Abstract
Breast cancer is the most prevalent cancer among women, and diagnosis and treatment represent a substantial challenge due to the lack of adequate molecular targets. It has been shown that long noncoding RNAs (lncRNAs) serve pivotal roles in regulating gene expression in tumors. The roles of long intervening noncoding RNA (Linc)-OIP5 has been demonstrated in different types of cancer; however, its function in breast cancer has not been determined. In the present study, expression of Linc-OIP5, YAP1 (Hippo signaling component) and JAG1 (Notch signaling component) in breast cancer cells with different degrees of malignancy were determined. To assess whether Linc-OIP5 regulated the malignant biological behaviors of MDA-MB-231 cells, its expression was knocked down using a specific small interfering RNA (siRNA), and cell proliferation was determined using a CCK-8 assay, apoptosis was evaluated using an Annexin V-FITC apoptosis detection kit, migration was assessed using a wound healing and transwell migration assays, and cell invasion examined using a transwell invasion assays. The effect of Linc-OIP5 knockdown on YAP1 and JAG1 expression was quantified using reverse transcription-quantitative PCR and immunoblotting. Cell proliferation, migration and invasion were reduced, while apoptosis was increased in MDA-MB-231 cells transfected with Linc-OIP5-specific siRNA. Mechanistic investigations showed that Linc-OIP5 knockdown downregulated YAP1 and JAG1 expression. The results of the present study suggest that Linc-OIP5 affects the malignant biological behaviors of MDA-MB-231 cells, at least partly through its effects on YAP1/JAG1 signaling. Whilst there are a number of mechanisms underlying the pathogenesis of breast cancer, the results of the present study highlight Linc-OIP5 as a potential therapeutic target in breast cancer.
Keywords: Linc-OIP5, breast cancer, hippo pathway, notch pathway, angiogenesis
Introduction
Breast cancer is the most commonly diagnosed malignancy and one of the leading causes of cancer-associated death in women worldwide (1–5). Breast tumors are heterogeneous neoplasms, composed of multiple subtypes, which exhibit distinct morphologies and clinical features (6,7). Clinically, although great advances have been made in the treatment of breast cancer over the last decade, recurrence and metastasis remain the principal causes of mortality in patients with this disease (2,8–11). Therefore, elucidating the pathogenic mechanisms underlying breast cancer development and progression to identify prognostic biomarkers may provide potentially novel therapeutic targets.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs >200 nucleotides in length which are transcribed from across the genome and participate in a variety of physiological and pathological processes as posttranscriptional regulators of gene expression (12–16). Long intervening noncoding RNAs (lincRNAs) are a type of lncRNA that have been demonstrated to be transcript units between protein coding genes, and display distinct tissue- and cell-specific expression (12,13). Accumulating evidence indicates that the abnormal expression of specific lincRNAs is closely associated with tumor initiation, progression, and metastasis (17,18). Linc-OIP5, a novel cancer-associated lincRNA, is dysregulated in various types of cancer (12,19); for example, Deng et al (12) demonstrated that Linc-OIP5 functions as an oncogene in lung adenocarcinoma. Additionally, Linc-OIP5 contributes to carcinogenic potential by controlling multiple myeloma cell proliferation and apoptosis (19). Therefore, Linc-OIP5 is considered to be an oncogene involved in tumorigenesis and progression. Although a number of reports have functionally characterized Linc-OIP5 in several tumors, the functional significance of Linc-OIP5 in breast cancer is largely unknown.
Yes-associated protein 1 (YAP1) and Jagged 1 (JAG1) are key components of the Hippo and Notch pathways, respectively, which participate in several biological processes, including maintenance of tissue homeostasis, regulation of stem cells in adults and progression of various tumors (6,7,20–25). YAP is a transcriptional coactivator which controls the activity of Hippo signaling through its phosphorylation and dephosphorylation, and binds with the TEA domain (TEAD1) transcription factor to activate target genes downstream of Hippo signaling (25–27). JAG1 is a key ligand of the Notch pathway, implicated in tumorigenesis and vascularization (28–30). YAP1 has been demonstrated to regulate oncogenic phenotypes of breast cancer cells (31–34), and JAG1 is associated with recurrence and poor prognosis in patients with breast cancer (35–37). Interestingly, it has previously been shown that YAP1 acts upstream of the Notch pathway and upregulates JAG1 expression (20,25,38). Based on unpublished data from our laboratory, it has been demonstrated that Linc-OIP5 knockdown influences the proliferation, migration, and tube formation of endothelial cells when cocultured with breast cancer cells. Furthermore, the expression of YAP1 and JAG1 in breast cancer cells with high-grade malignancy is significantly higher compared with breast cancer cells with moderate-grade malignancy, highlighting their potential as breast tumor markers, which warrant further investigation (unpublished data). In the present study, it was demonstrated that YAP1 and JAG1 can synergistically regulate the tumorigenesis and progression of breast cancer cells and the effects of Linc-OIP5 on this regulation was determined.
Linc-OIP5 may regulate the proliferation, apoptosis, migration and invasion of breast cancer cells, at least partly, by modulating the signaling pathways involving YAP1/JAG1, highlighting the therapeutic potential of targeting Linc-OIP5 for treating patients with breast cancer.
Materials and methods
Cell lines and culture conditions
The human breast cancer cell lines (MDA-MB-231 and MCF-7) were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and cultured in DMEM (Thermo Fisher Scientific Inc.) supplemented with 10% FBS (Thermo Fisher Scientific Inc.) and 1% penicillin-streptomycin (Hyclone; GE Healthcare Life Sciences) in a humidified atmosphere at 37°C with 5% CO2.
Reverse transcription-quantitative (RT-q)PCR and RT-PCR
Total RNA from cultured cells was isolated using a miRNA kit (Omega Bio-Tek Inc.), according to the manufacturer's protocol. Total RNA concentration was evaluated by measuring the absorbance at 260/280 nm using a NanoDrop-2000 (Thermo Fisher Scientific Inc.). RNA samples were reverse transcribed using a Reverse Transcription kit (Roche Diagnostics GmbH) to synthesize cDNA, according to the manufacturer's protocol. The reverse transcription temperature protocol was: 65°C for 10 min, 25°C for 10 min, 50°C for 1 h and 85°C for 5 min.
RT-PCR was performed on a thermal cycler (Bio-Rad Laboratories Inc.) with 2× Taq PCR Master mix (KT201-01; Tiangen Biotech Co., Ltd.), according to the manufacturer's protocols. The thermocycling conditions for Linc-OIP5 and YAP1 were: Pre-denaturation at 95°C for 5 min; followed by 30 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 2 min; with a final extension step at 72°C for 5 min. The thermocycling conditions for JAG1 mRNA was: Pre-denaturation at 95°C for 5 min; followed by 30 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec; with a final extension at 72°C for 7 min. The thermocycling conditions for GAPDH was: Pre-denaturation at 94°C for 3 min; followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min; with a final extension step at 72°C for 5 min. The following primer pairs were used to detect the mRNA levels of the indicated genes by RT-PCR: Linc-OIP5 forward, 5′-GCTGCGAAGATGGCGGAGTAAG-3′ and reverse, 5′-GCACGGACGCGCCTAACAC-3′; YAP1 forward, 5′-ACTCGGCTTCAGGTCCTCTTCC-3′ and reverse, 5′-TGGCTACGCAGGGCTAACTCC-3′; JAG1 forward, 5′-CAGTGCTACAACCGTGCCAGTG-3′ and reverse, 5′-CCCTCCCAGCCGTCACTACAG-3′; and GAPDH forward, 5′-CAGGAGGCATTGCTGATGAT-3′ and reverse, 5′-GAAGGCTGGGGCTCATTT-3′. The RT-PCR products were detected using agarose gel electrophoresis, and the images were sequentially scanned using a gel imaging system (Bio-Rad Laboratories Inc.).
RT-qPCR was performed on a StepOne™ PCR system (Thermo Fisher Scientific Inc.) with a SYBR® Premix Ex Taq™ (Roche Diagnostics GmbH), according to the manufacture's protocols. The RT-qPCR thermocycling conditions were: Prevention of cross-contamination at 50°C for 2 min; 95°C pre-denaturation for 10 min; followed by 40 cycles of 95°C denaturation for 15 sec, 60°C annealing for 1 min and 95°C extension for 15 sec; with a final extension step at 72°C for 5 min. Fluorescent signals were collected at 72°C. The following primer pairs were used to detect the mRNA levels of the indicated genes by RT-qPCR: Linc-OIP5 forward, 5′-GCTGCGAAGATGGCGGAGTAAG-3′ and reverse, 5′-CACGGTCCAACAGATGCACTCG-3′; YAP1 forward, 5′-CCTGCGTAGCCAGTTACCAACAC-3′ and reverse, 5′-GCTGCTCATGCTTAGTCCACTGTC-3′; JAG1 forward, 5′-TGTGGCTTGGATCTGTTGCTTGG-3′ and reverse, 5′-ACGTTGTTGGTGGTGTTGTCCTC-3′; and GAPDH forward, 5′-CAGGAGGCATTGCTGATGAT-3′ and reverse, 5′-GAAGGCTGGGGCTCATTT-3′. The relative expression of the genes was calculated using the 2−∆∆Cq method and the experiments were performed three times (39). The relative abundance of specific mRNA molecules was calculated using GAPDH mRNA for normalization. All the primers for RT-PCR and RT-qPCR were purchased from Sangon Biotech (Shanghai Sangon).
Immunofluorescence
A total of 7×104 cells were seeded into 24-well plates with a coverslip on the bottom and incubated at 37°C in 5% CO2. After 8 h, the cells on the coverslips were fixed with 4% paraformaldehyde (Wuhan Boster Biological Technology, Ltd.) for 10 min at room temperature and permeabilized for 20 min in 0.1% Triton X-100 (Beijing Solarbio Science & Technology Co., Ltd.). After blocking in goat serum (Wuhan Boster Biological Technology, Ltd.) for 30 min, slides were incubated with a primary antibody overnight at 4°C. Subsequently, slides were incubated with a goat anti-rabbit immunoglobulin G (IgG) FITC-conjugated secondary antibody (1:50 dilution; cat. no. SA00003-2; ProteinTech Group, Inc.) for 1 h at room temperature. Slides were subsequently counterstained with DAPI (cat. no. C1002; Beyotime Institute of Biotechnology) in the dark for 5 min at room temperature. The following primary antibodies were used for immunofluorescence staining: YAP1 (1:200 dilution; cat. no. GTX129151; GeneTex, Inc.) and JAG1 (1:100 dilution; cat. no. GTX48691; GeneTex, Inc.). Immunofluorescence images were acquired using an inverted fluorescence microscope (Ti-SR; Nikon Corporation; magnification, ×100).
Western blotting
Cells were lysed in RIPA lysis buffer supplemented with protease inhibitors (Applygen Technologies, Inc.). Protein concentrations were quantified using a bicinchoninic acid Protein assay kit (Applygen Technologies, Inc.) and equivalent quantities of protein (20 µg) from each sample were loaded on a 10% SDS gel and resolved using SDS-PAGE and transferred to PVDF membranes (EMD Millipore). Membranes were blocked with 5% non-fat milk in TBS-Tween (TBST; OriGene Technologies, Inc.) for 1 h at room temperature, and subsequently incubated overnight at 4°C with the indicated primary antibodies. The following day, PVDF membranes were washed three times with TBST and incubated with the appropriate horseradish peroxidase-conjugated Affinipure Goat anti-mouse/rabbit IgG secondary antibodies (1:10,000 dilution; cat. no. SA00001-1/SA00001-2, respectively; ProteinTech Group, Inc.) for 1 h at room temperature. Immunoreactive bands were visualized using an enhanced chemiluminescence kit (Biosharp Life Sciences). β-actin was used as the loading control. The following primary antibodies were used: YAP1 (1:200 dilution; cat. no. GTX129151; GeneTex, Inc.), JAG1 (1:100 dilution; cat. no. GTX48691; GeneTex, Inc.) and β-actin (1:1,800 dilution; cat. no. RPB340Mi01; Beyotime Institute of Biotechnology). An Amersham Imager 600 was used to image the blots (GE Healthcare), Image-pro plus (version 7; Media Cybernetics, Inc.) was used for densitometric analyses of immunoblots and quantification results were normalized to those of the loading control.
Small interfering RNAs (siRNAs)
Small interfering RNA duplexes targeting Linc-OIP5 and negative control siRNA duplexes were synthesized and purchased from Shanghai GenePharma Co., Ltd. The siRNA sequences were as follows: Negative control (NC) siRNA duplexes sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′; siLinc-OIP5 Duplex 1 sense, 5′-CCUACUGCCUUGUAAGAAUTT-3′ and antisense, 5′-AUUCUUACAAGGCAGUAGGTT-3′; siLinc-OIP5 Duplex 2 sense, 5′-CCAGCUGUCUUUGUGUCUUTT-3′ and antisense, 5′-AAGACACAAAGACAGCUGGTT-3′; siLinc-OIP5 Duplex 3 sense, 5′-CCAGUUAUCCUGCUAACAUTT-3′ and antisense, 5′-AUGUUAGCAGGAUAACUGGTT-3′.
A mixture of the three siRNAs targeting Linc-OIP5 were used, in a 1:1:1 ratio. MDA-MB-231 cells when they had reached 70–80% confluence using Lipofectamine® 3000 Transfection Reagent (Thermo Fisher Scientific Inc.) according to the manufacturer's protocol. Cells were seeded at a concentration density of 2.7×105 cells per well in 6-well plates. At 48 h post-transfection, the effectiveness of siRNA knockdown was assessed by RT-qPCR.
Cell proliferation assays
MDA-MB-231 cells were collected 48 h after transfection with Linc-OIP5 siRNA or NC siRNA, and analyzed using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.), according to the manufacturer's protocols. Cells were seeded in 96-well plates at a density of 3×103 cells/well and CCK-8 reagent (10 µl/well) was added into medium without serum (90 µl/well) and incubated for 3 h at 37°C. The amount of formazan dye generated by cellular dehydrogenase redox was measured by absorbance at 450 nm using a microplate reader (Molecular Devices, LLC), with the amount produced being proportional to the number of living cells. Cell proliferation was measured every 24 h for 3 days, with the optical density values of each well representing the survival/proliferation of cells. These experiments were repeated at least three times independently.
Wound healing assays
Wound healing assays were used to analyze the migratory ability of MDA-MB-231 cells transfected with Linc-OIP5 siRNA. A total of 3×105 cells per well were seeded into 24-well plates and cultured in DMEM with 1% FBS at 37°C in 5% CO2 for 48 h to allow the cells to adhere and form a confluent monolayer. Subsequently, the monolayers were scratched using the tip of a 10 µl pipette tip. The scratched wound was rinsed three times with PBS to remove debris. Cells were incubated at 37°C in 5% CO2 and monitored for 24 h. Wound healing was monitored by taking digital images from three different fields of view and from three independent samples at 0, 12, and 24 h after scratching. Scratch-wound images were captured using an inverted light microscope (TE2000-S; Nikon Corporation; magnification, ×40).
Transwell migration and invasion assays
Transwell assays were performed using 24-well Transwell plates (Corning Inc.). A total of 5×104 cells in serum-free DMEM (200 µl) were added to the upper chamber of each insert, while the medium in the lower chambers (600 µl) was supplemented with 10% FBS. After a 24 h incubation at 37°C, cells on the upper surface of the membrane were removed with a cotton tip, while those on the lower surface were stained for 20 min at room temperature with 0.1% crystal violet (Beijing Solarbio Science & Technology Co., Ltd.). For Transwell invasion assays, cells (1×105) were seeded in the upper chamber of inserts precoated with 40 µl Matrigel (BD Biosciences) prior to adding the 200 µl serum-free DMEM to the upper chamber. In the lower chamber, 600 µl DMEM supplemented with 10% FBS was added. Cells were incubated overnight at 37°C, after which the cells on the upper surface of the upper chamber were removed, whereas the invasive cells on the lower surface were fixed with methanol and stained with 0.1% crystal violet for 20 min at room temperature. Images were captured of three different fields of view, from three independent samples, using an inverted light microscope (TE2000-S; Nikon Corporation; magnification, ×100). Quantitative analysis of migratory and invasive cells was performed using ImageJ software (version 1.48 v; NIH Inc., Bethesda, Md, USA).
Flow cytometry
Apoptotic cells were determined using an Annexin V-FITC apoptosis detection kit (cat. no. 556547; BD Biosciences, Franklin Lakes, NJ, USA), according to the manufacturer's protocols. The assay was performed with two-color analysis of FITC (green)-labeled Annexin V binding and PI (red) uptake. Cells (5×105) were harvested and resuspended in 100 µl 1× binding buffer and stained with 5 µl FITC-Annexin V and 5 µl PI for 15 min in the dark at room temperature. Cell apoptosis was quantified using Cell Quest version 0.9.13 alpha (BD Biosciences) and data were analyzed using FlowJo version 10.1 (FlowJo LLC.). Results were calculated from three independent experiments.
Confocal laser-scanning microscopy
The annexin V-FITC apoptosis detection kit uses double staining to identify apoptotic cells using a confocal laser-scanning microscope (Olympus Corporation). The staining procedure was the same as described above for flow cytometry, and a drop of the cell suspension was placed on a glass slide and observed under a confocal microscope. Confocal images were obtained from three different fields of view from three independent samples (magnification, ×200).
Statistics and repeatability of experiments
Data are presented as the mean ± standard deviation (s.d.) of at least three repeats, and all error bars indicate s.d. SPSS version 21.0 (IBM Corp.) and GraphPad Prism version 7.0 (GraphPad Software Inc.) were used to evaluate statistical significance. A one-way ANOVA was used to compare the mean values between three or more data sets, with a post-hoc Student-Newman-Keuls test or the non-parametric Tamhane T2 test to compare the mean of each data set with that of every other data set. Statistical comparisons of the means of two data sets were performed using an unpaired Student's two-tailed t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of Linc-OIP5, YAP1 and JAG1 is upregulated in MDA-MB-231 cells
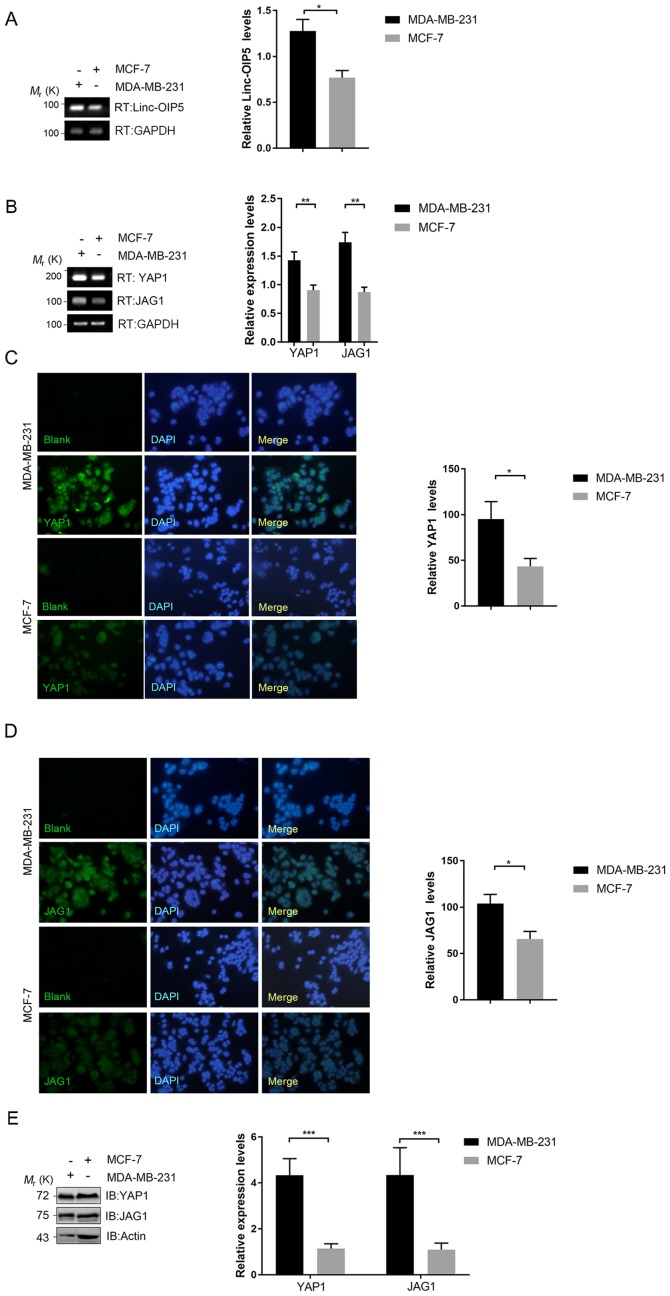
To understand the associations between Linc-OIP5, YAP1 and JAG1 in human breast cancer, their expression in breast cancer cells with different degrees of malignancy was determined using RT-qPCR. Linc-OIP5, YAP1 and JAG1 were all expressed in all breast cancer cells assessed at the mRNA level (Fig. 1A and B). The expression levels of Linc-OIP5 (P<0.05), YAP1 (P<0.01) and JAG1 (P<0.01) in MDA-MB-231 cells, which exhibit the highest degree of malignancy, were significantly higher compared with MCF-7 cells which are typically less malignant (Fig. 1A and B). Immunofluorescence and western blotting analyses further demonstrated the differential expression of YAP1 and JAG1 in the breast cancer cell lines at the protein level (Fig. 1C-E). The expression of YAP1 and JAG1 in MDA-MB-231 cells at the protein level was significantly higher compared with MCF-7 cells (P<0.05 and P<0.001, respectively) (Fig. 1C-E). Based on these data, MDA-MB-231 cells were used for further experimental analyses.
Figure 1.
Differential expression of Linc-OIP5, YAP1, and JAG1 in breast cancer cells with different degrees of malignancy. (A) Relative expression of Linc-OIP5 in MDA-MB-231 and MCF-7 cells. *P<0.05. (B) YAP1 and JAG1 expression levels were significantly higher in MDA-MB-231 cells compared with the MCF-7 cells. Expression was normalized to GAPDH. **P<0.01. Immunofluorescence analysis confirming that expression of (C) YAP1 and (D) JAG1 were significantly higher in MDA-MB-231 cells compared with MCF-7 cells. Immunofluorescence staining for YAP1 and JAG1 proteins (green) and nuclei (blue). *P<0.05; Magnification ×100. (E) Western blots showing that YAP1 and JAG1 expression levels cells were significantly upregulated in MDA-MB-231 cells compared with MCF-7 cells. Protein expression was normalized against β-actin. ***P<0.001.
Knockdown of Linc-OIP5 inhibits cell proliferation and increases apoptosis in MDA-MB-231 cells
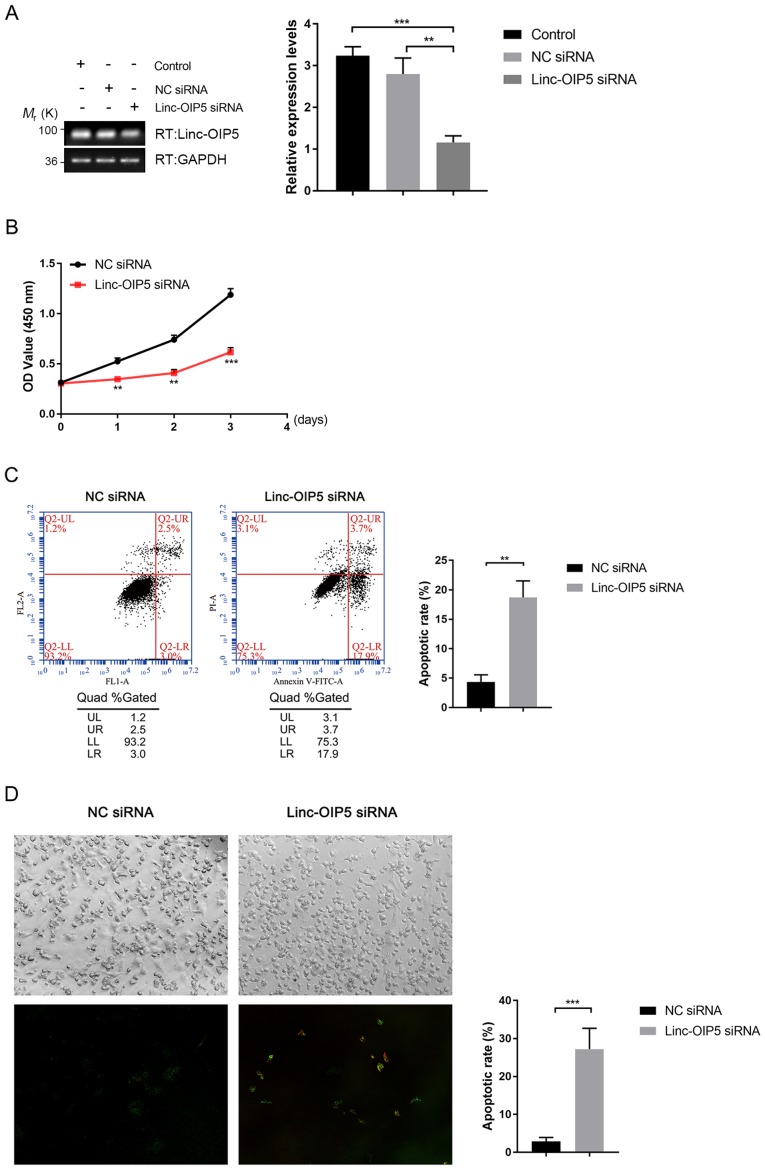
To determine the biological effects of Linc-OIP5 during the initiation and progression of breast cancer, its expression was knocked down in MDA-MB-231 cells using siRNA and the effects of knockdown on proliferation and apoptosis were investigated. RT-qPCR analysis showed that the mRNA expression levels of Linc-OIP5 was significantly decreased following transfection of Linc-OIP5 siRNA compared with NC-siRNA or mock-treated controls (P<0.01; Fig. 2A). The results of the cell proliferation (CCK-8) assays showed that Linc-OIP5 knockdown significantly decreased proliferation of MDA-MB-231 cells compared with the NC siRNA group after 24 h (P<0.01; Fig. 2B). Additionally, the rate of apoptosis in MDA-MB-231 cells following knockdown of Linc-OIP5 was detected using Annexin V-FITC/PI double staining combined with flow cytometry (Fig. 2C) and confocal laser-scanning microscopy (Fig. 2D). The results showed a significant increase in the rate of apoptosis and Annexin V staining when Linc-OIP5 was knocked down in MDA-MB-231 cells compared with the NC siRNA group (flow cytometry, P<0.01; confocal microscopy, P<0.001). These results suggest that Linc-OIP5 knockdown significantly inhibited the proliferation and promoted apoptosis of MDA-MB-231 cells.
Figure 2.
Knockdown of Linc-OIP5 decreases proliferation and promotes apoptosis in MDA-MB-231 cells. (A) Relative expression levels of Linc-OIP5 in cells transfected with Linc-OIP5 siRNA were significantly decreased compared with cells transfected with the NC siRNA. **P<0.01, ***P<0.001. (B) Knockdown of Linc-OIP5 significantly decreased the proliferative capacity of MDA-MB-231 cells compared with the NC siRNA transfected cells. **P<0.01, ***P<0.001. (C) Flow cytometry of Annexin V-FITC/PI double stained cells to determine the effect of Linc-OIP5 knockdown on apoptosis. **P<0.01. (D) Confocal laser-scanning microscopy showed a significant increase in the apoptotic rates of MDA-MB-231 cells transfected with Linc-OIP5 siRNA compared with NC siRNA, determined by FITC-labeled Annexin V (green) and PI (red) staining. Magnification, ×200. Values were normalized against the NC siRNA group. ***P<0.001. FL, fluorescence parameter; FITC, fluorescein isothiocyanate; PI, propidium iodide; NC, negative control; si, small interfering; UL, upper left, UR, upper right; LL, lower left; lower right.
Knockdown of Linc-OIP5 decreases cell migration and invasion in MDA-MB-231 cells
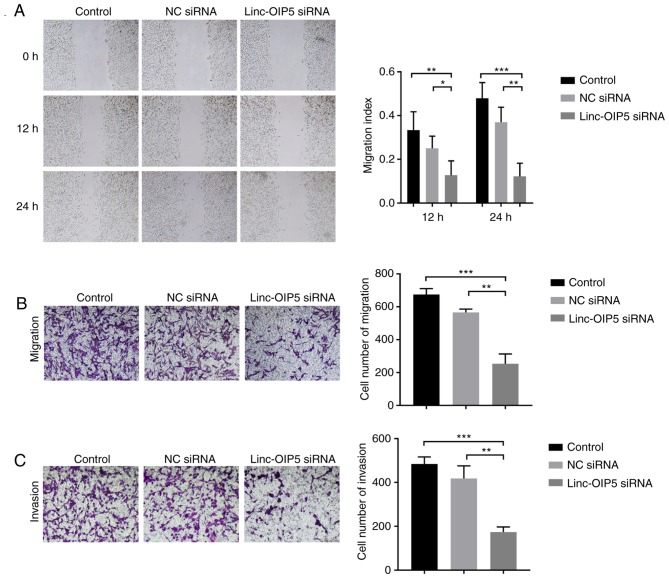
To determine the role of Linc-OIP5 in regulating the malignancy of MDA-MB-231 cells, cell migration and invasion were measured following Linc-OIP5 knockdown in vitro. Wound healing and transwell migration assays demonstrated that the migratory ability of MDA-MB-231 cells was reduced significantly following knockdown of Linc-OIP5. MDA-MB-231 cells transfected with Linc-OIP5 siRNA exhibited a decreased migratory capacity in the wound healing assay compared with the control group after 12 and 24 h (P<0.05 and P<0.01, respectively; Fig. 3A). Similarly, the results of the transwell migration assays showed that the number of cells which had migrated through the membrane was significantly decreased in cells transfected with Linc-OIP5 siRNA compared with the control group (P<0.05, P<0.01, respectively; Fig. 3B). Additionally, a transwell invasion assay was used to elucidate the effects of Linc-OIP5 knockdown on the invasive ability of MDA-MB-231 cells. The results showed that the number of cells which had invaded through the Matrigel was significantly decreased in the MDA-MB-231 cells transfected with Linc-OIP5 siRNA (P<0.01; Fig. 3C). These data demonstrate that Linc-OIP5 enhances the metastatic capacity of MDA-MB-231 breast cancer cells.
Figure 3.
Knockdown of Linc-OIP5 decreases migration and invasion of MDA-MB-231 cells. (A) Wound healing assays demonstrated a significant reduction in the migratory capacity of cells when Linc-OIP5 was knocked down. *P<0.05, **P<0.01, ***P<0.001; Magnification, ×40. (B) Transwell migration assays demonstrating that the number of cells which had migrated were significantly decreased in cells transfected with Linc-OIP5 siRNA. **P<0.01, ***P<0.001. Magnification, ×100. (C) Transwell invasion assays demonstrated that the number of cells which had invaded were significantly decreased in cells transfected with Linc-OIP5 siRNA. **P<0.01, ***P<0.001. Magnification, ×100. NC, negative control; si, small interfering.
Linc-OIP5 knockdown downregulates the expression of YAP1 and JAG1 in MDA-MB-231 cells
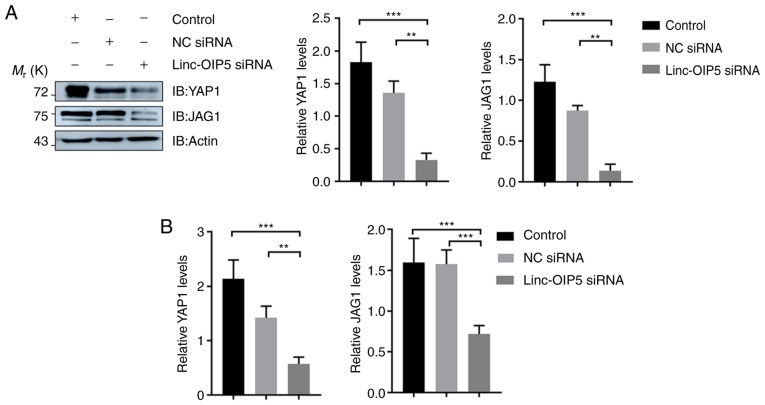
To determine the mechanisms underlying the effects of Linc-OIP5 in increasing the malignant potential of MDA-MB-231 breast cancer cells, the expression levels of YAP1 and JAG1 were measured in MDA-MB-231 cells transfected with Linc-OIP5 siRNA, relative to NC-siRNA- or mock-treated controls. The results of western blotting showed that Linc-OIP5 knock down significantly reduced the protein expression levels of YAP1 and JAG1 in MDA-MB-231 cells compared with the control group (P<0.01; Fig. 4A). Consistent with these findings, RT-qPCR analysis demonstrated that the YAP1 (P<0.01) and JAG1 (P<0.001) mRNA expression levels were also significantly reduced by knocking down Linc-OIP5 in MDA-MB-231 cells (Fig. 4B). Therefore Linc-OIP5 knockdown decreased YAP1 and JAG1 expression levels in MDA-MB-231 cells.
Figure 4.
YAP1 and JAG1 expression are downregulated in MDA-MB-231 cells transfected with Linc-OIP5 siRNA. (A) Western blotting confirmed that Linc-OIP5 knockdown significantly decreased the expression of YAP1 and JAG1 in MDA-MB-231 cells. Expression levels were normalized to β-actin. **P<0.01, ***P<0.001. (B) Reverse transcription-quantitative PCR demonstrated that the mRNA expression levels of YAP1 and JAG1 were prominently decreased in MDA-MB-231 cells treated with Linc-OIP5 siRNA. mRNA expression levels were normalized to GAPDH. **P<0.01, ***P<0.001. NC, negative control; si, small interfering.
Discussion
Several lncRNAs have emerged as important regulators of gene expression in mammary tumors. For example, loss of lncRNA Malat1 reduces the differentiation and metastatic capacity of mammary tumors (14), and lncRNA HOTAIR contributes to the development and tumorigenesis of breast cancer (3). Furthermore, Linc-OIP5 is closely associated with tumor initiation, progression and metastasis (17,18). Previous studies have shown that Linc-OIP5 contributes to the carcinogenic potential of lung adenocarcinoma and multiple myeloma (12,19). Based on unpublished data from our lab, Linc-OIP5 knockdown regulates the proliferation, migration and tube formation of endothelial cells when cocultured with breast cancer cells, suggesting that this molecule may be a promising marker for breast tumors. Additionally, Zeng et al (40) demonstrated that Linc-OIP5 was aberrantly expressed in breast cancer.
In the present study, to elucidate the functional effects of Linc-OIP5 on the development and progression of breast cancer, and the link between Linc-OIP5, YAP1 and JAG1, the expression levels of Linc-OIP5, YAP1, and JAG1 in breast cancer cell lines with different degrees of malignancy were determined. Expression of Linc-OIP5, YAP1, and JAG1 was shown to be highest in MDA-MB-231 cells, and thus, they were used for all subsequent experiments. Given that dysregulation of cell proliferation and apoptosis are hallmarks of cancer progression, the effects of Linc-OIP5 knockdown on cell proliferation and apoptosis in MDA-MB-231 cells were evaluated. It was demonstrated that Linc-OIP5 knockdown significantly increased proliferation and promoted apoptosis. Therefore, Linc-OIP5 may serve as an oncogene in breast cancer and its effects on cell proliferation may be associated with regulation of apoptosis. It should be noted however, that the green staining observed in the immunofluorescence experiments was not typical of Annexin-V staining. A possible explanation for this may be the uneven distribution of apoptotic protein (Annexin V) in MDA-MB-231 breast cancer cells. Furthermore, there is a misalignment between the bright-field and fluorescence images. This misalignment was the result of using two separate microscopes, as the confocal laser-scanning microscope used does not possess bright-field optics. Migration and invasion are considered to mediate the malignancy and metastatic capacity of tumors (2). To determine the role of Linc-OIP5 in metastasis of MDA-MB-231 breast cancer cells, the migratory and invasive capacities of the cells following knockdown of Linc-OIP5 were examined. The results showed that Linc-OIP5 knockdown resulted in a significant reduction of cell migration and invasion, suggesting that Linc-OIP5 may serve as an oncogenic regulator which increases the malignant behaviors of MDA-MB-231 breast cancer cells, and that inhibiting Linc-OIP5 function may suppress the progression and metastasis of breast cancer.
The molecular mechanism underlying the effects of Linc-OIP5 on breast cancer cells were investigated in this study. It has been suggested that YAP1 and JAG1 are associated with progression and recurrence in breast cancer (31–37). YAP1 acts upstream of the Notch pathway, as well as upregulating JAG1 expression (20,25,38). Based on the above studies, it was hypothesized that Linc-OIP5 may influence the proliferation, migration, invasion and apoptosis of MDA-MB-231 breast cancer cells, and may act upstream of YAP1/JAG1 signaling. The expression of YAP1 and JAG1 were determined in MDA-MB-231 breast cancer cells treated with or without Linc-OIP5 siRNA. The results suggested that both the mRNA and protein expression levels of YAP1 and JAG1 were significantly lower in MDA-MB-231 cells treated with Linc-OIP5 siRNA compared with the control, indicating that the effect of Linc-OIP5 on the malignant behaviors of MDA-MB-231 cells was partially associated with YAP1/JAG1 signaling. Together, the results indicate that Linc-OIP5 may act upstream of YAP1/JAG1 signaling to affect the proliferation, apoptosis, invasion and migration of MDA-MB-231 breast cancer cells.
In order to demonstrate a causal link between silencing of Linc-OIP5 and the lower expression levels of YAP1 and JAG1, rescue experiments were performed (data not shown). Unfortunately, the experimental results were not as expected. After consulting with a Plasmid Construction Company, the unexpected results may have been due to the use of a DNA construct >8,000 bp for Linc-OIP5 overexpression. This may have been too long, and ideally a <1,000 bp should be used as this typically leads to a higher success rate (Personal communication with the manufacturer). Therefore, establishing a causal link between Linc-OIP5 and YAP1/JAG1 will be the aim of future experiments. Future experiments should also investigate the function of Linc-OIP5 in EMT and in in vivo studies, possibly using xenograft mice models. EMT refers to the transformation of epithelial cells to mesenchymal cells under certain conditions, providing these cells with increased invasive and migratory capabilities (41,42). EMT is generally viewed as one of the primary mechanisms by which invasion and metastasis is increased (43). Therefore future research should establish the association between Linc-OIP5 and EMT to ascertain whether the effects of Linc-OIP5 observed in vitro translates to in vivo.
In conclusion, the results of the present study demonstrate that Linc-OIP5 is upregulated in MDA-MB-231 breast cancer cells. In addition, Linc-OIP5 knockdown inhibits MDA-MB-231 cell proliferation, migration and invasion, while inducing apoptosis, at least in part, through YAP1/JAG1 signaling. Linc-OIP5 may regulate JAG1 signaling through YAP1 signaling. Regulation of YAP1 and JAG1 by Linc-OIP5 may be a novel mechanism of oncogenic signal regulation in breast cancer. Therefore, Linc-OIP5 may serve as a breast cancer oncogene and may be a suitable therapeutic target for treatments aimed at preventing tumor progression and metastasis in patients with breast cancer.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CCK-8
Cell Counting Kit-8
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- lincRNAs
long intervening noncoding RNAs
- lncRNAs
long noncoding RNAs
- NC
negative control
- RT-qPCR
reverse transcription-quantitative PCR
- siRNA
small interfering RNA
- s.d.
standard deviation
Funding
The present study was supported by funding from the Project on Basic Scientific Research for Higher Education Institutions affiliated to Heilongjiang Province (Heilongjiang, China; grant no. 2018-KYYWFMY-0006).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SG and QZ designed the study. QZ and YY performed the experiments. QZ, XD, YL and HW analyzed the data. QZ wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lobba ARM, Carreira ACO, Cerqueira OLD, Fujita A, DeOcesano-Pereira C, Osorio CAB, Soares FA, Rameshwar P, Sogayar MC. High CD90 (THY-1) expression positively correlates with cell transformation and worse prognosis in basal-like breast cancer tumors. PLoS One. 2018;13:e0199254. doi: 10.1371/journal.pone.0199254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan S, Fan C, Liu N, Huang K, Fang X, Wang K. Downregulation of the long non-coding RNA ZFAS1 is associated with cell proliferation, migration and invasion in breast cancer. Mol Med Rep. 2018;17:6405–6412. doi: 10.3892/mmr.2018.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7:842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Wang P, Wang F, Wang L, Pan JH. Proprotein convertase subtilisin/kexin type 6 activates the extracellular signal-regulated kinase 1/2 and Wnt family member 3A pathways and promotes in vitro proliferation, migration and invasion of breast cancer MDA-MB-231 cells. Oncol Lett. 2018;16:145–150. doi: 10.3892/ol.2018.8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang Y, Chen H, Ji L, Du J, Xie X, Li X, Lou Y. Talin2 regulates breast cancer cell migration and invasion by apoptosis. Oncol Lett. 2018;16:285–293. doi: 10.3892/ol.2018.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen Y, Ji Y, Zhang Y, Jiang B, Tang C, Wang Q, Chen X, Jia L, Gu W, Xu X. Knockdown of Yes-associated protein induces the apoptosis while inhibits the proliferation of human periodontal ligament stem cells through crosstalk between Erk and Bcl-2 signaling pathways. Int J Med Sci. 2017;14:1231–1240. doi: 10.7150/ijms.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang N, Ke B, Hjortjensen K, Iglesiasgato D, Wang Z, Chang PC, Zhao Y, Niu XD, Wu T, Peng B, et al. YAP1 regulates prostate cancer stem cell-like characteristics to promote castration resistant growth. Oncotarget. 2017;8:115054–115067. doi: 10.18632/oncotarget.23014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Lin X, Xu J, Jing H, Qin Y, Li Y. SULT1E1 inhibits cell proliferation and invasion by activating PPARγ in breast cancer. J Cancer. 2018;9:1078–1087. doi: 10.7150/jca.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 10.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 11.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:e359–e386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 12.Deng J, Deng H, Liu C, Liang Y, Wang S. Long non-coding RNA OIP5-AS1 functions as an oncogene in lung adenocarcinoma through targeting miR-448/Bcl-2. Biomed Pharmacother. 2018;98:102–110. doi: 10.1016/j.biopha.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Mendell JT. Targeting a long noncoding RNA in breast cancer. N Engl J Med. 2016;374:2287–2289. doi: 10.1056/NEJMcibr1603785. [DOI] [PubMed] [Google Scholar]
- 14.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergmann JH, Spector DL. Long non-coding RNAs: Modulators of nuclear structure and function. Curr Opin Cell Biol. 2014;26:10–18. doi: 10.1016/j.ceb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Ann Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey GK, Kanduri C. Long noncoding RNAs and neuroblastoma. Oncotarget. 2015;6:18265–18275. doi: 10.18632/oncotarget.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang N, Chen J, Zhang H, Wang X, Yao H, Peng Y, Zhang WG. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017;8:e2975. doi: 10.1038/cddis.2017.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu N, Nguyen Q, Wan Y, Zhou T, Venter J, Frampton GA, DeMorrow S, Pan D, Meng F, Glaser S, et al. The Hippo signaling functions through the Notch signaling to regulate intrahepatic bile duct development in mammals. Lab Invest. 2017;97:843–853. doi: 10.1038/labinvest.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-King H, García NA, Ontoria-Oviedo I, Ciria M, Montero JA, Sepúlveda P. Hypoxia Inducible Factor-1α potentiates Jagged1-mediated angiogenesis by mesenchymal stem cell-derived exosomes. Stem Cells. 2017;35:1747–1759. doi: 10.1002/stem.2618. [DOI] [PubMed] [Google Scholar]
- 22.Sheldon H, Heikamp E, Turley H, Dragovic R, Thomas P, Oon CE, Leek R, Edelmann M, Kessler B, Sainson RC, et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;116:2385–2394. doi: 10.1182/blood-2009-08-239228. [DOI] [PubMed] [Google Scholar]
- 23.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 24.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, Camargo FD. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos A, Camargo FD. The Hippo signaling pathway and stem cell biology. Trends Cell Biol. 2012;22:339–346. doi: 10.1016/j.tcb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mammoto A, Muyleart M, Kadlec A, Gutterman D, Mammoto T. YAP1-TEAD1 signaling controls angiogenesis and mitochondrial biogenesis through PGC1α. Microvas Res. 2018;119:73–83. doi: 10.1016/j.mvr.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oon CE, Bridges E, Sheldon H, Sainson RCA, Jubb A, Turley H, Leek R, Buffa F, Harris AL, Li JL. Role of Delta-like 4 in Jagged1-induced tumour angiogenesis and tumour growth. Oncotarget. 2017;8:40115–40131. doi: 10.18632/oncotarget.16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benedito R, Roca C, Sörensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 30.Aspalter IM, Gordon E, Dubrac A, Ragab A, Narloch J, Vizán P, Geudens I, Collins RT, Franco CA, Abrahams CL, et al. Alk1 and Alk5 inhibition by Nrp1 controls vascular sprouting downstream of Notch. Nat Comm. 2015;17:7264. doi: 10.1038/ncomms8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin C, Xu X. YAP1-TEAD1-Glut1 axis dictates the oncogenic phenotypes of breast cancer cells by modulating glycolysis. Biomed Pharmacother. 2017;95:789–794. doi: 10.1016/j.biopha.2017.08.091. [DOI] [PubMed] [Google Scholar]
- 32.Wang T, Mao B, Cheng C, Zou Z, Gao J, Yang Y, Lei T, Qi X, Yuan Z, Xu W, Lu Z. YAP promotes breast cancer metastasis by repressing growth differentiation factor-15. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1744–1753. doi: 10.1016/j.bbadis.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 33.Wu Q, Li J, Sun S, Chen X, Zhang H, Li B, Sun S. YAP/TAZ-mediated activation of serine metabolism and methylation regulation is critical for LKB1-deficient breast cancer progression. Biosci Rep. 2017;37(pii):BSR20171072. doi: 10.1042/BSR20171072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou L, Chen L, Fang L. Scutellarin inhibits proliferation, invasion, and tumorigenicity in human breast cancer cells by regulating HIPPO-YAP signaling pathway. Med Sci Monit. 2017;23:5130–5138. doi: 10.12659/MSM.904492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selcuklu SD, Donoghue MT, Kerin MJ, Spillane C. Regulatory interplay between miR-21, JAG1 and 17beta-estradiol (E2) in breast cancer cells. Biochem Biophys Res Commun. 2012;423:234–239. doi: 10.1016/j.bbrc.2012.05.074. [DOI] [PubMed] [Google Scholar]
- 36.Dickson BC, Mulligan AM, Zhang H, Lockwood G, O'Malley FP, Egan SE, Reedijk M. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 37.Reedijk M, Pinnaduwage D, Dickson BC, Mulligan AM, Zhang H, Bull SB, O'Malley FP, Egan SE, Andrulis IL. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008;111:439–448. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 38.Slemmons KK, Crose LES, Riedel S, Sushnitha M, Belyea B, Linardic CM. A novel Notch-YAP circuit drives stemness and tumorigenesis in embryonal rhabdomyosarcoma. Mol Cancer Res. 2017;15:1777–1791. doi: 10.1158/1541-7786.MCR-17-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Zeng H, Wang J, Chen T, Zhang K, Chen J, Wang L, Li H, Tuluhong D, Li J, Wang S. Downregulation of long non-coding RNA Opa interacting protein 5-antisense RNA 1 inhibits breast cancer progression by targeting sex-determining region Y-box 2 by microRNA-129-5p upregulation. Cancer Sci. 2019;110:289–302. doi: 10.1111/cas.13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 42.Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: A cancer researcher's conceptual friend and foe. Am J Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ansieau S, Courtois-Cox S, Morel AP, Puisieux A. Failsafe program escape and EMT: A deleterious partnership. Semin Cancer Biol. 2011;21:392–396. doi: 10.1016/j.semcancer.2011.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.