routinely confirmed on the basis of clinical signs and symptoms, cerebrospinal fluid (CSF) cytology, and/or neuroimaging (contrast-enhanced brain MRI and/or CT) findings [2]. The presence of neoplastic cells in the CSF is the most useful finding to confirm the diagnosis and CSF cytology remains the gold diagnostic standard, but 25–30% of suspected clinical cases with MC diagnosed based on clinical picture and neuroimaging findings will not be confirmed by this method [3, 4]. Although the positive rate of CSF cytology for neoplastic cells will increase with the number of punctures, the false negative rate is still high, due to the similarity between tumor cells and ependymal cells, contamination by blood brought about by multiple punctures, difficulty of the distinction lymphoma cells caused by viral infections of central nervous system (CNS), few tumor cells in the collected specimen and inadequate preparation of the sample [5]. Measurement of tumor markers in the CSF may be convenient and of value in the adjunctive diagnosis of MC but lack sensitivity and specificity [6–9]. Amplification of tumor specific gene sequences by polymerase chain reaction may be used to detect minimal quantities of neoplastic cells in the CSF, which can be used when the gene mutations in the tumor are known but many cases of MC can be the first presentation of an as yet undetected primary cancer [10]. Contrast-enhanced brain MRI and/or CT is the technique of choice to evaluate patients with suspected MC but still has a 30% incidence of false-negative results [11]. Therefore, it is important to evaluate an alternative method for diagnosis of MC especially for cases with persistently negative CSF cytology and/or persistently negative neuroimaging results.
Circulating tumor DNA (ctDNA) consists of short, double stranded DNA fragments that are released by tumors [12]. The ctDNA implies the entire spectrum of tumor genome aberrations. Current approaches for detection of tumor genome aberrations in ctDNA include PCR-based methods targeting specific mutations and next-generation sequencing (NGS)-based options allowing detection of all possible aberrations in DNA. Improvements in read-length, sequence quality, and throughput allowed NGS to become a more promising method for quantifying ctDNA than PCR-based methods [13]. The tumor-derived ctDNA of plasma has been applied to detect multiple different types of cancer [14, 15]. However, plasma ctDNA from tumors confined to central nervous system was infrequently detectable given that physical obstacles such the blood-brain barrier could prevent ctDNA from entering the blood circulation [14, 16, 17]. Many studies have shown that CSF ctDNA could be an important method of liquid biopsy in patients with CNS cancers [17, 18]. However, only a single tumor type or single primary tumor type was included in these studies. Given that MC is involvement of the leptomeninges by metastatic tumors and can be observed in various kinds of solid tumors, we hope to evaluate the amounts of ctDNA among different primary tumor types and evaluate the clinical value of CSF ctDNA as liquid biopsy medium in the diagnosis of MC. Although the liquid biopsy approach has been shown to be promising, the sensitivity of CSF ctDNA with respect to conventional diagnosis of CSF cytology and neuroimaging findings have not been evaluated in patients with MC. Here, we evaluated the sensitivity of CSF ctDNA with NGS technology as liquid biopsy medium in the diagnosis of patients with MC.
Methods
Study subjects
Written informed consent has been obtained from all patients or their legal surrogates. The study protocol has been approved by the Ethics Committee of the Second Hospital of Hebei Medical University, Hebei, China. The patients with MC were enrolled from Department of Neurology in the Second Hospital of Hebei Medical University. In total, 35 patients with MC who underwent lumbar puncture for CSF cytology examination, CSF ctDNA extraction and cancer-associated gene mutations detection by next-generation sequencing (NGS) at the same time and underwent contrast-enhanced brain MRI and/or CT were enrolled in this study between October 2014 and September 2017. The diagnosis of MC had been established by clinical signs and symptoms in addition to positive CSF cytology and/or neuroimaging findings (contrast-enhanced brain MRI or CT) consistent with MC. The clinical signs and symptoms included headache, nausea and vomiting, convulsion, lower back pain, cranial nerve paralysis, paresthesia, gait disturbances, vertigo and defects in mental functioning. The positive CSF cytology result was defined by the morphology of neoplastic cells such as different size with irregular-shape, big nucleus with malignant signs such as lobulated state and malformed buds, increasing chromatin with basophilic coarse particles, mitotic activity with aberrant mitosis, obvious and polymorphic nucleolus accounting for the majority of chromatin and thickening nuclear membrane with saw-tooth-shaped and wear edge. The positive neuroimaging finding was defined as the presence of leptomeningeal enhancement.
Next-generation sequencing
Sample processing
Cerebrospinal fluid samples were collected from each patient. CSF samples in EDTA tubes were centrifuged for 5 min at 1000 g. The pellet was stored at − 20 °C, while the supernatant was centrifuged at 10,000 g for another 30 min [19]. The supernatant was aseptically transferred to pre-labeled Cryotubes stored at − 80 °C. The ctDNA was extracted from at least 5 ml CSF supernatant using the QIAamp Circulating Nucleic Acid kit (QIAGEN) as per the manufacturer’s instructions. Finally, ctDNA was quantified with the Qubit2.1 Fluorometer and Qubit dsDNA HS Assay kit (Life Technologies, Carlsbad, CA) following the recommended protocol.
Ion proton library preparation and sequencing
Preparation of the Ion Proton library and DNA sequencing were performed as described in our previous publications [20–22]. For each sample type, an adapter-ligated library was generated with the Ion AmpliSeq Library Kit 2.0 (Life Technologies) according to the manufacturer’s protocol. Briefly, pooled amplicons made from 10~20 ng ctDNA were end-repaired and ligated to Ion Adapters X and P1. AMPure beads (Beckman Coulter, Brea, CA) were used to purify adapter-ligated products, followed by nick-translation and PCR-amplification for a total of 5 cycles. AMPure beads were used to purify the resulting library. And the Agilent 2100 Bioanalyzer and Agilent Bioanalyzer DNA High-Sensitivity LabChip (Agilent Technologies) were used to determine the concentration and size of the library. Sample emulsion PCR and emulsion breaking were performed using the Ion OneTouch™2 system (Life Technologies) with the Ion PI Template OT2 200 Kit v3 (Life Technologies) as per the manufacturer’s instructions. Ion Sphere Particles (ISPs) were recovered, and template-positive ISPs were enriched with Dynabeads MyOne Streptavidin C1 beads (Life Technologies) on the Ion One Touch ES (enrichment system) (Life Technologies). ISPs enrichment was confirmed using the Qubit 2.0 Fluorometer (Life Technologies). The Ion Proton System using Ion PI v2 Chips (Life Technologies) were used to sequence barcoded samples for 100 cycles and the Ion PI Sequencing 200 Kit v3 (Life Technologies) was used for sequencing reactions following the recommended protocol.
We used the SV-OCP143-ctDNA panel (San Valley Biotech Inc., Beijing, China), which is capable of detecting somatic mutations from plasma or tissue samples on 143 cancer-related genes. Since the ctDNA in CSF is comprised of short DNA fragments, amplicons in the panel are specially designed for efficient amplification of ctDNA. For CSF samples, the total read count was more than 25 million to ensure the average base coverage depth over 10,000 x. Additionally, the average uniformity of base coverage was 95.5%. These strict quality control criteria ensured the reliability of sequencing.
Variant calling
Initial data from the sequencing runs were processed with the Ion Proton platform-specific pipeline software Torrent Suite v5.0 including generating sequencing reads, trim adapter sequences, filtering and removing poor signal-profile reads as described in our previous publications [20–22]. Initial variant calling from the sequencing data was generated with the TorrentSuite Software with a plug-in ‘variant caller v5.0’. Three filtering steps were used to eliminate erroneous base calling and generate final variant calling. For the first-step, the following filtering criteria were defined for CSF ctDNA: the average total coverage depth>10,000; each variant coverage>10; a variant frequency of each sample>0.1%; and p value<0.01. The second-step utilized the Integrative Genomics Viewer (IGV) software (http//www.broadinstitute.org/igv) or Samtools software (http://samtools.sourceforge.net) to eliminate possible DNA strand-specific errors after visual examination of called mutations.
Results
Characteristics of patients with MC
The clinical characteristics of 35 patients with MC were summarized in Table 1. There were 16 male and 19 female patients. The date of MC diagnosis was defined as the date of the first positive cytological study or of the first positive neuroimaging findings consistent with MC. The median age at the time of diagnosis of MC was 52 (range, 23–70) years. In 33 of 35 cases, the primary tumors were determined while no primary tumor could be found in the remaining 2 patients in spite of using various inspection methods. Of 33 patients who had clear and definite primary tumor, primary tumor included lung cancer (26 cases), gastric cancer (2 cases), breast cancer (2 cases), prostatic cancer (1 case), parotid carcinoma (1 case), and lymphoma (1 case). Of the 26 patients with lung cancer, a large majority of the patients were found to have adenocarcinoma histology (21/26, 81%). Of 33 patients who had clear and definite primary tumor, 7 patients (7/33, 21%) were diagnosed as MC before the initial presentation of primary tumor. The median time of the remaining 26 patients with MC from the initial diagnosis of the primary tumor to the diagnosis of MC was 355 (range, 1–2566) days. Headache, nausea and vomiting were the most common The baseline Karnofsky Performance Scores (KPS) was 60~80 in the majority of patients (30/35, 86%).
Table 1.
Clinical features of patients with MC
| No. | Primary tumor | Clinical manifestation | KPS |
|---|---|---|---|
| N01 | Unknown | Headache, nausea and vomiting | 60 |
| N02 | Breast cancer | Headache, nausea and vomiting | 60 |
| N03 | Unknown | Nausea and blurred vision | 30 |
| N04 | Gastric cancer | Headache, dizziness and paralysis | 80 |
| N05 | Gastric cancer | Headache and nausea | 70 |
| N06 | Lymphoma | Headache | 70 |
| N07 | Prostatic cancer | Headache | 70 |
| N08 | Parotid carcinoma | Headache, nausea and vomiting | 80 |
| N09 | Lung cancer | Headache, nausea and vomiting | 70 |
| N10 | Lung cancer | Headache and decreased vision | 80 |
| N11 | Lung cancer | Headache, nausea and vomiting | 80 |
| N12 | Lung cancer | Headache, nausea and vomiting | 70 |
| N13 | Lung cancer | Headache and dizziness | 70 |
| N14 | Lung cancer | Headache, nausea, vomiting and back pain | 70 |
| N15 | Lung cancer | Headache and neck pain | 70 |
| N16 | Lung cancer | Headache, nausea and vomiting | 60 |
| N17 | Lung cancer | Headache and dysarthria | 40 |
| N18 | Lung cancer | Headache, nausea and vomiting | 80 |
| N19 | Lung cancer | Leg pain | 70 |
| N20 | Lung cancer | Headache, nausea, vomiting, dizziness and neck pain | 70 |
| N21 | Lung cancer | Headache, nausea, vomiting and back pain | 60 |
| N22 | Lung cancer | Headache, nausea, vomiting and dizziness | 60 |
| N23 | Lung cancer | Headache, nausea, vomiting, dizziness and neck pain | 40 |
| N24 | Lung cancer | Headache, nausea, vomiting, dizziness and hearing loss | 60 |
| N25 | Lung cancer | Headache, nausea, vomiting and dizziness | 70 |
| N26 | Lung cancer | Headache, nausea, vomiting, confusion and decreased vision | 30 |
| N27 | Lung cancer | Blurred vision | 70 |
| N28 | Lung cancer | Headache and dizziness | 50 |
| N29 | Lung cancer | Headache, nausea, vomiting and back pain | 70 |
| N30 | Lung cancer | Headache, nausea, vomiting, confusion and back pain | 70 |
| N31 | Lung cancer | Headache, nausea, vomiting, and diplopia | 70 |
| N32 | Lung cancer | Headache, nausea and vomiting | 70 |
| N33 | Breast cancer | Headache and paralysis | 80 |
| N34 | Lung cancer | Headache, nausea and vomiting | 60 |
| N35 | Lung cancer | Speech disorder | 80 |
Sensitivity of all methods analyzed
The sensitivity of all methods analyzed was presented in Table 3. A total of 35 CSF samples were collected from 35 patients with MC for CSF cytology examination, CSF ctDNA extraction and cancer-associated gene mutations detection by NGS at the same time. Twenty-five CSF samples (25/35; 71%) were CSF cytology positive while all of the 35 CSF samples (35/35; 100%) were revealed having detectable tumor-derived ctDNA in which cancer-associated gene mutations were detected by NGS. The CSF immunocytochemistry investigations were performed in 14 CSF specimens with positive CSF cytology. All of 35 patients with MC in the study underwent contrast-enhanced brain MRI and/or CT and 22 neuroimaging features (22/35; 63%) were consistent with MC, while of 22 patients with leptomeningeal enhancement, 5 patients combined parenchymal brain metastasis. (Table 2 and Table 3).
Table 3.
Sensitivity of all methods analyzed
| % Sensitivity (no. of positive patients) of each method | |||
|---|---|---|---|
| CSF cytology | Neuroimaging | ctDNA | |
| Patients (35) | 71.43 (25) | 62.86 (22) | 100 (35) |
Table 2.
Concurrent result of CSF cytology, CSF ctDNA and neuroimaging findings in patients with MC
| No. | CSF immunocytochemistry | Neuroimaging findings | Concurrent result of CSF cytology | Concentration of CSF ctDNA (ng/ul) | CSF ctDNA gene mutations (mutation frequency) | Parenchymal brain metastasis |
|---|---|---|---|---|---|---|
| N01 | CK7(+), Ki-67(+), ER(−), PR(−) | Leptomeningeal enhancement | + | 5.36 |
TP53 R196* (68.55%) VHL R200W (5.19%) |
_ |
| N02 | Ki-67(+), CK7(+), ER(−), FR(−) | Leptomeningeal enhancement | + | 0.512 |
PTEN K13Q (0.455%) NOTCH1 (38.86%) |
_ |
| N03 | Ki-67(+), CEA(−), CK20(−), CK7(−), CK20(−) | Leptomeningeal enhancement | + | 0.204 | PTEN K13Q (0.311%) | + |
| N04 | Ki-67(+), CEA(+) | Leptomeningeal enhancement | + | 2.08 | TP53 L194R (45.69%) | _ |
| N05 | Not available | Leptomeningeal enhancement | _ | 1.17 | TP53R248Q (1.464%) | _ |
| N06 | Not available | Leptomeningeal enhancement | _ | 1.36 | PTEN R14G (0.204%) | _ |
| N07 | Not available | Leptomeningeal enhancement | _ | 0.29 | TP53 P301Q (0.761%) | _ |
| N08 | CK7 (+) | Leptomeningeal enhancement | + | 6.82 | TP53 C242G (72.914%) | _ |
| N09 | NapsinA (+), CK7(+) | Negative | + | 0.144 | TP53 I195F (89.921%) | _ |
| N10 | Not available | Negative | + | 0.238 | TP53 F113 V (55.412%) | _ |
| N11 | NapsinA (+), TTF1(+), CK7(+) | Negative | + | 5.36 | TP53 R337C (53.924%) | _ |
| N12 | CK7(+), NapsinA(+), Ki-67(−), TTF1(−), CDX2(−), CD20(−) | Leptomeningeal enhancement | + | 1.74 | EGFR L858R (82.178%) | + |
| N13 | TTF1(+), NapsinA (+) | Negative | + | 3.46 | EGFR L858R (29.903%) | _ |
| N14 | Not available | Leptomeningeal enhancement | _ | 3.52 | EGFR L858R (33.003%) | _ |
| N15 | Not available | Negative | + | 3.08 | EGFR L858R (14.159%) | _ |
| N16 | Not available | Negative | _ | 25.6 | EGFR L858R (0.229%) | _ |
| N17 | Not available | Leptomeningeal enhancement | + | 1.65 | TP53 P301Q (0.482%) | _ |
| N18 | Not available | Negative | + | 2.26 | EGFR E746_A750del (38.178%) | _ |
| N19 | Not available | Leptomeningeal enhancement | _ | 30.6 | EGFR T790 M (22.61%) | _ |
| N20 | Not available | Leptomeningeal enhancement | + | 2.58 | TP53 G245S (0.497%) | _ |
| N21 | Not available | Negative | + | 26.6 | EGFR L858R (78.401%) | _ |
| N22 | NapsinA(+), TTF1(−), Ki-67(−), CEA(+) | Negative | + | 1.42 | EGFR L858R (12.359%) | _ |
| N23 | Not available | Negative | + | 2.76 | EGFR L858R (24.468%) | _ |
| N24 | Not available | Negative | + | 3.58 | EGFR L858R (5.535%) | _ |
| N25 | NapsinA(+) | Leptomeningeal enhancement | + | 2.92 | PTEN C136Y (1.431%) | _ |
| N26 | Not available | Leptomeningeal enhancement | + | 7.54 | EGFR T790 M (6.286%) | _ |
| N27 | CK7(−), Ki-67(−), TTF1(−), NapsinA(−), CK20(−), CDX20(−), CDX2(−), CEA(−), FAP(−) | Leptomeningeal enhancement | + | 1.23 | EGFR L858R (79.739%) | + |
| N28 | Not available | Leptomeningeal enhancement | – | 4.1 | EGFR L858R (1.606%) | + |
| N29 | Not available | Negative | + | 3.36 | PTEN K13Q (0.387%) | _ |
| N30 | Not available | Negative | + | 4.16 | EGFR L858R (24.22%) | _ |
| N31 | CK7(+), NapsinA(+), Ki-67(−), PR(−), TTF1(−) | Leptomeningeal enhancement | + | 4.04 | EGFR T790 M (5.233%) | _ |
| N32 | Ki-67(−), CEA(+), CK7(+), TTF1(+), CDX2(−), CK20(−), NapsinA(+), ER(−), PR(−) | Leptomeningeal enhancement | + | 3.8 | EGFR L858R (3.74%) | _ |
| N33 | Not available | Leptomeningeal enhancement | _ | 1.51 | PTEN K13Q (0.464%) | + |
| N34 | Not available | Leptomeningeal enhancement | _ | 1.09 | TP 53 P250L (84.24%) | _ |
| N35 | Not available | Leptomeningeal enhancement | _ | 1.42 | FLT3 Y842C (0.404%) | _ |
The sensitivity of the neuroimaging was 88% (95% confidence intervals [95% CI], 75 to 100) (p = 22/25) and 63% (95% CI, 47 to 79) (p = 22/35) compared to those of CSF cytology and CSF ctDNA, respectively. The sensitivity of the CSF cytology was 71% (95% CI, 56 to 86) (n = 25/35) compared to that of CSF ctDNA. The specificity of CSF ctDNA in all cases was 100%. These data suggest a higher sensitivity of CSF ctDNA than those of CSF cytology and neuroimaging findings.
Nine cases were CSF cytology negative but neuroimaging positive while CSF ctDNA with cancer-associated gene mutations could be detected in them. Interestingly, neoplastic cells were found in CSF of patient N33 after 1 year from the first lumber puncture (Fig. 1).
Fig. 1.
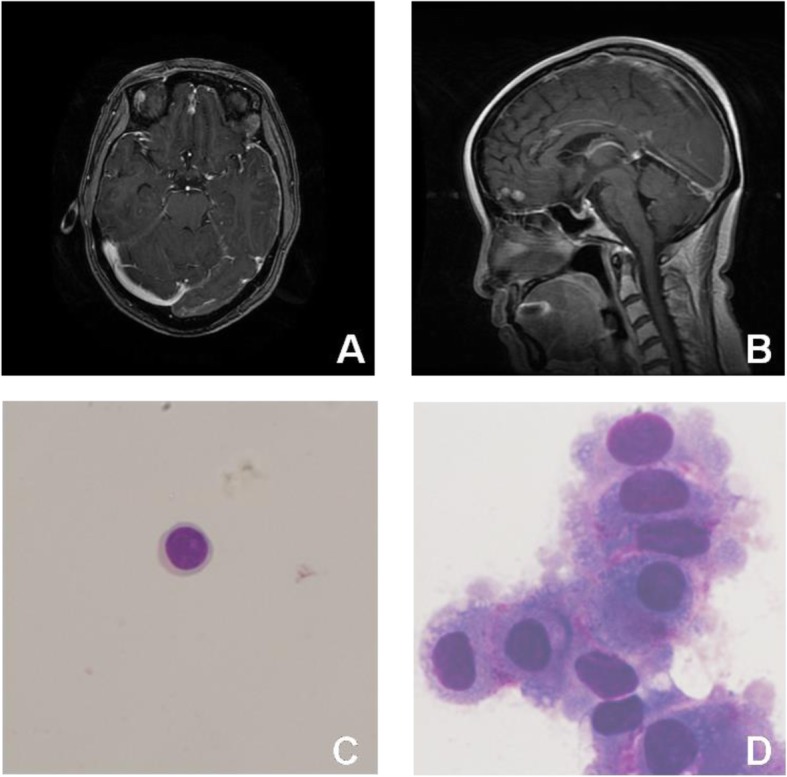
Head contrast enhanced MRI (a and b) of one patient showed abnormally uniform leptomeningeal enhancement. At the same time, no neoplastic cells were found in CSF by CSF cytology (c) while CSF sample revealed ctDNA and showed cancer-associated gene mutations of PTEN K13Q (0.464%), TP53 P301Q (0.809%), VHL R161Q (0.213%), CDKN2A A68T(0.235%) and FGFR2 Y375C (0.207%) by NGS. CSF cytology (d) 1 year later showed a large number of neoplastic cells
Concordance of EGFR activating mutations in primary lung adenocarcinomas and MC CSF samples
The comparisons between primary tumors and MC CSF samples were available only in 6 MC patients with lung adenocarcinoma (Table 4). We collected CSF samples from N10, N11, N15 and N31 during the tyrosine kinase inhibitor (TKI) therapy and N16 and N24 samples several months after the TKI. All patients, as carriers of EGFR mutations, of which 2 patients were found to contain exon 19 deletion and 4 patients to contain L858R. Additionally, CSF sample N11 also exhibited T790 M and E709A while CSF sample N31 also contained T790 M.
Table 4.
The comparisons of EGFR activating mutations between primary lung adenocarcinomas and MC CSF samples
| CSF sample No. | Primary lung adenocarcinoma tissues | CSF samples |
|---|---|---|
| N10 | EGFR 19Del | EGFR 19Del |
| N11 | EGFR L858R | EGFR L858R, E709A, T790 M |
| N15 | EGFR L858R | EGFR L858R |
| N16 | EGFR L858R | EGFR L858R |
| N24 | EGFR L858R | EGFR L858R |
| N31 | EGFR 19Del | EGFR 19Del, T790 M |
Discussion
In the present study, we compared the sensitivity of three methods including CSF cytology, neuroimaging findings and CSF ctDNA in the diagnosis of MC. The study revealed that CSF ctDNA had a higher sensitivity than the CSF cytology and neuroimaging findings. Most studies of ctDNA published to date have demonstrated the importance of plasma ctDNA as a liquid biopsy medium for various kinds of cancer given that benign tumors and non-neoplastic conditions do not generally give rise to ctDNA [23]. Similarly, CSF ctDNA may be useful in complementing the diagnosis of MC especially for cases with persistently negative CSF cytology and/or negative neuroimaging findings.
Currently, the presence of neoplastic cells in the CSF is the most useful criteria to confirm the diagnosis of MC and CSF cytology remains the gold diagnostic standard although the false negative rate of CSF cytology is still high. In our study, we found the sensitivity of CSF ctDNA is higher than that of CSF cytology. Previous study [14] had presented that ctDNA was often present in patients without detectable circulating tumor cells. Contrast-enhanced brain MRI is the technique of choice to evaluate patients with suspected MC but still has a 30% incidence of false-negative results [11]. The sensitivity of the neuroimaging was 63% compared to that of CSF-derived ctDNA by next-generation sequencing in our study. Neuroimaging studies are often noninformative or slow to reflect progression. Repeated neuroimaging also subjects patients to radiation, whereas monitoring ctDNA is noninvasive. However, other studies demonstrate that a cancer containing ~ 50 million malignant (rather than benign) cells releases sufficient DNA for detection in the circulation [24]. A cancer of this size is well below that required for definitive imaging at present. Measurement of tumor markers in the CSF may be convenient and of value in the adjunctive diagnosis of MC but lack sensitivity and specificity [6–9]. Unlike tumor markers such as CA19–9 or CEA, which are expressed in normal cells as well as in neoplastic cells, gene mutations of a clonal nature are only found in neoplasms.
In cancer patients, ctDNA are thought to be released in plasma as a result of tumor cells apoptosis and/or necrosis [14, 25]. ctDNA has played an important role in monitoring disease status of advanced cancer patients as a promising blood-based biomarker [26]. Many studies showed that ctDNA analysis can be utilized in early diagnosis of human malignancies including pancreatic, renal, advanced ovarian, colorectal, bladder, gastroesophageal, breast, melanoma, hepatocellular, head and neck cancers [14, 19, 27–29]. Previous study [14] showed that less than 10% of patients with gliomas harbored detectable ctDNA in the plasma while Yuxuan Wang et al. [18] demonstrated they identified detectable levels of CSF ctDNA in 74% of patients with primary tumors of the brain or spinal cord whereas no ctDNA was detected in patients whose tumors were not directly adjacent to a CSF reservoir. We attribute the 100% detection rate of CSF-derived ctDNA in our study to the reason that MC disseminates over the leptomeningeal surface, neoplastic cells shed into CSF and CSF is in direct contact with neoplastic cells and meninges while owing to physical obstacles such the blood–brain barrier, CSF ctDNA is unable to circulate fully within the blood system, resulting in a limited amount of ctDNA from CNS being released to plasma circulation.
Elena et al. [19] used next-generation sequencing to reveal somatic alterations in tumor-derived DNA from CSF in patients with CNS metastases mainly brain parenchyma metastasis while the positive rate is not 100 %. Our study showed a good result due to the patients who were recruited in this study were all definite MC cases. Tumor cells of MC cases were circulating in the CSF, which led to be easier to be caught. Consequently, this technique is more suitable for the diagnosis of MC than brain parenchyma metastasis.
As per the analysis for concordance of EGFR activating mutations between primary lung adenocarcinomas and MC, on the basis of patients’ CSF samples, the results showed that, EGFR activating mutations in CSF samples were consistent with those in primary adenocarcinomas. The showing of T790 M in two CSF samples, is possibly attributed to the fact that both two CSF samples were sequentially collected during the process of TKI therapy.
Our study find DNA mutation in CSF of patients with MC at 100% of our cohort, and it may give additional information to diagnose MC with negative CSF cytology. Furthermore, the type of tumor-associated gene mutations can provide many clues to the primary tumor type especially for patients whose primary tumor wouldn’t be found in spite of using various inspection methods. Additionally, looking for and knowing the primary tumor is the rather important diagnostic dependency.
However, our current study does have some limitations; for example, the cohort has had relative small numbers of patients and we used a smaller 143 gene panel. Though it was the most comprehensive gene panel at that time, including hot-spot mutation, copy number variants, gene fusion, and gene therapy information, it could not detect all possible aberrations in DNA. Thus, future study with larger size of samples from other departments and multiple institutions and using larger gene panel could help us to solve these issues.
Conclusions
This study suggests a higher sensitivity of CSF ctDNA than those of CSF cytology and neuroimaging findings. We find cancer-associated gene mutations in ctDNA from CSF of patients with MC at 100% of our cohort, and utilizing CSF ctDNA as liquid biopsy technology based on the detection of cancer-associated gene mutations may give additional information to diagnose MC with negative CSF cytology and/or negative neuroimaging findings. At the same time, cancer-associated gene mutations provide a reliable theory for MC targeted therapy.
Acknowledgements
The authors sincerely thank the MC patients for their help and willingness to participate in this study.
Abbreviations
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- CT
Computed tomography
- ctDNA
Circulating tumor DNA
- KPS
Karnofsky Performance Scores
- MC
Meningeal carcinomatosis
- MRI
Magnetic resonance imaging
- NGS
Next-generation sequencing
- NM
Neoplastic meningitis
- TKI
Tyrosine kinase inhibitor
Authors’ contributions
Study concept and design (JZC, LG and HB); acquisition and interpretation of the data (YZ, JYH, YLZ, XSG, LG, JZC and HB); drafting of the manuscript (YZ, LG and HB); critical revision of the manuscript for important intellectual content (LG and HB); acquisition of funding (HB); administrative, technical, or material support (administrative, technical, or material support); study supervision (LG and HB). All the authors read and approved the final manuscript.
Funding
This work was supported by a grant from the National key research and Development Program of China [No.2016YFC0904503]. The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Study data are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
This study has been approved by the Ethics Committee of the Second Hospital of Hebei Medical University. Written informed consent has been obtained from all patients or their legal surrogates.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yue Zhao, Email: zhaoyue_sunroad@163.com.
Jun-Ying He, Email: hjy_zn@126.com.
Yue-Li Zou, Email: sunnygirlzyl@126.com.
Xiao-Su Guo, Email: guoxiaosu1@163.com.
Jun-Zhao Cui, Email: 1097709288@qq.com.
Li Guo, Email: guolimed@126.com.
Hui Bu, Email: buhuisci@163.com.
References
- 1.Chamberlain MC. Neoplastic meningitis. Curr Neurol Neurosci Rep. 2008;8(3):249–258. doi: 10.1007/s11910-008-0038-6. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlain MC, Glantz M, Groves MD, Wilson WH. Diagnostic tools for neoplastic meningitis: detecting disease, identifying patient risk, and determining benefit of treatment. Semin Oncol. 2009;36(2):S35–S45. doi: 10.1053/j.seminoncol.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Azevedo CRASD, Cruz MRS, Chinen LTD, Peres SV, Peterlevitz MA, Pereira AEDA, et al. Meningeal carcinomatosis in breast cancer: prognostic factors and outcome. J Neuro-Oncol. 2011;104(2):565–572. doi: 10.1007/s11060-010-0524-y. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain MC. Leptomeningeal metastasis. Curr Opin Oncol. 2010;22(6):627–635. doi: 10.1097/CCO.0b013e32833de986. [DOI] [PubMed] [Google Scholar]
- 5.Martins SJ, Azevedo CR, Chinen LT, Cruz MR, Peterlevitz MA, Gimenes DL. Meningeal carcinomatosis in solid tumors. Arq Neuropsiquiatr. 2011;69(6):973–980. doi: 10.1590/S0004-282X2011000700024. [DOI] [PubMed] [Google Scholar]
- 6.Stockhammer G, Poewe W, Burgstaller S, Deisenhammer F, Muigg A, Kiechl S, et al. Vascular endothelial growth factor in CSF: a biological marker for carcinomatous meningitis. Neurol. 2000;54(8):1670–1676. doi: 10.1212/WNL.54.8.1670. [DOI] [PubMed] [Google Scholar]
- 7.Gauchez AS, Pez E, Boutonnat J, Bourre JC, Pelletier L, Payan R, et al. Early detection of leptomeningeal metastasis in patients with metastatic breast carcinoma: validation of CA 15–3 measurement in cerebrospinal fluid. Ann Biol Clin. 2007;65:6–653. [PubMed] [Google Scholar]
- 8.Groves MD, Hess KR, Puduvalli VK, Colman H, Conrad CA, Gilbert MR, et al. Biomarkers of disease: cerebrospinal fluid vascular endothelial growth factor (VEGF) and stromal cell derived factor (SDF)-1 levels in patients with neoplastic meningitis (NM) due to breast cancer, lung cancer and melanoma. J Neuro-Oncol. 2009;94(2):229–234. doi: 10.1007/s11060-009-9819-2. [DOI] [PubMed] [Google Scholar]
- 9.Orphanos G, Ioannidis G, Michael M, Kitrou G. Prostate-specific antigen in the cerebrospinal fluid: a marker of local disease. Med Oncol. 2009;26:2–143. doi: 10.1007/s12032-008-9096-5. [DOI] [PubMed] [Google Scholar]
- 10.Rhodes CH, Glantz MJ, Glantz L, Lekos A, Sorenson GD, Honsinger C, et al. A comparison of polymerase chain reaction examination of cerebrospinal fluid and conventional cytology in the diagnosis of lymphomatous meningitis. Cancer. 1996;77(3):543–548. doi: 10.1002/(SICI)1097-0142(19960201)77:3<543::AID-CNCR17>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.RJFM FRACP, George Krol MD, DLM D. Neuroimaging and cerebrospinal fluid cytology in the diagnosis of leptomeningeal metastasis. Ann Neurol. 1995;38(1):51–57. doi: 10.1002/ana.410380111. [DOI] [PubMed] [Google Scholar]
- 12.Guo X, Cui J, Zhao Y, Han W, Zou Y, Gao R, et al. The therapeutic value of cerebrospinal fluid ctDNA detection by next-generation sequencing for meningeal carcinomatosis: a case report. BMC Neurol. 2019;19:1–38. doi: 10.1186/s12883-018-1232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): clinical significance and utility in Cancer shaped by emerging technologies. Mol Cancer Res. 2016;14(10):898–908. doi: 10.1158/1541-7786.mcr-16-0044. [DOI] [PubMed] [Google Scholar]
- 14.Bettegowda C, Sausen M, Leary R, Kinde I, Agrawal N, Bartlett B, et al. DETECTION OF CIRCULATING TUMOR DNA IN EARLY AND LATE STAGE HUMAN MALIGNANCIES. Sci Transl Med. 2014;6:224–224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piccioni DE, Achrol AS, Kiedrowski LA, Banks KC, Boucher N, Barkhoudarian G, et al. Analysis of cell-free circulating tumor DNA in 419 patients with glioblastoma and other primary brain tumors. CNS Oncol. 2019;8:2–CNS34. doi: 10.2217/cns-2018-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De ML, Mayor R, Ng CK, Weigelt B, Martínezricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YS, Jiang BY, Yang JJ, Zhang XC, Zhang Z, Ye JY, et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in Leptomeningeal metastases of EGFR-mutant non-small cell lung Cancer: a new medium of liquid biopsy. Ann Oncol Official J Eur Soc Med Oncol. 2018;29:11. doi: 10.1200/jco.2011.29.27_suppl.11. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Springer S, Zhang M, Mcmahon KW, Kinde I, Dobbyn L, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112(31):9704–9709. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pentsova EI, Shah RH, Tang J, Boire A, You D, Briggs S, et al. Evaluating Cancer of the central nervous system through next-generation sequencing of cerebrospinal fluid. J Clin Oncol. 2016;34(20):2404–2415. doi: 10.1200/jco.2016.66.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai X, Sheng J, Tang C, Nandakumar V, Ye H, Ji H, et al. Frequent mutations in EGFR, KRAS and TP53 genes in human lung cancer tumors detected by ion torrent DNA sequencing. Plos One. 2014;9:4–e95228. doi: 10.1371/journal.pone.0095228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Huo X, Tang C, Ye H, Nandakumar V, Lou F, et al. Frequent KIT mutations in human gastrointestinal stromal tumors. Sci Rep. 2014;4:5907. doi: 10.1038/srep05907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai X, Zhang E, Ye H, Nandakumar V, Wang Z, Chen L, et al. PIK3CA and TP53 Gene Mutations in Human Breast Cancer Tumors Frequently Detected by Ion Torrent DNA Sequencing. Plos One. 2014;9:6–e99306. doi: 10.1371/journal.pone.0099306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jr LAD, Williams R, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nat. 2012;486(7404):537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canzoniero VL, Park BH. Use of cell free DNA in breast oncology. Biochim Biophys Acta. 2016;1865(2):266–274. doi: 10.1016/j.bbcan.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Calapre Leslie, Warburton Lydia, Millward Michael, Ziman Mel, Gray Elin S. Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Letters. 2017;404:62–69. doi: 10.1016/j.canlet.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Newman AM, Bratman SV, To J. Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:5–548. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creemers A, Krausz S, Strijker M, Mj VDW, Soer EC, Reinten RJ, et al. Clinical value of ctDNA in upper-GI cancers: a systematic review and meta-analysis. Biochim Biophys Acta. 2017;1868(2):394–403. doi: 10.1016/j.bbcan.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Corrò C, Hejhal T, Poyet C, Sulser T, Hermanns T, Winder T, et al. Detecting circulating tumor DNA in renal cancer: an open challenge. Exp Mol Pathol. 2017;102(2):255–261. doi: 10.1016/j.yexmp.2017.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Study data are available from the corresponding author upon reasonable request.


