Abstract
Breast cancer is the most common malignant tumor in women in the majority of countries, such as China, Britain and Australia, and its morbidity and mortality rates remain very high. Long non-coding RNAs (lncRNAs) are non-coding RNAs (ncRNAs) >200 nucleotides in length that lack open reading frames. LncRNA H19 is a transcription product of the H19 gene, and the aberrant expression of H19 can be demonstrated in various types of tumor cell. The purpose of the present review was to elaborate the role of H19 in breast cancer. H19 can regulate gene expression in breast cancer at multiple levels, including epigenetic, transcriptional and posttranscriptional. The abnormal expression of H19 is closely associated with the tumorigenesis and progression of breast cancer via different underlying molecular mechanisms, such as encoding microRNA-675, competing endogenous RNA regulation and interacting with MYC. A large number of clinical studies have suggested that H19 can serve as a potential biomarker for the diagnosis of breast cancer. High expression levels of H19 increases the drug resistance of breast cancer cells and is associated with poor prognosis within patients with breast cancer. Therefore, serum H19 levels may have momentous significance in the clinical setting.
Keywords: breast cancer, long non-coding RNA, H19, biological markers, drug resistance
1. Introduction
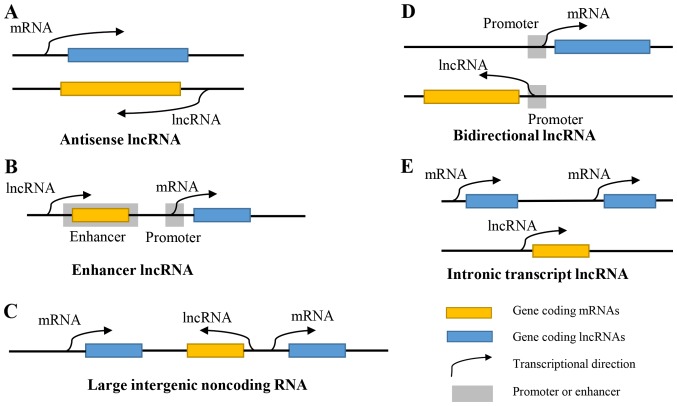
Approximately 93% of human genome DNA is transcribed into RNAs, but <2% of these nucleotide sequences can code for proteins, while the other 98% are non-coding RNAs (ncRNAs) that partially or completely lack the ability to be coded into proteins. The majority of these ncRNAs are known as long non-coding RNAs (lncRNAs) whose length exceeds 200 nucleotides (1). LncRNAs were at first regarded as the ‘noise’ of gene transcription. According to the position where they are relative to the protein-coding genes, lncRNAs can be roughly divided into antisense lncRNAs, enhancer lncRNAs, large intergenic non-coding RNAs, bidirectional lncRNAs and intronic transcript lncRNAs (2) (Fig. 1). With the existence and development of next-generation sequencing, lncRNAs have been demonstrated to affect multiple biological processes, including housekeeping functions and specialized functions, such as genomic imprinting and dosage compensation (3). LncRNAs can regulate gene expression via multiple mechanisms, including at the epigenetic, transcriptional and post-transcriptional levels (4). They participate in the regulation of a variety of cell activities, such as cell differentiation, proliferation, invasion, apoptosis and autophagy. through interacting with RNAs, DNAs or proteins (5).
Figure 1.
Classification and transcriptional processes of lncRNAs. (A) Antisense lncRNAs are endogenous lncRNAs that partly or completely complement other transcriptional sequences. (B) Enhancer lncRNAs are produced by the enhancers of protein-coding genes. (C) Large intergenic non-coding RNAs are transcribed independently by sequences located between protein-coding genes. (D) Bidirectional lncRNAs share the same promoter region as the protein-coding genes, but the transcriptional directions of the gene are reversed. (E) Intronic transcript lncRNAs are produced from the introns of genes. lncRNA, long non-coding RNA.
The H19 gene is located in 11p15.5, an imprinted region of chromosome 11, being adjacent to the insulin-like growth factor 2 (IGF2) gene, and is expressed only from the maternally inherited chromosome, while IGF2 is expressed only from the paternally inherited chromosome. H19, a lncRNA, is the transcription product of the H19 gene, and diversified transcript variants exist due to alternative splicing (6). Although H19 RNA molecules can be detected in both the cytoplasm and nucleus, H19 RNA primarily exists in cytoplasm. H19 functions in the form of regulatory RNA or ribosome regulators. H19 promotes biological processes such as apoptosis, angiogenesis, inflammation and cell death (7). Furthermore, Gene Ontology (GO) analyses predicted that H19 is connected with neurogenesis, angiogenesis and inflammation through DNA transcription, RNA folding, methylation and gene imprinting (8). The aberrant expression of H19 is associated with multiple diseases, including carcinoma, sarcoma, type 2 diabetes and hypertrophic cardiomyopathy (9–12). The present review primarily summarizes the role of H19 in the carcinogenesis and development of breast cancer.
2. H19 and cancer
Abnormal expression of H19 has been demonstrated in a variety different types of cancer cells, such as gastric cancer (13), pancreatic cancer (14), liver cancer (15) and breast cancer (16), affecting the development and progression of cancer through various mechanisms. H19 is an estrogen-regulated transcript, and a wide array of studies have confirmed that H19 regulates cell differentiation and proliferation (6). In addition, research into its application in the clinical setting revealed that H19 had the potential to serve as a biomarker of cancer, and had promise as a novel therapeutic option in oncotherapy (6,7). However, whether H19 functions as a tumor promoter or a tumor suppressor remains controversial (17). It is possible that H19 plays differential roles depending on the tissue type and developmental stage (6). Therefore, the clear-cut mechanisms underlying how H19, as an lncRNA, regulates the biological progression of cancer requires further investigation. The various roles of H19 in different kinds of cancers are briefly summarized in Table I.
Table I.
Expression and functional mechanisms of H19 in different types of cancer.
| Cancer type | Expression level | Mechanisms | (Refs.) |
|---|---|---|---|
| Lung cancer | Increased | i) H19 depresses miR-196b to elevate LIN28B; ii) H19 serves as a ceRNA and attaches miR-17 to modulate STAT3 expression; induces iii) H19 promotes EMT by downregulating miR-484; iv) H19 cisplatin resistance in patients | (81–84) |
| Gastric cancer | Increased | i) H19 functions as a ceRNA to regulate HER2 expression by suppressing let-7c expression; ii) H19-PEG10/IGF2BP3 axis promotes EMT in gastric cancer; iii) H19/miR-675 axis inhibits the expression of FADD and the downregulation of FADD inhibits the caspase cleavage cascades including caspase 8 and caspase 3 | (13,85,86) |
| Pancreatic cancer | Increased | i) H19/miR-675/E2F-1 regulatory loop affects the cell cycle; ii) H19 increases HMGA2-mediated EMT through antagonizing let-7 | (14,87) |
| Liver cancer | Increased | i) H19 targets miR-193a-3p and regulates PSEN1 expression, which influences the survival rates and proliferative abilities of HCC cells; ii) aberrant TGF-β/H19 signaling axis via Sox2 in TICs that regulates hepatocarcinogenesis; iii) abnormal regulation of H19 results in biallelic expression of IGF2, leading to exceptional cell proliferation | (15,88,89) |
| Colorectal cancer | Increased | i) Overexpression of H19 activates the RAS-MAPK signaling pathway, promoting invasion and metastasis of colorectal cancer; ii) H19 induces the EMT process in colon cancer cells; iii) H19 sponges miR-138 to upregulate the expression of HMGA1, enhancing the invasion and migration of colon cancer; iv) H19 competitively binds to miR-200a and depresses the expression of β-catenin in colorectal cancer | (90–93) |
| Endometrial cancer | Increased | i) Overexpression of H19 regulates the expression of HOXA10 via targeting miR-612, promoting cell proliferation of endometrial cancer; ii) H19 modulates EMT process, reinforcing the aggressiveness of endometrial cancer; iii) H19 acts as a sponge to bind let-7, leading to high expression of IGF1R and therefore promotes endometrial stromal hyperplasia | (94–96) |
| Breast cancer | Increased | i) H19 derives miR-675 and contributes to the development of breast cancer; ii) H19 functions as a ceRNA of various miRNAs and regulates the expression of corresponding target genes; iii) association between MYC and H19 plays an important role in the progression of breast cancer | (20–44,53–58) |
| Bladder cancer | Increased | i) High expression of H19 inhibits E-cadherin expression and strengthens metastasis of bladder cancer; ii) H19 acts as a ceRNA to sponge miR-29b-3p and promotes the expression of DNMT3B, resulting in metastasis and EMT of bladder cancer; iii) H19 increases miR-675 expression, which can inhibit the activation of p53 and reduce the expression of Bax/Bcl-2 and cyclin D1, leading to bladder cancer cell proliferation; iv) H19 increases the expression of ID2 and promotes bladder cancer growth | (97–100) |
| Prostate cancer | Decreased/increased | i) H19 and miR-675 is significantly downregulated in metastatic prostate cancer cells. miR-675 binds with 3′UTR of TGFBI mRNA and decreases the translation of TGFBI; ii) High expression of H19 is associated with the process of neuroendocrine transdifferentiation (NEtD) in prostate cancer | (101–102) |
miR, microRNA; ceRNA, competing endogenous RNA; EMT, epithelial-to-mesenchymal transition; HER2, human epidermal growth factor receptor 2; FADD, fas-associated via death domain; HMG, high-mobility group; PSEN1, presenilin-1; HCC, hepatocellular carcinoma; TGF-β, tumor growth factor-β; TICs, tumor-initiating hepatocytes; IGF2, insulin-like growth factor 2; RAS, rat sarcoma; MAPK, mitogen-activated protein kinase; DNMT3B, DNA-methylase 3β; ID2, inhibitor of DNA binding/differentiation 2; 3′UTR, 3′untranslated region; NEtD, neuroendocrine transdifferentiation.
3. H19 is involved in the tumorigenesis and progression of breast cancer
Breast cancer is a malignant tumor deriving from the epithelium of the mammary gland, and >99% of breast cancer cases occur in women worldwide; less than 1% (0.02%) of all breast cancer cases develop in men (18). According to data released by Globocan in 2012, the standardized mortality rate of breast cancer in the developed world was 149/100,000, and that in the less developed world was 115/100,000 (19). The global incidence and mortality rate of breast cancer increased in the late 1970s, while the mortality rate indicated signs of decline in the 1990s due to the introduction of breast cancer screening and comprehensive treatment. Metastasis is the leading cause of mortality in patients with breast cancer. The H19 gene is an oncogene in breast cancer and is highly expressed in cancer tissues compared with normal tissues. The expression level of H19 is associated with the oncogenesis, proliferation, invasion, metastasis and drug resistance of breast cancer, but the underlying molecular mechanisms are very different (7). Thus, serum H19 levels may possess clinical significance in the early diagnosis, treatment and prognosis of breast cancer.
H19 promotes breast tumorigenesis
A recent case-control study in China revealed that high expression levels of H19 were associated with an increased risk of breast carcinogenesis in both codominant and dominant models, and the association was more apparent in patients with estrogen receptor-positive (ER+), human epidermal growth factor receptor 2-negative (HER2-), and ER+-HER2-negative (HER2-) molecular subtypes (20). In addition, two single nucleotide polymorphisms (SNPs) were identified, and another study suggested that the expression levels of H19 in breast cancer cells with the rs2071095 CC genotype were significantly higher than those with the AA genotype, and that the CC genotype was markedly associated with an increased risk of breast cancer compared with the CA + AA genotypes (21). Furthermore, animal experiments demonstrated that the probability of breast carcinogenesis was increased in severe combined immunodeficiency mice injected with cells overexpressing the H19 gene (22).
The molecular mechanisms underlying H19-associated carcinogenesis may involve several aspects. The H19 promoter was activated by E2F transcription factor 1 (E2F1), which promoted cell cycle progression (particularly in the S-phase) of MCF-7 cells (23). Furthermore, H19 contributed to the epigenetic regulation of gene expression in breast cancer. H19 bound to and inhibited S-adenosylhomocysteine hydrolase, the sole enzyme that can hydrolyze S-adenosylhomocysteine (SAH) in humans. SAH can markedly suppress S-adenosylmethionine-dependent methyltransferases, which can methylate multiple cellular components, including DNA, RNAs and proteins, through a feedback mechanism. H19 knockdown increased the DNMT3B-mediated methylation of Nctc1, a gene encoding lncRNAs, within the Igf2-H19-Nctc1 locus. Thus, H19 altered DNA methylation and led to breast tumorigenesis (24). A new lncRNA within the H19/IGF2 locus named 91H is an antisense gene to H19. The 91H lncRNA also regulated the expression levels of H19 and IGF2 by epigenetic modifications and increased the tumorigenic properties of MDA-MB-231 cells both in vitro and in vivo (25). In summary, H19 plays an oncogenic role in breast tumorigenesis.
H19 encoding microRNA (miR)-675 contributes to the development of breast cancer
H19 is a precursor for miR-675 and generates two functional miRNAs, miR-675-5p and miR-675-3p (26). miR-675 is harbored in the first exon of H19 and miR-675 regulated by H19 was originally demonstrated to restrict the growth of the placenta prior to birth (27). A study of miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues demonstrated that the expression of miR-675 was significantly higher in the FFPE tissues of patients with breast cancer compared with the control group. However, the expression levels were not associated with age, lymph node stage, ER status or progestrogen receptor (PR) status. The frequency of miR-675 overexpression was higher in patients with low histological grade (histological grade I–II) cancer. In addition, there was a strong association between miR-675 and miR-24/93/98/378 levels in patients with breast cancer (28). In a prospective cohort study, DNA methylation levels of 517 miRNA-encoding genes were analyzed in the prediagnostic peripheral leukocytes of patients with colorectal cancer (n=159) or breast cancer (n=166). In the subjects who developed breast cancer, 8 miRNAs, including miR-675, were aberrantly methylated, while there were no significant associations in those with colorectal cancer. This aberrant methylation significantly affected the development of breast cancer (29). A luciferase assay revealed that H19/miR-675-5p decreased the expression levels of the ubiquitin ligase E3 family (c-Cbl and Cbl-b), and activated epidermal growth factor receptor (EGFR) and c-Met, enhancing the proliferation and migration of MDA-MB-231 breast cancer cells through the protein kinase B (Akt) and extracellular signal-regulated kinase (Erk) pathways (30). Furthermore, miR-675 was also involved in the mechanism through which H19 promoted Slug expression accompanied by the suppression of E-cadherin, and thereby increased the potential of cell invasion and metastasis in breast cancer (31). Huaier extract, a traditional Chinese medicine, decreased the expression levels of H19 and miR-675-5p and increased the expression levels of calcineurin B-like (CBL) protein. Huaier extract decreased the viability and increased the apoptosis rates of MDA-MB-231 and MDA-MB-468 cells by regulating the approach of the H19/miR-675-5p/CBL axis. Huaier extract was expected to be a novel clinical treatment for breast cancer (32). In summary, the ectopic expression of H19/miR-675 reinforced the aggressive phenotype of breast cancer cells. This abnormal expression aggrandized cell proliferation, cell migration, tumor growth and metastasis.
Competing endogenous RNA (ceRNA) regulatory network of H19 plays a vital role in the development of breast cancer
Multitudinous RNA transcripts have miRNA binding sites, which are also known as miRNA response elements (MREs). These RNA transcripts include mRNAs, pseudogene transcripts, lncRNAs and circular RNAs (circRNAs). They can compete with each other to bind to a shared miRNA through a common MRE, functioning as ceRNAs. There are multiple MREs on mRNA transcripts of protein-coding genes, and miRNAs can bind to corresponding mRNAs through MREs, leading to the degradation of mRNAs or inhibition of mRNA translation. Thus, RNA transcripts acting as ceRNAs can weaken the negative regulation of miRNA on target genes, promoting the expression of target genes. The example of lncRNAs as ceRNAs was first described in plants. LncRNAs indirectly regulate the expression level and translation of mRNAs through the bridge of MREs, so as to regulate the biological process of cells (33–36). H19 acted as a sponge for miR-200b/c and let-7b and promoted the expression of miRNA targets GIT2 and CYTH3, which motivated breast cancer cell migration through the epithelial-to-mesenchymal transition (EMT) and dissemination of breast cancer cells (37). In addition, H19 functioned as a ceRNA and bound to miR-152, which directly targeted DNA methyltransferase 1 (DNMT1). The overexpression of H19 diminished the inhibitory effect of miR-152 on DNMT1 expression and thereby promoted the proliferation and invasion of MCF-7 and MDA-MB-231 cells (38). H19 was indicated to be a target of miR-93-5p, and this molecule upregulated the expression levels of signal transducers and activators of transcription 3 (STAT3), a target of miR-93-5p. STAT3 significantly promotes the proliferation, migration and invasion of MCF-7 and MDA-MB-231 cells (39). H19 may also function as a ceRNA to sponge miRNA let-7a/b, resulting in the high expression level of the transcriptional factor LIN28. H19, let-7a/b and LIN28 formed a double-negative feedback loop that profoundly influenced the migration of breast cancer stem cells (BCSCs) (40). This ceRNA regulating effect released hypoxia-inducible factor 1α, leading to an increase of pyruvate dehydrogenase kinase 1, which influenced BCSC reprogramming and malignancy (41). In addition, let-7-regulated-Wnt signaling pathway activity was enhanced by the inhibitory effect of H19 on miR-146, which decreased asymmetric division and increased the self-renewal of BCSCs. H19 had a positive feedback regulatory effect on let-7 (42). Inversely, the knockdown of H19 reactivated let-7c, restraining the oestrogen receptor activated Wnt signaling pathway and inhibited the symmetric division of BCSCs (43).
Certain pseudogene transcripts also act as ceRNAs and affect H19 expression in breast cancer cells. For instance, HMGA1P7 mRNA, a transcription product of the HMGA1 gene, may increase the expression levels of H19 and IGF2 by a ceRNA mechanism and consequently accelerate cancer progression (44).
Similar regulatory mechanisms exist in other types of tumor, such as H19/miR-29b-3p in lung adenocarcinoma (45) and multiple myeloma (46), H19/miR-324-5p in ovarian carcinoma (47), H19/miR-194 in pancreatic carcinoma (48), H19/miR-106b-5p in seminoma (49), H19/miR-3126-5p in papillary thyroid carcinoma (50), H19/miR-326 in hepatocellular carcinoma (51) and H19/miR-194-5p in colorectal adenocarcinoma (52).
Thus, the ceRNA regulatory network of H19 plays a pivotal role in the proliferation, invasion and metastasis of breast cancer.
Interaction between MYC and H19 has profound impacts on the development of breast cancer
The oncogene MYC, including c-Myc, n-Myc and l-Myc, is deregulated in various types of tumor cell, including breast cancer cells. The overexpression of H19 influenced the let-7-mediated regulation of certain metastasis-promoting genes, including c-Myc (53). c-Myc protein bound to evolutionarily conserved E-boxes close to the imprinting control region to promote the histone acetylation and transcriptional initiation of the H19 promoter, distinctly inducing H19 expression (54). Furthermore, the c-Myc mRNA coding region instability determinant binding protein (CRD-BP) may bind to c-Myc and H19 and influence RNA stability, localization and translation. CRD-BP has been demonstrated to be associated with tumorigenesis, and experimental evidence suggested that CRD-BP was a proto-oncogene (55). Vigilin, a multi-K homology-domain protein, was present at several target sites of a zinc finger protein named CCCTC-binding factor, including promoter regions of c-Myc and several regions within the Igf2/H19 locus (56). Apart from vigilin, double-strand-break repair protein RAD21 and structural maintenance of chromosome protein 1 also bound to such regions and were associated with complicated chromatin organization and gene regulation (57). Furthermore, n-Myc has been demonstrated as a forceful glycogen synthase kinase-3-dependent regulator of DNA methyltransferase 3α 2 expression, changing the expression of H19 and the DNA methylation of corresponding imprinted loci via influencing signal transduction (58), which altered the biological process of breast cancer cells. In conclusion, the association between oncogene MYC and H19 expression plays a large role in the oncogenesis, proliferation, invasion and metastasis of breast cancer.
4. Potential clinical value of H19 in breast cancer
H19 serves as a potential biomarker for the diagnosis of breast cancer
The overexpression of H19 is associated with cells exhibiting higher tumorigenic phenotypes (22), which indicates that H19 expression levels can be used in the clinical diagnosis of breast cancer. A statistical analysis of clinical data revealed that H19 expression was significantly increased in breast cancer biopsies and plasma compared with that in the control group, and there was a significant correlation between the levels of plasma H19 and ER, PR, c-erbB-2 and lymph node metastasis in breast cancer. The diagnostic value of H19 for breast cancer was higher than carbohydrate antigen 153 and carcinoembryonic antigen (59). Reverse transcription-quantitative PCR was conducted in 12 lncRNAs that were confirmed as abnormally expressed in breast cancer cells, and the experimental results demonstrated that H19 was one of three lncRNAs (H19, HOTAIR and RP11-445H22.4) that notably increased in patients with breast cancer compared with the other nine lncRNAs (60). Furthermore, chromogenic in situ hybridization (CISH) was performed using the breast cancer tissues of 52 patients. The expression levels of H19 were disparate in different types of breast cancer. H19 was expressed in invasive breast carcinoma, ductal carcinoma in situ and normal adjacent tissue from higher to lower level, respectively (61). These data suggested that CISH with H19 is a potential clinical diagnostic assay with the ability to accurately classify breast cancer. In short, H19 possesses marked diagnostic significance and may serve as a potential biomarker for the early diagnosis of breast cancer.
H19 expression is negatively correlated with the prognosis of breast cancer
The expression levels of H19 were associated with tumor size, lymph node status and hormone negativity. Patients with breast cancer who exhibited high H19 expression levels had relatively unfavorable disease-free survival and overall survival (OS). The most prominent effect of H19 expression on clinical cancer prognosis was observed in triple-negative breast cancer (TNBC) (62). Another study examined the association between SNPs tagging the low-risk breast cancer loci in or near eight lncRNAs and the prognosis of breast cancer. The results revealed that only SNP rs2107425 near H19 were significantly associated with shorter metastasis-free survival (P=0.006 in univariate analysis; P=0.004 in multivariate analysis) (63). The postsurgical expression levels of H19 were significantly decreased in 82.1% of patients with breast cancer (32/39) compared with those in paired preoperative plasma (64), which indicated that the expression levels of H19 may reflect the curative effect of radical mastectomy. In summary, a negative correlation exists between the expression levels of H19 and the prognosis of patients with breast cancer.
Overexpression of H19 increases the drug resistance of breast cancer cells
H19 is a downstream target molecule of ERα, and the expression of ERα has been demonstrated to alter H19 levels, which decreases cell apoptosis in response to paclitaxel (PTX) treatment by suppressing the expression of the pro-apoptotic genes BIK and NOXA (65). Concrete mechanisms may include the inhibiting effect of H19 on the promoter activity of BIK via recruiting enhancer of zeste homolog 2 and trimethylating the lysine 27 of histone H3 (65). Endocrine therapies are widely used as an effective treatment for ERα+ breast cancer, such as Luminal A and Luminal B. However, the frequent occurrence of endocrine therapy resistance (ETR) leads to increased disease relapse and decreased OS. The expression levels of H19 were positively correlated with the expression of ERα in primary breast tumors (66). H19 regulated ERα expression at the transcript and protein levels and H19/ERα levels could be significantly downregulated by blocking the Notch and c-MET signaling pathway. Pharmacological inhibitors against this signaling pathway helped reverse the resistance to endocrinotherapy such as Tamoxifen and Fulvestrant (67). In addition, the expression levels of H19 were significantly increased in PTX-resistant TNBC cells compared with that in PTX-sensitive TNBC cells. Inversely, the knockdown of H19 can recover the chemosensitivity of PTX-resistant TNBC by mediating the Akt signaling pathway (16). Furthermore, overexpression of H19 enhanced breast cancer cell resistance to doxorubicin (Dox). The major effector proteins of H19-induced Dox resistance may include multidrug resistance protein 1 (MDR1) and multidrug resistance protein 4. The ubiquitin ligase component cullin 4A (CUL4A) has been regarded as an important factor that connects H19 and MDR1 expression, and the high expression levels of CUL4A compromised the cell chemical sensitivity to Dox and led to lower survival rates of patients with breast cancer undergoing chemotherapy (68). In summary, H19 affects the drug resistance and chemosensitivity of breast cancer cells. As a consequence, the response of breast cancer cells to chemotherapeutic drugs weakens, which leads to a decrease in the curative effect for the tumor.
5. Conclusion
Breast cancer is the most common malignant tumor in women in the majority of countries and usually occurs in women at 35–70 years of age (69,70). In recent years, breast cancer incidence has demonstrated a younger trend (71). The incidence of breast cancer had been demonstrated to be associated with multiple factors, such as a family history of breast cancer, early menarche, not being married, not having had children, radiographic exposure, long-term use of exogenous estrogen, postmenopausal obesity and alcohol consumption (72). Therefore, the tertiary prevention of breast cancer appears to be particularly important. In addition, primary and secondary prevention, including decreasing risk factors, early diagnosis and timely treatment, are key ways to decrease the mortality rate of patients with breast cancer.
According to a number of studies, many lncRNAs can be used as potential biomarkers in the diagnosis and prognostic evaluation of tumors. Excluding H19, there are a number of lncRNAs recently investigated in breast cancer, including NORAD (73), MIR2052HG (74), NAMPT-AS (75), HISLA (76), TROJAN (77), TINCR (78) and LINC00511 (79), some of which have also been reported as potential biomarkers. Among these lncRNAs, H19 is abnormally expressed in various cancer cells and is involved in the regulation of multiple physiological and pathological processes, affecting the occurrence and development of different types of cancer, including breast cancer. Likewise, the development of human breast cancer is influenced by polygenes and multifactor regulation, including H19 regulation (6,7). The regulatory mechanisms involve numerous aspects, such as encoding miR-675, regulating ceRNA, regulating the EMT and regulating tumor angiopoiesis.
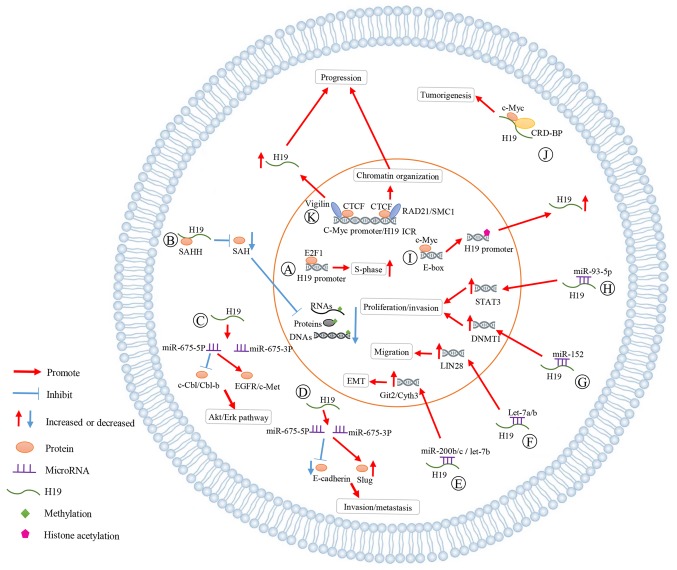
The primary mechanisms underlying the effect that H19 has on breast cancer are summarized in Fig. 2. A: As indicated, E2F1 activated H19 promoter to accelerate cell cycle progression (particularly the S-phase) of breast cancer cells. B: H19 bound SAHH to increase SAH levels and suppressed the methylation of DNA, RNAs and proteins. C: H19 generated miR-675-5p and miR-675-3p. H19/miR-675-5p decreased the expression levels of c-Cbl/Cbl-b and activated EGFR/c-Met, enhancing the proliferation and migration of breast cancer via Akt and Erk pathways. D: miR-675 was involved in the process in which H19 increased Slug expression and inhibited E-cadherin, promoting the cell invasion and metastasis in breast cancer. E-H: H19 acted as a ceRNA for miR-200b/c, let-7a/b, miR-152 and miR-93-5p, increasing the expression of related genes. The interaction between MYC and H19 has great influences on the development of breast cancer. I: C-Myc bound evolutionarily conserved E-boxes close to ICR to promote histone acetylation and transcriptional initiation of the H19 promoter, increasing the H19 expression. J: The complex of CRD-BP, c-Myc and H19 promoted tumorigenesis of breast cancer. K: Vigilin was present at several CTCF target sites, such as c-Myc promoter regions and IGF2/H19 locus, upregulating the H19 expression, and affecting the chromatin organization.
Figure 2.
Molecular mechanisms underlying lncRNA H19 regulation in breast cancer. lncRNA, long non-coding RNA; SAHH, s-adenosylmethionine; SAH, s-adenosylhomocysteine; miR, microRNA; EGFR, epidermal growth factor receptor; Akt, protein kinase B; Erk, extracellular signal-regulated kinase; ceRNA, competing endogenous RNA; DNMT1, DNA methyltransferase 1; STAT3, signal transducer and activator of transcription 3; ICR, imprinting control region; CRD-BP, cMyc mRNA coding region instability determinant; CTCF, CCCTC-binding factor; IGF2, insulin-like growth factor 2.
The diagnostic sensitivity and specificity of H19 in breast cancer is high (59,80), indicating that H19 is of high value in the diagnosis of breast cancer. Furthermore, clinical studies have demonstrated that high H19 expression is one of the factors leading to poor prognosis. Nevertheless, it remains difficult to develop clinical therapy targeting H19 due to certain adversities in its feasibility and possibility. In addition, drug resistance is a major difficulty in the treatment of breast cancer. With the development of research, there may be other antineoplastic drugs, as well as PTX, Dox and endocrine drugs, whose therapeutic effect may be impaired due to the high expression of H19.
Currently, certain deficiencies and problems remain in the studies that focus on the association between H19 and breast cancer. For instance, the potential mechanisms underlying H19/miR-675 regulation in gene expression and any specific molecular mechanisms underlying the interaction between H19 and MYC remain to be elucidated. In addition, the combination of basic research into H19 and the clinical diagnosis of breast cancer could be more proficient, and a unified standard for judging the expression level of H19 in the clinical setting should be available. Furthermore, to the best of our knowledge, there are few studies that have investigated whether H19 can be used in the treatment of breast cancer. In order to apply the studies on H19 to the clinical treatment of breast cancer, further research is required to comprehensively understand the complicated underlying mechanisms through which H19 regulates the biological characteristics of human breast cancer. As research on H19 progresses, anticancer drugs targeting H19 will be used to treat breast cancer more safely and effectively.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 31401094) and the Natural Science Foundation of Jiangsu Province, China (grant no. BK20181365).
Availability of data and materials
Not applicable.
Authors' contributions
JW and JS conceived the study and drafted the manuscript. FY critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 2.Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol Immunol. 2019;112:82–92. doi: 10.1016/j.molimm.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Charles Richard JL, Eichhorn PJA. Platforms for investigating LncRNA functions. SLAS Technol. 2018;23:493–506. doi: 10.1177/2472630318780639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia B, Xie T, Qiu X, Sun X, Chen J, Huang Z, Zheng X, Wang Z, Zhao J. Long noncoding RNA FALEC inhibits proliferation and metastasis of tongue squamous cell carcinoma by epigenetically silencing ECM1 through EZH2. Aging (Albany NY) 2019;11:4990–5007. doi: 10.18632/aging.102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim LJ, Wong S, Huang F, Lim S, Chong SS, Ooi LL, Kon OL, Lee CG. Roles and regulation of long non-coding RNAs in hepatocellular carcinoma. Cancer Res. 2019;79:5131–5139. doi: 10.1158/0008-5472.CAN-19-0255. [DOI] [PubMed] [Google Scholar]
- 6.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis-a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshimura H, Matsuda Y, Yamamoto M, Kamiya S, Ishiwata T. Expression and role of long non-coding RNA H19 in carcinogenesis. Front Biosci (Landmark Ed) 2018;23:614–625. doi: 10.2741/4608. [DOI] [PubMed] [Google Scholar]
- 8.Bao MH, Szeto V, Yang BB, Zhu SZ, Sun HS, Feng ZP. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018;9:281. doi: 10.1038/s41419-018-0282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Y, Li TL, Zhang HB, Deng JL, Zhang R, Sun H, Wan ZR, Liu YZ, Zhu YS, Wang G. Polymorphisms in IGF2/H19 gene locus are associated with platinum-based chemotherapeutic response in Chinese patients with epithelial ovarian cancer. Pharmacogenomics. 2019;20:179–188. doi: 10.2217/pgs-2018-0153. [DOI] [PubMed] [Google Scholar]
- 10.Ghaedi H, Zare A, Omrani MD, Doustimotlagh AH, Meshkani R, Alipoor S, Alipoor B. Genetic variants in long noncoding RNA H19 and MEG3 confer risk of type 2 diabetes in an Iranian population. Gene. 2018;675:265–271. doi: 10.1016/j.gene.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Gomez J, Lorca R, Reguero JR, Martín M, Morís C, Alonso B, Iglesias S, Díaz-Molina B, Avanzas P, Coto E. Genetic variation at the long noncoding RNA H19 gene is associated with the risk of hypertrophic cardiomyopathy. Epigenomics. 2018;10:865–873. doi: 10.2217/epi-2017-0175. [DOI] [PubMed] [Google Scholar]
- 12.Tarnowski M, Tkacz M, Czerewaty M, Poniewierska-Baran A, Grymuła K, Ratajczak MZ. 5-Azacytidine inhibits human rhabdomyosarcoma cell growth by downregulating insulin-like growth factor 2 expression and reactivating the H19 gene product miR-675, which negatively affects insulin-like growth factors and insulin signaling. Int J Oncol. 2015;46:2241–2250. doi: 10.3892/ijo.2015.2906. [DOI] [PubMed] [Google Scholar]
- 13.Wei Y, Liu Z, Fang J. H19 functions as a competing endogenous RNA to regulate human epidermal growth factor receptor expression by sequestering let7c in gastric cancer. Mol Med Rep. 2018;17:2600–2606. doi: 10.3892/mmr.2017.8184. [DOI] [PubMed] [Google Scholar]
- 14.Ma L, Tian X, Guo H, Zhang Z, Du C, Wang F, Xie X, Gao H, Zhuang Y, Kornmann M, et al. Long noncoding RNA H19 derived miR-675 regulates cell proliferation by down-regulating E2F-1 in human pancreatic ductal adenocarcinoma. J Cancer. 2018;9:389–399. doi: 10.7150/jca.21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Han C, Ungerleider N, Chen W, Song K, Wang Y, Kwon H, Ma W, Wu T. A transforming growth factor-β and H19 signaling axis in tumor-initiating hepatocytes that regulates hepatic carcinogenesis. Hepatology. 2019;69:1549–1563. doi: 10.1002/hep.30153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J, Han B, Wu X, Hao J, Dong X, Shen Q, Pang H. Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol Appl Pharmacol. 2018;359:55–61. doi: 10.1016/j.taap.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 17.El Hajj J, Nguyen E, Liu Q, Bouyer C, Adriaenssens E, Hilal G, Ségal-Bendirdjian E. Telomerase regulation by the long non-coding RNA H19 in human acute promyelocytic leukemia cells. Mol Cancer. 2018;17:85. doi: 10.1186/s12943-018-0835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 19.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Fu F, Chen Y, Qiu W, Lin S, Yang P, Huang M, Wang C. Genetic variants in long noncoding RNA H19 contribute to the risk of breast cancer in a southeast China Han population. Onco Targets Ther. 2017;10:4369–4378. doi: 10.2147/OTT.S127962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui P, Zhao Y, Chu X, He N, Zheng H, Han J, Song F, Chen K. SNP rs2071095 in LincRNA H19 is associated with breast cancer risk. Breast Cancer Res Treat. 2018;171:161–171. doi: 10.1007/s10549-018-4814-y. [DOI] [PubMed] [Google Scholar]
- 22.Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, Dugimont T, Curgy JJ. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–1895. doi: 10.1093/carcin/23.11.1885. [DOI] [PubMed] [Google Scholar]
- 23.Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280:29625–29636. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, Xie J, Giang K, Chung H, Sun X, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221. doi: 10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vennin C, Spruyt N, Robin YM, Chassat T, Le Bourhis X, Adriaenssens E. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett. 2017;385:198–206. doi: 10.1016/j.canlet.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Collette J, Le Bourhis X, Adriaenssens E. Regulation of human breast cancer by the long non-coding RNA H19. Int J Mol Sci. 2017;18(pii):E2319. doi: 10.3390/ijms18112319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai LL, Wang P, Zhou LY, Yin JY, Tang Q, Zhang TJ, Wang YX, Yang DQ, Lin J, Deng ZQ. Over-expression of miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues of breast cancer patients. Int J Clin Exp Med. 2015;8:11195–11201. [PMC free article] [PubMed] [Google Scholar]
- 29.Cordero F, Ferrero G, Polidoro S, Fiorito G, Campanella G, Sacerdote C, Mattiello A, Masala G, Agnoli C, Frasca G, et al. Differentially methylated microRNAs in prediagnostic samples of subjects who developed breast cancer in the European Prospective Investigation into Nutrition and Cancer (EPIC-Italy) cohort. Carcinogenesis. 2015;36:1144–1153. doi: 10.1093/carcin/bgv102. [DOI] [PubMed] [Google Scholar]
- 30.Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, Chassat T, Bourette RP, Le Bourhis X, Adriaenssens E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6:29209–29223. doi: 10.18632/oncotarget.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matouk IJ, Raveh E, Abu-Lail R, Mezan S, Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta. 2014;1843:1414–1426. doi: 10.1016/j.bbamcr.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Wang X, Chen T, Jiang L, Yang Q. Huaier extract inhibits breast cancer progression through a lncRNA-H19/miR-675-5p pathway. Cell Physiol Biochem. 2017;44:581–593. doi: 10.1159/000485093. [DOI] [PubMed] [Google Scholar]
- 33.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li LY, Guan GH, Liu Q, Qian YH, Xie D. The lncRNA H19 mediates breast cancer cell plasticity during EMT and MET plasticity by differentially sponging miR-200b/c and let-7b. Sci Signal. 2017;10(pii):eaak9557. doi: 10.1126/scisignal.aak9557. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Li Y, Li Y, Ren K, Li X, Han X, Wang J. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol. 2017:31. doi: 10.1002/jbt.21933. doi: 10.1002/jbt.21933. [DOI] [PubMed] [Google Scholar]
- 39.Li JP, Xiang Y, Fan LJ, Yao A, Li H, Liao XH. Long noncoding RNA H19 competitively binds miR-93-5p to regulate STAT3 expression in breast cancer. J Cell Biochem. 2019;120:3137–3148. doi: 10.1002/jcb.27578. [DOI] [PubMed] [Google Scholar]
- 40.Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8:e2569. doi: 10.1038/cddis.2016.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng F, Wang JH, Fan WJ, Meng YT, Li MM, Li TT, Cui B, Wang HF, Zhao Y, An F, et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene. 2018;37:1062–1074. doi: 10.1038/onc.2017.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang R, Li Y, Wang M, Tang SC, Xiao G, Sun X, Li G, Du N, Liu D, Ren H. MiR-146a promotes the asymmetric division and inhibits the self-renewal ability of breast cancer stem-like cells via indirect upregulation of Let-7. Cell Cycle. 2018;17:1445–1456. doi: 10.1080/15384101.2018.1489176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M, Li Y, Xiao GD, Zheng XQ, Wang JC, Xu CW, Qin S, Ren H, Tang SC, Sun X. H19 regulation of oestrogen induction of symmetric division is achieved by antagonizing Let-7c in breast cancer stem-like cells. Cell Prolif. 2019;52:e12534. doi: 10.1111/cpr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Martino M, Forzati F, Marfella M, Pellecchia S, Arra C, Terracciano L, Fusco A, Esposito F. HMGA1P7-pseudogene regulates H19 and Igf2 expression by a competitive endogenous RNA mechanism. Sci Rep. 2016;6:37622. doi: 10.1038/srep37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L, Liu L, Lu S. lncRNA H19 promotes viability and epithelial-mesenchymal transition of lung adenocarcinoma cells by targeting miR-29b-3p and modifying STAT3. Int J Oncol. 2019;54:929–941. doi: 10.3892/ijo.2019.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan Y, Zhang Y, Liu W, Huang Y, Shen X, Jing R, Pu J, Wang X, Ju S, Cong H, Chen H. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019;10:106. doi: 10.1038/s41419-018-1219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X, Zhou Y, Chen W, Chen L, Lu J, He F, Li X, Zhao L. Ginsenoside 20(S)-Rg3 prevents PKM2-targeting miR-324-5p from H19 sponging to antagonize the warburg effect in ovarian cancer cells. Cell Physiol Biochem. 2018;51:1340–1353. doi: 10.1159/000495552. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Zhu Q, Yang W, Shan Y, Yu Z, Zhang Q, Wu H. LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. J Cell Biochem. 2019;120:3874–3886. doi: 10.1002/jcb.27669. [DOI] [PubMed] [Google Scholar]
- 49.Wei J, Gan Y, Peng D, Jiang X, Kitazawa R, Xiang Y, Dai Y, Tang Y, Yang J. Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer Med. 2018;7:6247–6257. doi: 10.1002/cam4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li M, Chai HF, Peng F, Meng YT, Zhang LZ, Zhang L, Zou H, Liang QL, Li MM, Mao KG, et al. Estrogen receptor β upregulated by lncRNA-H19 to promote cancer stem-like properties in papillary thyroid carcinoma. Cell Death Dis. 2018;9:1120. doi: 10.1038/s41419-018-1077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei LQ, Li L, Lu C, Liu J, Chen Y, Wu H. Involvement of H19/miR-326 axis in hepatocellular carcinoma development through modulating TWIST1. J Cell Physiol. 2019;234:5153–5162. doi: 10.1002/jcp.27319. [DOI] [PubMed] [Google Scholar]
- 52.Li CF, Li YC, Wang Y, Sun LB. The Effect of LncRNA H19/miR-194-5p axis on the Epithelial-mesenchymal transition of colorectal adenocarcinoma. Cell Physiol Biochem. 2018;50:196–213. doi: 10.1159/000493968. [DOI] [PubMed] [Google Scholar]
- 53.Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S, Yang Y, Liu N, Zhao X, Santin AD, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076–3084. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 54.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, Tsao MS, Penn LZ. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 55.Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 2004;64:209–214. doi: 10.1158/0008-5472.CAN-03-2927. [DOI] [PubMed] [Google Scholar]
- 56.Liu Q, Yang B, Xie X, Wei L, Liu W, Yang W, Ge Y, Zhu Q, Zhang J, Jiang L, et al. Vigilin interacts with CCCTC-binding factor (CTCF) and is involved in CTCF-dependent regulation of the imprinted genes Igf2 and H19. FEBS J. 2014;281:2713–2725. doi: 10.1111/febs.12816. [DOI] [PubMed] [Google Scholar]
- 57.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popkie AP, Zeidner LC, Albrecht AM, D'Ippolito A, Eckardt S, Newsom DE, Groden J, Doble BW, Aronow B, McLaughlin KJ, et al. Phosphatidylinositol 3-kinase (PI3K) signaling via glycogen synthase kinase-3 (Gsk-3) regulates DNA methylation of imprinted loci. J Biol Chem. 2010;285:41337–41347. doi: 10.1074/jbc.M110.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, Yang J, Song X, Liu J. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016;17:187–194. doi: 10.3233/CBM-160630. [DOI] [PubMed] [Google Scholar]
- 60.Jiao ZY, Tian Q, Li N, Wang HB, Li KZ. Plasma long non-coding RNAs (lncRNAs) serve as potential biomarkers for predicting breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:1994–1999. doi: 10.26355/eurrev_201804_14727. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z, Weaver DL, Olsen D, DeKay J, Peng Z, Ashikaga T, Evans MF. Long non-coding RNA chromogenic in situ hybridisation signal pattern correlation with breast tumour pathology. J Clin Pathol. 2016;69:76–81. doi: 10.1136/jclinpath-2015-203275. [DOI] [PubMed] [Google Scholar]
- 62.Shima H, Kida K, Adachi S, Yamada A, Sugae S, Narui K, Miyagi Y, Nishi M, Ryo A, Murata S, et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res Treat. 2018;170:507–516. doi: 10.1007/s10549-018-4793-z. [DOI] [PubMed] [Google Scholar]
- 63.Riaz M, Berns EM, Sieuwerts AM, Ruigrok-Ritstier K, de Weerd V, Groenewoud A, Uitterlinden AG, Look MP, Klijn JG, Sleijfer S, et al. Correlation of breast cancer susceptibility loci with patient characteristics, metastasis-free survival, and mRNA expression of the nearest genes. Breast Cancer Res Treat. 2012;133:843–851. doi: 10.1007/s10549-011-1663-3. [DOI] [PubMed] [Google Scholar]
- 64.Han L, Ma P, Liu SM, Zhou X. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour Biol. 2016;37:6847–6854. doi: 10.1007/s13277-015-4568-7. [DOI] [PubMed] [Google Scholar]
- 65.Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang E, Shao L, Li A, Yang N, Han X, et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016;7:81452–81462. doi: 10.18632/oncotarget.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Basak P, Chatterjee S, Weger S, Bruce MC, Murphy LC, Raouf A. Estrogen regulates luminal progenitor cell differentiation through H19 gene expression. Endocr Relat Cancer. 2015;22:505–517. doi: 10.1530/ERC-15-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Basak P, Chatterjee S, Bhat V, Su A, Jin H, Lee-Wing V, Liu Q, Hu P, Murphy LC, Raouf A. Long non-coding RNA H19 acts as an estrogen receptor modulator that is required for endocrine therapy resistance in ER+ breast cancer cells. Cell Physiol Biochem. 2018;51:1518–1532. doi: 10.1159/000495643. [DOI] [PubMed] [Google Scholar]
- 68.Zhu QN, Wang G, Guo Y, Peng Y, Zhang R, Deng JL, Li ZX, Zhu YS. LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. Oncotarget. 2017;8:91990–92003. doi: 10.18632/oncotarget.21121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdel Hadi M. Breast cancer in developing countries: The shrinking age gap. Breast J. 2019;25:795–797. doi: 10.1111/tbj.13335. [DOI] [PubMed] [Google Scholar]
- 70.Sinaga ES, Ahmad RA, Shivalli S, Hutajulu SH. Age at diagnosis predicted survival outcome of female patients with breast cancer at a tertiary hospital in Yogyakarta, Indonesia. Pan Afr Med J. 2018;31:163. doi: 10.11604/pamj.2018.31.163.17284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fazel A, Hasanpour-Heidari S, Salamat F, Rajaie S, Kazeminezhad V, Naeimi-Tabiei M, Jahangirrad A, Sedaghat S, Hosseinpoor R, Ghasemi-Kebria F, et al. Marked increase in breast cancer incidence in young women: A 10-year study from Northern Iran, 2004–2013. Cancer Epidemiol. 2019;62:101573. doi: 10.1016/j.canep.2019.101573. [DOI] [PubMed] [Google Scholar]
- 72.Kaminska M, Ciszewski T, Łopacka-Szatan K, Miotla P, Staroslawska E. Breast cancer risk factors. Prz Menopauzalny. 2015;14:196–202. doi: 10.5114/pm.2015.54346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan BS, Yang MC, Singh S, Chou YC, Chen HY, Wang MY, Wang YC, Chen RH. LncRNA NORAD is repressed by the YAP pathway and suppresses lung and breast cancer metastasis by sequestering S100P. Oncogene. 2019;38:5612–5626. doi: 10.1038/s41388-019-0812-8. [DOI] [PubMed] [Google Scholar]
- 74.Cairns J, Ingle JN, Kalari KR, Shepherd LE, Kubo M, Goetz MP, Weinshilboum RM, Wang L. The lncRNA MIR2052HG regulates ERα levels and aromatase inhibitor resistance through LMTK3 by recruiting EGR1. Breast Cancer Res. 2019;21:47. doi: 10.1186/s13058-019-1130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, Zhang N, Liu Y, Su P, Liang Y, Li Y, Wang X, Chen T, Song X, Sang Y, et al. Epigenetic regulation of NAMPT by NAMPT-AS drives metastatic progression in triple-negative breast cancer. Cancer Res. 2019;79:3347–3359. doi: 10.1158/0008-5472.CAN-18-3418. [DOI] [PubMed] [Google Scholar]
- 76.Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, Tu Q, Yin D, Lin D, Wong PP, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21:498–510. doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 77.Jin X, Xu XE, Jiang YZ, Liu YR, Sun W, Guo YJ, Ren YX, Zuo WJ, Hu X, Huang SL, et al. The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci Adv. 2019;5:eaat9820. doi: 10.1126/sciadv.aat9820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong H, Hu J, Zou K, Ye M, Chen Y, Wu C, Chen X, Han M. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, Qin Q, Zhao L, Huang Q, Luo Z, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37:289. doi: 10.1186/s13046-018-0945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu G, Zhang W, Zhu L, Xia L. Upregulated long non-coding RNAs demonstrate promising efficacy for breast cancer detection: A meta-analysis. Onco Targets Ther. 2018;11:1491–1499. doi: 10.2147/OTT.S152241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren J, Fu J, Ma T, Yan B, Gao R, An Z, Wang D. LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle. 2018;17:1372–1380. doi: 10.1080/15384101.2018.1482137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Z, Lei W, Hu HB, Zhang H, Zhu Y. H19 promotes non-small-cell lung cancer (NSCLC) development through STAT3 signaling via sponging miR-17. J Cell Physiol. 2018;233:6768–6776. doi: 10.1002/jcp.26530. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Q, Li X, Li X, Li X, Chen Z. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J Cell Biochem. 2018;119:4447–4457. doi: 10.1002/jcb.26537. [DOI] [PubMed] [Google Scholar]
- 84.Wang Q, Cheng N, Li X, Pan H, Li C, Ren S, Su C, Cai W, Zhao C, Zhang L, Zhou C. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget. 2017;8:2558–2567. doi: 10.18632/oncotarget.13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yan J, Zhang Y, She Q, Li X, Peng L, Wang X, Liu S, Shen X, Zhang W, Dong Y, et al. Long noncoding RNA H19/miR-675 axis promotes gastric cancer via FADD/caspase 8/caspase 3 signaling pathway. Cell Physiol Biochem. 2017;42:2364–2376. doi: 10.1159/000480028. [DOI] [PubMed] [Google Scholar]
- 86.Ishii S, Yamashita K, Harada H, Ushiku H, Tanaka T, Nishizawa N, Yokoi K, Washio M, Ema A, Mieno H, et al. The H19-PEG10/IGF2BP3 axis promotes gastric cancer progression in patients with high lymph node ratios. Oncotarget. 2017;8:74567–74581. doi: 10.18632/oncotarget.20209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 88.Ma H, Yuan L, Li W, Xu K, Yang L. The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J Cell Biochem. 2018;119:8325–8335. doi: 10.1002/jcb.26883. [DOI] [PubMed] [Google Scholar]
- 89.Pope C, Mishra S, Russell J, Zhou Q, Zhong XB. Targeting H19, an imprinted long non-coding RNA, in hepatic functions and liver diseases. Diseases. 2017;5:E11. doi: 10.3390/diseases5010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang W, Redpath RE, Zhang C, Ning N. Long non-coding RNA H19 promotes the migration and invasion of colon cancer cells via MAPK signaling pathway. Oncol Lett. 2018;16:3365–3372. doi: 10.3892/ol.2018.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang W, Ning N, Jin X. The lncRNA H19 promotes cell proliferation by competitively binding to miR-200a and derepressing β-catenin expression in colorectal cancer. Biomed Res Int. 2017;2017:2767484. doi: 10.1155/2017/2767484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Q, Wang X, Tang C, Chen X, He J. H19 promotes the migration and invasion of colon cancer by sponging miR-138 to upregulate the expression of HMGA1. Int J Oncol. 2017;50:1801–1809. doi: 10.3892/ijo.2017.3941. [DOI] [PubMed] [Google Scholar]
- 93.Chen SW, Zhu J, Ma J, Zhang JL, Zuo S, Chen GW, Wang X, Pan YS, Liu YC, Wang PY. Overexpression of long non-coding RNA H19 is associated with unfavorable prognosis in patients with colorectal cancer and increased proliferation and migration in colon cancer cells. Oncol Lett. 2017;14:2446–2452. doi: 10.3892/ol.2017.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L, Wang DL, Yu P. LncRNA H19 regulates the expression of its target gene HOXA10 in endometrial carcinoma through competing with miR-612. Eur Rev Med Pharmacol Sci. 2018;22:4820–4827. doi: 10.26355/eurrev_201808_15617. [DOI] [PubMed] [Google Scholar]
- 95.Zhao L, Li Z, Chen W, Zhai W, Pan J, Pang H, Li X. H19 promotes endometrial cancer progression by modulating epithelial-mesenchymal transition. Oncol Lett. 2017;13:363–369. doi: 10.3892/ol.2016.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ghazal S, McKinnon B, Zhou J, Mueller M, Men Y, Yang L, Mueller M, Flannery C, Huang Y, Taylor HS. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med. 2015;7:996–1003. doi: 10.15252/emmm.201505245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu Z, Xu L, Wan Y, Zhou J, Fu D, Chao H, Bao K, Zeng T. Inhibition of E-cadherin expression by lnc-RNA H19 to facilitate bladder cancer metastasis. Cancer Biomark. 2018;22:275–281. doi: 10.3233/CBM-170998. [DOI] [PubMed] [Google Scholar]
- 98.Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta Mol Cell Res. 2017;1864:1887–1899. doi: 10.1016/j.bbamcr.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 99.Liu C, Chen Z, Fang J, Xu A, Zhang W, Wang Z. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 2016;37:263–270. doi: 10.1007/s13277-015-3779-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013;280:1709–1716. doi: 10.1111/febs.12185. [DOI] [PubMed] [Google Scholar]
- 101.Ramnarine VR, Alshalalfa M, Mo F, Nabavi N, Erho N, Takhar M, Shukin R, Brahmbhatt S, Gawronski A, Kobelev M, et al. The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. Gigascience. 2018 Jun 1; doi: 10.1093/gigascience/giy050. (Epub ahead of print). doi: 10.1093/gigascience/giy050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.