Abstract
The underlying mechanisms of resistance to chemoradiotherapy of human papilloma virus (HPV)-negative patients with oropharyngeal cancer (OPC) remain unclear. The present study aimed to characterize cancer stem cells (CSC) of the HPV-negative OPC cell line in terms of chemotherapy resistance. CSCs were isolated through magnetic activated cell sorting using the CSC specific marker aldehyde dehydrogenase 1 antibody, and characterized by sphere formation capacity, immunofluorescence staining, and CSC marker expression. CSC response to cisplatin treatment was evaluated via XTT-assays. Spheres of CSCs of the HPV-negative UTSCC-60A cell line were highly dark holospheres. RNA expression levels of CSC markers OCT4, SOX2, Kruppel-like factor 4 and BMI1 were significantly higher in CSC. CSCs were significantly resistant to cisplatin treatment at various dosages compared with nonCSC. The present study suggested that the proportion of CSCs is very low in the tumor bulk, CSCs are resistant to cisplatin in HPV-negative OPC, which requires further investigation to define their mechanism.
Keywords: oropharyngeal cancer, head and neck squamous cell carcinoma, human papilloma virus, cancer stem cell, chemotherapy resistance
Introduction
Head and neck squamous cell carcinoma (HNSCC) originates from the oral cavity, oropharynx, hypopharynx, nasopharynx, and larynx and is one of the most fatal among cancers. human papilloma virus (HPV) is believed to play role in the pathogenesis of HNSCC, specifically in oropharyngeal cancer (OPC), along with tobacco and alcohol. Despite HPV driven OPC increment in western countries (1), HPV-positive OPC responds well to chemoradiotherapy. In contrast, HPV-negative patients are generally resistant to these treatment modalities. Besides, advanced OPC requires multimodal approaches like surgery with adjuvant irradiation or concurrent irradiation with salvage surgery (2). As pharynx has important role in basic functions such as speech, chewing, and swallowing, complications and morbidity after treatment are the additional consequences that patients suffer together with the main problem of survival (3).
HPV-negative HNSCC patients are usually elder, the lesions are larger and overall survival is less when compared to HPV-positive HNSCC patients (4). It is also suggested that HPV-positive and HPV-negative cases' molecular background are different. According to Cancer Genome Atlas Network data in HPV-negative cases, TP53 84%, CCND1 31%, CDKN2A 58% and, FGFR1 10% have been altered. On the other hand, TP53 and CCND1 have been altered only in 3% and CDKN2A and FGFR1 had no alteration in HPV-positive cases (5). Although there has been improvement in understanding the background of HPV-negative cases, resistance to chemo/radiotherapy is still the dead end indicating the need of further studies. Cancer stem cell (CSC) model (hierarchic model) of cancer suggests that only a subpopulation of cells in the tumor bulk, CSC, have the potential to form the cancer cells that is different than the stochastic model of cancer in which every cell in the tumor bulk has the potential to form cancer cells. Besides, clonal evolution of CSC puts forwards another difficulty in revealing CSC mechanism and role in cancer because of subgroups and genetic heterogeneity, which means different genetic backgrounds and different phenotypes in CSC (6,7). Like stem cells, cancer stem cells (CSCs) have the potential of differentiation and self-renewal. First defined in leukemia (8), later in other cancers like breast (9), brain tumor (10), colon (11), and over (12), in 2007 CSC were defined in HNSCC (13). CSCs have recently been a hot spot in HNSCC research as they are postulated to play important role in initiation, progression, invasion, metastasis, and as well they are supposed to be resistant to chemo/radio therapy and responsible for recurrence (14). Their resistance to chemotherapy and radiation is through mechanisms like dormancy, DNA repair, multidrug-resistance-type membrane transporters, and escaping apoptosis (15). Although CSCs are very rare in the tumor bulk, sometimes even less than 1%, they are considered to be important to evaluate new therapeutic approaches (16). Not only by biological evidence but also via mathematical modeling it is also shown that a successful therapy should target CSCs as well (17). Additionally, CSC is an important target in cancer that still needs to be clarified. There is the necessity of understanding CSC better to elucidate their role in carcinogenesis of HPV-negative OPC. Therefore, in this study we focused on characterization of CSC in HPV-negative OPC cell line UTSCC-60A to carry our understanding to a higher level for possible treatment of OPC via CSC.
Materials and methods
Cell culture
UTSCC-60A (University of Turku- Squamous Carcinoma Cell 60A) cell line, which was used for the experiments, is HPV-negative oropharyngeal squamous cell carcinoma generated from left tonsil cancer, kindly provided by Reidar Grenman, Department of Otorhinolaryngology, Turku University, Finland (18–20). UTSCC-60A was cultured in Dulbecco's modified Eagle's medium (DMEM, Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (HyClone), 1% antibiotics (Thermo Fisher Scientific, Inc.) and, 1% GlutaMax supplement (Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere of 5% CO2 in air. Cells were passaged at 4–5 days.
Isolation of CSC
CSCs were isolated through magnetic activated cell sorting. Based on the cell number tyrpsinized (0.25% Trypsin-EDTA, Thermo Fisher Scientific, Inc.), magnetic beads (goat anti-mouse IgG magnetic beads, NEB) ratio was calculated. Trypsinized cells were re-suspended in 1 ml of cold PBS and kept in 4°C until combined with the magnetic beads. For 1×107 cells 0.25 µl of magnetic beads were washed with 1 ml of cold phosphate buffered saline (PBS, Thermo Fisher Scientific, Inc.) three times. This wash step included 2 min on the rotator followed by 2 min on the magnetic rack, discarding the PBS without disturbing the magnetic bead pellet every time. Then, magnetic beads were incubated with the first antibody mouse anti-ALDH1A1 (ab105920; Abcam) at 4°C for one and a half hour. Magnetic beads and antibody ratio were 1:1. After antibody incubation, magnetic beads were washed with cold PBS four times as same as mentioned above. Cells that were trypsinized previously and kept at 4°C were combined with magnetic beads and incubated at 4°C for half an hour. Cell and magnetic bead suspension were carried to magnetic rack for separation and kept on magnetic rack for 15 min. Without disturbing the cell-magnetic bead pellet supernatant was carried to a clean centrifuge tube representing nonCSC. Cell-magnetic bead pellet was re-suspended in CSC medium displaying the CSC population.
Sphere formation assay
Isolated CSCs were plated to ultra-low attachment surface 96U-well plate (Thermo Fisher Scientific, Inc.) in CSC medium: DMEM, B27 2%, N2 1%, 10 ng/ml human epidermal growth factor (EGF), 20 ng/ml human fibroblast growth factor (FGF), and antibiotics 1%. Everyday cells were supplemented with fresh EGF and FGF. Spheroids were evaluated under microscope.
RT-qPCR
TriPure (Roche, Basel, Switzerland) was applied directly to the isolated CSC and nonCSC followed by the manufacturer's protocol. Before cDNA synthesis from CSC and nonCSC samples, samples were treated with DNase (Thermo Fisher Scientific, Inc.) following manufacturer's specifications. 100 ng of total RNA for each sample was converted to first strand cDNA (RevertAid First Strand cDNA Synthesis kit, Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. All samples including controls; reverse transcriptase negative, no template negative, positive control; were amplified to confirm the cDNA synthesis without DNA contamination. SyberGreen (Thermo Fisher Scientific, Inc.) was used for RT-qPCR reaction on Applied Biosystems 7500 Real Time PCR system. CSC markers OCT4, SOX2, BMI1, CD133, and KLF4 RNA levels, quadruplicate samples for each marker were evaluated for 3 times. Gene specific amplification was normalized to β-actin expression. Relative quantification results were calculated with ΔΔCq method (21) through manufacturer's Sequence Detection System (SDS) software protocol.
Immunofluorescence staining
Isolated CSCs were cultured in CSC medium for 3 days for immunofluorescence staining. Then spheres were carried to chamber slides and incubated at 37°C in a humidified atmosphere of 5% CO2 in air, overnight. After spheres' attachment to the surface of the chamber slide were confirmed under microscope CSCs were fixed with 4% PFA/PBS and incubated at room temperature for 20 min. After wash steps, blocking buffer was applied, which was followed by first antibody, mouse monoclonal Anti-ALDH1A1 (abcam ab105920), 1:600 dilution application overnight at 4°C. Next day slides were washed with PBS than incubated with 1:1,000 dilution second antibody (goat anti-mouse IgG; Abcam) 1 h in dark at room temperature. Twice washed with PBS, nuclei staining (DAPI, Thermo Fisher Scientific, Inc.) were performed. After last wash step, chambers were removed from the slides and slides were mounted. CSCs were examined under microscope (Nikon Eclipse, Ts2). nonCSC immunofluorescence staining was performed as the same way as mentioned above for CSC, except the sphere formation step.
Cell proliferation assay
Triplicate samples for each dosage of Cisplatin (Sigma) for CSCs and nonCSCs plated to 96 well tissue culture plates as 5×103 cells/well and incubated at 37°C in a humidified atmosphere of 5% CO2 in air. After CSCs and nonCSCs attachment to the wells confirmed under microscope, cells were treated with Cisplatin for 24 h. Cisplatin dosage was used between 40 µg/ml-0.3125 µg/ml. 50 µl of XTT (Roche Cell Proliferation kit II; Roche) assay was added to the each well and absorbance was read at 490–660 nm using microplate reader Corona SH-9000Lab, following 4 h of incubation. Background was measured through average of wells containing only medium. Cell proliferation experiments were repeated twice with triplicate samples each time. For statistical significance the Student's t-test was determined.
Statistical analysis
Student's t-test was used to calculate the significance of RT-qPCR and cell proliferation assay results between CSC and nonCSC. P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated at least twice with at least triplicate samples.
Results
CSC spheres of UTSCC-60A cell line
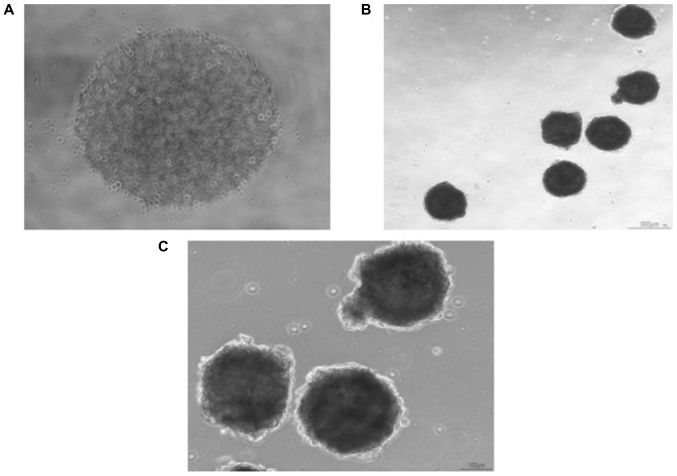
Before CSC isolation, cancer cells were counted (Invitrogen) and after isolation CSCs were counted. In UTSCC-60A cell-line there was only 0.6% of CSC which represented a small population compared to nonCSC. Isolated CSCs were plated as 2×103 in each well of 96 well-plate U-bottom with CSC medium. Spheres in each well could be observed under microscope (Fig. 1A). At the 3rd-4th day CSC spheres could be pictured after carrying to a flat-bottom well plates (Fig. 1B and C). Morphological images of spheres generated from UTSCC-60A were highly dense and dark.
Figure 1.
Sphere formation of CSCs of the UTSCC-60A cell line. (A) First day of sphere formation in ultra-low attachment 96 well-plate. Magnification, ×4. (B) CSC spheres cultured for 3 days in 96 well-plate were carried to 12 well-plate ultra-low attachment. Highly dark holoclones were observed. Magnification, ×4. (C) CSC spheres of UTSCC-60A cell line. Magnification, ×10. CSC, cancer stem cell.
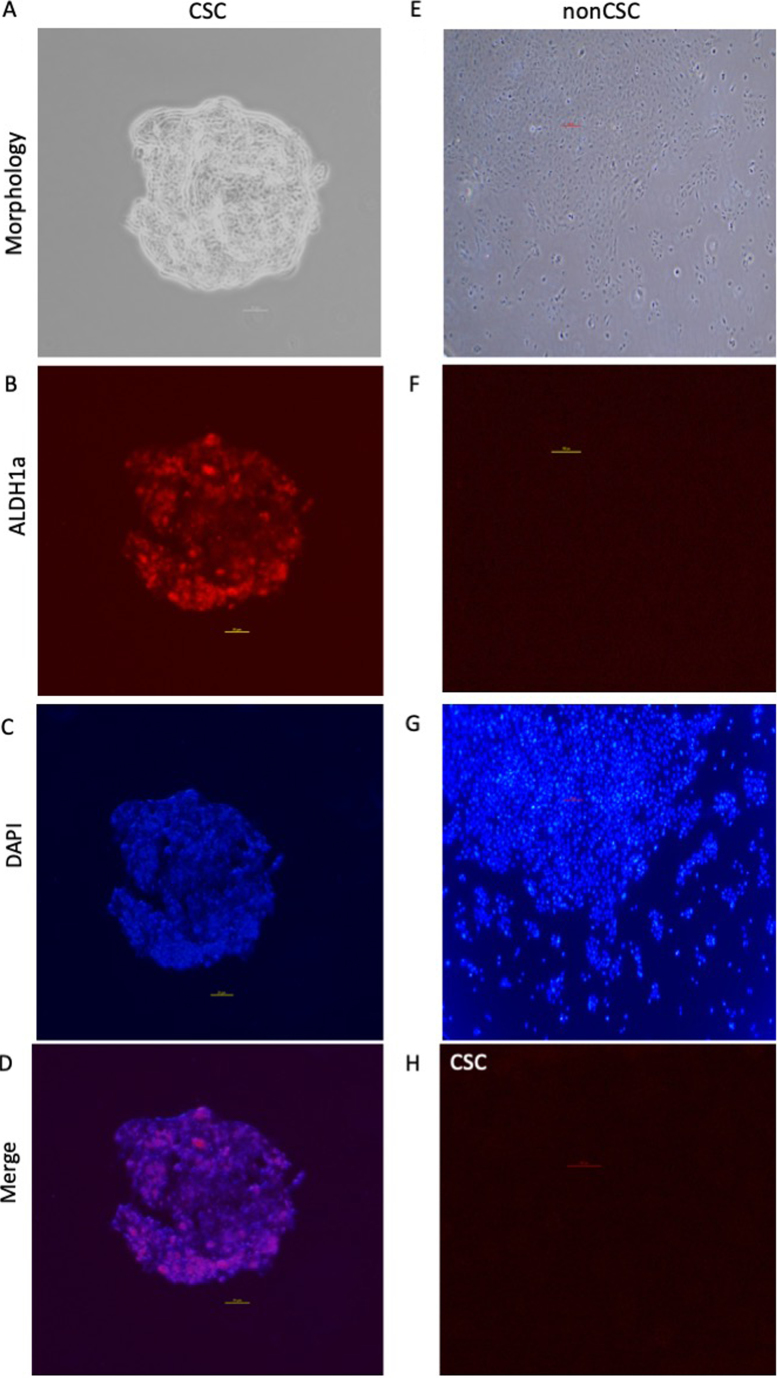
Immunofluorescence staining of spheres
For both cell type mouse Anti-ALDH1A1 (ab105920; Abcam) antibody was used for immunofluorescence staining. As expected, while ALDH1a expression could be observed in CSC spheres, there was no ALDH1a expression in nonCSC. This result confirmed the successful isolation of CSCs. In CSC ALDH1a expression was specific to spheroid formation. Negative control (no primary antibody) didn't display any ALDH1a expression for both cell lines representing clearance of undesired background staining. DAPI could be detected for both cell type, displaying different patterns for CSC and nonCSC (Fig. 2).
Figure 2.
Representative images from immunofluorescence staining of CSC spheres and non-CSC UTSCC-60A cells. Left column represents CSC, right column represents non-CSCs, except in (H). Magnification, ×10 (A-D) or ×4 (E-H). (A) Phase contrast image of CSC sphere. (B) ALDH1a expression in CSC sphere. (C) Nucleic staining of CSC sphere. (D) Merged image of ALDH1a and DAPI staining of CSC sphere. (E) Phase contrast image of non-CSC. (F) Control of non-CSC stained with ALDH1a antibody. (G) Nucleic staining of non-CSC. (H) Negative control of CSC (no ALDH1a antibody). ALDH1, aldehyde dehydrogenase 1; CSC, cancer stem cell.
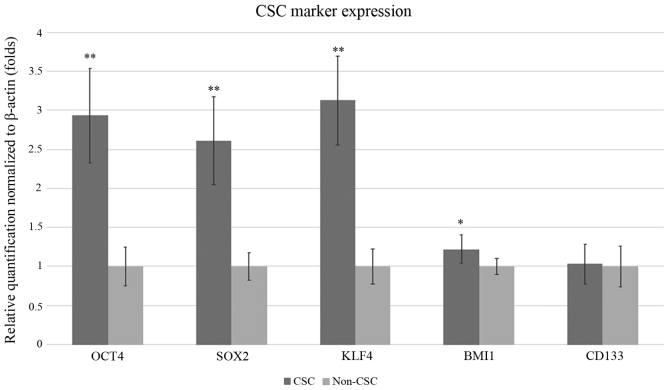
CSC marker expression by RT-qPCR
CSC markers OCT4, SOX2, KLF4 and BMI1were expressed significantly higher in CSC when compared to nonCSC. OCT4, SOX2, KLF4 and BMI1 were respectively 2.33, 2.53, 2.68, and 1.34 folds higher in CSC. For OCT4, SOX2 and KLF4 were all P<0.01, respectively P=0.0009, P=0.0001, P=0.0043, P=0.0013. CD133 expression was almost at the same level in both cell type (Fig. 3). CD133 gene expression was only 1.03-fold higher and P=0.91.
Figure 3.
Reverse transcription-quantitative PCR results of CSC marker expression normalized to β-actin expression. Relative expression of CSC and non-CSC for markers CD133, OCT4, SOX2, KLF4 and BMI1 are presented as fold changes. CSC expression levels for markers CD133, OCT4, SOX2, KLF4 and BMI1 were compared to non-CSC expression levels of the same genes. Error bars represent the mean ± SD of at least quadruplicate samples of three independent experiments. *P<0.05 and **P<0.01 vs. non-CSC. CSC, cancer stem cell; KLF4, Kruppel-like factor; BMI1, BMI1 proto-oncogene, polycomb ring finger.
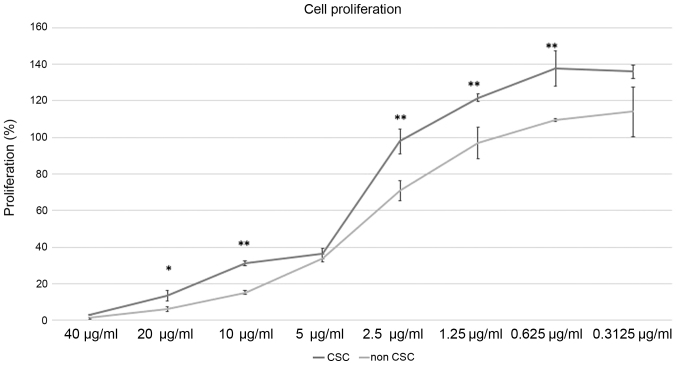
Effect of Cisplatin treatment to CSC and nonCSC
As CSC are thought to be resistant to chemotherapeutics we confirmed their proliferation compared to nonCSC after cisplatin treatment. We applied different doses of cisplatin between 40 to 0.3125 µg/ml. Our results displayed CSC to be more resistant to cisplatin (Fig. 4). For 20, 10, 2.5, 1.25, 0.625 µg/ml cisplatin treatment, cell proliferation percentages were significantly higher in CSC. P-values for these dosages were respectively P=0.017, P=0.0006, P=0.006, P=0.008, P=0073. For dosages of 5 and 0.3125 µg/ml cell proliferation rates weren't significant, P=0.32 and P=0.06 respectively.
Figure 4.
XTT-assay results of CSC and non-CSC after 24-h cisplatin treatment. Cell proliferation percentages are shown for 40–0.3125 µg/ml. Cell proliferation percentages for CSC and non-CSC were compared for the indicated dosages following 24-h cisplatin treatment. Error bars represent the mean ± SD of at least triplicate samples of two independent experiments. *P<0.05 and **P<0.01 vs. non-CSC. CSC, cancer stem cell.
Discussion
HPV-negative cases still remain problem for the treatment of OPC. It is known that they have different genetic background but their resistance to chemoradiation needs to be elucidated. Recent literature supports CSCs might be the reason of resistance to therapy in various kinds of cancers including HNSCC (22–25). As current anticancer therapies don't target CSCs specifically, instead of eradicating them they may even be expanding resistant CSC clones (16). Ionizing radiation itself can induce CSC properties (26–28). Cisplatin as well, is thought to be whether a chemotherapeutic that CSCs are resistant to or induces CSC properties in various types of cancer (29–32).CSC proportion in HPV- negative and HPV-positive HNSCC is challenging. Based on the data of their study Tang et al (33) stated that HPV status does not correlate with the CSC proportion of HNSCC and concluded that CSCs are more resistant to cisplatin than nonCSCs, which is irrelevant to HPV status. In another study investigating the effect of irradiation on CSC subject to HPV status, it was demonstrated that HPV-negative HNSCC cell line UM-SCC-1 had lower CSC proportion than the HPV-positive HNSCC cell line UM-SCC-47 and after irradiation both cell lines had elevated CSC proportion varied in course of time, but only HPV-positive cell line displayed significant plasticity in repopulating CSC phenotype in depleted cultures (34). Additionally, Zhang et al (35) demonstrated that HPV-positive HNSCC has higher CSC population than HPV-negative HNSCC and proposed that rather than absolute CSC number CSC phenotype might be more important for disease aggressiveness. Contradictorily, Vlashi et al (36) supported that HPV-negative cell lines have a higher proportion of CSC and radiation induced dedifferentiation of head and neck cancer cells into CSC depending on the HPV status. In the present study we detected coherent data of low CSC proportion in HPV-negative OPC cell line. Together with the low proportion of CSC our data displayed significantly high expression of CSC markers OCT4, SOX2, KLF4 and BIM1 compared to nonCSC indicating CSC phenotype.
Recently it is known that CSC may also originate from differentiated cells and nonCSC, and there are different phenotypes of CSC, displaying genetic heterogeneity (37,38). Almeida et al (39) defined holospheres, merospheres and paraspheres in HNSCC cell lines and suggested that CSCs with enhanced stemness and invasive potential are a specific population that exist in holospheres. Our morphological data also displayed highly dark and dense holospheres of CSCs.
Aldehyde dehydrogenase 1 (ALDH1) is an informative marker of CSC of HNSCC (40–43). In a systematic review of HNSCC, CSC markers used in many of the studies are defined as ALDH1, CD44 and BMI1, and in which ALDH1 is correlated tumor aggressiveness, tumor size, presence of lymph node metastasis and tumor staging (44). Likewise, a meta-analysis declared that ALDH1 positive patients had worse prognosis associated with common clinicopathological features and poor prognostic factors (45). Also, Yata et al showed that increased ALDH1 activity HNSCC cells represent CSC (46). We have displayed the expression of ALDH1a expression in tumor spheres of UTSCC-60A cell line while confirming CSC isolation.
CSC is a promising target for cancer treatment. Macha et al (47) targeted CSC via afitinib, a pan-EGFR-TKI, in HNSCC cell lines and showed that afitinib inhibited formation and growth of tumor spheres which also demonstrated significant radio-sensitization. Setúbal Destro Rodrigues et al (48) examined effects of Cetuximab and Erlotinib on the cell sub-populations in HNSCC cell lines and showed that EGFR blocking reduced cell proliferation, reduced motility of EMT-CSCs and increased sensitivity of Epi-CSCs to chemoradiation by inducing their differentiation. Also, the decrease of CSC marker expression, downregulation of migration and invasion, and quiescence of epithelial to mesenchymal transition was shown by targeting CSC in HNSCC through novel C-terminal Hsp90 inhibitors (KU711 and KU757) (49). Our data displayed that CSC were more resistant to cisplatin treatment than nonCSC (Fig. 4) and for all doses except 5 and 0.3125 µg/ml cell proliferation rate was significantly higher for CSC.
CSC studies shoulder great importance on understanding CSC mechanism in carcinogenesis and how will this information be translated to therapy. Even though this is a comprehend step because of the underlying complex mechanisms and genetic heterogeneity, recent improvements in CSC information is promising. Eventually it can be proposed that not only targeting cancer cells but also targeting CSC in cancer specifically is probably an important advancement in cancer treatment.
In this study, CSC of HPV-negative OPC cell line is characterized. It's been shown that CSC proportion is low but CSC markers are highly expressed and CSCs are more resistant to cisplatin treatment than nonCSC. The present study suggests that even the proportion of CSC may be low in HPV-negative OPC, CSC properties may be an important factor in resistance to cisplatin, which needs further investigation to define the underlying mechanisms.
Acknowledgements
The authors would like to thank Professor Reidar Grenman (Department of Otorhinolaryngology-Head and Neck Surgery, Turku University and Turku University Hospital, Turku, Finland) for providing the UTSCC-60A cell line.
Funding
The present study was supported by a grant to MG from the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant no. 17K11401).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
MG designed the study, acquired the data, analyzed the data and prepared the manuscript. EG acquired the data, analyzed the data, performed the statistical analysis and prepared the manuscript. ST designed the study and edited the manuscript. KE performed the statistical analysis and edited the manuscript. MH designed the study, controlled the quality of the data and reviewed the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Božinović K, Sabol I, Dediol E, Milutin Gašperov N, Manojlović S, Vojtechova Z, Tachezy R, Grce M. Genome-wide miRNA profiling reinforces the importance of miR-9 in human papillomavirus associated oral and oropharyngeal head and neck cancer. Sci Rep. 2019;9:2306. doi: 10.1038/s41598-019-38797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohan DM, Popat S, Kaplan SE, Rigual N, Loree T, Hicks WL., Jr Oropharyngeal cancer: Current understanding and management. Curr Opin Otolaryngol Head Neck Surg. 2009;17:88–94. doi: 10.1097/MOO.0b013e32832984c0. [DOI] [PubMed] [Google Scholar]
- 3.Preuss SF, Quante G, Semrau R, Mueller RP, Klussmann JP, Guntinas-Lichius O. An analysis of surgical complications, morbidity, and cost calculation in patients undergoing multimodal treatment for operable oropharyngeal carcinoma. Laryngoscope. 2007;117:101–105. doi: 10.1097/01.mlg.0000246191.90021.5e. [DOI] [PubMed] [Google Scholar]
- 4.Biktasova A, Hajek M, Sewell A, Gary C, Bellinger G, Deshpande HA, Bhatia A, Burtness B, Judson B, Mehra S, et al. Demethylation therapy as a targeted treatment for human papillomavirus-associated head and neck cancer. Clin Cancer Res. 2017;23:7276–7287. doi: 10.1158/1078-0432.CCR-17-1438. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network, corp-author. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 7.Xie X, Teknos TN, Pan Q. Are all cancer stem cells created equal? Stem Cells Transl Med. 2014;3:1111–1115. doi: 10.5966/sctm.2014-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identificationof human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 11.Ricci-Vitiani L, Pagliuca A, Palio E, Zeuner A, De Maria R. Colon cancer stem cells. Gut. 2008;57:538–548. doi: 10.1136/gut.2007.127837. [DOI] [PubMed] [Google Scholar]
- 12.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, Dombkowski D, Preffer F, Maclaughlin DT, Donahoe PK. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci USA. 2006;103:11154–11159. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albers AE, Chen C, Köberle B, Qian X, Klussmann JP, Wollenberg B, Kaufmann AM. Stem cells in squamous head and neck cancer. Crit Rev Oncol Hematol. 2012;81:224–240. doi: 10.1016/j.critrevonc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Morrison R, Schleicher SM, Sun Y, Niermann KJ, Kim S, Spratt DE, Chung CH, Lu B. Targeting the mechanisms of resistance to chemotherapy and radiotherapy with the cancer stem cell hypothesis. J Oncol. 2011;12011:941876. doi: 10.1155/2011/941876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, Los M. Cancer stem cell markers in common cancers-therapeutic implications. Trends Mol Med. 2008;14:450–460. doi: 10.1016/j.molmed.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Dingli D, Michor F. Successful therapy must eradicate cancer stem cells. Stem Cells. 2006;24:2603–2610. doi: 10.1634/stemcells.2006-0136. [DOI] [PubMed] [Google Scholar]
- 18.Laban S, Steinmeister L, Gleißner L, Grob TJ, Grénman R, Petersen C, Gal A, Knecht R, Dikomey E, Kriegs M. Sorafenib sensitizes head and neck squamous cell carcinoma cells to ionizing radiation. Radiother Oncol. 2013;109:286–292. doi: 10.1016/j.radonc.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Maushagen R, Reers S, Pfannerstill AC, Hahlbrock A, Stauber R, Rahmanzadeh R, Rades D, Pries R, Wollenberg B. Effects of paclitaxel on permanent head and neck squamous cell carcinoma cell lines and identification of anti-apoptotic caspase 9b. J Cancer Res Clin Oncol. 2016;142:1261–1271. doi: 10.1007/s00432-016-2150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamagawa S, Beder LB, Hotomi M, Gunduz M, Yata K, Grenman R, Yamanaka N. Role of miR-200c/miR-141 in the regulation of epithelial-mesenchymal transition and migration in head and neck squamous cell carcinoma. Int J Mol Med. 2014;33:879–886. doi: 10.3892/ijmm.2014.1625. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJS, chmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott SC, Rodriguez-Ramirez C, McDermott SP, Wicha MS, Nör JE. FGFR signaling regulates resistance of head and neck cancer stem cells to cisplatin. Oncotarget. 2018;9:25148–25165. doi: 10.18632/oncotarget.25358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018:5416923. doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prieto-Vila M, Takahashi RU, Usuba W, Kohama I, Ochiya T. Drug resistance driven by cancer stem cells and their niche. Int J Mol Sci. 2017;18(pii):E2574. doi: 10.3390/ijms18122574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghisolfi L, Keates AC, Hu X, Lee DK, Li CJ. Ionizing radiation induces stemness in cancer cells. PLoS One. 2012;7:e43628. doi: 10.1371/journal.pone.0043628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-Casal R, Bhattacharya C, Ganesh N, Bailey L, Basse P, Gibson M, Epperly M, Levina V. Non-small cell lung cancer cells survived ionizing radiation treatment display cancer stem cell and epithelial-mesenchymal transition phenotypes. Mol Cancer. 2013;12:94. doi: 10.1186/1476-4598-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Kim KH, Kim DG, Cho HJ, Kim Y, Rheey J, Shin K, Seo YJ, Choi YS, Lee JI, Lee J, Joo KM, Nam DH. FoxM1 promotes stemness and radio-resistance of glioblastoma by regulating the master stem cell regulator sox2. PLoS One. 2015;10:e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barr MP, Gray SG, Hoffmann AC, Hilger RA, Thomale J, O'Flaherty JD, Fennell DA, Richard D, O'Leary JJ, O'Byrne KJ. Generation and characterization of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PLoS One. 2013;8:e54193. doi: 10.1371/journal.pone.0054193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh RD, Ghuwalewala S, Das P, Mandloi S, Alam SK, Chakraborty J, Sarkar S, Chakrabarti S, Panda CK, Roychoudhury S. MicroRNA profiling of cisplatin-resistant oral squamous cell carcinoma cell lines enriched with cancer-stem-cell-like and epithelial-mesenchymal transition-type features. Sci Rep. 2016;6:23932. doi: 10.1038/srep23932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki H, Takahashi RU, Prieto-Vila M, Kawamura Y, Kondo S, Shirota T, Ochiya T. CD44 exerts a functional role during EMT induction in cisplatin-resistant head and neck cancer cells. Oncotarget. 2018;9:10029–10041. doi: 10.18632/oncotarget.24252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang P, Xu C, Zhou M, Zhou H, Dong W, Wu X, Chen A, Feng Q. RXRα-enriched cancer stem cell-like properties triggered by CDDP in head and neck squamous cell carcinoma (HNSCC) Carcinogenesis. 2018;39:252–262. doi: 10.1093/carcin/bgx138. [DOI] [PubMed] [Google Scholar]
- 33.Tang AL, Owen JH, Hauff SJ, Park JJ, Papagerakis S, Bradford CR, Carey TE, Prince ME. Head and neck cancer stem cells: The effect of HPV-an in vitro and mouse study. Otolaryngol Head Neck Surg. 2013;149:252–60. doi: 10.1177/0194599813486599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid P, Wilson P, Li Y, Marcu LG, Staudacher AH, Brown MP, Bezak E. In vitro investigation of head and neck cancer stem cell proportions and their changes following X-ray irradiation as a function of HPV status. PLoS One. 2017;12:e0186186. doi: 10.1371/journal.pone.0186186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Kumar B, Piao L, Xie X, Schmitt A, Arradaza N, Cippola M, Old M, Agrawal A, Ozer E, et al. Elevated intrinsic cancer stem cell population in human papillomavirus-associated head and neck squamous cell carcinoma. Cancer. 2014;120:992–1001. doi: 10.1002/cncr.28538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlashi E, Chen AM, Boyrie S, Yu G, Nguyen A, Brower PA, Hess CB, Pajonk F. Radiation-induced dedifferentiation of head and neck cancer cells into cancer stem cells depends on human papillomavirus status. Int J Radiat Oncol Biol Phys. 2016;94:1198–1206. doi: 10.1016/j.ijrobp.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, Costea DE, Mackenzie IC. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71:5317–5326. doi: 10.1158/0008-5472.CAN-11-1059. [DOI] [PubMed] [Google Scholar]
- 39.Almeida LO, Guimarães DM, Squarize CH, Castilho RM. Profiling the behavior of distinct populations of head and neck cancer stem cells. Cancers (Basel) 2016;8(pii):E7. doi: 10.3390/cancers8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophy Res Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 41.Sayed SI, Dwivedi RC, Katna R, Garg A, Pathak KA, Nutting CM, Rhys-Evans P, Harrington KJ, Kazi R. Implications of understanding cancer stem cell (CSC) biology in head and neck squamous cell cancer. Oral Oncol. 2011;47:237–243. doi: 10.1016/j.oraloncology.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 43.Qian X, Wagner S, Ma C, Coordes A, Gekeler J, Klussmann JP, Hummel M, Kaufmann AM, Albers AE. Prognostic significance of ALDH1A1-positive cancer stem cells in patients with locally advanced, metastasized head and neck squamous cell carcinoma. J Cancer Res Clin Oncol. 2014;140:1151–1158. doi: 10.1007/s00432-014-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curtarelli RB, Gonçalves JM, Dos Santos LGP, Savi MG, Nör JE, Mezzomo LAM, Rodríguez Cordeiro MM. Expression of cancer stem cell biomarkers in human head and neck carcinomas: A systematic review. Stem Cell Rev. 2018;14:769–784. doi: 10.1007/s12015-018-9839-4. [DOI] [PubMed] [Google Scholar]
- 45.Zhou C, Sun B. The prognostic role of the cancer stem cell marker aldehyde dehydrogenase 1 in head and neck squamous cell carcinomas: A meta-analysis. Oral Oncol. 2014;50:1144–1148. doi: 10.1016/j.oraloncology.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 46.Yata K, Beder LB, Tamagawa S, Hotomi M, Hirohashi Y, Grenman R, Yamanaka N. MicroRNA expression profiles of cancer stem cells in head and neck squamous cell carcinoma. Int J Oncol. 2015;47:1249–1256. doi: 10.3892/ijo.2015.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macha MA, Rachagani S, Qazi AK, Jahan R, Gupta S, Patel A, Seshacharyulu P, Lin C, Li S, Wang S, et al. Afatinib radiosensitizes head and neck squamous cell carcinoma cells by targeting cancer stem cells. Oncotarget. 2017;8:20961–20973. doi: 10.18632/oncotarget.15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Setúbal Destro Rodrigues MF, Gammon L, Rahman MM, Biddle A, Nunes FD, Mackenzie IC. Effects of cetuximab and erlotinib on the behaviour of cancer stem cells in head and neck squamous cell carcinoma. Oncotarget. 2018;9:13488–13500. doi: 10.18632/oncotarget.24416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian C, Kovatch KJ, Sim MW, Wang G, Prince ME, Carey TE, Davis R, Blagg BSJ, Cohen MS. Novel c-terminal Heat shock protein 90 inhibitors (KU711 and Ku757) are effective in targeting head and neck squamous cell carcinoma cancer stem cells. Neoplasia. 2017;19:1003–1011. doi: 10.1016/j.neo.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.