Abstract
The epigenetic regulation of gene expression (via DNA methylation, histone modification and microRNA interference) contributes to a variety of diseases, particularly cancer. Protein deubiquitination serves a key role in the mechanism underlying histone modification, and consequently influences tumor development and progression. Improved characterization of the role of ubiquitinating enzymes has led to the identification of numerous deubiquitinating enzymes (DUBs) with various functions. Gastric cancer (GC) is a highly prevalent cancer type that exhibits a high mortality rate. Latest analysis about cancer patient revealed that GC is sixth deadliest cancer type, which frequently occur in male (7.2%) than female (4.1%). Complex associations between DUBs and GC progression have been revealed in multiple studies; however, the molecular mechanism underpinning the metastasis and recurrence of GC is yet to be elucidated. Generally, DUBs were upregulated in gastric cancer. The relation of DUBs and tumor size, classification and staging was observed in GC. Besides, 5-yar survival rate of patients with GC is effeccted by expression level of DUBs. Among the highly expressed DUBs, specifically six DUBs namely UCHs, USPs, OTUs, MJDs, JAMMs and MCPIPs effect on this survival rate. Consequently, the association between GC and DUBs has received increasing attention in recent years. Therefore, in the present review, literature investigating the association between DUBs and GC pathophysiology was analyzed and critically appraised.
Keywords: deubiquitinating enzymes, gastric cancer, survival
1. Introduction
Epigenetic mechanisms are implicated in tumorigenesis and cancer progression, and are defined as heritable changes that do not affect the DNA sequence. Examples include DNA methylation, histone modification and microRNA (miR) interference (1). Histone modification serves an important role in transcriptional regulation, DNA repair and replication, and chromosomal condensation. Several studies have indicated that histone modifications typically occur at the N-terminus, primarily in the form of acetylation, methylation, phosphorylation or ubiquitination (2,3).
Ubiquitination describes a post-translational modification of proteins under the conditions of normal homeostasis or disease, which involves the addition of the evolutionarily conserved small protein ubiquitin (4) or UBLs (ubiquitin-like proteins) (5) that tag the protein for proteasomal degradation or non-degradative processes (6). Ubiquitin is a 76-amino acid polypeptide that can covalently conjugate with protein substrates via a mechanism involving three enzymes: Ubiquitin-activating (E1), ubiquitin-conjugating (E2) and ubiquitin ligase (E3). The ubiquitination of a specific protein substrate requires the selective recruitment of E1, E2 and E3 (7–9). In eukaryotic cells, the structure of ubiquitin is highly conserved and the protein responds to certain chemical signals (such as phosphorylation, oxidation, misfolding and damage to the ubiquitinated protein) to induce the ubiquitin-proteasome degradation pathway (10). Notably, the activity of deubiquitinating enzymes (DUBs) directly influences the turnover rate, activity, regeneration and localization of various proteins in cells. In addition, DUBs serve an important role in homeostasis (11), the stabilization and degradation of proteins, and signal transduction pathways (11). Changes in protein structure, abnormal spatial and temporal expression, and uncontrolled activity can result in the development of certain conditions, including arthritis, neurodegenerative and cardiovascular diseases, and tumors. In humans, DUBs can serve a role in the genesis of tumors as either oncogenes or tumor suppressor genes.
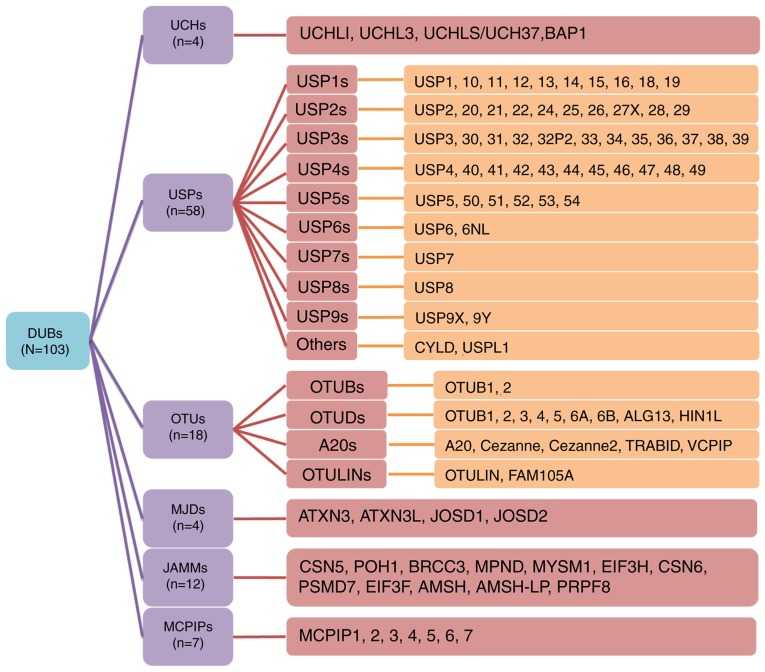
The DUB protein family reportedly comprises 103 members, the majority of which are cysteine proteases. As a result of similarities in their amino acid sequences and molecular structures, these proteins can be divided into the following six families: Ubiquitin C-terminal hydrolases (UCHs) (12), ubiquitin-specific proteases (USPs), ovarian tumor-related proteases (OTUs), Machado-Joseph disease protein domain proteases (MJDs), Jab1/MPN domain-associated metalloisopeptidase (JAMM) domain proteins and monocyte chemotactic protein-induced proteins (MCPIPs) (13). To further summarize and stratify the aforementioned proteins, subfamilies are also detailed in Fig. 1. USP16 (14), USP6NL (15), ubiquitin thioesterase otulin (OTULIN) (16) and family with sequence similarity 105 member A (FAM105) (17) were also recently discovered. The majority of these DUBs are associated with tumor progression and several studies have revealed the association between DUBs and gastric cancer (GC) (18,19). Of note, GC is the second leading cause of cancer-associated mortality and the fourth most common cancer type worldwide (20,21).
Figure 1.
Members of the DUB family. The DUB family contains numerous members, which have been divided into subfamilies. The USPs are the largest subfamily of DUBs, and were further divided into 9 subfamilies (USP1-9) in the present study; CYLD lysine 63 deubiquitinase and USPL1 have been listed as other USP members. The first digits indicates the subfamily, for example, USP14 belongs to the USP1 subfamily. The subfamily classification of the ovarian tumor-related protease family refers to existing taxonomies (102). DUB, deubiquitinating enzyme; USP, ubiquitin-specific protease.
The lack of a comprehensive understanding of the molecular mechanism underpinning GC metastasis and recurrence suggests that further investigation is required. Thus, DUBs and their potential association with the progression of GC were the primary focus of the present review. Within the present study, subsequent data analyses were performed using the Gene Expression Profiling Interactive Analysis (GEPIA) website (http://gepia.cancer-pku.cn), which primary collates data from The Cancer Genome Atlas and the Genotype-Tissue Expression project databases. The name of a each target gene was input into the GEPIA website and the corresponding data was extracted (22).
2. UCHs and GC
The enzymes of the UCH protein family contain a conserved catalytic domain known as the UCH domain, which comprises ~230 amino acids (23). This protein family includes four members, including UCHL1/protein gene product 9.5, UCHL3, UCHL5/UCH37 and BRCA1 associated protein-1 (BAP1) (24–27). The activities of these proteins have been associated with the occurrence and development of cancer, and several studies have identified that UCHL1, UCHL5 and BAP1 are specifically involved in the formation of GC (24–26).
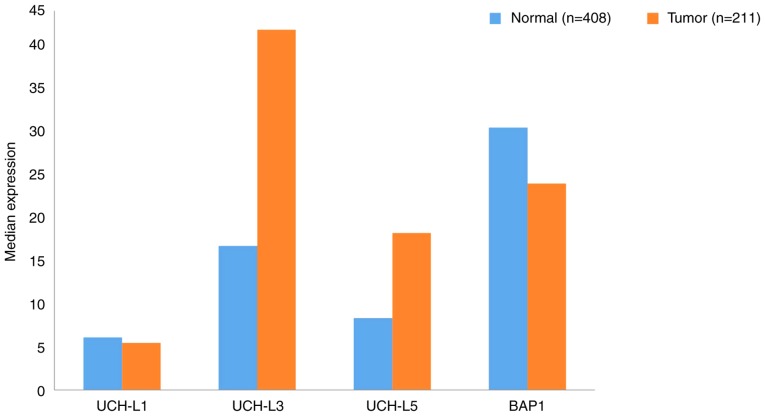
Using data extracted from GEPIA, the gene expression profiles of UCHs between GC samples and the paired normal tissues are presented in Fig. 2. The gene expression levels of UCHL3 and UCHL5 in tumor tissues were upregulated >2-fold compared with normal tissues. To the best of our knowledge, no studies have reported the link between UCHL3 and GC; however, UCHL5 has been identified as a potential biomarker of GC with novel prognostic value. For elderly patients with dysregulated protein homeostasis, high levels of UCHL5 inhibited proteasome activity, and were determined to promote the apoptosis of cancer cells (28). Regarding UCHL1, research has shown that it could be a candidate biomarker and therapeutic target for GC metastasis, as UCHL1 promoted this process via the Akt and Erk1/2 pathways (29). BAP1 expression is downregulated in gastric carcinoma and its decreased expression was associated with a malignant phenotype (histological grade) and a more advanced TNM stage (30). Furthermore, low BAP1 expression was revealed to be associated with poor prognosis in patients with gastric adenocarcinoma and GC (30).
Figure 2.
Gene expression profile of ubiquitin C-terminal hydrolases between gastric cancer samples and paired normal tissues. Data were extracted using the Gene Expression Profiling Interactive Analysis website. UCH, Ubiquitin C-terminal hydrolase; BAP1, BRCA1 associated protein-1.
Associations between UCHs and certain clinicopathological features, and the 5-years survival rates of patients with GC are presented in Table I. High expression levels of UCHs in patients with GC were predominantly associated with tumor size and TNM stage, but not sex or age. Analysis of these expression levels indicated that the higher the degree of positive BAP1 and UCHL5 expression in GC, the higher the 5-year survival rate of patients. Conversely, increased expression of UCHL1 was revealed to reduce the survival rate of patients.
Table I.
Association between DUBs and clinicopathological variables and survival in patients with gastric cancer.
| Age, years | Sex | Tumor size, cm | Lauren classification | TNM stage | 5-year survival | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DUB | Expression | Cases | <60 | ≥60 | M | F | <5 | ≥5 | Intestinal | Diffuse | I and II | III and IV | Patients, n | Rate (%) |
| BAP1 (30) | Positive | 211 | 124 | 87 | 145 | 66 | 42 (<3) | 169 (≥3) | 51 | 117 | 91 | 74 | 136 | 65 |
| Negative | 263 | 144 | 119 | 180 | 83 | 20 (<3) | 243 (≥3) | 43 | 174 | 120 | 189 | 100 | 38 | |
| P-value | 0.381 | 0.948 | 0.001 | 0.042 (Mix=43/46) | 0.01 | 0.001 | ||||||||
| UCHL1 (29) | Positive | 87 | 37 | 50 | 67 | 20 | 23 (<3) | 64 (≥3) | NF | NF | 25 | 62 | 26 | 30 |
| Negative | 109 | 59 | 50 | 80 | 29 | 43 (<3) | 66 (≥3) | NF | NF | 53 | 56 | 66 | 61 | |
| P-value | 0.107 | 0.561 | 0.038 | NF | 0.004 | 0.001 | ||||||||
| UCHL5 (28) | Positive | 379 | 181 (<66) | 198 (≥66) | 188 | 191 | 139 | 240 | 181 | 198 | 150 | 229 | 258 | 65 |
| Negative | 111 | 51 (<66) | 60 (≥66) | 54 | 57 | 50 | 61 | 34 | 77 | 40 | 71 | 42 | 38 | |
| P-value | 0.747 | 0.914 | 0.173 | 0.004 | 0.001 | 0.108 | ||||||||
| USP10 (45) | Positive | 198 | 95 (≤57) | 103 (>57) | 137 | 61 | 83 (<4) | 115 (≥4) | 111 | 87 | 108 | 90 | 106 | 64 |
| Negative | 167 | 85 (≤57) | 82 (>57) | 118 | 49 | 60 (<4) | 107 (≥4) | 72 | 95 | 69 | 98 | 79 | 40 | |
| P-value | 0.578 | 0.761 | 0.182 | 0.060 | 0.004 | 0.003 | ||||||||
| USP14 (44) | Positive | 62 | 36 | 26 | 49 | 13 | NF | NF | NF | NF | 17 | 45 | NF | NF |
| Negative | 51 | 32 | 19 | 44 | 7 | NF | NF | NF | NF | 21 | 30 | NF | NF | |
| P-value | 0.613 | 0.316 | NF | NF | 0.124 | NF | ||||||||
| USP20 (57) | Positive | 52 | 21 | 31 | 37 | 15 | 39 | 13 | NF | NF | 30 | 22 | 23 | 45 |
| Negative | 37 | 18 | 19 | 24 | 13 | 11 | 26 | NF | NF | 13 | 24 | 8 | 20 | |
| P-value | 0.439 | 0.529 | 0.001 | NF | 0.036 | 0.003 | ||||||||
| USP22 (58) | Positive | 125 | 55 | 70 | 88 | 37 | 56 | 69 | NF | NF | 23 | 102 | 23 | 18 |
| Negative | 94 | 42 | 52 | 74 | 20 | 39 | 45 | NF | NF | 59 | 35 | 45 | 48 | |
| P-value | 0.920 | 0.165 | 0.283 | NF | 0.004 | 0.001 | ||||||||
| USP3 (71) | Positive | 67 | NF | NF | 42 | 25 | NF | NF | 37 | 30 | 16 | 51 | 13 | 20 |
| Negative | 80 | NF | NF | 25 | 27 | NF | NF | 61 | 19 | 50 | 30 | 48 | 60 | |
| P-value | NF | 0.653 | NF | 0.007 | 0.001 | 0.001 | ||||||||
| USP33 (72) | Positive | 58 | 24 (≤55) | 34 (>55) | 32 | 26 | 38 | 20 | NF | NF | 34 | 24 | 40 | 68 |
| Negative | 63 | 23 (≤55) | 40 (>55) | 19 | 32 | 32 | 31 | NF | NF | 12 | 51 | 26 | 42 | |
| P-value | 0.583 | 0.095 | 0.182 | NF | 0.001 | 0.001 | ||||||||
| USP39 (74) | Positive | 26 | NF | NF | NF | NF | NF | NF | NF | NF | NF | NF | 6 | 24 |
| Negative | 27 | NF | NF | NF | NF | NF | NF | NF | NF | NF | NF | 18 | 62 | |
| P-value | NF | NF | NF | NF | NF | 0.019 | ||||||||
| USP42 (85) | Positive | 45 | 15 (<55) | 30 (≥55) | 36 | 9 | 25 | 20 | NF | NF | 49 | 41 | NF | NF |
| Negative | 45 | 11 (<55) | 34 (≥55) | 31 | 14 | 14 | 31 | NF | NF | 18 | 27 | NF | NF | |
| P-value | 0.486 | 0.334 | 0.033 | NF | 0.006 | NF | ||||||||
| USP44 (86) | Positive | 90 | NF | NF | 60 | 30 | NF | NF | NF | NF | 32 | 58 | 33 | 36.8 |
| Negative | 117 | NF | NF | 78 | 39 | NF | NF | NF | NF | 51 | 66 | 59 | 50.5 | |
| P-value | NF | 1 | NF | NF | 0.440 | 0.033 | ||||||||
| USP9X (97) | Positive | 43 | 31 | 12 | 34 | 9 | 29 | 14 | NF | NF | 11 | 32 | 4 | 10 |
| Negative | 25 | 15 | 10 | 18 | 7 | 12 | 13 | NF | NF | 16 | 9 | 8 | 32 | |
| P-value | 0.304 | 0.508 | 0.114 | NF | 0.006 | 0.003 | ||||||||
| OTUB1 (105) | Positive | 78 | 33 | 45 | 56 | 22 | 35 | 43 | NF | NF | 10 | 68 | 20 | 25 |
| Negative | 78 | 40 | 38 | 64 | 14 | 56 | 22 | NF | NF | 22 | 56 | 33 | 42 | |
| P-value | 0.261 | 0.128 | 0.001 | NF | 0.017 | 0.027 | ||||||||
| EIF3F (133) | Positive | 66 | 33 | 33 | 49 | 17 | 34 | 32 | NF | NF | 53 | 13 | 56 | 85 |
| Negative | 129 | 51 | 78 | 94 | 36 | 80 | 49 | NF | NF | 46 | 90 | 90 | 70 | |
| P-value | 0.170 | 0.870 | 0.160 | NF | 0.020 | 0.040 | ||||||||
The expression of DUBs in GC and normal tissues was not significantly associated with sex or age, but was associated with tumor size, tumor stage, grading and 5-year-survival. Positive expression refers to the positive expression of a certain protein in gastric cancer. All expression was detected by immunohistochemistry. BAP1, BRCA1 associated protein-1; DUBs, deubiquitinating enzymes; EIF3F, eukaryotic translation initiation factor 3 subunit F; NF, not found; OTUB1, otubain 1; TNM, tumor-node-metastasis; UCH, ubiquitin C-terminal hydrolase; USP, ubiquitin-specific protease. *P<0.05.
3. USPs and GC
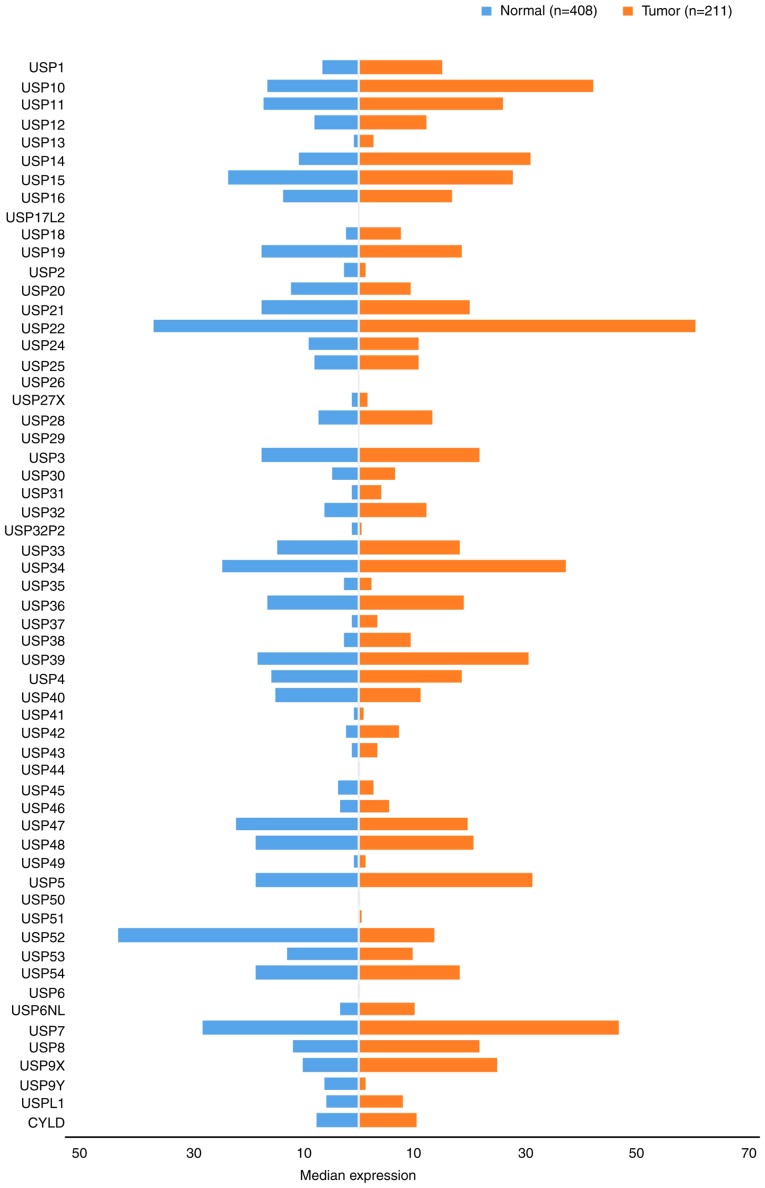
USPs are the most diverse family of DUBs and the USP subclass represents the majority of DUBs encoded by the human genome. Consequently, numerous studies have investigated their function, substrates and mechanisms of action in various diseases. The discovery of gene mutations and the upregulation of USPs in various types of cancer, and their potential for targeted small molecule-mediated inhibition, indicates USPs as a promising therapeutic target. There is also increasing interest in the development of USP-specific inhibitors as antiviral and anticancer agents (31). In the present study, the USP family was stratified into 10 subgroups comprising USPs 1–10. The gene expression profile of USPs was compared between GC and paired normal tissues (Fig. 3).
Figure 3.
Gene expression profiles of ubiquitin-specific proteases between gastric cancer samples and paired normal tissues. Data were extracted using the Gene Expression Profiling Interactive Analysis website. USP, ubiquitin-specific protease. CYLD, CYLD lysine 63 deubiquitinase.
USP1s and GC
The USP1 subfamily includes 11 members: USP1 (32), USP10-13 (33–36), USP14-16 (37–39), USP17L2 (40), USP18 (41) and USP19 (42). Of the USP1 subfamily members investigated, the expression levels in CG tissues was higher compared with those in the adjacent normal tissues (Fig. 3). The expression levels of USP13 and USP18 were <10 Transcripts Per Million (TPM), which were relatively low compared with the other USP1s investigated (which were expressed at levels >10 TPM). USP17L2 expression was not detected. The highest levels of expression were noted for USP10, followed by USP14 and USP11, which exhibited expression levels >20 TPM.
The expression of the majority of USP1s has been associated with tumor growth, though studies into GC have predominantly investigated USP10, USP14 and USP15 (43). USP10 and USP14 are independent predictors of prognosis for patients with GC, and the increased expression of USP10 in GC has been associated with the 5-year survival rate of patients. A previous study demonstrated that the downregulation of USP10, as well as the absence of USP14 expression in GC cells had significant effects on gastric wall invasion and lymph node metastasis, increased malignant biological behavior and reduced survival rate, as determined from a large number of clinical samples (44,45). Conversely, vimentin expression was upregulated in human GC tissues and cell lines as a result of deubiquitination, following interactions with USP14 and miR-320a, which may contribute to the aggressiveness of GC cells (46). It was also reported that the direct targeting of USP14 and vimentin by miR-320a inhibited GC cell proliferation, migration and invasion. miR-320a not only directly suppresses vimentin expression, but also binds to USP14, indirectly downregulating vimentin in GC tissues. Therefore, a high positive expression rate of USP14 correlates with a high recurrence rate in patients with GC (44,45).
USP2s and GC
The 10 USP2 family members are: USP2 (47), USP20 (48), USP21 (49), USP22 (50), and USP24-29 (26,51–55). As depicted in Fig. 3, the protein expression level of the USP2 subfamily in GC tissues were typically higher compared with those in normal gastric tissues in 9/10 cases, although the opposite was true for USP20. Notably, USP26 and 29 were not detected in gastric tissues, yet USP22 was expressed at levels as high as 60.31 TPM.
USP20, USP22 and USP28 have been previously determined to be associated with GC. Compared with normal tissues, high expression levels of USP28 were detected in GC tissues, and were also associated with the distant metastasis of tumors. Conversely, USP28 downregulation may significantly inhibit the proliferation and migration of GC cells; however, the effects of USP28 expression on the proliferation and migration of gastric epithelial mucosal cell lines were not significant (46). The aforementioned findings provide a novel insight for the development of therapeutic strategies to treat GC via the regulation of USP28 (56). USP20 also serves an important role in gastric tumorigenesis and progression. A negative association between USP20 expression and tumor size, tumor invasion and TNM stage has previously been reported (Table I). It was revealed that USP20 expression negatively correlated with patient prognosis and its anti-tumor activity. The mechanism underlying the effects of USP20 included the positive regulation of claspin stabilization in GC, thus, USP20 represents a promising molecular target for the development of novel therapeutic drugs (57). USP22-mediated protein stabilization of B cell-specific Moloney murine leukemia virus integration site 1 promotes the stemness of GC stem cells as well as GC progression, and its expression may also serve an important role in gastric carcinoma (58–60). Yang et al discussed that USP22 expresion is correlate with cancer progression. Where they found that around 57% of gastric cancer tisues showed high expression of USP22 comparing with normal connective tissue. This overexpression of USP22 consequentially effecct on tumor size, inavsion and metastasis (60). Additionally, both USP20 and USP22 expression are positively correlated with the 5-year survival rate of patients with GC (57,60).
USP3s and GC
The USP3 subfamily represents the largest family of USPs and consists of the following 12 members: USP3 (61), USP 30 (62), USP31 (63,64), USP32, USP32P2 (22), USP33 (64), USP34 (65), USP35 (66), USP36 (67), USP37 (68), USP38 (69) and USP39 (70). As shown in Fig. 3, excluding USP32P2 and USP35, the expression of each member was upregulated in GC tissues compared with normal gastric tissues. The highest expression levels were exhibited by USP34 (37.21), while the lowest were reported for USP32P2 (0.33). The expression levels of USP30, USP31, USP32P2, USP35, USP37 and USP38 were <10 TPM.
USP3 s may also serve as useful biomarkers to predict the prognosis of patients with GC. Studies investigating USP3s revealed their ability to influence cell proliferation, cell cycle regulation and transfer-related protein expression (61). In vivo experiments revealed that USP3s promoted the growth and metastasis of GC. Additionally, the high expression levels of these proteins imparted a lower survival rate in patients (71). Studies have also discovered that tumor location, tumor infiltration depth and TNM stage are all associated with USP33 upregulation, and affect the overall survival rate and prognosis of patients with GC. USP33 may also be linked with the prognosis of GC (62), and its high expression levels indicated longer survival times in patients (72).
It was also determined that short hairpin RNA-mediated downregulation of USP39, another member of the USP3 subfamily, inhibited GC cell proliferation and colony formation. USP39 inhibition also induced G2/M phase arrest and increased poly (ADP-ribose) polymerase cleavage (Asp214) suggesting that USP39 is critical for GC cell proliferation. As USP39 is upregulated in certain types of cancer, and hyperproliferation is a hallmark of cancer, USP39 may represent a potential therapeutic target for the treatment of several cancer types (73). By contrast, miR-133a expression was inversely correlated with USP39, which it directly targets by binding at the 3′-untranslated region; the high expression rate of USP39 indicated a longer survival time for patients (74).
USP4s and GC
The USP4 subfamily has a total of 11 members: USP4 (75), USP40 (76), USP41 (77), USP42 (78), USP43 (79), USP44, USP45 (80), USP46 (80), USP47 (80), USP48 (81) and USP49 (82). Generally, the expression of USP4 s in GC tissues was increased compared with those in normal adjacent tissues; however, USP40, USP44, USP45 and USP47 were downregulated (Fig. 3). The expression levels of the seven upregulated members were all <10.
Studies into GC have investigated USP42, USP44 and USP47. It has been reported that USP47 may represent a drug resistant target for GC. Additionally, it was determined that miR-204-5p was downregulated in GC, and may inhibit the proliferation of GC cells by targeting USP47 and RAB22A, thus serving a role in suppressing cancer development. Therefore, the recovery of miR-204-5p expression may be a potential therapeutic strategy for the treatment of GC (83,84). In vitro analyses also demonstrated that USP42 silencing suppressed cell proliferation by inducing G0/G1 arrest, and inhibited cellular invasion via matrix metalloprotease and epithelial-mesenchymal transition regulation. The increased expression of USP42 may be important in tumor progression and the metastasis of GC, and may serve as a prognostic marker (85). The combination of USP44 expression and DNA ploidy status may also serve as an independent prognostic marker in GC. Notably, the expression rate of USP44 in GC is negatively correlated with the 5-year survival rate of patients (86).
USP5s and GC
The USP5 subfamily comprises six members, including USP5 (87), USP50 (88), USP51 (89), USP52 (90), USP53 (91) and USP54 (92). The expression of USP5s in GC and normal tissues differed (Fig. 3). Notably, the expression of USP52 in CG tissues was upregulated 3-fold compared with that in normal gastric tissues, and USP52 was differentially expressed compared with USP5. The expression levels of USP50 and USP51 were <1 in normal and gastric tumor tissues, and although USP5s have been associated with multiple cancer types (93), no studies have reported the association between USP5s and GC.
USP6s and GC
At present, the USP6 subfamily comprises only two members, USP6 (94) and USP6NL (15). As presented in Fig. 3, the two members, particularly USP6, were not highly expressed in either GC or normal tissues. However, USP6 has been reported to contribute to the progression of colon cancer and may therefore represent a valuable prognostic biomarker for patients. USP6NL (also known as RN-tre) is a GTPase-activating protein involved in the regulation of endocytosis and signal transduction. USP6NL upregulation results in increased glycolysis in breast cancer cells and highlights a point of metabolic vulnerability for the targeting of certain therapeutic agents in a subset of aggressive basal-like breast tumors. The association between the USP6 subfamily and GC progression requires further investigation (15,95).
USP7s and GC
USP7 is currently the only member of the USP7 subfamily. Its expression levels in GC tissues are higher than those in normal tissue (46.63 TPM and 28.04 TPM, respectively; Fig. 3). Studies investigating USP7 and its relation to GC are yet to be performed; however, H. pylori was reported to affect the expression of the USP family via alternative H. pylori-specific mechanisms distinct from the conserved signaling pathways, during the activation of the innate immune response (18).
USP8s and GC
The roles of USP8 and its substrate (epidermal growth factor) have been evaluated in cancer therapy, and their possible targeting for the treatment of Cushing's disease has been investigated (96); USP8 is the only member of the USP8 subfamily. As shown in Fig. 3, USP8 expression in GC tissues was increased ~2-fold compared with that in normal tissues.
USP9s and GC
The USP9 subfamily comprises two members, USP9X and Y. As presented in Fig. 3, the expression of USP9X in GC tissues was upregulated 2-fold compared with that of normal tissues. By contrast, the expression of USP9Y in normal tissues was higher than that of cancerous tissues, though its overall expression was notably lower than that of USP9X. Upregulation of the deubiquitinating enzyme USP9X in GC suggested that it may be associated with certain oncogenes, and it was also significantly associated with reduced survival rate (97). A link between USP9Y and GC has not yet been confirmed, although its expression has been revealed to correlate with certain breast cancer characteristics (98).
Other proteins and GC
Additional USP family members include CYLD lysine 63 deubiquitinase (CYLD) and USPL1. Their expression in GC tissues was notably increased compared with normal tissues (Fig. 3). The CYLD signaling pathway serves a biological function similar to that of the oncogenes in gastrointestinal tumors, and has been associated with the occurrence and development of GC (99,100). Moreover, genetic variations affecting USPL1 expression have been linked to breast cancer (101).
Analysis of USP gene expression in GC and normal tissues (Fig. 3) revealed that their expression in tumor tissues is markedly upregulated compared with that in normal tissues; the majority of GC and normal tissues exhibited detectable basal levels of USP expression. Of note, the expression of certain genes was upregulated >2-fold; increased expression of USP52 (tumor=43.5; normal=13.6) was reported in normal tissues compared with GC samples. However, whether USPs may be considered as reliable prognostic indicators of GC requires further investigation. As presented in Table I, associations between USPs, and the clinicopathological features and prognosis of GC were reported. In addition, the increased expression of the majority of USPs in GC tissues was associated with poor prognosis. It was also revealed that the expression profiles of USP10, USP20 and USP33 were the opposite of those aforementioned.
4. OTUs and GC
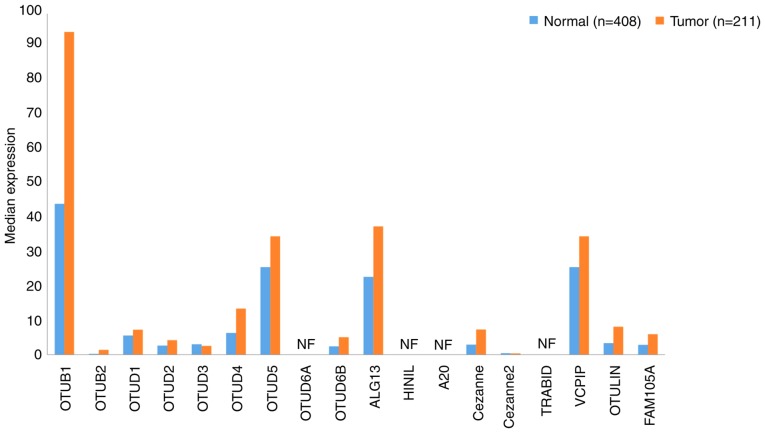
A total of 18 OTU family DUBs exist in humans, the majority of which have been associated with the prognosis of patients with tumors. OTUs can be divided into four categories: OTUBs, OTUDs, A20s and OTULINs (102). The gene expression profiles of OTUs in GC samples and paired normal tissues ware presented in Fig. 4.
Figure 4.
Gene expression profiles of ovarian tumor-related proteases between gastric cancer samples and paired normal tissues. Data were extracted using the Gene Expression Profiling Interactive Analysis website. NF, not found.
OTUBs and GC
The OTUB family comprises two members, OTUB1 (103) and OTUB2 (104), which are expressed in the majority of tissues. In the present study, their expression was determined to be increased 2-fold compared with that of normal tissues. Additionally, the expression of OTUB1 in gastric tissues was markedly higher than that of OTUB2 (which was almost undetectable), yet the expression levels of OTUB1 in GC tissues were as high as 93.09, which was twice that exhibited in normal tissues (Fig. 4). OTUB1-isoform 2 was reported to be a predictor of poor prognosis and to promote tumor progression in patients with GC (95). However, its potential clinical application as a marker of tumor invasiveness requires further investigation. Poor prognosis of patients with GC was revealed to correlate with high expression of OTUB1-isoform 2 (105); the association between OTUB2 and GC remains to be further studied.
OTUDs and GC
OTUDs are the largest class of DUBs, which comprises the following nine members: OTUD1-5 (106–110), OTUD6A (111) and B (112), UDP-N-acetylglucosamine transferase subunit ALG13 homolog (ALG13) and hematological and neurological expressed 1 protein (HIN1 L) (102). Using data extracted from GEPIA, the expression levels of both of the OTUD subfamily members in GC and normal tissues were determined to be relatively low. OTUD6A and HIN1L were undetectable, although the expression levels of OTUD4 and OTUD5 were >10 in GC tissues. Further investigation is required to determine the pathophysiological role of OTUDs in GC progression.
A20s and GC
The A20 subfamily contains five members: A20, Cezanne (113), Cezanne2 (114), Ubiquitin thioesterase ZRANB1 (TRABID) (115) and ubiquitinating protein VCIP135 (VCPIP) (102). As exhibited in Fig. 4, the expression of VCPIP in GC and normal tissues was notably high, yet low expression levels of other A20s were detected. Additionally, to the best of our knowledge, no data regarding the expression of TRABID has yet been reported. Inhibition of A20 expression or overexpression of miR-200a may prevent the polydiallylation of receptor interacting serine/threonine kinase 1, and promote caspase-8 lysis and tumor necrosis factor-related apoptosis inducing ligand-associated apoptosis (102). A20 is able to induce apoptosis in GC cells, thus may be considered as a potential therapeutic target for GC (116). In the current study, the expression levels of A20s in GC tissues were not high, yet notable levels of VCPIP were detected, suggesting that further study into the prognostic value of A20s in GC is required.
OTULINs and GC
The OTULIN subfamily comprises only two members, OTULIN and FAB105A (102). The role of OTULIN in immune homeostasis and inflammation has been reported to result in certain autoimmune and cancer-associated defects (117). Data analysis in the present study indicated that OTULIN and FAB105A expression in GC tissues was increased compared with that in normal tissues; however, the levels of expression remained low (Fig. 4). Conversely, the expression levels of OTUB1, OTUD5, ALG13 and VCPIP were markedly increased. In particular, OTUB1 expression in GC tissues was 93.09, which is >2-fold higher than the expression level observed in healthy tissues. Moreover, certain studies have revealed that the high expression rate of OTUB1 in GC tissues was associated with poor prognosis (105,118). The association between the prognosis of patients with GC and the expression of other members of the OTU subfamily remains unclear; thus further investigation is required.
5. MJDs and GC
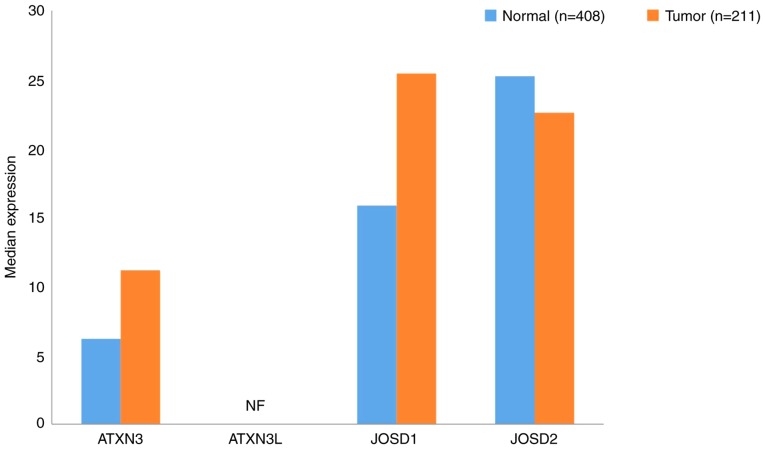
Ataxin (ATXN)3, ATXN3L, Josephin domain containing (JOSD)1 (119) and JOSD2 (120) all belong to the MJD subfamily. In the present study, JOSD1 and 2 were revealed to be expressed in both GC and normal tissues; however, the expression of ATXN3L was not detected (Fig. 5). The expression levels of ATXN3 and JOSD1 in GC tissues were increased compared with normal tissues. Notably, the expression level of JOSD2 was downregulated in GC samples. Furthermore, the expression of ATXN3 in GC was determined to be associated with tumor cell proliferation and infiltration (121). Therefore, the association between MJDs and the prognosis of patients with GC requires further analysis.
Figure 5.
Gene expression profiles of Machado-Joseph disease protein domain proteases between gastric cancer samples and paired normal tissues. Data were extracted using the Gene Expression Profiling Interactive Analysis website. NF, not found. ATXN, ataxin; JOSD, Josephin domain containing.
6. JAMMs and GC
The JAMM subfamily comprises 12 members, including COP9 signalsome subunit (CSN)5, 26S proteasome non-ATPase regulatory subunit 14 (POH1) (122), BRCA1/BRCA2-containing complex subunit 3 (BRCC3) (123), MPN domain containing (MPND) (124), myb-like SWIRM and MPN domains 1 (MYSM1) (125), eukaryotic translation initiation factor 3 subunit (EIF3)H, CSN6 (126), 26S proteasome non-ATPase regulatory subunit 7 (PSMD7) (127), EIF3F, anti-Müllerian hormone (AMSH) (128), AMSH-LP (129) and pre-mRNA-processing-splicing factor 8 (PRPF8) (130). The data presented in Fig. 6 demonstrate that the expression levels of JAMMs in GC tissues were upregulated compared with those in normal tissues, particularly EIF3H and EIF3F, in which the expression levels were >120. The expression of BRCC3 was not detected. These findings suggest that the inhibition of CSN5 may result in a significant increase in p53 levels, indicating that CSN5 may be a crucial regulator of p53 and its associated intracellular signaling pathway, via CSN5-mediated cell activity.
Figure 6.
Gene expression profiled of the Jab1/MPN domain-associated metalloisopeptidases between gastric cancer samples and paired normal tissues. Data were extracted using the Gene Expression Profiling Interactive Analysis website. NF, not found.
Moreover, upregulation of CSN5 has been significantly associated with the progression of GC; therefore, CSN5 may represent a novel target for the treatment of this disease (131). EIF3H was also reported to influence the progression of GC (132), and therefore, may serve as a potential therapeutic target. In particular, the strategy of inhibiting EIF3H expression may suppress the progression of GC and improve patient prognosis (131). Furthermore, EIF3F was determined to serve an important role in the recurrence of GC; increased expression rates of EIF3F in GC were associated with higher 5-year survival rates of patients (133).
7. MCPIPs and GC
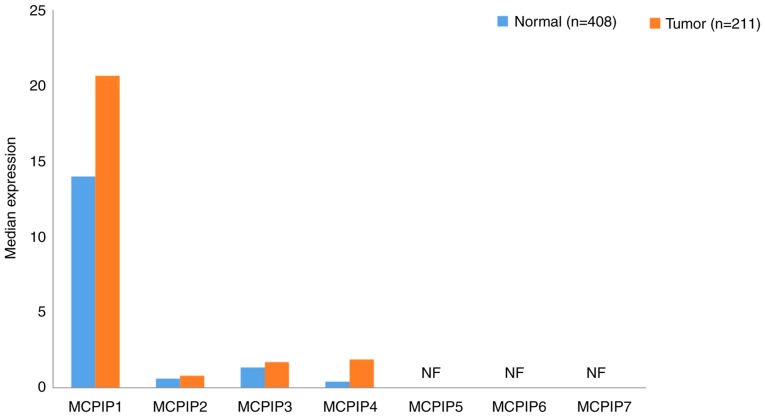
The MCPIP subfamily includes MCPIP1 (134), MCPIP2-4 (135), MCPIP5 (136), MCPIP6 and 7 (137). The expression data of the MCPIP subfamily in GC and normal tissues are presented in Fig. 7. MCPIPs were expressed at markedly low levels in GC and normal tissues; and only MCPIP1 was expressed at levels >10. It has been demonstrated that MCPIP3 serves a negative role in the migration of human colorectal cancer cells (138). In the same study, researchers demonstrated that overexpression of MCPIP3 inhibit cell migration, which confirmed by downregulation of E-cadherin (Marker of EMT). Alternately, mutated MCPIP3 was responsible for enhancing cancer cell migration; However, MCPIP3 expression could not inhibit the cell growth and proliferation (138). Though none of the MCPIP family members were determined to be associated with the 5-year survival rate of patients with GC. In addition, the expression profiles of MCPIP5-7 in GC and normal tissues have not yet been determined. Therefore, the association between MCPIPs and GC should be further evaluated in the future.
Figure 7.
Gene expression profiles of monocyte chemotactic protein-induced proteins between gastric cancer samples and paired normal tissues. Data were extracted using the Gene Expression Profiling Interactive Analysis website. NF, not found. MCPIP, monocyte chemotactic protein-induced proteins.
8. Conclusions and future perspectives
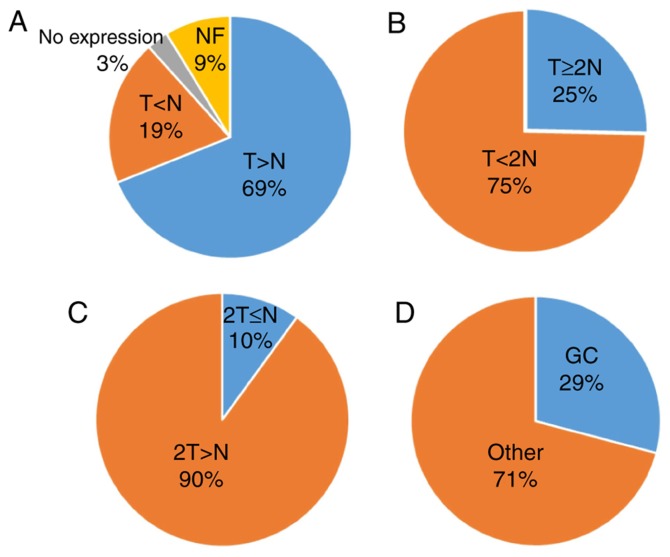
According to global cancer statistics in 2018 (125), 18.1 million new cancer cases and 9.6 million cancer-associated mortalities were reported worldwide, and the incidence of GC was ranked sixth; the incidence of GC was 5.7% (18.1 million) and the mortality rate was 8.2% (9.6 million) of the total cancer cases. Furthermore, the incidence of GC in males was 7.2% (9.5 million), and the mortality rate was 9.5% (5.4 million), compared with an incidence of 4.1% (8.6 million) and mortality rate of 6.5% (4.2 million) in female patients (139). The high prevalence and mortality rates suggest that novel therapeutic strategies are required to treat this disease. The present review focused on the association between GC and DUBs. The present study reported that DUBs are typically upregulated in the majority of GC tissues (79%). A total of 25% of the reported GC cases exhibited a ≥2-fold increase in DUB expression compared with that of normal tissues. Only 19% of healthy tissues exhibited enhanced USP32P2 and USP52 expression, in which this expression was twice the level of that in GC tissues (Fig. 8D). Notably, USP17L2, USP26 and USP29 expression was detected in both GC and normal tissues.
Figure 8.
Expression of DUBs in GC. Data were extracted using the Gene Expression Profiling Interactive Analysis website. (A) T>N (n=71); T<N (n=20); Not expressed (n=3); NF (n=9). No data found for T=N≠0. (B) T≥2N (n=18). (C) 2T≤N (n=2). (D) GC-related DUBs (n=30). AMSH, anti-Müllerian hormone; BAP1, BRCA1 associated protein-1; DUBs, DUB, deubiquitinating enzymes; EIF3, eukaryotic translation initiation factor 3 subunit; FAB105A, family with sequence similarity 105 member A; GC, gastric cancer; JOSD, Josephin domain containing; MCPIP, monocyte chemotactic protein-induced proteins; OUT, ovarian tumor-related protease; PRPF8, pre-mRNA-processing-splicing factor 8; UCH, ubiquitin C-terminal hydrolase; USP, ubiquitin-specific protease.
On the contrary, the number of DUBs associated with GC was determined to be 29%. Following analysis of data from previously published studies (Table I), the expression of DUBs in GC and normal tissues was not determined to be associated with either sex or age; however, an association between DUBs and tumor size, classification and staging was observed. In addition, the expression level of DUBs was significantly associated with the 5-year survival rate of patients with GC. Among the upregulated genes in GC, six DUBs were linked to a high 5-year survival rate, though the difference between the two was not significant. Thus, DUBs may serve a dual role in the prognosis of GC. However, further investigation is required. Providing that DUBs can be divided into two categories according to the prognosis of GC, the common features associated with this disease and DUBs may be identified, in which DUBs may be considered in the development of treatments for GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JS, XS and YG conducted literature searching and wrote this review. JS conducted the data analysis. The language of the review was edited by MAAM.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pidsley R, Lawrence MG, Zotenko E, Niranjan B, Statham A, Song J, Chabanon RM, Qu W, Wang H, Richards M, et al. Enduring epigenetic landmarks define the cancer microenvironment. Genome Res. 2018;28:625–638. doi: 10.1101/gr.229070.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zentner GE, Henikoff S. High-resolution digital profiling of the epigenome. Nat Rev Genet. 2014;15:814–827. doi: 10.1038/nrg3798. [DOI] [PubMed] [Google Scholar]
- 3.Onder O, Sidoli S, Carroll M, Garcia BA. Progress in epigenetic histone modification analysis by mass spectrometry for clinical investigations. Expert Rev Proteomics. 2015;12:499–517. doi: 10.1586/14789450.2015.1084231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A. Ubiquitin: Roles in protein modification and breakdown. Cell. 1983;34:11–12. doi: 10.1016/0092-8674(83)90131-9. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 9.Finley D, Ciechanover A, Varshavsky A. Ubiquitin as a central cellular regulator. Cell. 2004;116(Suppl 2):S29–S32. doi: 10.1016/S0092-8674(03)00971-1. [DOI] [PubMed] [Google Scholar]
- 10.Zhou MJ, Chen FZ, Chen HC. Ubiquitination involved enzymes and cancer. Med Oncol. 2014;31:93. doi: 10.1007/s12032-014-0093-6. [DOI] [PubMed] [Google Scholar]
- 11.Johnston SC, Riddle SM, Cohen RE, Hill CP. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18:3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y, Shen X. Ubiquitin carboxyl-terminal hydrolases: Involvement in cancer progression and clinical implications. Cancer Metastasis Rev. 2017;36:669–682. doi: 10.1007/s10555-017-9702-0. [DOI] [PubMed] [Google Scholar]
- 13.McDonough M, Sangan P, Gonda DK. Characterization of novel yeast RAD6 (UBC2) ubiquitin-conjugating enzyme mutants constructed by charge-to-alanine scanning mutagenesis. J Bacteriol. 1995;177:580–585. doi: 10.1128/jb.177.3.580-585.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu JC, Dawson VL, Dawson TM. Usp16: Key controller of stem cells in Down syndrome. EMBO J. 2013;32:2788–2789. doi: 10.1038/emboj.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avanzato D, Pupo E, Ducano N, Isella C, Bertalot G, Luise C, Pece S, Bruna A, Rueda OM, Caldas C, et al. High USP6NL levels in breast cancer sustain chronic AKT phosphorylation and GLUT1 stability fueling aerobic glycolysis. Cancer Res. 2018;78:3432–3444. doi: 10.1158/0008-5472.CAN-17-3018. [DOI] [PubMed] [Google Scholar]
- 16.Weber A, Elliott PR, Pinto-Fernandez A, Bonham S, Kessler BM, Komander D, El Oualid F, Krappmann D. A linear diubiquitin-based probe for efficient and selective detection of the deubiquitinating enzyme OTULIN. Cell Chem Biol. 2017;24:1299–1313.e7. doi: 10.1016/j.chembiol.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taneera J, Fadista J, Ahlqvist E, Atac D, Ottosson-Laakso E, Wollheim CB, Groop L. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum Mol Genet. 2015;24:1945–1955. doi: 10.1093/hmg/ddu610. [DOI] [PubMed] [Google Scholar]
- 18.Coombs N, Sompallae R, Olbermann P, Gastaldello S, Goppel D, Masucci MG, Josenhans C. Helicobacter pylori affects the cellular deubiquitinase USP7 and ubiquitin-regulated components TRAF6 and the tumour suppressor p53. Int J Med Microbiol. 2011;301:213–224. doi: 10.1016/j.ijmm.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Saldana M, VanderVorst K, Berg AL, Lee H, Carraway KL. Otubain 1: A non-canonical deubiquitinase with an emerging role in cancer. Endocr Relat Cancer. 2019;26:R1–R14. doi: 10.1530/ERC-18-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20:4483–5890. doi: 10.3748/wjg.v20.i16.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 22.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Todi SV, Paulson HL. Balancing act: Deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011;34:370–382. doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang Y, Fu D, Shen XZ. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim Biophys Acta. 2010;1806:1–6. doi: 10.1016/j.bbcan.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Kim HJ, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- 26.Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- 27.Case A, Stein RL. Mechanistic studies of ubiquitin C-terminal hydrolase L1. Biochemistry. 2006;45:2443–2452. doi: 10.1021/bi052135t. [DOI] [PubMed] [Google Scholar]
- 28.Arpalahti L, Laitinen A, Hagström J, Mustonen H, Kokkola A, Böckelman C, Haglund C, Holmberg CI. Positive cytoplasmic UCHL5 tumor expression in gastric cancer is linked to improved prognosis. PLoS One. 2018;13:e0193125. doi: 10.1371/journal.pone.0193125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu YY, Yang M, Zhao M, Luo Q, Yang L, Peng H, Wang J, Huang SK, Zheng ZX, Yuan XH, et al. The de-ubiquitinase UCHL1 promotes gastric cancer metastasis via the Akt and Erk1/2 pathways. Tumour Biol. 2015;36:8379–8387. doi: 10.1007/s13277-015-3566-0. [DOI] [PubMed] [Google Scholar]
- 30.Yan S, He F, Luo R, Wu H, Huang M, Huang C, Li Y, Zhou Z. Decreased expression of BRCA1-associated protein 1 predicts unfavorable survival in gastric adenocarcinoma. Tumour Biol. 2016;37:6125–6133. doi: 10.1007/s13277-015-3983-0. [DOI] [PubMed] [Google Scholar]
- 31.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Das DS, Das A, Ray A, Song Y, Samur MK, Munshi NC, Chauhan D, Anderson KC. Blockade of deubiquitylating enzyme USP1 inhibits DNA repair and triggers apoptosis in multiple myeloma cells. Clin Cancer Res. 2017;23:4280–4289. doi: 10.1158/1078-0432.CCR-16-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapadia B, Nanaji NM, Bhalla K, Bhandary B, Lapidus R, Beheshti A, Evens AM, Gartenhaus RB. Fatty Acid Synthase induced S6Kinase facilitates USP11-eIF4B complex formation for sustained oncogenic translation in DLBCL. Nat Commun. 2018;9:829. doi: 10.1038/s41467-018-03028-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aron R, Pellegrini P, Green EW, Maddison DC, Opoku-Nsiah K, Wong JS, Daub AC, Giorgini F, Finkbeiner S. Publisher correction: Deubiquitinase Usp12 functions noncatalytically to induce autophagy and confer neuroprotection in models of Huntington's disease. Nat Commun. 2018;9:4333. doi: 10.1038/s41467-018-05653-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Zhang M, Jing Y, Yin X, Ma P, Zhang Z, Wang X, Di W, Zhuang G. Deubiquitinase USP13 dictates MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat Commun. 2018;9:215. doi: 10.1038/s41467-017-02693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, Aura C, Barba I, Peg V, Prat A, et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 39.Adorno M, Sikandar S, Mitra SS, Kuo A, Nicolis Di Robilant B, Haro-Acosta V, Ouadah Y, Quarta M, Rodriguez J, Qian D, et al. Usp16 contributes to somatic stem-cell defects in Down's syndrome. Nature. 2013;501:380–384. doi: 10.1038/nature12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw JA, Page K, Blighe K, Hava N, Guttery D, Ward B, Brown J, Ruangpratheep C, Stebbing J, Payne R, et al. Genomic analysis of circulating cell-free DNA infers breast cancer dormancy. Genome Res. 2012;22:220–231. doi: 10.1101/gr.123497.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 42.Combaret L, Adegoke OA, Bedard N, Baracos V, Attaix D, Wing SS. USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. Am J Physiol Endocrinol Metab. 2005;288:E693–E700. doi: 10.1152/ajpendo.00281.2004. [DOI] [PubMed] [Google Scholar]
- 43.Xie L, Wei J, Qian X, Chen G, Yu L, Ding Y, Liu B. CXCR4, a potential predictive marker for docetaxel sensitivity in gastric cancer. Anticancer Res. 2010;30:2209–2216. [PubMed] [Google Scholar]
- 44.Fu Y, Ma G, Liu G, Li B, Li H, Hao X, Liu L. USP14 as a novel prognostic marker promotes cisplatin resistance via Akt/ERK signaling pathways in gastric cancer. Cancer Med. 2018;7:5577–5588. doi: 10.1002/cam4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng Z, Wu HX, Zhan N, Huang YB, Wang ZS, Yang GF, Wang P, Fu GH. Prognostic significance of USP10 as a tumor-associated marker in gastric carcinoma. Tumour Biol. 2014;35:3845–3853. doi: 10.1007/s13277-013-1509-1. [DOI] [PubMed] [Google Scholar]
- 46.Zhu Y, Zhang Y, Sui Z, Zhang Y, Liu M, Tang H. USP14 de-ubiquitinates vimentin and miR-320a modulates USP14 and vimentin to contribute to malignancy in gastric cancer cells. Oncotarget. 2017;8:48725–48736. doi: 10.18632/oncotarget.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renatus M, Parrado SG, D'Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, Kroemer M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berthouze M, Venkataramanan V, Li Y, Shenoy SK. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 2009;28:1684–1796. doi: 10.1038/emboj.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Lubin A, Chen H, Sun Z, Gong F. The deubiquitinating protein USP24 interacts with DDB2 and regulates DDB2 stability. Cell Cycle. 2012;11:4378–4384. doi: 10.4161/cc.22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. Possible role of USP26 in patients with severely impaired spermatogenesis. Eur J Hum Genet. 2005;13:336–340. doi: 10.1038/sj.ejhg.5201335. [DOI] [PubMed] [Google Scholar]
- 53.Weber A, Heinlein M, Dengjel J, Alber C, Singh PK, Häcker G. The deubiquitinase Usp27× stabilizes the BH3-only protein Bim and enhances apoptosis. EMBO Rep. 2016;17:724–738. doi: 10.15252/embr.201541392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 55.Liu J, Chung HJ, Vogt M, Jin Y, Malide D, He L, Dundr M, Levens D. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011;30:846–858. doi: 10.1038/emboj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao LJ, Zhang T, Feng XJ, Chang J, Suo FZ, Ma JL, Liu YJ, Liu Y, Zheng YC, Liu HM. USP28 contributes to the proliferation and metastasis of gastric cancer. J Cell Biochem. 2018 Nov 28; doi: 10.1002/jcb.28040. (Epub ahead of print). doi: 10.1002/jcb.28040. [DOI] [PubMed] [Google Scholar]
- 57.Wang C, Yang C, Ji J, Jiang J, Shi M, Cai Q, Yu Y, Zhu Z, Zhang J. Deubiquitinating enzyme USP20 is a positive regulator of Claspin and suppresses the malignant characteristics of gastric cancer cells. Int J Oncol. 2017 Mar 8; doi: 10.3892/ijo.2017.3904. (Epub ahead of print). doi: 10.3892/ijo.2017.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Y, Fu HL, Wang Z, Huang H, Ni J, Song J, Xia Y, Jin WL, Cui DX. USP22 maintains gastric cancer stem cell stemness and promotes gastric cancer progression by stabilizing BMI1 protein. Oncotarget. 2017;8:33329–33342. doi: 10.18632/oncotarget.16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y, Jin YJ, Zhang YH, Meng HX, Zhao BS, Jiang Y, Zhu JW, Liang GY, Kong D, Jin XM. Ubiquitin-specific peptidase 22 overexpression may promote cancer progression and poor prognosis in human gastric carcinoma. Transl Res. 2015;16:407–416. doi: 10.1016/j.trsl.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Yang DD, Cui BB, Sun LY, Zheng HQ, Huang Q, Tong JX, Zhang QF. The co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinoma. Cell Biochem Biophys. 2011;61:703–710. doi: 10.1007/s12013-011-9229-x. [DOI] [PubMed] [Google Scholar]
- 61.Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 62.Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 63.Tzimas C, Michailidou G, Arsenakis M, Kieff E, Mosialos G, Hatzivassiliou EG. Human ubiquitin specific protease 31 is a deubiquitinating enzyme implicated in activation of nuclear factor-kappaB. Cell Signal. 2006;18:83–92. doi: 10.1016/j.cellsig.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 64.Akhavantabasi S, Akman HB, Sapmaz A, Keller J, Petty EM, Erson AE. USP32 is an active, membrane-bound ubiquitin protease overexpressed in breast cancers. Mamm Genome. 2010;21:388–397. doi: 10.1007/s00335-010-9268-4. [DOI] [PubMed] [Google Scholar]
- 65.Sy SM, Jiang J, O WS, Deng Y, Huen MS. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 2013;41:8572–8580. doi: 10.1093/nar/gkt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Serricchio M, Jauregui M, Shanbhag R, Stoltz T, Di Paolo CT, Kim PK, McQuibban GA. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy. 2015;11:595–606. doi: 10.1080/15548627.2015.1034408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Endo A, Matsumoto M, Inada T, Yamamoto A, Nakayama KI, Kitamura N, Komada M. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J Cell Sci. 2009;122:678–686. doi: 10.1242/jcs.044461. [DOI] [PubMed] [Google Scholar]
- 68.Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, Kirkpatrick DS, Jackson PK, Fang G, Dixit VM. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell. 2011;42:511–523. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 69.Lin M, Zhao Z, Yang Z, Meng Q, Tan P, Xie W, Qin Y, Wang RF, Cui J. USP38 Inhibits type I interferon signaling by editing TBK1 Ubiquitination through NLRP4 Signalosome. Mol Cell. 2016;64:267–281. doi: 10.1016/j.molcel.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 70.van Leuken RJ, Luna-Vargas MP, Sixma TK, Wolthuis RM, Medema RH. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle. 2008;7:2710–2719. doi: 10.4161/cc.7.17.6553. [DOI] [PubMed] [Google Scholar]
- 71.Fang CL, Lin CC, Chen HK, Hseu YC, Hung ST, Sun DP, Uen YH, Lin KY. Ubiquitin-specific protease 3 overexpression promotes gastric carcinogenesis and is predictive of poor patient prognosis. Cancer Sci. 2018;109:3438–3449. doi: 10.1111/cas.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Pang X, Ji L, Sun Y, Ji Y. Reduced expression of deubiquitinase USP33 is associated with tumor progression and poor prognosis of gastric adenocarcinoma. Med Sci Monit. 2018;24:3496–505. doi: 10.12659/MSM.908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Yu Q, Huang L, Yu P. Lentivirus-mediated inhibition of USP39 suppresses the growth of gastric cancer cells via PARP activation. Mol Med Rep. 2016;14:301–306. doi: 10.3892/mmr.2016.5252. [DOI] [PubMed] [Google Scholar]
- 74.Dong X, Su H, Jiang F, Li H, Shi G, Fan L. miR-133a, directly targeted USP39, suppresses cell proliferation and predicts prognosis of gastric cancer. Oncol Lett. 2018;15:8311–3818. doi: 10.3892/ol.2018.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang L, Zhou F, Drabsch Y, Gao R, Snaar-Jagalska BE, Mickanin C, Huang H, Sheppard KA, Porter JA, Lu CX, ten Dijke P. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat Cell Biol. 2012;14:717–726. doi: 10.1038/ncb2522. [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Schrodi S, Rowland C, Tacey K, Catanese J, Grupe A. Genetic evidence for ubiquitin-specific proteases USP24 and USP40 as candidate genes for late-onset Parkinson disease. Hum Mutat. 2006;27:1017–1023. doi: 10.1002/humu.20382. [DOI] [PubMed] [Google Scholar]
- 77.Pinilla-Vera M, Xiong Z, Zhao Y, Zhao J, Donahoe MP, Barge S, Horne WT, Kolls JK, McVerry BJ, Birukova A, et al. Full spectrum of LPS activation in alveolar macrophages of healthy volunteers by whole transcriptomic profiling. PLoS One. 2016;11:e0159329. doi: 10.1371/journal.pone.0159329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J. 2011;30:4921–4930. doi: 10.1038/emboj.2011.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.He L, Liu X, Yang J, Li W, Liu S, Liu X, Yang Z, Ren J, Wang Y, Shan L, et al. Imbalance of the reciprocally inhibitory loop between the ubiquitin-specific protease USP43 and EGFR/PI3K/AKT drives breast carcinogenesis. Cell Res. 2018;28:934–951. doi: 10.1038/s41422-018-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borrero J, Jimenez JJ, Gutiez L, Herranz C, Cintas LM, Hernandez PE. Use of the usp45 lactococcal secretion signal sequence to drive the secretion and functional expression of enterococcal bacteriocins in Lactococcus lactis. Appl Microbiol Biotechnol. 2011;89:131–143. doi: 10.1007/s00253-010-2849-z. [DOI] [PubMed] [Google Scholar]
- 81.Schweitzer K, Naumann M. CSN-associated USP48 confers stability to nuclear NF-kappaB/RelA by trimming K48-linked Ub-chains. Biochim Biophys Acta. 2015;1853:453–469. doi: 10.1016/j.bbamcr.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 82.Weinstock J, Wu J, Cao P, Kingsbury WD, McDermott JL, Kodrasov MP, McKelvey DM, Suresh Kumar KG, Goldenberg SJ, Mattern MR, Nicholson B. Selective dual inhibitors of the cancer-related deubiquitylating proteases USP7 and USP47. ACS Med Chem Lett. 2012;3:789–792. doi: 10.1021/ml200276j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang B, Yin Y, Hu Y, Zhang J, Bian Z, Song M, Hua D, Huang Z. MicroRNA-204-5p inhibits gastric cancer cell proliferation by downregulating USP47 and RAB22A. Med Oncol. 2015;32:331. doi: 10.1007/s12032-014-0331-y. [DOI] [PubMed] [Google Scholar]
- 84.Naghavi L, Schwalbe M, Ghanem A, Naumann M. Deubiquitinylase USP47 promotes RelA phosphorylation and survival in gastric cancer cells. Biomedicines. 2018;6(pii):E62. doi: 10.3390/biomedicines6020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hou K, Zhu Z, Wang Y, Zhang C, Yu S, Zhu Q, Yan B. Overexpression and biological function of ubiquitin-specific protease 42 in gastric cancer. PLoS One. 2016;11:e0152997. doi: 10.1371/journal.pone.0152997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishimura S, Oki E, Ando K, Iimori M, Nakaji Y, Nakashima Y, Saeki H, Oda Y, Maehara Y. High ubiquitin-specific protease 44 expression induces DNA aneuploidy and provides independent prognostic information in gastric cancer. Cancer Med. 2017;6:1453–1464. doi: 10.1002/cam4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J Biol Chem. 2009;284:5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aressy B, Jullien D, Cazales M, Marcellin M, Bugler B, Burlet-Schiltz O, Ducommun B. A screen for deubiquitinating enzymes involved in the G2/M checkpoint identifies USP50 as a regulator of HSP90-dependent Wee1 stability. Cell Cycle. 2010;9:3815–3822. doi: 10.4161/cc.9.18.13133. [DOI] [PubMed] [Google Scholar]
- 89.Wang Z, Zhang H, Liu J, Cheruiyot A, Lee JH, Ordog T, Lou Z, You Z, Zhang Z. USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 2016;30:946–959. doi: 10.1101/gad.271841.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang S, Liu L, Cao C, Song N, Wang Y, Ma S, Zhang Q, Yu N, Ding X, Yang F, et al. USP52 acts as a deubiquitinase and promotes histone chaperone ASF1A stabilization. Nat Commun. 2018;9:1285. doi: 10.1038/s41467-018-03588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kazmierczak M, Harris SL, Kazmierczak P, Shah P, Starovoytov V, Ohlemiller KK, Schwander M. Progressive hearing loss in mice carrying a mutation in Usp53. J Neurosci. 2015;35:15582–15598. doi: 10.1523/JNEUROSCI.1965-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fraile JM, Campos-Iglesias D, Rodriguez F, Espanol Y, Freije JM. The deubiquitinase USP54 is overexpressed in colorectal cancer stem cells and promotes intestinal tumorigenesis. Oncotarget. 2016;7:74427–74434. doi: 10.18632/oncotarget.12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Y, Wang WM, Zou LY, Li L, Feng L, Pan MZ, Lv MY, Cao Y, Wang H, Kung HF, et al. Ubiquitin specific peptidase 5 mediates Histidine-rich protein Hpn induced cell apoptosis in hepatocellular carcinoma through P14-P53 signaling. Proteomics. 2017:17. doi: 10.1002/pmic.201600350. doi: 10.1002/pmic.201600350. [DOI] [PubMed] [Google Scholar]
- 94.Oliveira AM, Perez-Atayde AR, Inwards CY, Medeiros F, Derr V, Hsi BL, Gebhardt MC, Rosenberg AE, Fletcher JA. USP6 and CDH11 oncogenes identify the neoplastic cell in primary aneurysmal bone cysts and are absent in so-called secondary aneurysmal bone cysts. Am J Pathol. 2004;165:1773–1780. doi: 10.1016/S0002-9440(10)63432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang A, Kumar JB, Thomas A, Bourke AG. A spontaneously resolving breast lesion: Imaging and cytological findings of nodular fasciitis of the breast with FISH showing USP6 gene rearrangement. BMJ Case Rep. 2015;2015(pii):bcr2015213076. doi: 10.1136/bcr-2015-213076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jian F, Cao Y, Bian L, Sun Q. USP8: A novel therapeutic target for Cushing's disease. Endocrine. 2015;50:292–296. doi: 10.1007/s12020-015-0682-y. [DOI] [PubMed] [Google Scholar]
- 97.Fu X, Xie W, Song X, Wu K, Xiao L, Liu Y, Zhang L. Aberrant expression of deubiquitylating enzyme USP9X predicts poor prognosis in gastric cancer. Clin Res Hepatol Gastroenterol. 2017;41:687–692. doi: 10.1016/j.clinre.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 98.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, Dong L, Tang E, Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- 99.Xia JT, Chen LZ, Jian WH, Wang KB, Yang YZ, He WL, Chen D, Li W. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-B signaling. J Transl Med. 2014;12:33. doi: 10.1186/1479-5876-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun B, Li L, Ma W, Wang S, Huang C. MiR-130b inhibits proliferation and induces apoptosis of gastric cancer cells via CYLD. Tumour Biol. 2016;37:7981–9787. doi: 10.1007/s13277-016-5221-9. [DOI] [PubMed] [Google Scholar]
- 101.Bermejo JL, Kabisch M, Dunnebier T, Schnaidt S, Melchior F, Fischer HP, Harth V, Rabstein S, Pesch B, Brüning T, et al. Exploring the association between genetic variation in the SUMO isopeptidase gene USPL1 and breast cancer through integration of data from the population-based GENICA study and external genetic databases. Int J Cancer. 2013;133:362–372. doi: 10.1002/ijc.28040. [DOI] [PubMed] [Google Scholar]
- 102.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kato K, Nakajima K, Ui A, Muto-Terao Y, Ogiwara H, Nakada S. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol Cell. 2014;53:617–630. doi: 10.1016/j.molcel.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 105.Wang YQ, Zhang QY, Weng WW, Wu Y, Yang YS, Shen C, Chen XC, Wang L, Liu KJ, Xu MD, Sheng WQ. Upregulation of the Non-coding RNA OTUB1-isoform 2 contributes to gastric cancer cell proliferation and invasion and predicts poor gastric cancer prognosis. Int J Biol Sci. 2016;12:545–557. doi: 10.7150/ijbs.13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carneiro AP, Reis CF, Morari EC, Maia YC, Nascimento R, Bonatto JM, de Souza MA, Goulart LR, Ward LS. A putative OTU domain-containing protein 1 deubiquitinating enzyme is differentially expressed in thyroid cancer and identifies less-aggressive tumours. Br J Cancer. 2014;111:551–558. doi: 10.1038/bjc.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Flierman D, van der Heden van Noort GJ, Ekkebus R, Geurink PP, Mevissen TE, Hospenthal MK, Komander D, Ovaa H. Non-hydrolyzable diubiquitin probes reveal linkage-specific reactivity of deubiquitylating enzymes mediated by S2 pockets. Cell Chem Biol. 2016;23:472–482. doi: 10.1016/j.chembiol.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, Xing G, Kong X, Wang L, Li Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17:1169–1181. doi: 10.1038/ncb3218. [DOI] [PubMed] [Google Scholar]
- 109.Zhao Y, Majid MC, Soll JM, Brickner JR, Dango S, Mosammaparast N. Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J. 2015;34:1687–1703. doi: 10.15252/embj.201490497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo J, Lu Z, Lu X, Chen L, Cao J, Zhang S, Ling Y, Zhou X. OTUD5 regulates p53 stability by deubiquitinating p53. PLoS One. 2013;8:e77682. doi: 10.1371/journal.pone.0077682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim SY, Kwon SK, Lee SY, Baek KH. Ubiquitin-specific peptidase 5 and ovarian tumor deubiquitinase 6A are differentially expressed in p53+/+ and p53−/− HCT116 cells. Int J Oncol. 2018 Mar 5; doi: 10.3892/ijo.2018.4302. (Epub ahead of print). doi: 10.3892/ijo.2018.4302. [DOI] [PubMed] [Google Scholar]
- 112.Santiago-Sim T, Burrage LC, Ebstein F, Tokita MJ, Miller M, Bi W, Braxton AA, Rosenfeld JA, Shahrour M, Lehmann A, et al. Biallelic variants in OTUD6B cause an intellectual disability syndrome associated with seizures and dysmorphic features. Am J Hum Genet. 2017;100:676–688. doi: 10.1016/j.ajhg.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Evans PC, Smith TS, Lai MJ, Williams MG, Burke DF, Heyninck K, Kreike MM, Beyaert R, Blundell TL, Kilshaw PJ. A novel type of deubiquitinating enzyme. J Biol Chem. 2003;278:23180–23186. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- 114.Xu Z, Pei L, Wang L, Zhang F, Hu X, Gui Y. Snail1-dependent transcriptional repression of Cezanne2 in hepatocellular carcinoma. Oncogene. 2014;33:2836–2845. doi: 10.1038/onc.2013.243. [DOI] [PubMed] [Google Scholar]
- 115.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 116.Guo T, Zhang Y, Qu X, Che X, Li C, Fan Y, Wan X, Ma R, Hou K, Zhou H, et al. miR-200a enhances TRAIL-induced apoptosis in gastric cancer cells by targeting A20. Cell Biol Int. 2018;42:506–514. doi: 10.1002/cbin.10924. [DOI] [PubMed] [Google Scholar]
- 117.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-B signaling and cell death: So similar, yet so different. Cell Death Differ. 2017;24:1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weng W, Zhang Q, Xu M, Wu Y, Zhang M, Shen C, Chen X, Wang Y, Sheng W. OTUB1 promotes tumor invasion and predicts a poor prognosis in gastric adenocarcinoma. Am J Transl Res. 2016;8:2234–2244. [PMC free article] [PubMed] [Google Scholar]
- 119.Wang X, Zhang L, Zhang Y, Zhao P, Qian L, Yuan Y, Liu J, Cheng Q, Xu W, Zuo Y, et al. JOSD1 negatively regulates type-I interferon antiviral activity by deubiquitinating and stabilizing SOCS1. Viral Immunol. 2017;30:342–349. doi: 10.1089/vim.2017.0015. [DOI] [PubMed] [Google Scholar]
- 120.Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H, Norberg E. PHGDH defines a metabolic subtype in lung adenocarcinomas with poor prognosis. Cell Rep. 2017;19:2289–2303. doi: 10.1016/j.celrep.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 121.Zhang J, Huang JY, Chen YN, Yuan F, Zhang H, Yan FH, Wang MJ, Wang G, Su M, Lu G, et al. Whole genome and transcriptome sequencing of matched primary and peritoneal metastatic gastric carcinoma. Sci Rep. 2015;5:13750. doi: 10.1038/srep13750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, Weekes D, Festy F, Beesley J, Morris JR. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 2012;31:3918–3934. doi: 10.1038/emboj.2012.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 124.Sun H, Guo D, Su Y, Yu D, Wang Q, Wang T, Zhou Q, Ran X, Zou Z. Hyperplasia of pericytes is one of the main characteristics of microvascular architecture in malignant glioma. PLoS One. 2014;9:e114246. doi: 10.1371/journal.pone.0114246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou L, Shi L, Guo H, Yao X. MYSM-1 suppresses migration and invasion in renal carcinoma through inhibiting epithelial-mesenchymal transition. Tumour Biol. 2015 Sep 27; doi: 10.1007/s13277-015-4138-z. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 126.Xiao D, Yang S, Huang L, He H, Pan H, He J. COP9 signalosome subunit CSN5, but not CSN6, is upregulated in lung adenocarcinoma and predicts poor prognosis. J Thorac Dis. 2018;10:1596–1606. doi: 10.21037/jtd.2018.02.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Niu Z, Lei R, Shi J, Wang D, Shou W, Wang Z, Wang Y, Wang Z, Huang W. A polymorphism rs17336700 in the PSMD7 gene is associated with ankylosing spondylitis in Chinese subjects. Ann Rheum Dis. 2011;70:706–907. doi: 10.1136/ard.2010.130039. [DOI] [PubMed] [Google Scholar]
- 128.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhu W, Liu Y, Ling B. Quantum mechanics and molecular mechanics study of the catalytic mechanism of human AMSH-LP domain deubiquitinating enzymes. Biochemistry. 2015;54:5225–5234. doi: 10.1021/acs.biochem.5b00527. [DOI] [PubMed] [Google Scholar]
- 130.Wickramasinghe VO, Gonzalez-Porta M, Perera D, Bartolozzi AR, Sibley CR, Hallegger M, Ule J, Marioni JC, Venkitaraman AR. Regulation of constitutive and alternative mRNA splicing across the human transcriptome by PRPF8 is determined by 5′ splice site strength. Genome Biol. 2015;16:201. doi: 10.1186/s13059-015-0749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sang MM, Du WQ, Zhang RY, Zheng JN, Pei DS. Suppression of CSN5 promotes the apoptosis of gastric cancer cells through regulating p53-related apoptotic pathways. Bioorg Med Chem Lett. 2015;25:2897–2901. doi: 10.1016/j.bmcl.2015.05.057. [DOI] [PubMed] [Google Scholar]
- 132.Wang X, Wang H, Zhao S, Sun P, Wen D, Liu T, Liu H, Yang Z, Ma Z. Eukaryotic translation initiation factor EIF3H potentiates gastric carcinoma cell proliferation. Tissue Cell. 2018;53:23–29. doi: 10.1016/j.tice.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 133.Cheng Y, Jia C, Li G, Li H. Expression of eukaryotic initiation factor 3f is associated with prognosis in gastric carcinomas. Oncol Res Treat. 2014;37:198–202. doi: 10.1159/000360779. [DOI] [PubMed] [Google Scholar]
- 134.Tahara H, Kay MA, Yasui W, Tahara E. MicroRNAs in cancer: The 22nd hiroshima cancer Seminar/the 4th Japanese Association for RNA interference joint international symposium, 30 August 2012, grand prince hotel Hiroshima. Jpn J Clin Oncol. 2013;43:579–582. doi: 10.1093/jjco/hyt037. [DOI] [PubMed] [Google Scholar]
- 135.Huang S, Liu S, Fu JJ, Tony Wang T, Yao X, Kumar A, Liu G, Fu M. Monocyte chemotactic protein-induced protein 1 and 4 form a complex but act independently in regulation of interleukin-6 mRNA degradation. J Biol Chem. 2015;290:20782–20792. doi: 10.1074/jbc.M114.635870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Roy A, Kolattukudy PE. Monocyte chemotactic protein-induced protein (MCPIP) promotes inflammatory angiogenesis via sequential induction of oxidative stress, endoplasmic reticulum stress and autophagy. Cell Signal. 2012;24:2123–2131. doi: 10.1016/j.cellsig.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 137.Mansour MA. Ubiquitination: Friend and foe in cancer. Int J Biochem Cell Biol. 2018;101:80–93. doi: 10.1016/j.biocel.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 138.Suk FM, Chang CC, Lin RJ, Lin SY, Chen YT, Liang YC. MCPIP3 as a potential metastasis suppressor gene in human colorectal cancer. Int J Mol Sci. 2018;19:E1350. doi: 10.3390/ijms19051350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.