Abstract
The homeobox (HOX) genes, a class of transcription factors, are known to promote embryonic development and induce tumor formation. To date, the HOXA and HOXB gene families have been reported to be associated with breast cancer. However, the expression and exact role of homeobox C13 (HOXC13) in breast cancer has not yet been investigated. In the present study, the HOXC13 expression in human breast cancer was evaluated using the Oncomine database and Cancer Cell Line Encyclopedia (CCLE). Next, the Gene expression-based Outcome for Breast cancer online database, cBioportal, University of California Santa Cruz Xena browser and bc-GenExMinerv were used to explore the specific expression of HOXC13 in breast cancer. The methylation and mutation status of HOXC13 in breast cancer was then validated using the CCLE and cBioportal databases. Finally, the co-expression of HOX transcript antisense RNA (HOTAIR) and HOXC13 in breast cancer were analyzed and their impact on clinical prognosis determined. It was found that the expression of HOXC13 was high in breast cancer compared with other types of cancer, such as gastric cancer and colon cancer. Following co-expression analysis, a significant positive association was identified between HOTAIR and HOXC13. An association between HOTAIR and HOXC13, and lymph node and distant metastasis recurrence was also revealed during the development of breast cancer. Of note, survival analysis showed that high expression of HOTAIR and HOXC13 predicted poor prognosis. These findings revealed that HOXC13 plays an important role in the progression of breast cancer. However, the specific mechanism needs to be confirmed by subsequent experiments.
Keywords: homeobox C13, breast cancer, bioinformatics, prognosis
Introduction
Breast cancer, the most common type of cancer in women, presents a challenge for global research (1). Molecular research on breast cancer was at a bottleneck, until the identification of a correlation between BRCA1/2 DNA repair associated and breast cancer was reported and marked great progress in the research, treatment and prognosis of breast cancer (2,3). Following this, genetic susceptibility has become the focus of breast cancer research (4). However, in the context of big data, with the development of high-throughput sequencing technology, it is not difficult to find susceptibility genes.
The Homeobox (HOX) genes is a large class of transcription factors that play an important role in embryogenesis and oncogenesis, as well as the distribution of fat and hair in body (5–7). In humans, the HOX gene family contains 39 HOX genes located on 4 different chromosomes (7p15, 17q21.2, 12q13 and 2q31) (8). The 39 HOX genes are divided into 4 clusters (HOXA, HOXB, HOXC and HOXD) (9). Each HOX gene contains a well-conserved DNA sequence, known as the homeobox (10). The unique expression pattern, including mutation, and dependent mechanism of the HOX genes regulates, to some extent, the embryonic development of vertebrates (11,12). When HOX protein expression is upregulated, it may lead to cancer (5). It has also been reported that the HOXC gene family is highly expressed in certain solid tumors, including lung, prostate and colon cancer (13,14). The HOXA and HOXB gene families have a similar expression in breast cancer, which is derived from the ectoderm. Whether the expression level of the HOX gene follows the origin of the germ layer in cancer requires further investigation. A study has reported that HOXC13, a member of the HOXC gene family, is highly expressed in the MCF-7 cell line (15). Thus, the present study aimed to explore the expression and significance of HOXC13 in breast cancer.
In the present study, the Oncomine and tumor public databases (bc-GenExMinerv 4.2; GOBO database; CCLE) were used to analyze the expression level of HOXC13 in different types of cancer including breast cancer. HOXC13 was then further investigated in breast cancer. The expression and co-expression of HOXC13 in breast cancer was re-analyzed using the University of California, Santa Cruz (UCSC) cancer genomics browser. Finally, the clinical significance of HOXC13 in breast cancer was further explored.
Materials and methods
Oncomine database verification
The Oncomine (www.oncomine.org) database is a public bioinformatics database containing gene expression data set that has become an industry-standard tool cited in >1,100 peer-reviewed journal articles (16,17). The Oncomine platform has been used as a foundation for ground-breaking discoveries with unique features that include scalability, high quality, consistency and standardized analysis (18). In order to screen out the most meaningful RNA probes, the paired Student's t-test was used to generate P-values to compare expression differences between cancer and healthy adjacent non-cancerous tissues. Relevant statistical indicators were used as follows: P<1×10−4, fold change >4 and gene ranking in the top 10%. Moreover, the Oncomine database was used to explore the co-expression analysis of HOXC13 in breast cancer. The search term ‘HOXC13’ was used, followed by coexpression analysis and selecting the cancer type as breast cancer. Lastly, the database of TCGA breast was chosen. Furthermore, the same cut-off values used as aforementioned.
Cancer cell line encyclopedia (CCLE) verification
The CCLE (portals.broadinstitute.org) provides public access to genomic data, analysis and visualization for >1,100 cell lines from various tumors, such as gastric cancer cell line AGS and intestinal cancer cell line SW480 (19,20). Each gene of the human genome has multiple datasets and data identifiers, obtained by high-throughput sequencing. The 5 major dataset types are copy number, mRNA expression (Affymetrix), reverse phase protein array, reduced representation bisulfite sequencing, and mRNA expression (RNA sequencing). The expression and methylation level of HOXC13 was analyzed in each tumor cell line using CCLE, using the search term ‘HOXC13’.
Gene expression-based Outcome for Breast cancer Online (GOBO) analysis
The GOBO database (version 1.0.3; co.bmc.lu.se/gobo/gsa_cellines.pl) is a user-friendly public database. It allows for rapid assessment of gene expression levels, identification of co-expressed genes and association with outcome for single genes, gene sets or gene signatures in an 1,881-sample breast cancer dataset (21). The most important functionality of the GOBO database is the possibility of investigating gene expression levels in breast cancer subgroups and cell lines for gene sets (22). Breast cancer subtypes are classified into basal A, basal B and luminal in the GOBO database. A correlation map is a square table where each line and column represent a gene. Each cell represents an interaction between two genes and is colored according to the value of the Pearson's correlation coefficient between these two genes, from dark blue (coefficient=−1) to dark red (coefficient=1). Cells from the diagonal of the correlation map represent the interaction of a gene with itself and are colored black.
UCSC cancer genomics browser analysis
UCSC (xena.ucsc.edu) is an online exploration tool for public and private multi-omics functional genomics and clinical/phenotype data (23). The Cancer Genome Atlas (TCGA; http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) were used in the present study. Using the UCSC database, a heat map of HOXC13 expression in various subtypes of breast cancer, such as LuminalA, luminalB, normal-like, Basal-like and Her2-riched was generated. At the same time, the co-expression of HOXC13 in breast cancer was analyzed.
cBioPortal database analysis
The public cBioPortal site (www.cbioportal.org; last entered, 28th March 2019) is hosted by the Center for Molecular Oncology at Memorial Sloan Kettering Cancer Center. The cBioportal database has been recognized as a way to verify gene mutations (24–26). This database can also analyze gene expression in different tumors and different studies (23,27). This database has been recognized by multiple studies (26,28). The online cBioPortal for Cancer Genomics was used to provide mutations of HOXC13 (including nonstart, missense, truncation and missense) and its expression in different studies (28). The cancer types and databases used were breast (TCGA2015), breast (TCGA), breast invasive carcinoma breast (TCGA PanCan) (29–34).
Breast cancer gene-expression miner (bc-GenExMiner) analysis
bc-GenExMinerv (version 4.2) is a statistical mining tool of published annotated genomic data (35,36). The statistical analyses are grouped in three modules: Expression, prognosis and correlation. The co-expression of HOXC13 in breast cancer was explored using this database; each study is validated across multiple databases to avoid discrepancies in individual databases. At the same time, the effects of HOXC13 and HOX transcript antisense RNA (HOTAIR) on the survival prognosis in breast cancer were also analyzed.
Statistical analysis
Pearson test and Spearman's rank test were used to evaluate coexpression. The analysis criteria selected were as follows: Gene, nodal and estrogen receptor status of the cohorts to be explored, event on which survival analysis will be based and splitting criterion of median HOXC13 expression. To analyze the prognostic value of HOXC13 and HOTAIR, the patient samples are split into two groups according to the median expressions of HOXC13 and HOTAIR. The two patient cohorts were compared by a Kaplan-Meier survival plot, and the hazard ratio with 95% confidence intervals and logrank P-value are calculated. Significance was determined by the P-value provided by each database.
Results
HOXC13 mRNA expression in human cancer
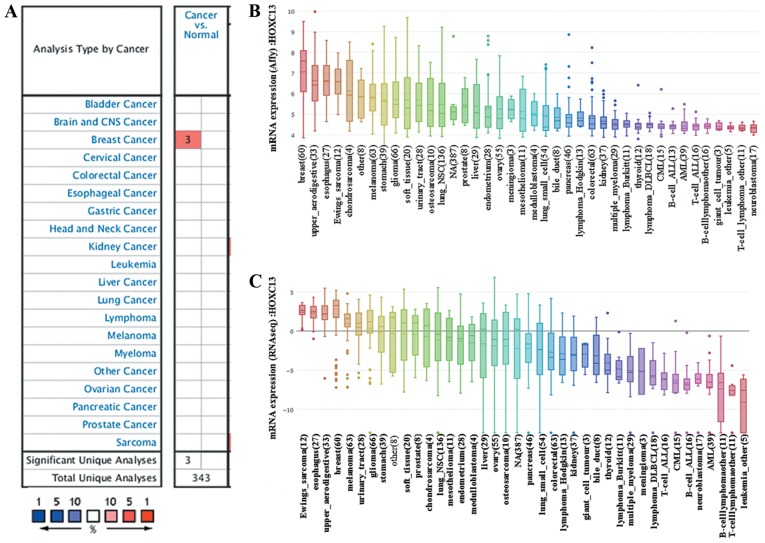
HOXC13 has been proven to be highly expressed in digestive tract-derived tumors (37–39). However, to the best of our knowledge, there have been no reports on the expression of HOXC13 in breast cancer. Therefore the expression levels of HOXC13 in various human tumors from the Oncomine database and CCLE were determined. A simultaneous fold change of >4, gene rank of >10% and P<1×10−4 was set as the threshold. To our surprise, HOXC13 was found by high-throughput sequencing and biological gene chip technology to be abnormally highly expressed in breast cancer (Fig. 1A). Only breast cancer revealed significant unique analysis in the Oncomine database when the fold change was >4.
Figure 1.
HOXC13 mRNA expression level in different types of human cancer. This image shows the expression of HOXC13 in human tumors. (A) Graph showing the number of datasets with a statistically significant mRNA HOXC13 overexpression, based on the cut-off value of P<1×10−4 and fold change >4 in the Oncomine database. The cell color is determined by the best gene rank percentile for the analyses within the cell. Blue represents low expression in tumors, red represents high expression in tumors and white represents no difference in tumor tissues and normal tissues. As shown in the figure, breast cancer has three data sets showing high expression, based on the cut-off value of P<1×10−4, fold change >4 and gene ranking in the top 10%. (B) mRNA expression of HOXC13 in different cancer cell lines. The expression of HOXC13 ranks highest in breast cancer using Affy gene chip data in the Cancer Cell Line Encyclopedia. (C) The mRNA expression of HOXC13 ranks fourth highest in different tumor cell lines RNA-seq data, behind that of Ewings sarcoma, esophagus and upper aerodigestive tract. Affy, Affymetrix; RNA seq, RNA sequencing; CNS, central nervous system; HOXC13, homeobox C13.
HOXC13 mRNA expression in breast cancer
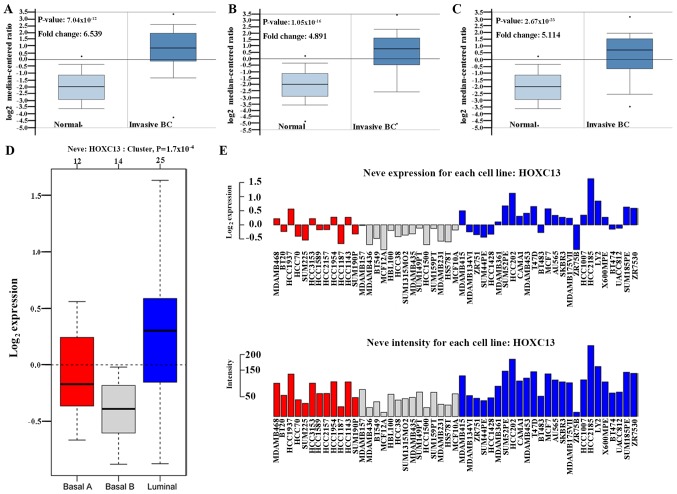
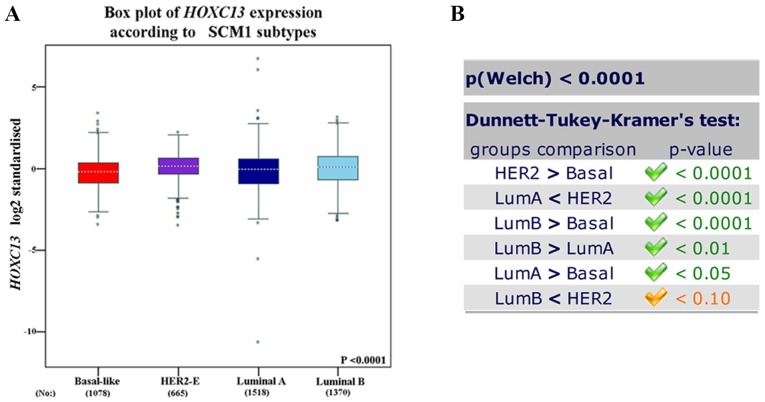
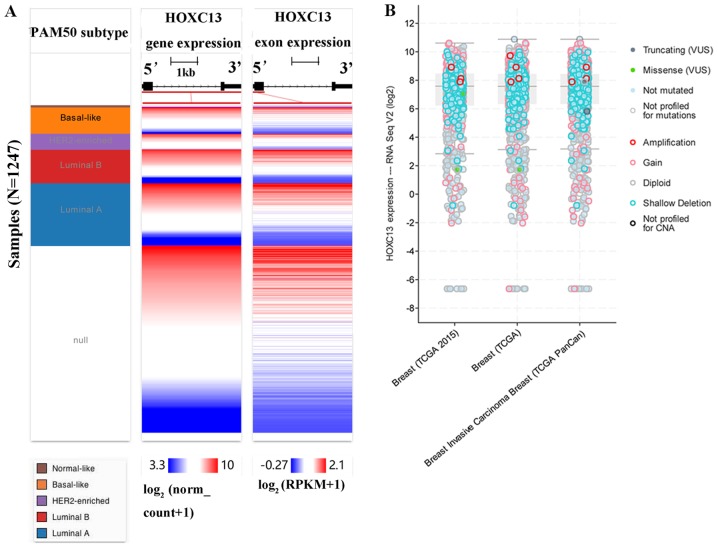
The high expression of HOXC13 in breast cancer has been preliminary confirmed in the Oncomine database, GOBO database and CCLE. From the CCLE database, it was found that at the RNAseq level, the expression level of HOXC13 in breast cancer ranked fourth and ranked first in the Affy level. However, there is no report on the specific expression of HOXC13 in breast cancer. Therefore, its specific expression in various subtypes of breast cancer was further explored. HOXC13 was analyzed in various tumor types via Oncomine database and GOBO database and explored HOXC13 in various breast cancer cells via GOBO database and CCLE. HOXC13 was found to be highly expressed in invasive and luminal-like breast cancer than in any other subtype (Figs. 2 and 3). Such expression characteristics were consistent with breast cancer cell lines and tissues (Figs. 2D and 3). Using the UCSC cancer genomics browser analysis, the heat map of the gene and exon expression of HOXC13 in various subtypes of breast cancer was obtained (Fig. 4A). Furthermore, the expression of HOXC13 in the different data sets, including breast (TCGA 2015), breast (TCGA), breast invasive carcinoma breast (TCGA PanCan), was explored using the cBioPortal database. The expression characteristics of breast cancer in multiple data sets such as amplification and missense mutations, were demonstrated (Fig. 4B).
Figure 2.
HOXC13 expression analysis in different breast cancer subtypes. Box plots represent the expression of HOXC13 in invasive breast cancer from the Oncomine database. (A) Invasive BC (invasive lobular breast carcinoma): P-value=7.04×10−12. (B) Invasive BC (invasive breast carcinoma): P-value=1.05×10−16. (C) Invasive BC (invasive ductal breast carcinoma): P-value=2.67×10−23. (D-E) Red represents breast cancer subtype basal A, gray represents breast cancer subtype basal B, and blue represents breast cancer subtype luminal. (D) Using GOBO analysis, in various subtypes of breast cancer, the HOXC13 expression was significantly higher in luminal-like breast cancer: P=0.00017. (E) GOBO analysis showing the expression of HOXC13 in each breast cancer cell line. BC, breast carcinoma; GOBO, Gene expression-based Outcome for Breast cancer Online; HOXC13, homeobox C13. The neve expression refers to the base two logarithm of the expression of the gene in each cell, and the neve intensity refers to the expression level of each breast cancer cell relative to the expression of the internal reference.
Figure 3.
HOXC13 expression analysis in breast cancer subtypes of SCM1 subtypes. (A) The box plot of HOXC13 expression according to SCM1 subtypes from bc-GenExMiner version 4.2. (B) Group comparison in each subgroup of breast cancer, and corresponding statistical values in SCM1 subtypes. HER2, human epidermal growth factor receptor 2; Lum, luminal; HOXC13, homeobox C13; SCM, subtype clustering model.
Figure 4.
Analysis of the expression of HOXC13 in breast cancer subtypes. (A) HOXC13 expression heatmap of each breast cancer subtype in TCGA from the University of California, Santa Cruz Xena browser. Null data indicates samples that have no gene expression data. (B) HOXC13 expression analysis and mutation status in breast cancer using cBioPortal. HOXC13 was significantly amplified in breast cancer subtypes, including breast invasive carcinoma. The present study included datasets on breast (TCGA2015), breast (TCGA) and breast invasive carcinoma breast (TCGA PanCan) datasets. TCGA, The Cancer Genome Atlas; RNA seq, RNA sequencing; HER2, human epidermal growth factor receptor 2; RPKM, reads per kilobase of transcript per million mapped reads; HOXC13, homeobox C13; VUS, variants of uncertain significance; CNA, copy number alteration.
HOXC13 methylation and mutation in human breast cancer
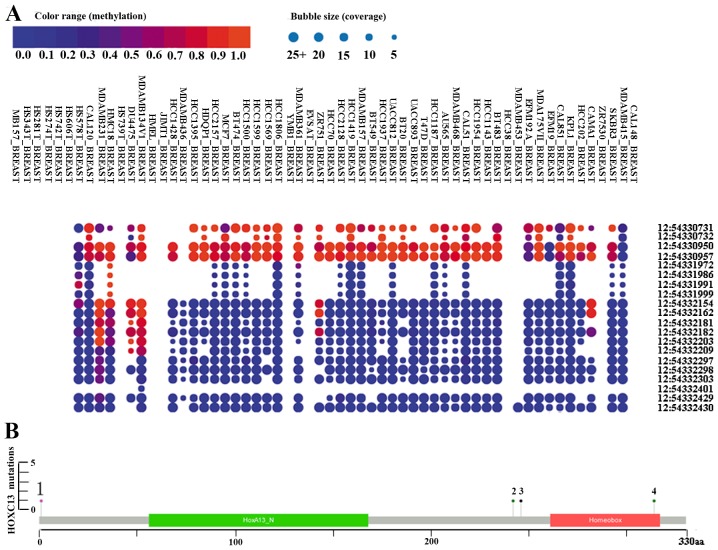
The bubble chart shows the methylation level of HOXC13 in breast cancer cell lines from the CCLE (Fig. 5A). HOXC13 is highly methylated at three sites (positions 54,330,731, 54,330,950 and 54,330,957) on chromosome 12 from methylation and coverage (Fig. 5A). The discovery of cpG island methylation further supported the high expression of HOXC13 in breast cancer. cBioPortal was used to assess the frequency of HOXC13 mutations in breast cancer (Figs. 4B and 5). HOXC13 contains multiple mutations in breast cancer such as amplification, gain, missense and truncation (Fig. 4B). Missense and truncation are two major forms of mutation (Fig. 5).
Figure 5.
Methylation and mutation status of HOXC13 in breast cancer. (A) Methylation status of HOXC13 in breast cancer cell lines. The x-axis displays the cell line in which methylation was measured. The y-axis displays the position of the methylation data. The number before the colon is the chromosome, and the number after the colon the position. Red, orange and blue represent high, medium and low levels of methylation. The size of the bubble represents coverage. (B) Mutation status of HOXC13 in breast cancer from cBioPortal. The mutation types include nonstart, missense and truncation, and missense is shown as 1, 2, 3 and 4. The boxes in green and red represent the open reading frame. The scale shows where the mutation site is located in the sequence. Corresponding changed protein name contains M1, S242Y, X_246splice and S242Y. The data used included studies on breast cancer (29–34).
Co-expression of HOXC13 mRNA in breast cancer
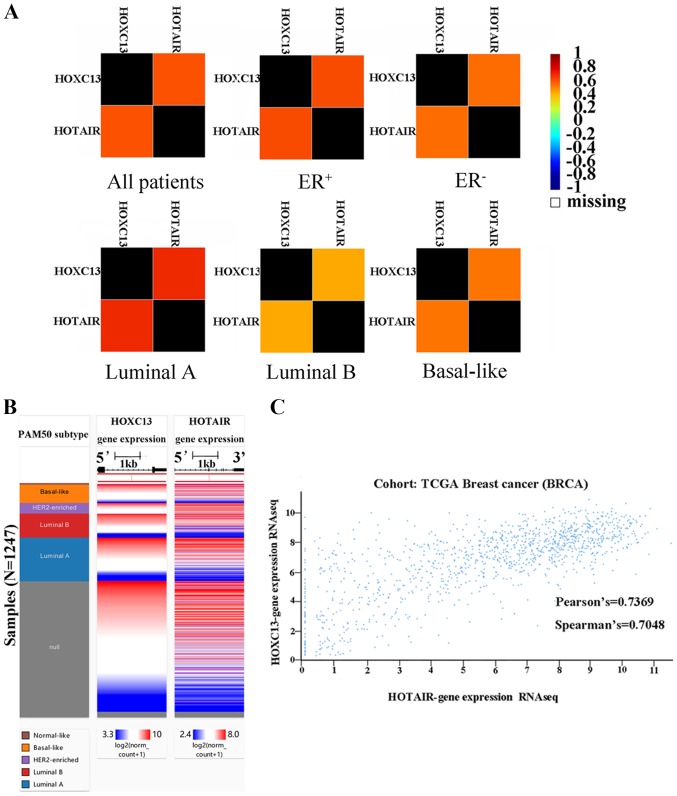
To investigate the reason for the high expression of HOXC13 in breast cancer, bc-GenExMiner version 4.2 was used to analyze the potential co-expression of HOXC13 in breast cancer. It was found that HOTAIR and HOXC13 are highly co-expressed in breast cancer (Fig. 6A). Furthermore, to verify the co-expression of HOXC13 and HOTAIR in breast cancer, their co-expression heat maps were obtained and mined (Fig. 6B) and correlation analysis was performed using the UCSC Xena browser (Fig. 6C).
Figure 6.
Co-expression analysis of HOXC13 and HOTAIR. (A) A correlation map illustrates pairwise correlations between HOXC13 and HOTAIR. They are all patients, ER+, ER-, luminal A, luminal B and basal-like respectively according to the group. (B) The co-expression heat map of HOXC13 and HOTAIR in TCGA derived from the UCSC Xena browser. (C) Association between HOXC13 and HOTAIR gene expression in TCGA breast cancer derived from the UCSC Xena browser. The Pearson's value was 0.7369 and the Spearman's value was 0.7048. HOXC13, homeobox C13; HOTAIR, HOX transcript antisense RNA; UCSC, University of California Santa Cruz; ER, estrogen receptor; -, negative; +, positive; TCGA, The Cancer Genome Atlas; RNA seq, RNA sequencing; HER2, human epidermal growth factor receptor 2; PAM, prediction analysis of microarray.
Impact of HOXC13 and HOTAIR on the prognosis of patients with breast cancer
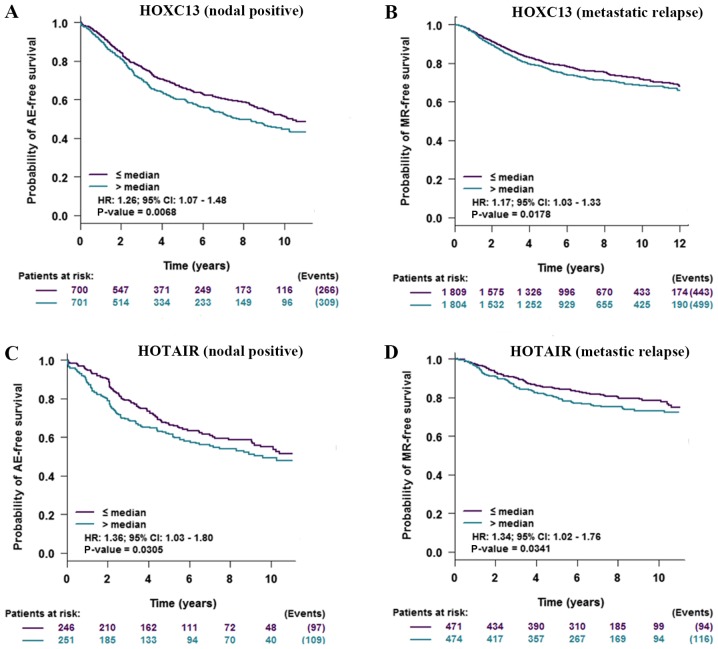
To verify the impact of the high expression of HOXC13 and HOTAIR on patients with breast cancer, prognostic analysis of HOXC13 and HOTAIR in breast cancer was performed and HOXC13 and HOTAIR were found to have a negative impact on the prognosis of patients with tumor and lymph node metastasis (Fig. 7).
Figure 7.
Prognostic value of HOXC13 and HOTAIR in breast cancer. (A) Prognostic analysis of HOXC13 with regards to positive nodal status. The higher the expression of HOXC13, the shorter the AE-free survival time. (B) Prognostic analysis for HOXC13 with regards to metastatic relapse. The higher the expression of HOXC13, the shorter the MR-free survival time. (C) Prognostic analysis for HOTAIR with regards to positive nodal status. The higher the expression of HOTAIR, the shorter the AE free survival time. (D) Prognostic analysis for HOTAIR with regards to metastatic relapse. The higher the expression of HOTAIR, the shorter the MR-free survival time. HOTAIR, HOX transcript antisense RNA; AE, any event; MR, metastatic relapse.
Discussion
The mammary gland is an appendage of the ectoderm, whose formation begins during embryonic development (40). Moreover, breast cancer cells remain highly associated with ectoderm cells (41). Studies have shown that the HOX gene plays an important role in embryonic development and tumor formation (9,42–44). However, to the best of our knowledge, no research has reported the exact role of HOXC13 in breast cancer so far.
The HOXC13 gene is considered an element of hair morphogenesis, and the HOXC13 protein is a member of the human replication complex in growing cells (45,46). This id the same for breast adenocarcinoma MCF7 lines (46). HOXC13 can promote the expression of a series of proto-oncogenes, including topoisomerase I and II, and allow these expression products to form a replication complex (47,48). Therefore, HOXC13 is involved in the development of tumors. In addition, HOXC13 increases the metastasis of ectodermal-derived melanoma (49). HOTAIR is the product of the transcription of the HOXC gene cluster antisense strand (50). HOTAIR is transcribed from the mammalian HOXC gene cluster on chromosome 12q13.13 (51,52). HOTAIR predicts poor prognosis in tumor cell cycle and metastasis (53). It has been reported that HOTAIR upregulates HOX in colon cancer (39). A strong co-expression of HOTAIR and HOXC13 was confirmed in proximal and distal colon cancer, suggesting that HOTAIR and HOXC13 could promote tumor and lymph node metastasis (39,54–56).
In the present study, it was first identified that HOXC13 is highly expressed in breast cancer both at the cellular and tissue levels. This was a finding from the co-verification of the data from TCGA's Oncomine or GSE's bc-GenExMiner. It was found for the first time that HOXC13 is most highly expressed in luminal-like subtype of breast cancer. This laid the foundation for the future study of the relationship between HOXC13 and breast cancer surface hormone receptors (estrogen receptor and progesterone receptor). In order to explore the high expression of HOXC13 in breast cancer, its methylation and mutation status were investigated. To our surprise, three sites on chromosome 12 were found to be consistently highly methylated in different breast cancer cell lines. Certain studies have reported that HOX gene methylation regulates hereditary breast cancer (57–59). In addition, the methylation level of HOXA11 is significantly higher in breast cancer compared with that in normal tissues, and is positively associated with family history and lymph node metastasis in breast cancer (59). Furthermore, the methylation level of HOXD13 in breast cancer is almost identical to that of HOXA11, and leads to shorter survival time (58). Therefore, the present findings and the HOX gene family have consistent trends in methylation levels and similar prognostic effects in breast cancer. However, further studies on the specific regulation mechanism of HOXC13 methylation in HOXC13 transcription is required. The present study is the first to report the mutation of HOXC13 in breast cancer. Missense may be an indispensable cause of the high expression of HOXC13 in breast cancer.
The lncRNA HOTAIR is derived from the region between HOXC11 and HOXC12 (51). It has been shown that HOXC10, HOXC11, HOXC12 and HOXC13 are adjacent to each other in the HOXC gene cluster (52). Simultaneously, the HOXC distal enhancer has non-specific enhancement of HOXC10 and HOTAIR enhancer activity, promoting the HOXC10 and HOTAIR expression (60). On the other hand, specific intergenic non-coding RNAs (including HOTAIR) in the HOX loci can directly modulate the expression of the HOX gene in normal and cancer status (61). This has been confirmed in colon cancer (39,55). Therefore the mechanism between HOXC13 and HOTAIR will be explored further in this respect. In the present study, the co-expression of HOTAIR and HOXC13 provided a new direction for studying the function of HOTAIR in breast cancer. In addition, a study has identified through meta-analysis that HOTAIR has a statistically significant effect on lymph node and distant metastasis in various types of cancer, including breast cancer, gastric cancer and colorectal cancer, which was consistent with our conclusion (62). The present data showed that HOTAIR and HOXC13 were significantly associated with lymph node metastasis and distant metastasis recurrence. Furthermore, they were shown to have a significant impact on survival period.
In conclusion, the present study was performed using public databases and revealed the expression and clinical significance of HOXC13 in breast cancer. However, the specific interactions and mechanisms involved require further experimental verification, which will be performed in future studies.
Acknowledgements
Not applicable.
Funding
This study was supported by the Shenzhen Science and Research Innovation Foundation (grant no. JCYJ20170815090309586).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WW contributed to the experimental design and fundraising. JWC and LZ contributed to the acquisition of data. LZZ assisted in the data processing. CL processed the data and wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Barrios CH, Reinert T, Werutsky G. Global breast cancer research: Moving forward. Am Soc Clin Oncol Educ Book. 2018;38:441–450. doi: 10.1200/EDBK_209183. [DOI] [PubMed] [Google Scholar]
- 2.Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 3.Nathanson KL, Domchek SM. Therapeutic approaches for women predisposed to breast cancer. Annu Rev Med. 2011;62:295–306. doi: 10.1146/annurev-med-010910-110221. [DOI] [PubMed] [Google Scholar]
- 4.Rousset-Jablonski C, Gompel A. Screening for familial cancer risk: Focus on breast cancer. Maturitas. 2017;105:69–77. doi: 10.1016/j.maturitas.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Luo Z, Rhie SK, Farnham PJ. The enigmatic HOX genes: Can we crack their code? Cancers (Basel) 2019;11(pii):E323. doi: 10.3390/cancers11030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turcotte M, Abadi A, Peralta-Romero J, Suarez F, Reddon H, Gomez-Zamudio J, Burguete-Garcia AI, Cruz M, Meyre D. Genetic contribution to waist-to-hip ratio in Mexican children and adolescents based on 12 loci validated in European adults. Int J Obes (Lond) 2019;43:13–22. doi: 10.1038/s41366-018-0055-8. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Chen Y, Zhu G, Hysi PG, Wu S, Adhikari K, Breslin K, Pospiech E, Hamer MA, Peng F, et al. Meta-analysis of genome-wide association studies identifies 8 novel loci involved in shape variation of human head hair. Hum Mol Genet. 2018;27:559–575. doi: 10.1093/hmg/ddx416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Wang Y, Song G, Zhang X, Gao S, Liu H. HOX cluster-embedded antisense long non-coding RNAs in lung cancer. Cancer Lett. 2019;450:14–21. doi: 10.1016/j.canlet.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 9.Bhatlekar S, Fields JZ, Boman BM. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018;2018:3569493. doi: 10.1155/2018/3569493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 11.Gordon J. Hox genes in the pharyngeal region: How Hoxa3 controls early embryonic development of the pharyngeal organs. Int J Dev Biol. 2018;62:775–783. doi: 10.1387/ijdb.180284jg. [DOI] [PubMed] [Google Scholar]
- 12.Mallo M. The vertebrate tail: A gene playground for evolution. Cell Mol Life Sci. 2019 Sep 26; doi: 10.1007/s00018-019-03311-1. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl) 2014;92:811–823. doi: 10.1007/s00109-014-1181-y. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y, Luo J, Sun Q, Xu T, Sun S, Chen M, Lin X, Qian Q, Zhang Y, Cao L, et al. HOXC13 promotes proliferation of lung adenocarcinoma via modulation of CCND1 and CCNE1. Am J Cancer Res. 2017;7:1820–1834. [PMC free article] [PubMed] [Google Scholar]
- 15.Svingen T, Tonissen KF. Altered HOX gene expression in human skin and breast cancer cells. Cancer Biol Ther. 2003;2:518–523. doi: 10.4161/cbt.2.5.441. [DOI] [PubMed] [Google Scholar]
- 16.Shi S, Zhang ZG. Role of Sp1 expression in gastric cancer: A meta-analysis and bioinformatics analysis. Oncol Lett. 2019;18:4126–4135. doi: 10.3892/ol.2019.10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng L, Shi L, Dai H. Bioinformatics prognostic biomarkers among Krüppel-like transcription factors (KLFs) in breast cancer. Cancer Biomark. 2019 Oct 12; doi: 10.3233/CBM-190199. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang K, Gao J, Luo M. Identification of key pathways and hub genes in basal-like breast cancer using bioinformatics analysis. Onco Targets Ther. 2019;12:1319–1331. doi: 10.2147/OTT.S158619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng L, Pandya PH, Liu E, Chandra P, Wang L, Murray ME, Carter J, Ferguson M, Saadatzadeh MR, Bijangi-Visheshsaraei K, et al. Integration of genomic copy number variations and chemotherapy-response biomarkers in pediatric sarcoma. BMC Med Genomics. 2019;12(Suppl 1):S23. doi: 10.1186/s12920-018-0456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Streit M, Gratzl S, Stitz H, Wernitznig A, Zichner T, Haslinger C. Ordino: A visual cancer analysis tool for ranking and exploring genes, cell lines and tissue samples. Bioinformatics. 2019;35:3140–3142. doi: 10.1093/bioinformatics/btz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández-Nogueira P, Bragado P, Almendro V, Ametller E, Rios J, Choudhury S, Mancino M, Gascón P. Differential expression of neurogenes among breast cancer subtypes identifies high risk patients. Oncotarget. 2016;7:5313–5326. doi: 10.18632/oncotarget.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zen g, Xiao Y, Zhu J, Peng C, Liang W, Lin H. Knockdown of nucleophosmin 1 suppresses proliferation of triple-negative breast cancer cells through activating CDH1/Skp2/p27kip1 pathway. Cancer Manag Res. 2018;11:143–156. doi: 10.2147/CMAR.S191176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klonowska K, Czubak K, Wojciechowska M, Handschuh L, Zmienko A, Figlerowicz M, Dams-Kozlowska H, Kozlowski P. Oncogenomic portals for the visualization and analysis of genome-wide cancer data. Oncotarget. 2016;7:176–192. doi: 10.18632/oncotarget.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen W, Gong J, Wu P, Zhao M, Wang M, Chen H, Sun J. Mutations in gliclazide-associated genes may predict poor bladder cancer prognosis. FEBS Open Bio. 2019;9:457–467. doi: 10.1002/2211-5463.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen E, Qin X, Peng K, Xu X, Li W, Cheng X, Tang C, Cui Y, Wang Z, Liu T. Identification of potential therapeutic targets among CXC chemokines in breast tumor microenvironment using integrative bioinformatics analysis. Cell Physiol Biochem. 2018;45:1731–1746. doi: 10.1159/000487782. [DOI] [PubMed] [Google Scholar]
- 26.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kline CLB, Ralff MD, Lulla AR, Wagner JM, Abbosh PH, Dicker DT, Allen JE, El-Deiry WS. Role of dopamine receptors in the anticancer activity of ONC201. Neoplasia. 2018;20:80–91. doi: 10.1016/j.neo.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martelotto LG, De Filippo MR, Ng CK, Natrajan R, Fuhrmann L, Cyrta J, Piscuoglio S, Wen HC, Lim RS, Shen R, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–189. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34:427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan J, Ong CK, Lim WK, Ng CC, Thike AA, Ng LM, Rajasegaran V, Myint SS, Nagarajan S, Thangaraju S, et al. Genomic landscapes of breast fibroepithelial tumors. Nat Genet. 2015;47:1341–1345. doi: 10.1038/ng.3409. [DOI] [PubMed] [Google Scholar]
- 35.Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 36.Jézéquel P, Frénel JS, Campion L, Guérin-Charbonnel C, Gouraud W, Ricolleau G, Campone M. bc-GenExMiner 3.0: New mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013;2013:bas060. doi: 10.1093/database/bas060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo J, Wang Z, Huang J, Yao Y, Sun Q, Wang J, Shen Y, Xu L, Ren B. HOXC13 promotes proliferation of esophageal squamous cell carcinoma via repressing transcription of CASP3. Cancer Sci. 2018;109:317–329. doi: 10.1111/cas.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcinkiewicz KM, Gudas LJ. Altered histone mark deposition and DNA methylation at homeobox genes in human oral squamous cell carcinoma. J Cell Physiol. 2014;229:1405–1416. doi: 10.1002/jcp.24577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatangelo F, Di Mauro A, Scognamiglio G, Aquino G, Lettiero A, Delrio P, Avallone A, Cantile M, Botti G. Posterior HOX genes and HOTAIR expression in the proximal and distal colon cancer pathogenesis. J Transl Med. 2018;16:350. doi: 10.1186/s12967-018-1725-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson GW. Identification of signaling pathways in early mammary gland development by mouse genetics. Breast Cancer Res. 2004;6:105–108. doi: 10.1186/bcr776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tickle C, Crawley A, Goodman M. Mechanisms of invasiveness of epithelial tumours: Ultrastructure of the interactions of carcinoma cells with embryonic mesenchyme and epithelium. J Cell Sci. 1978;33:133–155. doi: 10.1242/jcs.33.1.133. [DOI] [PubMed] [Google Scholar]
- 42.Morgan R, El-Tanani M. HOX genes as potential markers of circulating tumour cells. Curr Mol Med. 2016;16:322–327. doi: 10.2174/1566524016666160316145715. [DOI] [PubMed] [Google Scholar]
- 43.Kamkar F, Xaymardan M, Asli NS. Hox-mediated spatial and temporal coding of stem cells in homeostasis and neoplasia. Stem Cells Dev. 2016;25:1282–1289. doi: 10.1089/scd.2015.0352. [DOI] [PubMed] [Google Scholar]
- 44.Nunes FD, de Almeida FC, Tucci R, de Sousa SC. Homeobox genes: A molecular link between development and cancer. Pesqui Odontol Bras. 2003;17:94–98. doi: 10.1590/S1517-74912003000100018. [DOI] [PubMed] [Google Scholar]
- 45.Godwin AR, Capecchi MR. Hoxc13 mutant mice lack external hair. Genes Dev. 1998;12:11–20. doi: 10.1101/gad.12.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comelli L, Marchetti L, Arosio D, Riva S, Abdurashidova G, Beltram F, Falaschi A. The homeotic protein HOXC13 is a member of human DNA replication complexes. Cell Cycle. 2009;8:454–459. doi: 10.4161/cc.8.3.7649. [DOI] [PubMed] [Google Scholar]
- 47.La Starza R, Trubia M, Crescenzi B, Matteucci C, Negrini M, Martelli MF, Pelicci PG, Mecucci C. Human homeobox gene HOXC13 is the partner of NUP98 in adult acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36:420–423. doi: 10.1002/gcc.10182. [DOI] [PubMed] [Google Scholar]
- 48.Gurevich RM, Aplan PD, Humphries RK. NUP98-topoisomerase I acute myeloid leukemia-associated fusion gene has potent leukemogenic activities independent of an engineered catalytic site mutation. Blood. 2004;104:1127–1136. doi: 10.1182/blood-2003-10-3550. [DOI] [PubMed] [Google Scholar]
- 49.Cantile M, Scognamiglio G, Anniciello A, Farina M, Gentilcore G, Santonastaso C, Fulciniti F, Cillo C, Franco R, Ascierto PA, Botti G. Increased HOX C13 expression in metastatic melanoma progression. J Transl Med. 2012;10:91. doi: 10.1186/1479-5876-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo CJ, Kingston RE. HOTAIR lifts noncoding RNAs to new levels. Cell. 2007;129:1257–1259. doi: 10.1016/j.cell.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Wang J, Zhong Y, Guo R, Chu D, Qiu H, Yuan Z. HOTAIR: A key regulator in gynecologic cancers. Cancer Cell Int. 2017;17:65. doi: 10.1186/s12935-017-0434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, Yu SZ, Pu PY, Liu N, You YP, et al. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol. 2013;15:1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18:1930–1936. [PubMed] [Google Scholar]
- 55.Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Y, Zang Y, Yang Y, Xiang J, Chen Z. Candidate genes involved in metastasis of colon cancer identified by integrated analysis. Cancer Med. 2019;8:2338–2347. doi: 10.1002/cam4.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pilato B, Pinto R, De Summa S, Lambo R, Paradiso A, Tommasi S. HOX gene methylation status analysis in patients with hereditary breast cancer. J Hum Genet. 2013;58:51–53. doi: 10.1038/jhg.2012.118. [DOI] [PubMed] [Google Scholar]
- 58.Zhong Z, Shan M, Wang J, Liu T, Xia B, Niu M, Ren Y, Pang D. HOXD13 methylation status is a prognostic indicator in breast cancer. Int J Clin Exp Pathol. 2015;8:10716–10724. [PMC free article] [PubMed] [Google Scholar]
- 59.Xia B, Shan M, Wang J, Zhong Z, Geng J, He X, Vu T, Zhang D, Pang D. Homeobox A11 hypermethylation indicates unfavorable prognosis in breast cancer. Oncotarget. 2017;8:9794–9805. doi: 10.18632/oncotarget.14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milevskiy MJ, Al-Ejeh F, Saunus JM, Northwood KS, Bailey PJ, Betts JA, McCart Reed AE, Nephew KP, Stone A, Gee JM, et al. Long-range regulators of the lncRNA HOTAIR enhance its prognostic potential in breast cancer. Hum Mol Genet. 2016;25:3269–3283. doi: 10.1093/hmg/ddw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Botti G, De Chiara A, Di Bonito M, Cerrone M, Malzone MG, Collina F, Cantile M. Noncoding RNAs within the HOX gene network in tumor pathogenesis and progression. J Cell Physiol. 2018;234:395–413. doi: 10.1002/jcp.27036. [DOI] [PubMed] [Google Scholar]
- 62.Sun Z, Wu XY, Wu CL. The association between LncRNA HOTAIR and cancer lymph node metastasis and distant metastasis: A meta-analysis. Neoplasma. 2018;65:178–184. doi: 10.4149/neo_2018_170114N34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.