Abstract
The aim of the present study was to investigate the effect of Forkhead box transcription factor M1 (FoxM1)-silencing on the growth, migration and invasion of K1 human papillary thyroid carcinoma (PTC) cells. The effect of FoxM1-small interfering RNA (siRNA) in K1 cells was detected by western blot analysis. FoxM1-siRNA and control siRNA were transfected into K1 cells using Lipofectamine® 2000 (transfection group, T) and the non-meaning sequence group (NM). K1 cells exposed to PBS solution comprised the blank control group (CON). Cell proliferation ability was detected using an MTT assay. Cell migration and invasion was detected by the single cell scratch test and Transwell invasion assay, respectively. Western blot analysis indicated that FoxM1 siRNA downregulated the expression of FoxM1 protein. Cell proliferation, migration and invasion were significantly lower in the T group compared with the NM and CON groups (P<0.05). These results indicated that silencing of FoxM1 expression could block growth, invasion and migration of K1 cells. This study may provide a novel target gene for targeted therapy of PTC.
Keywords: Forkhead box transcription factor M1, invasion, migration, proliferation, thyroid papillary carcinoma
Introduction
Thyroid cancer ranks first among endocrine tumors and is increasing in prevalence in China, in particular papillary cancer of the thyroid gland (1). Thyroid-associated tests have been added to routine physical examinations and have increased the detection of thyroid cancers (2). Thyroid cancer-associated mortality is particularly prevalent in Asia and China. China accounts for 15.6% of thyroid cancer cases globally and 13.8% of global cancer-associated mortality (3). Thyroid cancer is a major disease threat in China.
Forkhead transcription factor M1 (FoxM1) possesses a ‘wing helix’ DNA binding domain, which is a common structural domain of the Forkhead family (4). The main function of FoxM1 is to regulate the transition G1 phase in cells, thereby regulating mitosis; thus, FoxM1 serves an important role in the cell growth cycle (5,6). FoxM1 enhances the proliferation of cells, particularly in fetal tissues (7). Elevated expression of FoxM1 in tumors influences tumor occurrence and development, including promoting the growth of tumor cells, neovascularization, and the invasive spread of tumor cells (8). Elevated production of FoxM1 can prevent apoptosis of tumor cells and delay their aging (9). Previous studies have reported an association between FoxM1 expression and paclitaxel resistance in tumors, including those of breast cancer (10), colon cancer (11), gastric cancer (12), and liver cancer (13). It has been reported that when FoxM1 expression is silenced in tumor cells, the cells become sensitive to paclitaxel (14).
The role of FoxM1 in papillary thyroid carcinoma (PTC) remains unclear. In the present study, the proliferation, migration and invasion rate was detected in K1 human PTC cells with FoxM1 silencing. The molecular mechanism of FoxM1 has been thoroughly studied in previous reports (15). The present study was not concerned with the molecular mechanism, but rather with the effect of FoxM1 on tumor cells. The results demonstrated an association of FoxM1 expression with a number of biological behaviors of tumor cells, suggesting that FoxM1 may serve as a novel therapeutic gene target for PTC, in particular for patients who cannot have surgery or do not respond to chemotherapy.
Materials and methods
Culture and passage of K1 human thyroid cancer cells
The K1 human thyroid cancer cell line was purchased from the Shanghai Cell Bank (Chinese Academy of Sciences), and preserved by the Central Laboratory of North China University of Science and Technology (Tangshan, China). K1 cells were cultured in RPMI-1640 complete medium (Biological Industries) containing penicillin, streptomycin, and 10% bovine embryo serum (Biological Industries) in an incubator maintained at 37°C with 5% CO2. K1 cell growth was monitored by optical microscopy, replenishing the medium every 24 to 48 h, according to cell growth. Following adherence of K1 cells to the wall of the culture flask and growth covering ~70% of the wall, the cells were passaged, and the shed cells and cell debris were removed gently by 2–3 washes with PBS. Trypsin (0.25%) was subsequently added to digest the cells. When cell morphology changed to a roughly spherical shape, and the cells had detached from the flask wall, complete medium was added to stop the digestion. The cells were evenly mixed in the medium and half were transferred to another culture flask, and cultured using the aforementioned conditions. As pancreatin affects cell growth, the medium was changed every 12 h.
Transfection of FoxM1-small interfering RNA (siRNA)
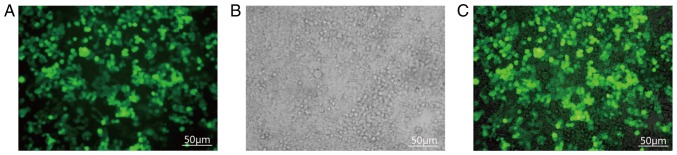
The FoxM1-siRNA target sequence 5′-GGCUGCACUAUCAACAAUATTUAUUGUUGAUAGUGCAGCCTT-3′ was synthesized by Suzhou Gema Gene Limited. Lipofectamine 2000 was purchased from Invitrogen (Thermo Fisher Scientific, Inc.). Aliquots of detached cells with the appropriate density (70%) were seeded into 6-well plates. The cells were transfected when they had grown to an appropriate state (to the fastest growth rate) to avoid the effect of penicillin and streptomycin prior to transfection. The penicillin/streptomycin-free culture medium was changed 24 h prior to transfection, and the cells in the 6-well plates were washed twice with PBS prior to transfection. Uninoculated RPMI-1640 medium (1 ml) was added to each well, and the cells were incubated for 1 h to produce a starvation condition. siRNA (5 µl) and Lipofectamine® 2000 (5 µl; Thermo Fisher Scientific, Inc.) were added to 500 µl blank RPMI-1640 medium and incubated for 5 min at 37°C. The two solutions were subsequently mixed and allowed to stand for 20 min at 37°C prior to being added into two wells with the starved cells forming the transfection group (group T). The above protocol was repeated with the replacement of the previous siRNA with the control siRNA (sequence 5′-CCAUGAGGAGUACUGCCAATT-3′) to prepare the non-meaning sequence group (group NM). The remaining two wells received only PBS and comprised the control (group CON). The 6-well plates were gently shaken to spread the liquid evenly on the bottom of the wells. After 6 h of transfection, the liquid was removed from each well, RPMI-1640 medium containing 10% serum was added, and culturing was continued for 48 h. The cells were then observed using fluorescence microscopy, and the proportion of fluorescent cells was recorded. Transfection was defined as successful when the proportion of cells with fluorescence exceeded 80% (Fig. 1).
Figure 1.
Fluorescence microscopy 48 h following transfection with small interfering RNA targeting FoxM1 (magnification, ×200). FoxM1, Forkhead box transcription factor M1. (A) Fluorescence microscopy detection. (B) Light transmittance detection. (C) Merge of (A) and (B) Scale bar, 50 µm.
Western blot analysis
To extract total protein, digested cells with appropriate density were prepared from a single cell suspension and centrifuged at 330 × g for 5 min at 37°C. A small volume of ice-cold PBS solution was subsequently added into the centrifugation bottle to resuspend and centrifuge the cells. This step was repeated twice. Following the final wash, the supernatant was removed and the cell pellet was resuspended in 200 µl RIPA pyrolysis working solution (Biological Industries) and transferred to an Eppendorf tube. The preparation was ultrasonicated for 10 sec to destroy the cells. After shaking 10 times, the cells were pipetted evenly on ice for 30 min, and centrifuged at 4°C for 15 min at 4,000 × g. The supernatant was stored at −20°C.
Protein was quantified using a bicinchoninic acid protein quantification kit, according to the manufacturer's protocols. The same volume of 2X protein loading buffer was added, followed by denaturation at 100°C for 5 min and preservation at −80°C for later use.
A volume containing 30 µg protein was used per well for SDS-PAGE with a 5% concentration gel and 10% separation gel. The previously prepared protein samples and markers were added sequentially to the wells for electrophoresis at 80 and 120 V, respectively. Following electrophoresis, the separation gel region, in which the proteins with different molecular weights became uniformly distributed, was removed and a PVDF membrane was soaked in methanol for about 5 sec, followed by rinsing with transmembrane solution. The transmembrane solution-soaked membrane was placed in a wet transmembrane apparatus in which the membrane was sandwiched between filter paper. Proteins were transferred from the gel to the membrane for 1 h at 90 V. The transmembrane solution on the membrane surface was washed away with Tris buffered saline-Tween 20 (TBST). The membrane was blocked using a solution of 10% skimmed milk for 2 h at 37°C.
The membrane surface was flushed with TBST, followed by shaking, and exposed to a primary antibody kit solution (Invitrogen; Thermo Fisher Scientific, Inc.) (FOXm1; cat. no. sc-271746; 1:500; Thermo Fisher Scientific, Inc.) at 4°C for 12 h. The membrane was rinsed two to three times (10 min each) with TBST, prior to shaking in the presence of pre-configured horseradish peroxidase-IgG secondary antibody kit (1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. 31430) at 37°C for 2 h. The membrane was rinsed two to three times (10 min each) with TBST. A BeyoECL Star super-sensitized electrochemiluminescence kit, was used to detect proteins according to the manufacturer's protocols. ImageJ software (version 1.8.0; National Institutes of Health) was used to analyze the gray scale values of the immunofluorescent bands. The expression (%) of associated protein was calculated as (gray value of target protein/gray value of internal reference) ×100.
Cell viability based on MTT assay
DMSO was purchased from Sigma-Aldrich; Merck KGaA. MTT was purchased from Amresco, LLC. Cells in each group were digested and single cell suspensions were prepared and evenly seeded in 96-well plates with appropriate cell densities (70%). An equal volume of PBS was added to the well to prevent a detrimental effect on cell growth, due to water evaporation. Each group comprised six wells, which were divided into 24, 48 and 72 h samples (two wells for each time). Each well received 20 µl MTT solution agent 4 h prior to each time-point to allow the formation of blue formazan. At 24, 48 or 72 h, the liquid in each well was removed and 150 µl DMSO was added, followed by shaking for 10 min. The optical density (OD) value of each well was measured at a wavelength of 490 mm, and blank wells were used for normalizing the spectrophotometer. The maximum and minimum values of each well were eliminated from the data, and the remaining data were averaged, with the experiment repeated three times. The inhibition rate (%) of FoxM1-siRNA on cell growth was calculated as (OD490Group CON-OD490 Group T)/OD490 Group CON ×100.
Migration scratch assay
The cell groups were cultured in 6-well plates in pairs (70%). When the cells had formed a monolayer covering the bottom of the well, a 200-µl pipette tip was used to make a scratch in each monolayer, with the width of the scratch ~0.6 mm. Each well was washed two to three times using PBS to remove non-adherent cells, cell debris and residual medium. RPMI-1640 medium was added (0 h). The wells were imaged at 0, 24, 48 and 72 h with a fluorescence microscope to observe the changes in the scratch area in the images.
Invasion assay
Prior to the experiment, the prepared Matrigel (BD Biosciences) was placed at 4°C for 12 h. The Matrigel and blank RPMI-1640 medium were dispensed at a ratio of 1:9, and 20 µl prepared Matrigel was evenly spread on each Transwell chamber. Care was taken not to produce bubbles when adding the Matrigel. The Transwell chambers were subsequently placed into one incubator for 12 h to allow coagulation of the Matrigel. The cell groups were cultured for 48 h, digested, and subsequently seeded into the upper chamber with a cell density of 2×105 cells/100 µl of serum-free, penicillin/streptomycin-free medium. A total of 600 µl RPMI-1640 medium containing only 10% serum was added to the lower chamber, which was subsequently transferred to an incubator for 18 h. The chamber was gently washed three times with PBS to remove the cells on the surface of the upper chamber. The chamber was soaked with 4% formaldehyde for 30 min at 37°C. The solution was removed and the chamber was rinsed with PBS and allowed to air-dry. The cells were then stained with 80% crystal violet for 15–20 min at 37°C and subsequently rinsed with distilled water. The polycarbonate membrane was cut from the chamber and placed on a glass slide. Subsequent to the membrane being air-dried, it was covered using neutral gum and with a cover glass to further to air-dry. Cells permeating the membrane were then observed by optical microscopy (magnification, ×200). Randomly selected visual fields (n=15) were examined to record the number of cells and the mean value was calculated. The experiment was repeated three times.
Statistical analyses
The data were processed using SPSS 17.0 software (SPSS Inc.). The results are expressed as mean ± standard deviation. The results of cell migration and invasion tests were analyzed by one-way ANOVA. The results of MTT were analyzed using the ANOVA of repeated measurements. P<0.05 considered to indicate a statistical significant difference.
Results
Expression of FoxM1 protein
The expression of FoxM1 protein in group T (cells transfected with siRNA targeting FoxM1) was lower compared with the other two groups (P<0.05); however, there was no statistical significance between the CON and NM groups (P>0.05; Fig. 2; Table I).
Figure 2.
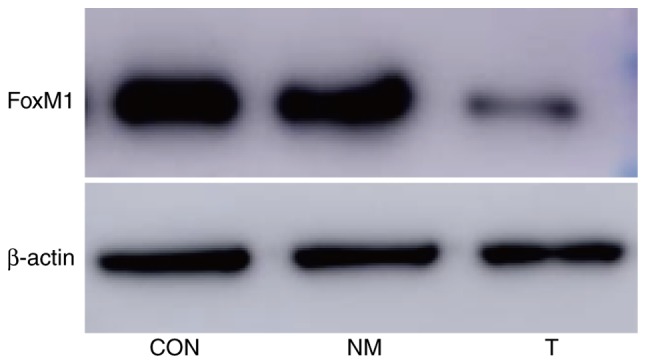
Relative FoxM1 protein expression in each group. FoxM1, Forkhead box transcription factor M1; CON, blank control group; NM, non-meaning sequence group; T, transfection group.
Table I.
Effect of FoxM1 silencing on protein expression (n=5).
P<0.05 vs. group T. FoxM1, Forkhead box transcription factor M1; CON, blank control group; NM, non-meaning sequence group; T, transfection group.
Cell proliferation
Cell growth in group T was significantly inhibited compared with the other two groups (P<0.05). There was no significant difference between group NM and group CON (P>0.05; Table II).
Table II.
Effect of FoxM1 silencing on K1 cell growth (n=6).
| Group | |||||
|---|---|---|---|---|---|
| Time-point (h) | CON | NM | T | F | P-value |
| 24 | 0.48±0.01a | 0.46±0.02a | 0.36±0.02 | 73.29 | <0.001 |
| 48 | 0.66±0.04a | 0.64±0.02a | 0.48±0.01 | 72.50 | <0.001 |
| 72 | 0.72±0.03a | 0.71±0.01a | 0.57±0.02 | 99.11 | <0.001 |
P<0.05 vs. group T. FoxM1, Forkhead box transcription factor M1; CON, blank control group; NM, non-meaning sequence group; T, transfection group.
Cell migration
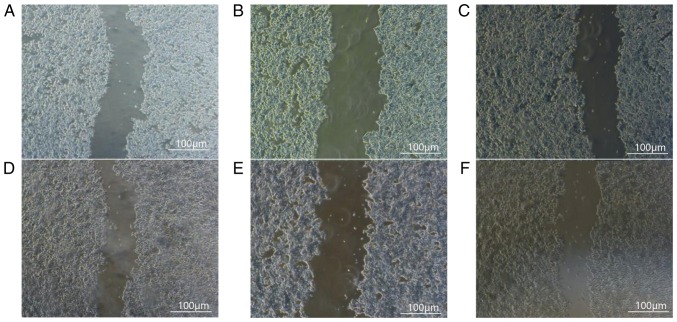
After 24 h, the relative distance between the scratches in group CON (0.83±0.01) was significantly shorter than in group T (0.93±0.01; P<0.05), indicating that the migration speed of the K1 cells was significantly slower following FoxM1 silencing compared with the CON and NM groups. Comparison of cell migration ability between the CON and NM groups indicated no statistically significant difference (P>0.05; Fig. 3; Table III).
Figure 3.
Changes in the migratory ability of cells following FoxM1-silencing. (A) Blank control group, (B) meaningless sequence group and (C) experimental group at 0 h. (D) Blank control group, (E) meaningless sequence group and (F) experimental group at 24 h. Scale bar, 100 µm. FoxM1, Forkhead box transcription factor M1.
Table III.
Effect of FoxM1 silencing on cell migration (n=9).
P<0.05 vs. group T. FoxM1, Forkhead box transcription factor M1; CON, blank control group; NM, non-meaning sequence group; T, transfection group.
Detection of in vitro invasive ability of tumor cells
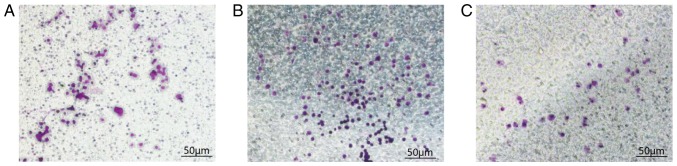
There was no significant difference in the number of cells penetrating the matrix membrane in the NM group compared with the meaningless sequence group. This experiment confirmed that silencing FoxM1 significantly reduced the invasive ability of K1 human thyroid cancer cells. The mean numbers of the cells that penetrated the upper chamber, and reached the lower chamber were 37.20±3.96, 85.40±5.13 and 92.40±3.05/field in the T, NM and CON groups, respectively. There was a statistical significance between the CON and T groups (P<0.001), indicating that compared with group CON, the number of cells penetrating the matrix membrane in group T was significantly reduced. There was no statistical significance between the CON and NM groups (P=0.20; Fig. 4 and Table IV).
Figure 4.
Detection of in vitro invasive ability of tumor cells. Cell number passing through the Transwell chamber (magnification, ×200). (A) Blank control group. (B) Meaningless sequence group. (C) Experimental group. Scale bar, 50 µm. FoxM1, Forkhead box transcription factor M1
Table IV.
Effect of FoxM1 silencing on invasion in a Transwell chamber assay (n=20).
| Group | Amount of cells (cells/field) | F | P-value |
|---|---|---|---|
| CON | 92.40±3.05a | 264.09 | <0.001 |
| NM | 85.40±5.13a | ||
| T | 37.20±3.96 |
P<0.05 vs. group T. FoxM1, Forkhead box transcription factor M1; CON, blank control group; NM, non-meaning sequence group; T, transfection group.
Discussion
The results of the present study indicated that the proliferation, migration, and invasion of PTC cells are suppressed following FoxM1-silencing. The results are consistent with prior observations that inhibition of FoxM1 expression can alter the biological changes in tumor cell proliferation, migration, invasion, and other biological changes (16). Therefore, FoxM1 appears to promote several cancer-associated functions of PTC cells. The incidence rate of thyroid cancers in developing countries are high, as it accounts for only 1% of all malignant tumors (17). They remain however, the most common endocrine tumor, as thyroid cancers currently rank as the 10th most frequent tumor disease in China (17), among which PTC is the most common, accounting for ~70% of all types of thyroid cancers. Furthermore, the incidence of PTC is increasing (18). Malignancy of PTC is less common and its growth is relatively slow, making it prone to lymph node metastasis (19). Surgery is the most effective treatment method for thyroid cancers. However, the complex anatomy, rich blood supply and the endocrine effects of PTC can result in a number of postoperative complications (20). The postoperative 10-year survival rate is high; however, the recurrence rate is also high, which results in an increased mortality rate over time (21). Therefore, the need for novel targeted therapeutic drugs has become urgent. Physiological processes, including proliferation and apoptosis, are abnormal in tumor cells (22). FoxM1 is a member of the Forkhead transcription factor family (23). FoxM1 can regulate a number of metabolic-associated processes to maintain the balance of tumor cell proliferation and energy metabolism. Furthermore, FoxM1 is also involved in the regulation of tumor cell apoptosis, metastasis, and other related processes, and is associated with the metastasis, angiogenesis and epithelial-mesenchymal transition of tumor cells (24,25). Abnormal expression of FoxM1 is associated with poor clinical classification and poor prognosis in patients with cancer (26).
Based on the aforementioned characteristics, a quantitative index diagnosis system of malignant tumors based on the FoxM1 gene was previously established (27). Subsequent studies have documented an accuracy of 94% against early oral, skin and neck cancers. Therefore, FoxM1 gene expression can be suggested as a reliable method for the early diagnosis of associated tumors and has great practical potential in the clinical diagnosis and treatment of tumors. FoxM1 has the same effect on other thyroid cancer cell lines (28) as its role in TPC-1 cell line has been demonstrated. Alvarez-Fernández and Medema (16) examined the underlying molecular mechanism of FoxM1, therefore this was not the focus of the present study; however, to the best of our knowledge, cell scratch test data have not been provided in earlier studies. The cell scratch test gauges the ability of cells, including cancer cells, to migrate. Metastasis of cancer often results in a poor prognosis. Therefore, controlling the spread of cancer by blunting metastasis is a prudent strategy for cancer control and prevention.
In summary, FoxM1 is crucial in the occurrence and growth of PTC, and may be a valuable target for treatment. This study demonstrated the effect of FoxM1 on the proliferation, migration and invasion ability of PTC cells, however it was not able to demonstrate the role of FoxM1 in PTC cells. Therefore, further examination of the other biological effects of FoxM1 on PTC cells is required, in order to verify the results of the present study.
Acknowledgements
Not applicable.
Funding
This study was funded by the Hebei Institute of Science and Technology Information (grant no. 20150523).
Availability of data and materials
All the data generated or analyzed in this study are included in this published article.
Authors' contributions
GZ and XW made substantial contributions to conception and design; GZ, YZ and MC acquired data; XW, JC and WC analysed and interpreted data; GZ, GY, YZ, WC, MC and QL were involved in drafting the manuscript and revising it critically for important intellectual content; GY and YZ were responsible for the preparation of experimental materials and equipment, WC and MC were responsible for the final data statistics and collation, QL, GY and YZ were responsible for performing the experiment, WC and YZ were responsible for reviewing the relevant literature. GZ gave final approval of the version to be published.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hedinger C, Williams ED, Sobin LH. The WHO histological classification of thyroid tumors: A commentary on the second edition. Cancer. 1989;63:908–911. doi: 10.1002/1097-0142(19890301)63:5<908::AID-CNCR2820630520>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 2.Zhu T, Xu J, Tian T, Niu F, An CM. Interpretation of the 2017 US Preventive Services Working Group Recommendations for Thyroid Cancer Screening. Cancer Res. 2018;45:710–714. [Google Scholar]
- 3.Stewart B, Wild C. World cancer report 2014. International Agency for Research on Cancer. 2014 [Google Scholar]
- 4.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, Shin B, Datta A, Raychaudhuri P, Costa RH. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, Park YY, Kim SH, Nguyen OT, Yoo YS, Chan GK, Sun X, Cho H. Human mitochondrial Fis1 links to cell cycle regulators at G2/M transition. Cell Mol Life Sci. 2014;71:711–725. doi: 10.1007/s00018-013-1428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Yuan C, Wu J, Elsayed Z, Fu Z. Polo-like kinase 1-mediated phosphorylation of Forkhead box protein M1b antagonizes Its SUMOylation and facilitates its mitotic function. J Biol Chem. 2015;290:3708–3719. doi: 10.1074/jbc.M114.634386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- 8.Cui J, Shi M, Xie D, Wei D, Jia Z, Zheng S, Gao Y, Huang S, Xie K. FOXM1 promotes the Warburg effect and pancreatic cancer progression via transactivation of LDHA expression. Clin Cancer Res. 2014;20:2595–2606. doi: 10.1158/1078-0432.CCR-13-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yung MM, Chan DW, Liu VW, Yao KM, Ngan HY. Activation of AMPK inhibits cervical cancer cell growth through AKT/FOXO3a/FOXM1 signaling cascade. BMC Cancer. 2013;13:327. doi: 10.1186/1471-2407-13-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW, Zhao F, Monteiro LJ, Coombes RC, Medema RH, Khoo US, Lam EW. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35:990–1002. doi: 10.1038/onc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song IS, Jeong YJ, Han J. Mitochondrial metabolism in cancer stem cells: A therapeutic target for colon cancer. BMB Rep. 2015;48:539–540. doi: 10.5483/BMBRep.2015.48.10.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin H, Park MH, Kim SM. 3,3′-Diindolylmethane potentiates paclitaxel-induced antitumor effects on gastric cancer cells through the Akt/FOXM1 signaling cascade. Oncol Rep. 2015;33:2031–2036. doi: 10.3892/or.2015.3758. [DOI] [PubMed] [Google Scholar]
- 13.Huang X, Qin J, Lu S. Up-regulation of miR-877 induced by paclitaxel inhibits hepatocellular carcinoma cell proliferation though targeting FOXM1. Int J Clin Exp Pathol. 2015;8:1515–1524. [PMC free article] [PubMed] [Google Scholar]
- 14.Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW, Zhao F, Monteiro LJ, Coombes RC, Medema RH, Khoo US, Lam EW. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2015;35:990–1002. doi: 10.1038/onc.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, Liao Y, He H, Xin Q, Tu Z, Kong S, Cui T, Wang B, Quan S, Li B, et al. FoxM1 Directs STAT3 expression essential for human endometrial stromal decidualization. Sci Rep. 2015;5:13735. doi: 10.1038/srep13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Fernández M, Medema RH. Novel functions of FoxM1: From molecular mechanisms to cancer therapy. Front Oncol. 2013;3:30. doi: 10.3389/fonc.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen WQ, Zheng RS, Zhang SW, Li N, Zhao P, Li GL, Wu LY, He J. Report of incidence and mortality in china cancer registries, 2008. Chin J Cancer Res. 2012;24:171–180. doi: 10.1007/s11670-012-0171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLeod DS, Sawka AM, Cooper DS. Controversies in primary treatment of low-risk papillary thyroid cancer. Lancet. 2013;381:1046–1057. doi: 10.1016/S0140-6736(12)62205-3. [DOI] [PubMed] [Google Scholar]
- 19.Xue S, Wang P, Liu J, Li R, Zhang L, Chen G. Prophylactic central lymph node dissection in cN0 patients with papillary thyroid carcinoma: A retrospective study in China. Asian J Surg. 2016;39:131–136. doi: 10.1016/j.asjsur.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Yun JS, Lee YS, Jung JJ, Nam KH, Chung WY, Chang HS, Park CS. The Zuckerkandl's tubercle: A useful anatomical landmark for detecting both the recurrent laryngeal nerve and the superior parathyroid during thyroid surgery. Endocr J. 2008;55:925–930. doi: 10.1507/endocrj.K08E-132. [DOI] [PubMed] [Google Scholar]
- 21.Tuttle RM, Ball DW, Byrd D, Dilawari RA, Doherty GM, Duh QY, Ehya H, Farrar WB, Haddad RI, Kandeel F, et al. Thyroid carcinoma. J Natl Compr Canc Netw. 2010;8:1228–1274. doi: 10.6004/jnccn.2010.0093. [DOI] [PubMed] [Google Scholar]
- 22.Dias RC, Marangoni DV, Riley LW, Moreira BM. Identification of uropathogenic Escherichia coli clonal group A (CgA) in hospitalised patients. Mem Inst Oswaldo Cruz. 2009;104:787–789. doi: 10.1590/S0074-02762009000500021. [DOI] [PubMed] [Google Scholar]
- 23.Laoukili J, Kooistra MR, Brás A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Zou Y, Yang H, Wang J, Pan H. Downregulation of FoxM1 inhibits proliferation, invasion and angiogenesis of HeLa cells in vitro and in vivo. Int J Oncol. 2014;45:2355–2364. doi: 10.3892/ijo.2014.2645. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad A, Wang Z, Kong D, Ali S, Li Y, Banerjee S, Ali R, Sarkar FH. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2016;158:607. doi: 10.1007/s10549-016-3904-y. [DOI] [PubMed] [Google Scholar]
- 26.Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, Sivanandan K, Coombes RC, Medema RH, Hartman J, et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. 2010;29:2983–2995. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The MT, Hutchison IL, Costea DE, Neppelberg E, Liavaag PG, Purdie K, Harwood C, Wan H, Odell EW, Hackshaw A, Waseem A. Exploiting FOXM1-orchestrated molecular network for early squamous cell carcinoma diagnosis and prognosis. Int J Cancer. 2013;132:2095–2106. doi: 10.1002/ijc.27886. [DOI] [PubMed] [Google Scholar]
- 28.Yuan H, Min Z, Chen J. Study on the relationship between the expression of FoxM1 gene and the activity and invasion of TPC-1 cells in papillary thyroid carcinoma. Chin J Endocrine Surg. 2014;8:33–37. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated or analyzed in this study are included in this published article.