Abstract
The prognostic value of the genotype of the PRSS1 gene in patients with pancreatic ductal adenocarcinoma (PDAC) remains poorly understood. The aim of the present study was to evaluate the association between the PRSS1 genotype and clinicopathological characteristics of patients with PDAC, as well as to explore the prognostic significance of the PRSS1 genotype in patients with PDAC. A total of 124 patients with PDAC patients were included in the current study and the PRSS1 genotype of the enrolled patients was determined by the polymerase chain reaction. Associations between the PRSS1 genotype and clinicopathological characteristics were subsequently analyzed using the Chi-square test. The impact of the PRSS1 genotype on patient prognosis was assessed using the Kaplan-Meier method, and predictive factors of overall survival (OS) time were analyzed by Cox regression. A total of 56 patients with PDAC (45.16%) had the T/C PRSS1 genotype, which was associated with large tumor sizes (P=0.027) and higher tumor node metastasis (TNM) stages (P=0.041). Following a median follow-up of 19 months, the T/C genotype of PRSS1 genotype was associated with a shorter OS time (P=0.037) compared with the C/C or T/T PRSS1 genotypes. Univariate and multivariate analyses revealed that PRSS1 genotype was identified to be an independent prognostic factor for the OS time of patients with PDAC. The results obtained in the current study suggested that the PRSS1 genotype, as well as factors such as the serum level of carbohydrate antigen 19-9 and the TNM stage, may act as independent prognostic factors for the OS time of patients with PDAC.
Keywords: serine protease 1, pancreatic cancer, prognosis
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer and has a 5-year survival rate of 6% was reported in year 2011 worldwide (1). Although advances have been made in the prevention of PDAC as well as in surgical approaches and adjuvant therapy, the mortality rates in patients with PDAC remain high (2,3). Several risk factors such as smoking (4), diabetes (5) and hereditary pancreatitis (6) have been identified. However, tissue biomarkers or gene signatures for the prognosis prediction of PDAC have not been well established (7). The identification of a reliable prognostic genetic determinant in PDAC is an area of ongoing research and may aid in individualized risk assessment and treatment decision-making. Furthermore, biomarkers might present novel therapeutic targets for the treatment of PDAC.
The inflammatory process is a key mediator of pancreatic cancer development and progression, and patients with chronic pancreatitis have an increased risk of developing pancreatic cancer (8–10). Trypsin-1, the main trypsinogen secreted by the pancreas and encoded by the serine protease 1 gene (PRSS1), has attracted attention in the field of PDAC research (11). Previous studies revealed that mutations in PRSS1 are associated with autoimmune (12) and chronic pancreatitis (13). Progressive mutation accumulation and clonal expansion, however, are required for the development of invasive PDAC (11). Mutations in the PRSS1 gene have been reported to be associated with an increased incidence of PDAC (10,14–16). The −409 C/T PRSS1 genotype has been found to protect against the development of pancreatic cancer in the Han Chinese population (17); however, the prognostic value of the aforementioned PRSS1 genotype in patients with PDAC remains poorly understood.
The aim of the current study was to investigate the association of the −409 C/T PRSS1 genotype with the clinicopathological characteristics and prognosis of patients with PDAC.
Materials and methods
Patient recruitment
Patients with PDAC from The First Affiliated Hospital of Fujian Medical University (Fuzhou, China) were enrolled between 2015 and 2017. The inclusion criteria were as follows: i) Patients with pathologically confirmed PDAC diagnosis; ii) patients who had received radical resection; and iii) patients who had not received any preoperative therapy (including radiotherapy, chemotherapy or targeted therapy). Patients with any comorbid or previous malignancies were excluded from the analysis. A total of 124 patients were eligible for inclusion in the current study, including 76 males and 48 females, with a median age of 60 years (range, 28–86 years).
The present study was performed in accordance with the ethical principles described in the Declaration of Helsinki and was approved by the Ethics and Research Committee of The First Affiliated Hospital of Fujian Medical University. Written informed consent was obtained from all patients.
Genotype analysis
Peripheral blood samples were collected from the peripheral vein. Following anticoagulation with EDTA and centrifugation in 1,509 × g at 4°C for 5 min, genomic DNA was extracted from each patient using a TIANamp Blood DNA kit (Tiangen Biotech Co., Ltd.), and was stored at −20°C. Following the manufacturer's instructions, the full-length PRSS1 gene was amplified, and following purification, sequencing was performed and analyzed by Sangon Biotech Co., Ltd. The 50 µl reaction mixture used to generate the fragments contained 200 ng genomic DNA, 10 mmol/l Tris-hydrochloric acid (pH 9.0), 50 mmol/l potassium chloride, 0.1% triton, 2 mmol/l magnesium chloride, 0.25 mmol/l deoxynucleotide mix, 100 ng upstream primer, 100 ng downstream primer and 3.0 units of Taq DNA-polymerase (Sangon Biotech Co., Ltd). The thermocycling conditions included an initial denaturing step at 95°C for 5 min, followed by 30 cycles at 95°C for 30 sec, 55°C for 30 sec and 72°C for 1 min, followed by a final extension step at 72°C for 10 min. PCR products were subsequently run on an 1% agarose gel in 1X TAE buffer and purified by a MinElute Gel Extraction kit (Qiagen GmbH). Following the manufacturer's instructions, a Perkin-Elmer Big Dye Sequencing kit (PerkinElmer, Inc.) and an ABI PRISM7700 sequencer (PerkinElmer, Inc.) were used for sequencing. The PRSS1 primers as follows: Forward, 5′-GGTCCTGGGTCTCATACCTT-3′ and reverse, 5′-GGGTAGGAGGCTTCACACTT-3′.
Statistical analysis
Statistical analysis was performed using SPSS software (version 20.0; IBM Corp.). Data are expressed as means ± standard deviation or number and percentage as appropriate. The association between the PRSS1 genotype and the clinicopathological parameters of the patients was evaluated using the c2 or Fisher's exact tests as appropriate. The overall survival (OS) was defined as the time period between surgery and mortality or the last follow-up. Survival outcomes were evaluated by the Kaplan-Meier method and compared using the log-rank test. A Cox proportional hazard regression model was used for univariate and multivariate analyses of prognosis predictive factors. P<0.05 was considered to indicate a statistically significant difference.
Results
PRSS1 genotype in PDAC
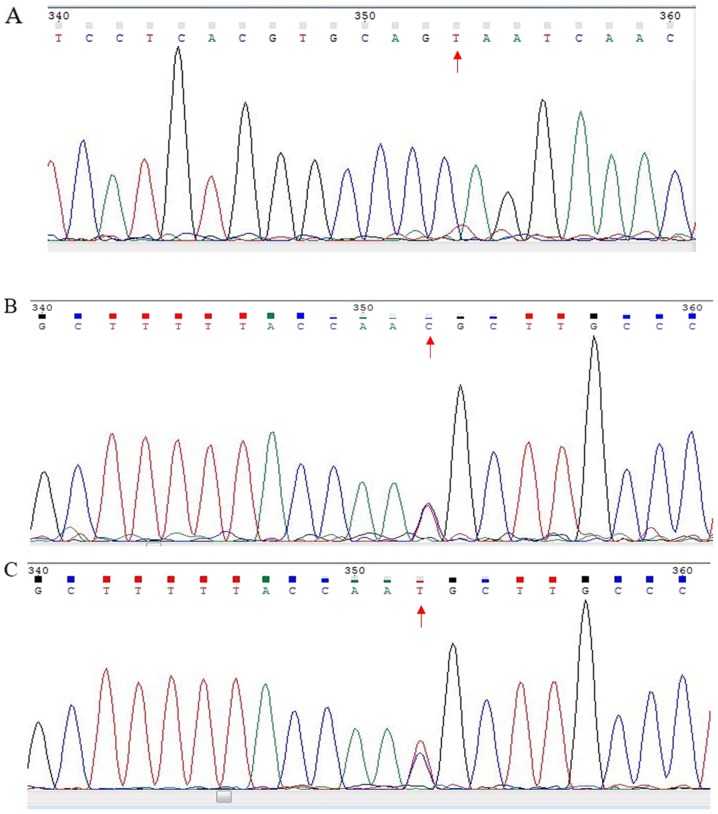
In the current study, PCR was used to determine the PRSS1 genotype in the enrolled patients. The results revealed that multiple loci contained one of three genotypes: C/C, C/T or T/T (Fig. 1), which is similar to results that have been previously reported (18,19). Moreover, 58 patients with PDAC (43.75%) had the T/C genotype (Table I), which was regarded as a mutation.
Figure 1.
Representative DNA sequencing analysis for rs181626272. Gene mutation sites of the serine protease 1 gene genotypes (A) T/T, (B) C/C and (C) C/T. The red arrows refer to the gene mutation sites.
Table I.
Prognostic impact of PRSS1 genotype on the prognosis of patients with pancreatic ductal adenocarcinoma.
| PRSS1 genotype | |||
|---|---|---|---|
| Variable | T/C | T/T and C/C | P-value |
| Age, years (mean ± SD) | 61.5±12.7 | 59.7±10.3 | 0.253 |
| Sex, n (%) | 0.534 | ||
| Male | 36 (64.3) | 40 (58.8) | |
| Female | 20 (35.7) | 28 (41.2) | |
| Tumor size, cm (mean ± SD) | 3.9±2.6 | 3.2±1.5 | 0.027 |
| Tumor location, n (%) | 0.688 | ||
| Head | 41 (73.2) | 53 (77.9) | |
| Body and tail | 15 (26.8) | 15 (22.1) | |
| CEA, ng/ml (mean ± SD) | 7.7±15.1 | 15.9±48.7 | 0.038 |
| CA19-9, U/ml (mean ± SD) | 291.6±375.9 | 302.2±361.5 | 0.717 |
| CA125, U/ml (mean ± SD) | 50.1±89.3 | 45.7±78.7 | 0.460 |
| TNM stage, n (%) | 0.041 | ||
| I | 11 (19.6) | 19 (27.9) | |
| II | 13 (23.2) | 27 (39.7) | |
| III | 27 (48.2) | 20 (29.4) | |
| IV | 5 (8.9) | 2 (2.9) | |
| Perineural invasion, n (%) | 0.874 | ||
| Yes | 19 (33.9) | 24 (35.3) | |
| No | 37 (66.1) | 44 (64.7) | |
| Vascular invasion, n (%) | 0.442 | ||
| Yes | 43 (76.8) | 56 (82.4) | |
| No | 13 (23.2) | 12 (17.6) | |
PRSS1, serine protease 1; TNM, tumor-node-metastasis; CEA, carcinoembryonic antigen; CA, carbohydrate antigen.
Associations between the PRSS1 genotype and clinicopathological characteristics
As shown in Table I, the T/C PRSS1 genotype was associated with larger tumor sizes (P=0.027), higher tumor-node-metastasis (TNM) stages (P=0.041) and higher levels of serum carcinoembryonic antigen (CEA) (P=0.038) compared with the C/C or T/T PRSS1 genotypes. However, no associations between the T/C PRSS1 genotype and age, sex, tumor location, serum carbohydrate antigen (CA) 19-9 and 12-5 levels, perineural invasion or lymphovascular invasion were observed (all P>0.05).
Prognostic impact of the PRSS1 genotype
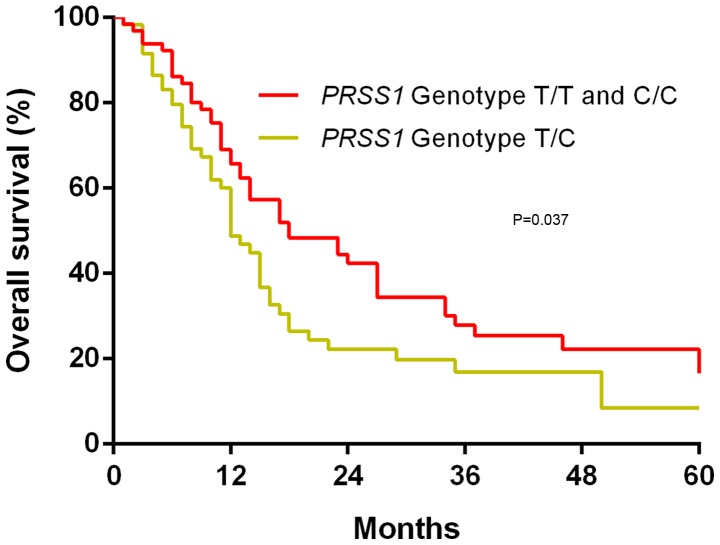
The possible association of the PRSS1 genotype with OS in patients with PDAC was investigated using Kaplan-Meier survival curves and log-rank tests. It was revealed that following a median follow-up period of 19 months, the T/C PRSS1 genotype was associated with a worse OS time compared with the T/T and C/C PRSS1 genotypes (P=0.037; Fig. 2).
Figure 2.
Overall survival time of patients with pancreatic ductal adenocarcinoma grouped by PRSS1 genotype. There was no significant difference between the T/T and C/C PRSS1 genotype (P=0.761). PRSS1, serine protease 1 gene.
Univariate and multivariate analyses of predictive factors of OS
Univariate analysis revealed that surgical type (P=0.032), serum CA19-9 level (P=0.001), serum CEA level (P=0.034), TNM stage III (P=0.005), TNM stage IV (P<0.001), and PRSS1 genotype (P=0.002) were significantly associated with the OS time of patients with PDAC (Table II).
Table II.
Univariate analysis of factors affecting the overall survival time in patients with pancreatic ductal adenocarcinoma.
| Univariate analysis | |||
|---|---|---|---|
| Variable | HR | 95% CI | P-value |
| Sex (male vs. female) | 0.785 | 0.510–1.208 | 0.271 |
| Age | 1.016 | 0.996–1.036 | 0.113 |
| Surgery type (Whipple surgery vs. pancreatectomy) | 1.626 | 1.042–2.537 | 0.032 |
| Resection margin (yes vs. no) | 1.049 | 0.476–2.315 | 0.905 |
| Tumor size | 1.155 | 1.038–1.285 | 0.008 |
| Serum CA19-9 level | 1.001 | 1.000–1.002 | 0.001 |
| Serum CEA level | 1.007 | 1.001–1.014 | 0.034 |
| Serum CA125 level | 1.001 | 0.999–1.004 | 0.194 |
| TNM stage | |||
| I | Reference value | 0.001 | |
| II vs. I | 1.691 | 0.931- 3.072 | 0.085 |
| III vs. I | 2.257 | 1.272–4.005 | 0.005 |
| IV vs. I | 5.320 | 2.186–12.947 | <0.001 |
| Perineural invasion (yes vs. no) | 1.303 | 0.841–2.020 | 0.237 |
| Vascular invasion (yes vs. no) | 1.487 | 0.921–2.398 | 0.104 |
| PRSS1 genotype (T/C vs. C/C or T/T) | 1.961 | 1.293–2.973 | 0.002 |
HR, hazard ratio; CEA, carcinoembryonic antigen; CA, carbohydrate antigen, PRSS1, serine protease 1 gene.
A multivariate analysis using the Cox regression model was subsequently performed, and the results revealed that the serum CA19-9 level [hazard ratio (HR)=1.001; P=0.001], TNM stage III (HR=2.078; P=0.031), TNM stage IV (HR=3.090; P=0.041), and PRSS1 genotype (HR=2.196; P=0.002) were independent predictors of the OS time in patients with PDAC (Table III).
Table III.
Multivariate analysis of factors affecting the overall survival time in patients with pancreatic ductal adenocarcinoma.
| Variable | Regression coefficient | SE | HR | 95% CI | P-value |
|---|---|---|---|---|---|
| Surgery type (Whipple surgery vs. pancreatectomy) | −0.009 | 0.274 | 0.991 | 0.579–1.695 | 0.973 |
| Tumor size | 0.081 | 0.067 | 1.085 | 0.952–1.236 | 0.223 |
| Serum CA19-9 level | 0.001 | <0.001 | 1.001 | 1.000–1.002 | 0.001 |
| Serum CEA level | 0.006 | 0.005 | 1.006 | 0.996–1.016 | 0.230 |
| TNM stage | |||||
| I | Reference value | 0.110 | |||
| II vs. I | 0.613 | 0.345 | 1.847 | 0.938–3.634 | 0.076 |
| III vs. I | 0.731 | 0.339 | 2.078 | 1.068–4.041 | 0.031 |
| IV vs. I | 1.128 | 0.551 | 3.090 | 1.049–9.104 | 0.041 |
| PRSS1 genotype (T/C vs. C/C or T/T) | 0.786 | 0.249 | 2.196 | 1.347–3.580 | 0.002 |
SE, standard error; HR, hazard ratio; CEA, carcinoembryonic antigen; CA, carbohydrate antigen, PRSS1, serine protease 1 gene.
Discussion
The current study revealed that the T/C PRSS1 genotype was statistically associated with larger tumor size and higher TNM stage compared with other genotypes. To the best of the authors' knowledge, the present study was the first to demonstrate that the T/C PRSS1 genotype was associated with poor OS, and that the aforementioned genotype may serve as an independent prognostic factor of OS in patients with PDAC, together with factors including serum CA19-9 level and TNM stage.
Previous studies revealed that the processes of genotype accumulation and clonal expansion are required for the development of invasive PDAC (11,16,20). Many efforts have been made to identify gene mutations associated with pancreatic cancer, including mutL homolog 1 and PMS1 homolog 1, mismatch repair system component (21), and BRCA1/2 DNA repair (22). Increasing numbers of studies have demonstrated that mutations in the PRSS1 gene are associated with an increased incidence of PDAC (10,14–16). Furthermore, the −409 C/T PRSS1 genotype was found to exert a protective effective against pancreatic cancer in the Han Chinese population (17). PRSS1 encodes trypsin-1, the main trypsinogen enzyme secreted by the pancreas. PRSS1 mutations have been associated with increased trypsin-1 serum levels as well as the trypsin and α1-antitrypsin imbalance observed in patients with pancreatic cancer, which may alter the pancreatic microenvironment and increase immunological escape and clonal proliferation of pancreatic cancer cells (19). However, the clinicopathological and prognostic significance of PRSS1 mutations in patients with PDAC remains unclear.
PRSS1 mutations have been previously shown to be associated with autoimmune (12) and chronic pancreatitis (13). Additionally, the mean serum trypsin level was increased in patients with pancreatic cancer compared with healthy controls (19). While the PRSS1 gene is not currently considered to be an oncogene, tumor-suppressor gene or genome-maintenance gene, PRSS1 genotypes were strongly correlated with the trypsin and α1-antitrypsin imbalance in patients with pancreatic cancer (13).
Traditional prognostic factors, including serum CA19-9 level and TNM stage, were identified as independent prognostic factors of OS in patients with PDAC in the current study, similarly to previously published studies (7,23). In the current study, the T/C PRSS1 genotype indicated a poor prognosis and a shorter OS time in patients with PDAC, as demonstrated by the Kaplan-Meier method. Furthermore, associations between the PRSS1 genotype and clinicopathological parameters such as large tumor size and higher TNM stage were identified in the current study, suggesting that the PRSS1 genotype may be involved in the progression and aggressiveness of pancreatic cancer. Moreover, by using univariate and Cox regression analysis, the results obtained in the current study demonstrated that the PRSS1 genotype was an independent prognostic factor of OS in patients with PDAC. The PRSS1 genotype may aid the selection of treatment plans and the identification of patients who may benefit from additional adjuvant therapy.
The current study had a number of limitations. Firstly, the present study was a single-centre retrospective study. Secondly, a limited sample size was included in the current study. Thirdly, patients with unresectable PDAC were not included in the current study due to insufficient data. Finally, all the patients enrolled in the current study were of Chinese descent, therefore the association between the mutation status of PRSS1 and prognosis of patients of different ethnicities was not investigated. Nevertheless, the current study contributed to the understanding of the association between the PRSS1 genotype and prognosis of patients with PDAC.
In conclusion, the current study demonstrated that the T/C PRSS1 genotype was associated with large tumor size, higher TNM stage and poor OS compared with other genotypes. The PRSS1 genotype, together with other factors including serum CA19-9 level and TNM stage, may therefore be used as an independent prognostic factor of OS in patients with PADC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HW, ZZ and YC conceived and designed the study. FD and XC acquired data. HW, SC, ZS and QL analysed the data. HW, ZZ and YC interpretated, drafted and revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics and Research Committee of The First Affiliated Hospital of Fujian Medical University (approval no. 2018-053). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ryan D, Hong T, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140–2141. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 2.Uesaka K, Boku N, Fukutomi A, Okamura Y, Konishi M, Matsumoto I, Kaneoka Y, Shimizu Y, Nakamori S, Sakamoto H, et al. Adjuvant chemotherapy of S-1 versus gemcitabine for resected pancreatic cancer: A phase 3, open-label, randomised, non-inferiority trial (JASPAC 01) Lancet. 2016;388:248–257. doi: 10.1016/S0140-6736(16)30583-9. [DOI] [PubMed] [Google Scholar]
- 3.Picozzi VJ, Oh SY, Edwards A, Mandelson MT, Dorer R, Rocha FG, Alseidi A, Biehl T, Traverso LW, Helton WS, et al. Five-year actual overall survival in resected pancreatic cancer: A contemporary single-institution experience from a multidisciplinary perspective. Ann Surg Oncol. 2017;24:1722–1730. doi: 10.1245/s10434-016-5716-z. [DOI] [PubMed] [Google Scholar]
- 4.Rulyak S, Lowenfels A, Maisonneuve P, Brentnall T. Risk factors for the development of pancreatic cancer in familial pancreatic cancer kindreds. Gastroenterology. 2003;124:1292–1299. doi: 10.1016/S0016-5085(03)00272-5. [DOI] [PubMed] [Google Scholar]
- 5.Huxley R, Ansary-Moghaddam A, Berrington De González A, Barzi F, Woodward M. Type-II diabetes and pancreatic cancer: A meta-analysis of 36 studies. Br J Cancer. 2005;92:2076–2083. doi: 10.1038/sj.bjc.6602619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu QC, Zhuang ZH, Zeng K, Cheng ZJ, Gao F, Wang ZQ. Prevalence of pancreatic diabetes in patients carrying mutations or polymorphisms of the PRSS1 gene in the Han population. Diabetes Technol Ther. 2009;11:799–804. doi: 10.1089/dia.2009.0051. [DOI] [PubMed] [Google Scholar]
- 7.Winter JM, Yeo CJ, Brody JR. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J Surg Oncol. 2013;107:15–22. doi: 10.1002/jso.23192. [DOI] [PubMed] [Google Scholar]
- 8.Hausmann S, Kong B, Michalski C, Erkan M, Friess H. The role of inflammation in pancreatic cancer. Adv Exp Med Biol. 2014;816:129–151. doi: 10.1007/978-3-0348-0837-8_6. [DOI] [PubMed] [Google Scholar]
- 9.Pinho A, Chantrill L, Rooman I. Chronic pancreatitis: A path to pancreatic cancer. Cancer Lett. 2014;345:203–209. doi: 10.1016/j.canlet.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Weiss F. Pancreatic cancer risk in hereditary pancreatitis. Front Physiol. 2014;5:70. doi: 10.3389/fphys.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitcomb DC. Genetic risk factors for pancreatic disorders. Gastroenterology. 2013;144:1292–1302. doi: 10.1053/j.gastro.2013.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao F, Li YM, Hong GL, Xu ZF, Liu QC, He QL, Lin LQ, Weng SH. PRSS1_p.Leu81Met mutation results in autoimmune pancreatitis. World J Gastroenterol. 2013;19:3332–3338. doi: 10.3748/wjg.v19.i21.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao F, Liu QC, Zhang S, Zhuang ZH, Lin CZ, Lin XH. PRSS1 intron mutations in patients with pancreatic cancer and chronic pancreatitis. Mol Med Rep. 2012;5:449–451. doi: 10.3892/mmr.2011.684. [DOI] [PubMed] [Google Scholar]
- 14.Hengstler JG, Bauer A, Wolf HK, Bulitta CJ, Tanner B, Oesch F, Gebhard S, Boettger T. Mutation analysis of the cationic trypsinogen gene in patients with pancreatic cancer. Anticancer Res. 2000;20:2967–2974. [PubMed] [Google Scholar]
- 15.Rebours V, Boutron-Ruault M, Schnee M, Férec C, Maire F, Hammel P, Ruszniewski P, Lévy P. Risk of pancreatic adenocarcinoma in patients with hereditary pancreatitis: A national exhaustive series. Am J Gastroenterol. 2008;103:111–119. doi: 10.1111/j.1572-0241.2007.01597.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Férec C. Chronic pancreatitis: Genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2009;10:63–87. doi: 10.1146/annurev-genom-082908-150009. [DOI] [PubMed] [Google Scholar]
- 17.Liu Q, Lin X, Liu J, Liu A, Gao F. The −409 C/T genotype of PRSS1 protects against pancreatic cancer in the Han Chinese population. Dig Dis Sci. 2012;57:573–579. doi: 10.1007/s10620-011-1893-6. [DOI] [PubMed] [Google Scholar]
- 18.Zeng K, Liu QC, Lin JH, Lin XH, Zhuang ZH, Gao F, Ou QS. Novel mutations of PRSS1 gene in patients with pancreatic cancer among Han population. Chin Med J (Engl) 2011;124:2065–2067. [PubMed] [Google Scholar]
- 19.Yi Q, Dong F, Lin L, Liu Q, Chen S, Gao F, He Q. PRSS1 mutations and the proteinase/antiproteinase imbalance in the pathogenesis of pancreatic cancer. Tumour Biol. 2016;37:5805–5810. doi: 10.1007/s13277-015-3982-1. [DOI] [PubMed] [Google Scholar]
- 20.Campa D, Pastore M, Capurso G, Hackert T, Di Leo M, Izbicki JR, Khaw KT, Gioffreda D, Kupcinskas J, Pasquali C, et al. Do pancreatic cancer and chronic pancreatitis share the same genetic risk factors? A PANcreatic Disease ReseArch (PANDoRA) consortium investigation. Int J Cancer. 2018;142:290–296. doi: 10.1002/ijc.31047. [DOI] [PubMed] [Google Scholar]
- 21.Gargiulo S, Torrini M, Ollila S, Nasti S, Pastorino L, Cusano R, Bonelli L, Battistuzzi L, Mastracci L, Bruno W, et al. Germline MLH1 and MSH2 mutations in Italian pancreatic cancer patients with suspected Lynch syndrome. Fam Cancer. 2009;8:547–553. doi: 10.1007/s10689-009-9285-1. [DOI] [PubMed] [Google Scholar]
- 22.Fogelman D, Wolff R, Kopetz S, Javle M, Bradley C, Mok I, Cabanillas F, Abbruzzese J. Evidence for the efficacy of Iniparib, a PARP-1 inhibitor, in BRCA2-associated pancreatic cancer. Anticancer Res. 2011;31:1417–1420. [PubMed] [Google Scholar]
- 23.Berger AC, Garcia M, Jr, Hoffman JP, Regine WF, Abrams RA, Safran H, Konski A, Benson AB, III, MacDonald J, Willett CG. Postresection CA 19-9 predicts overall survival in patients with pancreatic cancer treated with adjuvant chemoradiation: A prospective validation by RTOG 9704. J Clin Oncol. 2008;26:5918–5922. doi: 10.1200/JCO.2008.18.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.