Abstract
One of the main causes of cancer incidence and mortality worldwide is lung cancer. This study focused on the function of basic leucine zipper ATF-like transcription factor (BATF) in non-small cell lung cancer (NSCLC). Using NSCLC patient data from The Cancer Genome Atlas, the present study demonstrated that BATF expression in NSCLC tissues was significantly higher compared with that in adjacent non-tumor tissues (P=6.56×10−6). Lentivirus-mediated short hairpin RNA (shRNA) was used to knock down BATF expression in the human A549 NSCLC cell line and assessed by reverse transcription-quantitative PCR and western blotting. Cell proliferation was evaluated by MTT assay and Celigo imaging cytometry. Apoptosis was detected by fluorescence-activated cell sorting and caspase 3/7 activity analysis. The results revealed that knockdown of BATF inhibited the proliferation of A549 cells. Compared with that of the control group, the apoptosis rate of the BATF-shRNA group was significantly higher. In summary, knockdown of BATF inhibited the proliferation of A549 cells and promoted apoptosis. These results provide important information about the underlying mechanism of the pathogenesis of NSCLC.
Keywords: basic leucine zipper ATF-like transcription factor, non-small cell lung cancer, short hairpin RNA, proliferation, apoptosis
Introduction
One of the leading causes of cancer death worldwide is lung cancer (1). There are 1.8 million new cases of lung cancer each year, and 1.6 million people succumb to lung cancer (2). Nearly 70% of patients with lung cancer are in an advanced stage with localized spread and/or distant metastasis (3).
Among the types of diagnosed lung cancers, non-small cell lung cancer (NSCLC) accounts for ~80% (4). Despite advances in treatment, the 5-year survival rate of patients with lung cancer is still <15% (5). The presence of local recurrence of tumors and distant metastasis may be the most common cause of death (6). The carcinogenesis and progression of NSCLC involve both environmental and genetic factors and are multistep processes (7–9). Although chemotherapy, radiotherapy, surgery, molecular-targeted therapy and immunotherapy have greatly improved the survival and prognosis of patients with NSCLC to date, the underlying molecular mechanisms remain unclear and require further study (10). Therefore, it is important to study the genes involved in the metastasis, tumor progression and invasion, as well as to determine the molecular mechanisms underlying the effects of these genes in the treatment and prediction of NSCLC prognosis.
The basic leucine zipper ATF-like transcription factor (BATF) family comprises three members (BATF, BATF2 and BATF3) and belongs to the group of activator protein 1 (AP-1) transcription factors (11). BATF is located on human chromosome 14q24.3; there is only one transcript (NM_006399) of BATF (12). The protein encoded by this gene is a nuclear alkaline leucine zipper protein, which belongs to the AP-1/activating transcription factor (ATF) superfamily. The BATF protein is a negative regulator of AP-1/ATF transcription events, and AP-1 complexes activate or inhibit their target genes (13). Regulation of the AP-1 gene effects cell survival, differentiation and proliferation (14,15). An increasing number of studies suggest that BATF may affect cancer development (16–18). However, the functional mechanisms of BATF in NSCLC are poorly understood.
The present study used The Cancer Genome Atlas (TCGA) data to analyze BATF expression in NSCLC. Through a series of experiments, the role of BATF in NSCLC was explored. The results could provide important information to explain the pathogenesis of NSCLC and reveal that BATF may be a potential novel target for the treatment of non-small cell lung cancer.
Materials and methods
BATF expression analysis in TCGA database
TCGA is a publicly funded project and contains RNA expression data for various types of cancer, including NSCLC (http://cancergenome.nih.gov/). The mRNA expression profiling data of NSCLC and matched adjacent mucosa was downloaded from TCGA (datasets TCGA-38, TCGA-49 and TCGA-50) (19).
Cell lines and culture conditions
The human NSCLC cell line A549 was purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and cultured in RPMI-1640 medium (Thermo Fisher Scientific, Inc) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin (Sigma-Aldrich; Merck KGaA) and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA), at 37°C in a humidified incubator containing 5% CO2.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tissue samples and cultured cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. cDNA was synthesized from total RNA (500 ng) in a 10 µl reaction volume using a PrimeScript RT-PCR kit (Takara Bio, Inc.) using the following conditions: 5 min at 25°C, 25 min at 52°C and 10 min at 80°C. Then, qPCR was performed using the power SYBR Green master mix (Thermo Fisher Scientific, Inc.). GAPDH was used as an internal control. The primers used were: BATF forward, 5′-AGAAGAGTTCAGAGGAGGGA-3′ and reverse, 5′-CGTTCTGTTTCTCCAGGTCT-3′; GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. qPCR was performed using SYBR Premix Ex Taq II (Takara Biotechnology Co., Ltd.), and the thermocycling conditions used were: 95°C for 10 min, followed by 39 cycles of 95°C for 1 min, 55°C for 30 sec, and 72°C for 45 sec. The 2−ΔΔCq method was used to determine relative (BATF/GAPDH) mRNA expression (20).
Lentiviral shRNA vector construction and infection
For knockdown of BATF, a lentiviral shRNA vector targeting the human BATF gene was constructed by Shanghai GeneChem Co., Ltd. The targeting sequence of BATF was synthesized and cloned into the lentiviral vector pGVX115-GFP (Shanghai GeneChem Co., Ltd.) to produce pGV115-shBATF at 37°C for 2 h. For control cells, transfection was performed using an empty vector carrying GFP. To establish stable cell lines, 2 µg/ml puromycin was added to the medium for 1 week following transfection with the lentiviral vectors. Knockdown efficiency was verified using RT-qPCR and western blot analysis.
Western blot analysis
Cell lines were harvested 3 days post transfection and washed twice with cold PBS, and then cell lysates were prepared by adding radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) containing 1 mM phenylmethanesulfonyl. The lysate was stored at −20°C until further experimentation. Proteins (~30 µg each lane) were separated with 12% SDS-PAGE, and transferred onto PVDF membranes (EMD Millipore; Merck KGaA). Membranes were incubated with primary antibodies to BATF (1:500; cat. no. ab236876; Abcam), and β-actin (1:500; cat. no. sc-47778; Santa Cruz Biotechnology) overnight at 4°C. Next, the membrane was washed and incubated with the corresponding secondary antibodies [horseradish peroxidase-conjugated (HRP) goat anti-rabbit; 1:5,000; cat. no. P044801-2; Dako; Agilent Technologies or HRP goat anti-mouse IgG H&L; 1:1,000; cat. no. ab6708; Abcam or goat anti-rabbit IgG; 1:1,500; cat. no. bs-0295G; BIOSS] at room temperature for 1 h. Protein bands were quantified using a bio-imaging system (DNR Bio-Imaging Systems, Ltd.).
Cell proliferation and viability assays
Cell proliferation and viability were analyzed by the Celigo imaging cytometry system and MTT assay, respectively. The fluorescence intensity of entire wells, of transfected A549 cells was detected, and the number of cells was automatically calculated by the Celigo imaging cytometry system (Nexcelom Bioscience LLC). Cell viability was measured using a Cell Viability kit (MTT; Roche Diagnostics) solubilized in 150 µl dimethyl sulfoxide according to the manufacturer's instructions. The absorption of the solution was measured at 570 nm at various time points.
Apoptosis assay
A total of 1×105 cells/well were incubated in 6-well plates for 24 h and treated with shBATF and shCtrl for 48 h, both at 37°C. The cells were washed, and the Annexin V-APC Apoptosis Detection kit (eBioscience, Inc.) was used according to the manufacturer's instructions. Cells were analyzed by flow cytometry (FACScalibur; BD Biosciences), and the percentage of apoptotic cells was determined using ModFit LT v5.0 software (Verity Software House, Inc.). Each experiment was performed three times.
Caspase 3/7 activity was measured using a homogeneous luminescence-based assay with a Caspase 3/7 Glo Assay kit (Promega Corporation) according to the manufacturer's instructions. Following transfection with shCtrl and shBATF, A549 cells were treated with 0, 0.01, 0.1 and 1.0 µM cetuximab for 24 h. For luminosity, the cells were analyzed by an Infinite F500 multifunction microplate reader (Tecan Trading AG).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software, Inc.). Statistical differences were analyzed and calculated by unpaired two-tailed Student's t-test or one-way analysis of variance (ANOVA) followed by Bonferroni's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
The expression levels of BATF in TCGA dataset
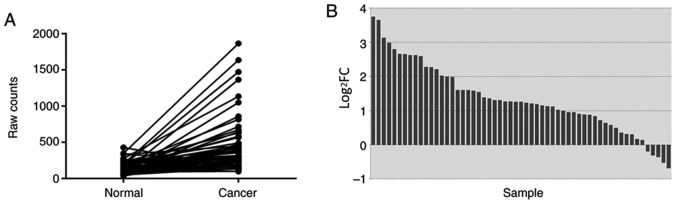
In TCGA database, a total of 57 pairs of mRNA expression profiles of NSCLC tissues and adjacent non-tumor tissues were screened. The differences in the expression levels of NSCLC and adjacent non-tumor tissues were analyzed and described using a paired samples linear graph (Fig. 1A) and a bar graph (Fig. 1B). Compared with that in adjacent non-tumor tissues, BATF expression was significantly upregulated in NSCLC (P=6.56×10−6).
Figure 1.
Expression levels of BATF in lung cancer compared with adjacent non-tumor tissues in TCGA data. (A) Expression levels of BATF in lung cancer compared with adjacent non-tumor tissues. (B) Log2FC of the expression level of BATF in lung cancer compared with adjacent non-tumor tissues. Normal, adjacent non-tumor tissues; TCGA, The Cancer Genome Atlas; BATF, basic leucine zipper ATF-like transcription factor; FC, fold-change.
Knockdown efficiency of BATF by shRNA lentivirus infection in A549 cells
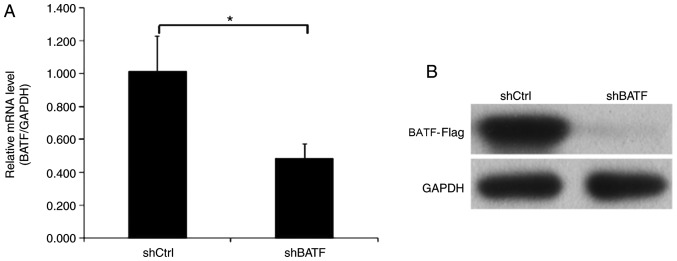
Endogenous BATF expression in human NSCLC A549 cells was detected by RT-qPCR. In the A549 cell line, BATF was moderately expressed, which indicated that A549 cells were suitable for BATF knockdown analysis. To investigate the role of BATF in A549 cells, lentiviruses carrying shBATF and shCtrl were used to infect A549 cells; the transfection efficiency was evaluated using RT-qPCR, and the results revealed that BATF mRNA levels were reduced by 50% in the shBATF group compared with the shCtrl group (Fig. 2A). Western blot analysis demonstrated similar results (Fig. 2B).
Figure 2.
Expression of BATF in A549 cell lines detected by RT-qPCR and western blot analysis following shBATF/shCtrl infection. (A) The mRNA expression level of BATF in the A549 cell line was detected by RT-qPCR analysis. (B) The protein expression level of BATF in the A549 cell line was detected by western blot analysis. *P<0.05. BATF, basic leucine zipper ATF-like transcription factor; shBATF, short hairpin RNA targeting BATF; shCtrl, control short hairpin RNA; RT-qPCR, reverse transcription-quantitative PCR.
Knockdown of BATF inhibits the proliferation of A549 cells
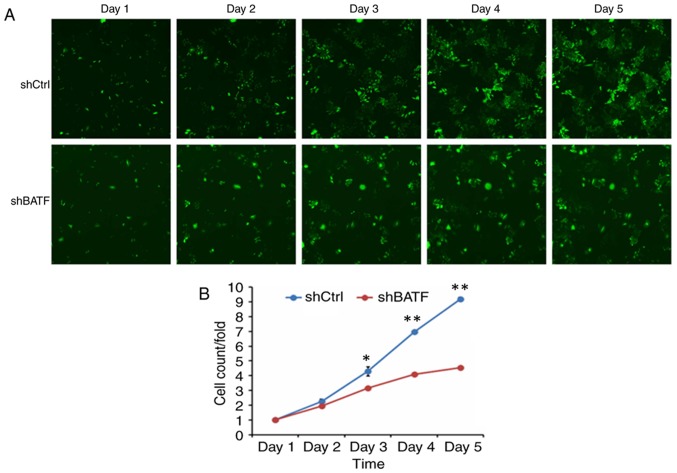
To generate the cell growth curve, a cell imaging system and an MTT assay were used. As shown in Fig. 3, the proliferation in the shBATF group was significantly inhibited compared with that in the shCtrl group according to the Celigo imaging cytometry system (P<0.01).
Figure 3.
A549 cell proliferation following shBATF/shCtrl infection detected by the Celigo imaging cytometry system. (A) A549 cell proliferation following shBATF/shCtrl infection between days 1 and 5. (B) Proliferation curves of A549 cells based on the Celigo imaging cytometry. *P<0.05; **P<0.01. BATF, basic leucine zipper ATF-like transcription factor; shBATF, short hairpin RNA targeting BATF; shCtrl, control short hairpin RNA; BATF, basic leucine zipper ATF-like transcription factor; shBATF, short hairpin RNA targeting BATF; shCtrl, control short hairpin RNA.
Cell viability was determined by MTT assay. The results demonstrated that in the A549 cell line, the cell viability of the shBATF group was significantly lower (P<0.001) between days 3 and 5 compared with that of the shCtrl group (Fig. 4).
Figure 4.
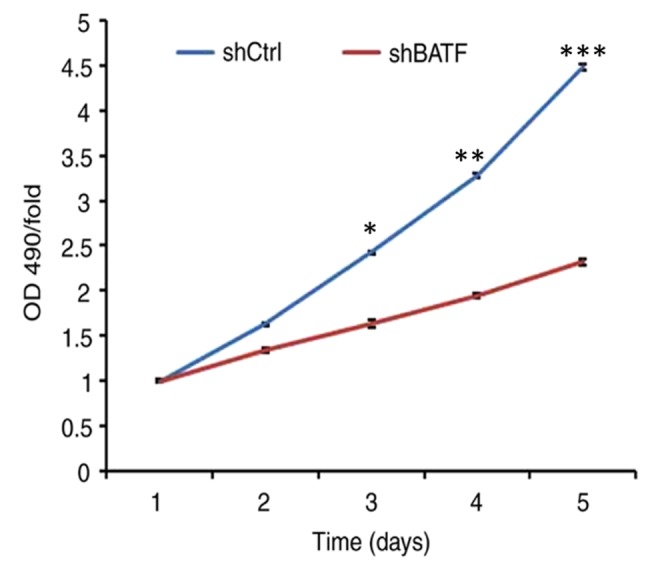
A549 cell viability following shBATF and shCtrl infection examined by MTT assay between days 1 and 5. *P<0.05; **P<0.01; ***P<0.001. BATF, basic leucine zipper ATF-like transcription factor; shBATF, short hairpin RNA targeting BATF; shCtrl, control short hairpin RNA; OD, optical density.
Knockdown of BATF induces apoptosis in A549 cells
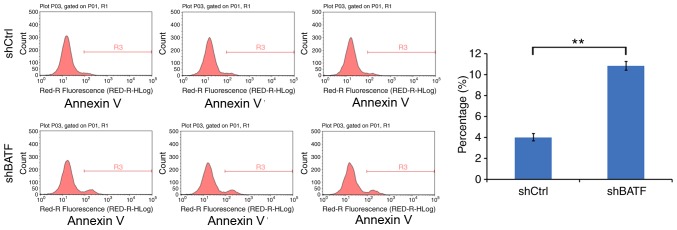
The effect of BATF knockdown on apoptosis was detected by annexin V staining. Compared with the control group, the apoptotic rate of the shBATF group was significantly increased in the A549 cell line (P<0.01; Fig. 5), suggesting that knockdown of BATF promoted apoptosis in A549 cells.
Figure 5.
Apoptosis of A549 cells following shBATF and shCtrl infection was examined by flow cytometry. (A) Apoptosis analysis in A549 cells with shBATF or shCtrl infection. (B) The percentage of apoptotic A549 cells. **P<0.01. BATF, basic leucine zipper ATF-like transcription factor; shBATF, short hairpin RNA targeting BATF; shCtrl, control short hairpin RNA.
Knockdown of BATF increases caspase 3/7 activity in A549 cells
As shown in Fig. 6, in the A549 cell line, caspase 3/7 activity was significantly increased following shBATF infection compared with shCtrl infection (P<0.01).
Figure 6.
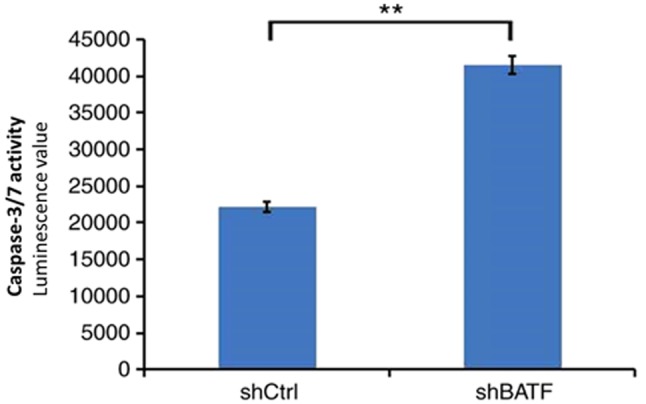
Caspase 3/7 activity of the A549 cell line was determined following shBATF/shCtrl infection in the shBATF and shCtrl groups. **P<0.01. BATF, basic leucine zipper ATF-like transcription factor; shBATF, short hairpin RNA targeting BATF; shCtrl, control short hairpin RNA.
Discussion
The main aims of this study were to investigate the biological roles of BATF in human NSCLC cells and to elucidate the role of BATF in the development of lung cancer. Lung cancer is often a multifactorial process involving processes such as ECM degradation, transmission, basement membrane, cell adhesion, vascular production and movement (21–25). Therefore, identification of novel biomarkers is crucial for the improvement of clinical outcomes for NSCLC patients.
Dorsey et al (19) first described BATF as a modulator of the AP-1 transcription complex in specific human tissues following identification in a cDNA library from Epstein-Barr virus-infected human B cells. Analysis of polyadenylated mRNA from different human tissues and established cell lines revealed a strong hybridization in Raji Burkitt's lymphoma cells and in the healthy lung tissue (26). BATF knockout mice have a defect in TH17 development based on its direct regulation of both RORγt and of RORγt target genes such as IL-17A. (27). BATF serves a crucial role in Th17 differentiation and exhibits distinct DNA binding specificity and protein-protein interactions with Th17-specific factors (28). Th17 cells serve a key role in the development of NSCLC (29).
In the present study, screening of TCGA database revealed that BATF was expressed at significantly higher levels in NSCLC tissues compared with adjacent non-tumor tissues. In addition, BATF was moderately expressed in the A549 NSCLC cell line. Therefore, lentivirus-mediated BATF shRNA was constructed and transfected into A549 cells; the results revealed that cell proliferation was inhibited in BATF-silenced A549 cells. Compared with that of the control group, the apoptotic rate of the BATF-shRNA group was significantly higher. These results suggested that BATF may regulate NSCLC cell proliferation and apoptosis in vitro. A number of studies provide support that BATF may play an important role in the development of different types of cancer, including colon cancer, lymphoma and multiple myeloma (16–18).
There were certain limitations to the present study. Only one lung cancer cell line was used; the results will be verified in other lung cancer cell lines and normal cell lines in a future study. In addition, a larger sample size is required to identify the association between BATF expression and clinical features of NSCLC in the future.
In conclusion, the results of the present study demonstrated that BATF was upregulated in NSCLC tissues in TCGA database. In the human NSCLC A549 cell line, BATF knockdown inhibited cell proliferation and promoted apoptosis. Although detailed mechanisms remain to be elucidated, these results suggested that BATF may serve an oncogenic role in the development of NSCLC. Therefore, BATF may be a potential target for the treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 81800279) and the Natural Science Foundation of Jiangsu Province (grant no. BK20180197).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in The Cancer Genome Atlas repository (http://cancergenome.nih.gov).
Authors' contributions
HM, YF and LP designed the experiments. HM wrote the manuscript. LP and BZ acquired the data, and wrote and revised the manuscript. HH was responsible for the analysis and discussion of the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Approval was obtained to review the data from Institutional Ethical Committee of The First Affiliated Hospital of Soochow University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr, Wu YL, Paz-Ares L. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 3.Sangodkar J, Katz S, Melville H, Narla G. Lung adenocarcinoma: Lessons in translation from bench to bedside. Mt Sinai J Med. 2010;77:597–605. doi: 10.1002/msj.20226. [DOI] [PubMed] [Google Scholar]
- 4.Vansteenkiste J, Crinò L, Dooms C, Douillard JY, Faivre-Finn C, Lim E, Rocco G, Senan S, Van Schil P, Veronesi G, et al. 2nd ESMO consensus conference on lung cancer: Early-stage non-small-cell lung cancer consensus on diagnosis, treatment and follow-up. Ann Oncol. 2014;25:1462–1474. doi: 10.1093/annonc/mdu089. [DOI] [PubMed] [Google Scholar]
- 5.Chabowski M, Polański J, Jankowska-Polanska B, Lomper K, Janczak D, Rosinczuk J. The acceptance of illness, the intensity of pain and the quality of life in patients with lung cancer. J Thorac Dis. 2017;9:2952–2958. doi: 10.21037/jtd.2017.08.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huo Y, Li A, Wang Z. LncRNA AWPPH participates in the metastasis of non-small cell lung cancer by upregulating TGF-β1 expression. Oncol Lett. 2019;18:4246–4252. doi: 10.3892/ol.2019.10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NSCLC Meta-analysis Collaborative Group Preoperative chemotherapy for non-small-cell lung cancer: A systematic review and meta-analysis of individual participant data. Lancet. 2014;383:1561–1571. doi: 10.1016/S0140-6736(13)62159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank L, Christodoulou E, Kazerooni EA. Radiation risk of lung cancer screening. Semin Respir Crit Care Med. 2013;34:738–747. doi: 10.1055/s-0033-1358615. [DOI] [PubMed] [Google Scholar]
- 10.Chalela R, Curull V, Enríquez C, Pijuan L, Bellosillo B, Gea J. Lung adenocarcinoma: From molecular basis to genome-guided therapy and immunotherapy. J Thorac Dis. 2017;9:2142–2158. doi: 10.21037/jtd.2017.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jabeen R, Goswami R, Awe O, Kulkarni A, Nguyen ET, Attenasio A, Walsh D, Olson MR, Kim MH, Tepper RS, et al. Th9 cell development requires a BATF-regulated transcriptional network. J Clin Invest. 2013;123:4641–4653. doi: 10.1172/JCI69489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 13.Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol. 2013;13:499–509. doi: 10.1038/nri3470. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez JM, Floyd DH, Weilbaecher KN, Green PL, Boris-Lawrie K. Multiple facets of junD gene expression are atypical among AP-1 family members. Oncogene. 2008;27:4757–4767. doi: 10.1038/onc.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 16.Schleussner N, Merkel O, Costanza M, Liang HC, Hummel F, Romagnani C, Durek P, Anagnostopoulos I, Hummel M, Jöhrens K, et al. The AP-1-BATF and -BATF3 module is essential for growth, survival and TH17/ILC3 skewing of anaplastic large cell lymphoma. Leukemia. 2018;32:1994–2007. doi: 10.1038/s41375-018-0045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai L, Cui X, Zhang X, Cheng L, Liu Y, Yang Y, Fan P, Wang Q, Lin Y, Zhang J, et al. SARI inhibits angiogenesis and tumour growth of human colon cancer through directly targeting ceruloplasmin. Nat Commun. 2016;7:11996. doi: 10.1038/ncomms11996. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Gil M, Pak HK, Park SJ, Lee AN, Park YS, Lee H, Lee H, Kim KE, Lee KJ, Yoon DH, et al. Engagement of CD99 reduces AP-1 activity by inducing BATF in the human multiple myeloma cell line RPMI8226. Immune Netw. 2015;15:260–267. doi: 10.4110/in.2015.15.5.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol. 2018;51:116–128. doi: 10.1016/j.semcancer.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Zarogoulidis P, Tsakiridis K, Karapantzou C, Lampaki S, Kioumis I, Pitsiou G, Papaiwannou A, Hohenforst-Schmidt W, Huang H, Kesisis G, et al. Use of proteins as biomarkers and their role in carcinogenesis. J Cancer. 2015;6:9–18. doi: 10.7150/jca.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, Chella A, Viola P, Pullara C, Mucilli F, Buttitta F. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 24.He L, Zhang Y, Sun H, Jiang F, Yang H, Wu H, Zhou T, Hu S, Kathera CS, Wang X, et al. Targeting DNA flap endonuclease 1 to impede breast cancer progression. EBioMedicine. 2016;14:32–43. doi: 10.1016/j.ebiom.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas C, Ji Y, Lodhi N, Kotova E, Pinnola AD, Golovine K, Makhov P, Pechenkina K, Kolenko V, Tulin AV. Non-NAD-like poly(ADP-ribose) polymerase-1 inhibitors effectively eliminate cancer in vivo. EBioMedicine. 2016;13:90–98. doi: 10.1016/j.ebiom.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorsey MJ, Tae HJ, Sollenberger KG, Mascarenhas NT, Johansen LM, Taparowsky EJ. B-ATF: A novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene. 1995;11:2255–2265. [PubMed] [Google Scholar]
- 27.Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, Murphy KM. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma QY, Huang DY, Zhang HJ, Wang S, Chen XF. Upregulation of bacterial-specific Th1 and Th17 responses that are enriched in CXCR5+CD4+ T cells in non-small cell lung cancer. Int Immunopharmacol. 2017;52:305–309. doi: 10.1016/j.intimp.2017.09.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in The Cancer Genome Atlas repository (http://cancergenome.nih.gov).