Abstract
Accumulating evidence has demonstrated that thioredoxin interacting protein (TXNIP) is abnormally expressed in a variety of malignant tumors and functions as a tumor suppressor. However, the association between TXNIP and clear cell renal cell carcinoma (CCRCC) has not yet been fully elucidated. The aim of the present study was to evaluate the role of TXNIP in CCRCC using The Cancer Genome Atlas (TCGA) database. The RNA sequencing data and corresponding clinical data were collected from TCGA database. The association between TXNIP and patient clinicopathological characteristics was analyzed using analysis of variance and logistic regression. The Kaplan-Meier method and Cox proportional hazards model were used to assess the association between TXNIP and overall survival. Gene Set Enrichment Analysis (GSEA) was used to explore the associated signaling pathways. TXNIP expression was identified to be decreased in CCRCC tissues compared with normal tissues. The decreased expression of TXNIP in CCRCC was significantly associated with clinical stage [OR=0.509 for III vs. I (P=0.002); OR=0.527 for IV vs. I (P=0.012)], T stage [OR=0.552 for T3 vs. T1 (P=0.002)] and grade [OR=0.261 for G4 vs. G1 (P=0.027)]. Kaplan-Meier survival analysis indicated that cases of CCRCC with low TXNIP expression were associated with poorer prognoses compared with those with a high expression level (P=0.002). Univariate and multivariate Cox analyses indicated that TXNIP was an independent prognostic factor in CCRCC. GSEA revealed that 6 pathways exhibited significant differential enrichment in the TXNIP high-expression phenotype, including the WNT signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, the phosphatidylinositol signaling system, the transforming growth factor-β (TGF-β) signaling pathway, autophagy and the Janus kinase (JAK)-STAT signaling pathway. Taken together, the results of the present study indicate that TXNIP expression may be a potential prognostic marker for patients with CCRCC. In addition, the WNT signaling pathway, MAPK signaling pathway, phosphatidylinositol signaling system, TGF-β signaling pathway, autophagy and the JAK-STAT signaling pathway may be the crucial pathways regulated by TXNIP in CCRCC.
Keywords: clear cell renal cell carcinoma, thioredoxin interacting protein, The Cancer Genome Atlas, prognosis, Gene Set Enrichment Analysis
Introduction
Renal cell carcinoma (RCC) is the most frequently occurring type of kidney cancer (85%). RCC has the highest malignancy and mortality rates among all types of urogenital cancer. Clear cell renal cell carcinoma (CCRCC) is the most common type of RCC, accounting for almost 85% of the reported cases of RCC (1). Although improvements in both chemotherapy and surgical techniques have been made, the prognosis of certain patients with CCRCC remains poor, and advanced CCRCC continues to be fatal due to tumor recurrence and metastasis (2). In addition, the pathogenesis of CCRCC remains unknown, and the underlying molecular mechanisms are complex. Therefore, investigating the pathogenesis and identifying novel biomarkers is of the utmost importance for the early detection and treatment of patients with CCRCC.
Thioredoxin interacting protein (TXNIP) is a small 38 kDa protein that was identified to be a vitamin D3-induced gene, and it was originally named vitamin D3 upregulated protein 1 (3). Another study indicated that TXNIP binds to and negatively modulates the activity of thioredoxin (TRX) (4). A previous study demonstrated that TXNIP deficiency stimulates transforming growth factor-β (TGF-β)-induced epithelial-mesenchymal transition, indicating that TXNIP may suppress tumor progression (5). In gastroesophageal adenocarcinoma, TXNIP has been identified to be an independent prognostic factor for distant metastasis-free survival and overall survival (6). Additionally, TXNIP has been demonstrated to serve a role as a tumor suppressor gene by directly inducing the expression of the tumor suppressor protein p53 (7). Accumulating evidence has demonstrated that TXNIP is abnormally expressed in a variety of malignant tumors, and functions as a tumor suppressor (8) in malignancies such as hepatoma (9), breast cancer (10), pancreatic ductal adenocarcinoma (11) and esophageal adenocarcinoma (12). However, the association between TXNIP and CCRCC is yet to be fully elucidated.
In the present study, the association between TXNIP expression and the clinicopathological characteristics of patients with CCRCC was evaluated, and the prognostic value of TXNIP in CCRCC was analyzed based on data obtained from The Cancer Genome Atlas (TCGA). Furthermore, the biological pathways associated with TXNIP were determined using Gene Set Enrichment Analysis (GSEA), which may provide further insight into the molecular mechanisms of CCRCC.
Patients and methods
RNA sequencing and clinical information
The RNA sequencing data and corresponding clinical data were collected from TCGA database (https://cancergenome.nih.gov/). A total of 539 CCRCC tissues and 72 normal tissues were obtained. Ultimately, 529 cases were selected for subsequent analysis, while cases without clinical prognostic information were excluded. Detailed clinical information, including age at diagnosis, sex, T stage, lymph-node status, metastasis and clinical stage was recorded. Any information that was unavailable or unknown was regarded as missing.
GSEA
GSEA was used to evaluate whether an a priori set of genes exhibited significant differential expression between the high- and low-TXNIP groups (13,14). The TXNIP mRNA expression level was used as a phenotype label. In total, 1,000 gene set permutations were performed for each analysis. The nominal P-value, false discovery rate (FDR) and normalized enrichment scores (NES) were used to classify the signaling pathways enriched in each phenotype.
Statistical analysis
All statistical analyses were performed using R (v.3.6.0) software and SPSS v.24.0 software (IBM Corp.). Comparison of the expression levels of TXNIP between CCRCC and normal groups was performed using an unpaired Student's t-test, and paracancerous groups with a paired t-test. Based on the median value for TXNIP expression (274.17), patients with CCRCC were divided into high- and low-risk groups. Analysis of variance followed by a Least Significance Difference post hoc test and logistic regression were used to analyze the TXNIP expression and pathological parameters of CCRCC. Kaplan-Meier survival analysis was used with the log-rank test to compare the overall survival between the high- and low-TXNIP expression groups. The univariate and multivariate Cox proportional hazards model was used to evaluate the prognostic value of TXNIP expression. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulated TXNIP expression in CCRCC
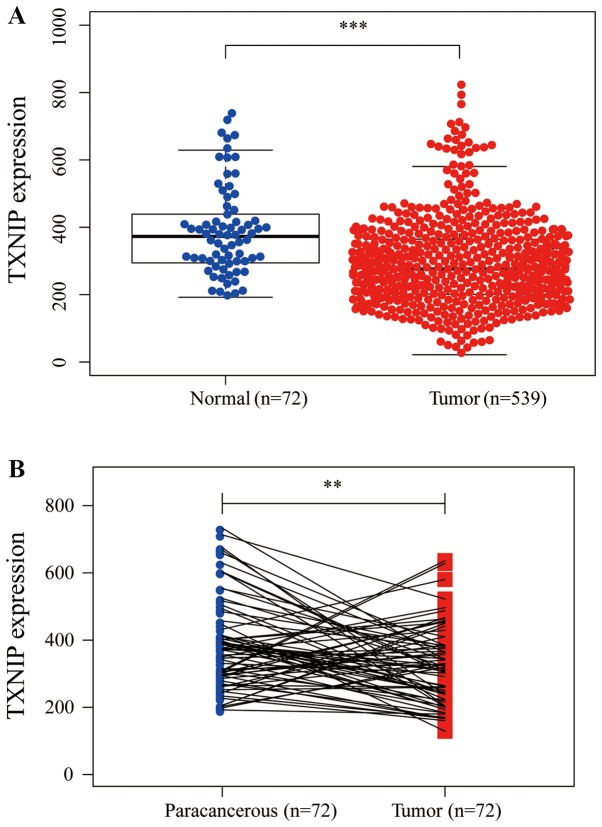
TXNIP expression was investigated in 539 CCRCC tissues and 72 normal tissues. The results indicated that TXNIP expression was decreased in the CCRCC tissues compared with in the normal tissues (P<0.001; Fig. 1A). Furthermore, the expression of TXNIP in 72 pairs of CCRCC tissue and paracancerous tissue were also investigated; the results revealed that TXNIP was downregulated in CCRCC tissues (P<0.01; Fig. 1B), demonstrating that TXNIP may suppress CCRCC tumorigenesis.
Figure 1.
TXNIP is markedly downregulated in CCRCC tissues compared with normal or paracancerous tissues. (A) TXNIP expression in cancer tissues was significantly decreased compared with normal tissues. ***P<0.001. (B) TXNIP expression was significantly decreased in CCRCC compared with 72 pairs of paracancerous tissues. **P<0.01. TXNIP, thioredoxin interacting protein; CCRCC, clear cell renal cell carcinoma.
Patient clinical characteristics
From TCGA database, 529 tumors with both gene expression data and clinical parameters were analyzed. The clinical characteristics of the patients including age, sex, metastasis, lymph-node status, clinical stage, T stage and grade are presented in Table I.
Table I.
Clinical characteristics of patients with renal clear cell carcinoma.
| Variable | Cases, n | Percentage |
|---|---|---|
| Age at diagnosis, years | ||
| ≤60 | 262 | 49.53 |
| >60 | 267 | 50.47 |
| Sex | ||
| Male | 341 | 64.46 |
| Female | 188 | 35.54 |
| Metastasis | ||
| M0 | 423 | 84.43 |
| M1 | 78 | 15.57 |
| Lymph-node status | ||
| N0 | 239 | 93.73 |
| N1 | 16 | 6.27 |
| Clinical stage | ||
| I | 265 | 50.09 |
| II | 56 | 10.59 |
| III | 125 | 23.63 |
| IV | 83 | 15.69 |
| T stage | ||
| T1 | 271 | 51.23 |
| T2 | 68 | 12.85 |
| T3 | 179 | 33.84 |
| T4 | 11 | 2.08 |
| Grade | ||
| G1 | 14 | 2.67 |
| G2 | 227 | 43.32 |
| G3 | 206 | 39.31 |
| G4 | 77 | 14.70 |
Associations between TXNIP expression and the clinical characteristics of patients with CCRCC
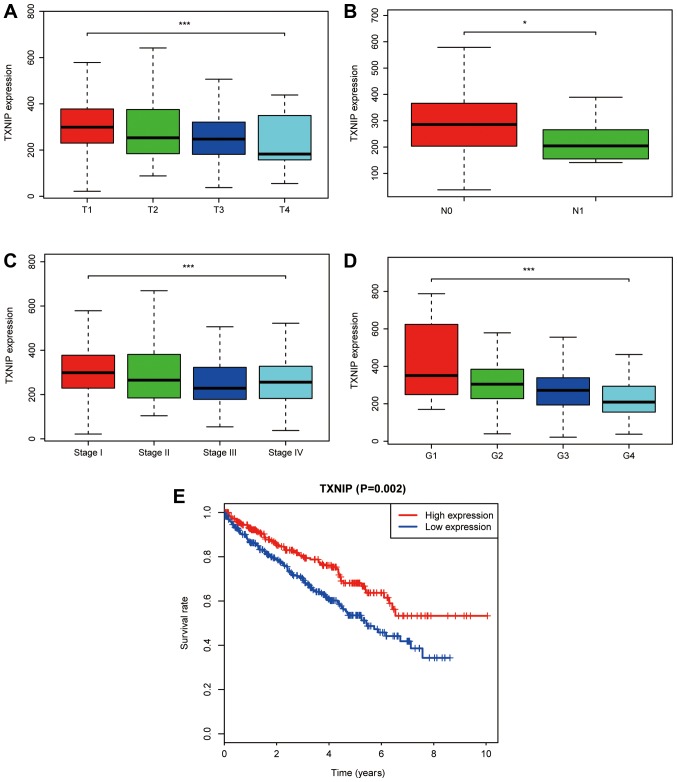
A total of 529 CCRCC samples with TXNIP expression data from TCGA database were analyzed. As shown in Fig. 2A-D, the decreased expression of TXNIP was significantly associated with T stage (P<0.001), lymph-node status (P<0.05), clinical stage (P<0.001) and grade (P<0.001).
Figure 2.
Association of TXNIP expression with patient clinicopathological characteristics. (A) T stage. (B) Lymph-node status. (C) Clinical stage. (D) Grade. (E) Effect of TXNIP expression on the overall survival of patients with CCRCC in The Cancer Genome Atlas datasets. TXNIP, thioredoxin interacting protein; CCRCC, clear cell renal cell carcinoma. *P<0.05 and ***P<0.001.
Univariate analysis using logistic regression indicated that decreased TXNIP expression was associated with poor prognostic features (Table II). The decreased expression of TXNIP in CCRCC was significantly associated with clinical stage [OR=0.509 for III vs. I (P=0.002); OR=0.527 for IV vs. I (P=0.012)], T stage [OR=0.552 for T3 vs. T1 (P=0.002)] and grade [OR=0.261 for G4 vs. G1 (P=0.027); Table II]. These results revealed that cases of CCRCC with a low TXNIP expression level were associated with poor clinical outcome.
Table II.
TXNIP expression is associated with patient clinical pathological characteristics.
| Clinical characteristics | Total | Odds ratio in TXNIP expression | P-value |
|---|---|---|---|
| Age (continuous) | 529 | 0.991 (0.977–1.005) | 0.250 |
| Sex (female vs. male) | 529 | 1.261 (0.883–1.806) | 0.203 |
| Metastasis ((M1 vs. M0) | 501 | 0.611 (0.357–1.037) | 0.069 |
| Lymph-node status (N1 vs. N0) | 255 | 0.737 (0.451–1.196) | 0.219 |
| Clinical stage (II vs. I) | 321 | 0.669 (0.375–1.188) | 0.170 |
| Clinical stage (III vs. I) | 390 | 0.509 (0.329–0.784) | 0.002 |
| Clinical stage (IV vs. I) | 348 | 0.527 (0.317–0.867) | 0.012 |
| T stage (T2 vs. T1) | 339 | 0.611 (0.357–1.037) | 0.069 |
| T stage (T3 vs. T1) | 450 | 0.552 (0.376–0.808) | 0.002 |
| T stage (T4 vs. T1) | 282 | 0.281 (0.060–0.994) | 0.065 |
| Grade (G2 vs. G1) | 241 | 0.745 (0.223–2.226) | 0.607 |
| Grade (G3 vs. G1) | 220 | 0.504 (0.151–1.511) | 0.234 |
| Grade (G4 vs. G1) | 91 | 0.261 (0.073–0.839) | 0.027 |
Data were analyzed using logistic regression. TXNIP, thioredoxin interacting protein.
Univariate and multivariate analysis of survival
As indicated in Fig. 2E, Kaplan-Meier survival analysis indicated that CCRCC with low TXNIP expression was associated with a poorer prognosis compared with CCRCC with high TXNIP expression (P=0.002). In univariate analysis, age (HR=1.033; P<0.001), metastasis (HR=4.284; P<0.001), clinical stage (HR=1.889; P<0.001), T stage (HR=1.941; P<0.001), grade (HR=2.293; P<0.001) and TXNIP expression (HR=0.652; P<0.001) were associated with the overall survival of patients with CCRCC. Using multivariate analysis, TXNIP expression was revealed to be an independent predictor of the prognosis of patients with CCRCC (HR=0.733; P=0.009; Table III).
Table III.
Univariate and multivariate cox regression analysis of patients with renal clear cell carcinoma.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age | 1.033 (1.019–1.047) | <0.001 | 1.035 (1.021–1.051) | <0.001 |
| Sex | 1.074 (0.779–1.481) | 0.663 | – | – |
| Metastasis | 4.284 (3.106–5.908) | <0.001 | 1.033 (0.664–2.555) | 0.442 |
| Lymph-node status | 0.865 (0.739–1.012) | 0.069 | – | – |
| Clinical stage | 1.889 (1.649–2.164) | <0.001 | 1.687 (1.078–2.640) | 0.022 |
| T stage | 1.941 (1.639–2.299) | <0.001 | 0.864 (0.574–1.301) | 0.485 |
| Grade | 2.293 (1.854–2.836) | <0.001 | 1.462 (1.148–1.861) | 0.002 |
| TXNIP | 0.652 (0.534–0.797) | <0.001 | 0.733 (0.581–0.926) | 0.009 |
TXNIP, thioredoxin interacting protein; HR, hazard ratio; CI, confidence interval.
Identification of TXNIP-associated signaling pathways using GSEA
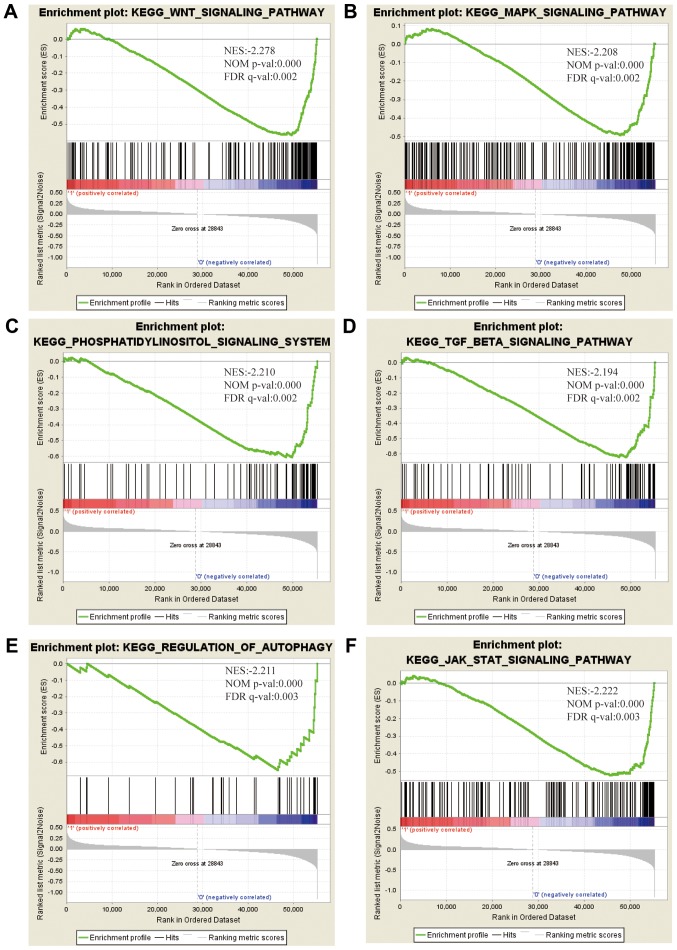
To elucidate the signaling pathway associated with TXNIP, GSEA was performed between the high- and low-TXNIP expression datasets. Significant differences were observed in the enrichment of the Molecular Signatures Database collection (FDR<0.05, and normalized P<0.05). Based on their NES, normalized (NOM) P-values and FDR q-values, 6 pathways that exhibited significant differential enrichment in the TXNIP-high expression phenotype were identified, including the WNT signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, the phosphatidylinositol signaling system, the TGF-β signaling pathway, autophagy and the Janus kinase (JAK)-STAT signaling pathway (Fig. 3; Table IV).
Figure 3.
Enrichment plots from GSEA. GSEA results indicated that the (A) WNT signaling pathway, (B) MAPK signaling pathway, (C) phosphatidylinositol signaling system, (D) TGF-β signaling pathway, (E) autophagy, and (F) the JAK-STAT signaling pathway were differentially enriched in CCRCC with high TXNIP expression. NES, normalized enrichment score; NOM p-val, normalized P-value; FDR q-val, false discovery rate q-value; GSEA, Gene Set Enrichment Analysis; TXNIP, thioredoxin interacting protein; CCRCC, clear cell renal cell carcinoma; MAPK, mitogen-activated protein kinase; TGF-β, transforming growth factor-β; JAK, Janus kinase.
Table IV.
Gene sets enriched in the high expression phenotype group.
| Gene set name | NES | NOM P-value | FDR q-value |
|---|---|---|---|
| KEGG_WNT_SIGNALING_PATHWAY | −2.278 | <0.001 | 0.002 |
| KEGG_MAPK_SIGNALING_PATHWAY | −2.208 | <0.001 | 0.002 |
| KEGG_PHOSPHATIDYLINOSITOL_SIGNALING_SYSTEM | −2.210 | <0.001 | 0.002 |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | −2.194 | <0.001 | 0.002 |
| KEGG_REGULATION_OF_AUTOPHAGY | −2.211 | <0.001 | 0.003 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | −2.222 | <0.001 | 0.003 |
MSigDB collection: c2. cp. kegg. v6. 2. symbols. gmt. MAPK, mitogen-activated protein kinase; TGF-BETA, transforming growth factor-β; JAK, janus kinase; NES, normalized enrichment score; NOM p-val, normalized P-value; FDR q-val, false discovery rate q-value; MSigDB, Molecular Signatures Database.
Discussion
Previously, a number of studies have indicated that TXNIP functions as a tumor-suppressor gene in various types of cancer (8,15). Numerous studies have confirmed that the overexpression of TXNIP results in an increase in the levels of apoptosis, the suppression of cell proliferation, decreased metastasis and suppressed tumor growth (16–20). However, little is known about the association between TXNIP and CCRCC. In the present study, the expression of TXNIP in CCRCC tissues and its potential prognostic value were investigated.
In addition, CCRCC high-throughput RNA-sequencing data from TCGA were also investigated. TXNIP was revealed to be significantly downregulated in CCRCC tissues compared with adjacent normal or normal tissues. In addition, the decreased expression of TXNIP in CCRCC was evidently associated with T stage, lymph-node status, clinical stage, grade and a poor prognosis. To further elucidate the signaling pathways associated with TXNIP, GSEA was performed between the high- and low-TXNIP expression datasets. A total of 6 pathways were identified, which exhibited significant differential enrichment in the TXNIP-high expression phenotype, including the WNT, MAPK, phosphatidylinositol and TGF-β signaling pathways, as well as autophagy and the JAK-STAT signaling pathway. This indicates that TXNIP may serve as a potential therapeutic and prognostic target in CCRCC.
TXNIP is considered to be a tumor suppressor and is essential for regulating the redox status of cells by interacting with TRX. Feingold et al (12) showed that the overexpression of TXNIP reduces the clonogenicity and proliferation of esophageal adenocarcinoma cells. TXNIP deficiency results in the high viability and estrogen-induced growth of breast tumors (21). In addition, the overexpression of TXNIP may lead to attenuated tumor growth and markedly diminished metastasis in orthotopic anaplastic thyroid cancer (19). Furthermore, TXNIP overexpression induces apoptosis and represses proliferation by triggering mitochondrial-mediated reactive oxygen species generation and MAPK signaling pathway activation in SMMC7221 cells (16). In addition, the overexpression of TXNIP has been demonstrated to be associated with the improved overall survival rate of patients with breast cancer (22). Similarly, the present study indicated that TXNIP is downregulated in CCRCC tissues, and is associated with T stage, lymph-node status, clinical stage, grade, survival and poor prognosis, highlighting the crucial role of TXNIP in the progression of CCRCC.
To further observe the molecular functions of TXNIP, GSEA was conducted, revealing that TXNIP was able to regulate a number of crucial signaling pathways, including the WNT signaling pathway, MAPK signaling pathway, phosphatidylinositol signaling system, the TGF-β signaling pathway, autophagy and the JAK-STAT signaling pathway. Wang et al (23) revealed that the Wnt/β-catenin signaling axis serves a key role in regulating cell proliferation in CCRCC. Hong et al (24) identified that the activation of the MAPK signaling pathway in RCC enhanced cell viability and invasiveness. TGF-β serves a crucial role in regulating various biological functions, including cell differentiation, proliferation, metastasis, angiogenesis, cellular microenvironment and immune surveillance (25). TGF-β has also been indicated to promote the aggressive behavior and invasion of CCRCC (26). In addition, there is evidence to indicate that autophagy is associated with the development of CCRCC, including cell death, proliferation and metastasis (27–29). It has also been suggested that the resistance of RCC to interferon-α is associated with defective JAK-STAT activation (30). However, further mechanistic studies are required to validate the regulatory mechanisms of TXNIP in CCRCC, which may provide novel therapeutic approaches for CCRCC.
Although the results of the present study improved understanding of the association between TXNIP and CCRCC, there were several limitations: Firstly, only TCGA data were analyzed, and clinical specimen verification was not performed. Secondly, the association between TXNIP mRNA and TXNIP protein expression was not assessed, and should be included in further studies.
In conclusion, the present study identified that TXNIP expression may be a potential prognostic marker for patients with CCRCC. However, further functional studies are required to explore the molecular mechanisms of TXNIP in CCRCC progression.
Acknowledgements
The results presented are in whole based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
All authors were involved in the conception and design of the study. JQ, XL, JS and HJ performed data curation and statistical analysis. YG and QL, contributed to data analysis and drafted and reviewed this manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11:517–525. doi: 10.1038/nrurol.2014.194. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Salamanca JI, Huang WC, Millan I, Bertini R, Bianco FJ, Carballido JA, Ciancio G, Hernández C, Herranz F, Haferkamp A, et al. Prognostic impact of the 2009 UICC/AJCC TNM staging system for renal cell carcinoma with venous extension. Eur Urol. 2011;59:120–127. doi: 10.1016/j.eururo.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Chen KS, DeLuca HF. Isolation and characterization of a novel cDNA from HL-60 cells treated with 1,25-dihydroxyvitamin D-3. Biochim Biophys Acta. 1994;1219:26–32. doi: 10.1016/0167-4781(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 4.Nishiyama A, Matsui M, Iwata S, Hirota K, Masutani H, Nakamura H, Takagi Y, Sono H, Gon Y, Yodoi J. Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274:21645–21650. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 5.Masaki S, Masutani H, Yoshihara E, Yodoi J. Deficiency of thioredoxin binding protein-2 (TBP-2) enhances TGF-β signaling and promotes epithelial to mesenchymal transition. PLoS One. 2012;7:e39900. doi: 10.1371/journal.pone.0039900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woolston CM, Madhusudan S, Soomro IN, Lobo DN, Reece-Smith AM, Parsons SL, Martin SG. Thioredoxin interacting protein and its association with clinical outcome in gastro-oesophageal adenocarcinoma. Redox Biol. 2013;1:285–291. doi: 10.1016/j.redox.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung H, Kim MJ, Kim DO, Kim WS, Yoon SJ, Park YJ, Yoon SR, Kim TD, Suh HW, Yun S, et al. TXNIP maintains the hematopoietic cell pool by switching the function of p53 under oxidative stress. Cell Metab. 2013;18:75–85. doi: 10.1016/j.cmet.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Yu Q, Chng WJ. TXNIP (VDUP-1, TBP-2): A major redox regulator commonly suppressed in cancer by epigenetic mechanisms. Int J Biochem Cell Biol. 2011;43:1668–1673. doi: 10.1016/j.biocel.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Kwon HJ, Won YS, Suh HW, Jeon JH, Shao Y, Yoon SR, Chung JW, Kim TD, Kim HM, Nam KH, et al. Vitamin D3 upregulated protein 1 suppresses TNF α induced NF-κB activation in hepatocarcinogenesis. J Immunol. 2010;185:3980–3989. doi: 10.4049/jimmunol.1000990. [DOI] [PubMed] [Google Scholar]
- 10.Cadenas C, Franckenstein D, Schmidt M, Gehrmann M, Hermes M, Geppert B, Schormann W, Maccoux LJ, Schug M, Schumann A, et al. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res. 2010;12:R44. doi: 10.1186/bcr2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu G, Zhou L, Liu H, Shan Y, Zhang X. MicroRNA-224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP-mediated HIF1α Pathway. Cell Physiol Biochem. 2018;48:1735–1746. doi: 10.1159/000492309. [DOI] [PubMed] [Google Scholar]
- 12.Feingold PL, Surman DR, Brown K, Xu Y, McDuffie LA, Shukla V, Reardon ES, Crooks DR, Trepel JB, Lee S, et al. Induction of thioredoxin-interacting protein by a histone deacetylase inhibitor, entinostat, is associated with DNA damage and apoptosis in esophageal adenocarcinoma. Mol Cancer Ther. 2018;17:2013–2023. doi: 10.1158/1535-7163.MCT-17-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 15.Sheth SS, Bodnar JS, Ghazalpour A, Thipphavong CK, Tsutsumi S, Tward AD, Demant P, Kodama T, Aburatani H, Lusis AJ. Hepatocellular carcinoma in Txnip-deficient mice. Oncogene. 2006;25:3528–3536. doi: 10.1038/sj.onc.1209394. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Yue Z, Xiong W, Sun P, You K, Wang J. TXNIP overexpression suppresses proliferation and induces apoptosis in SMMC7221 cells through ROS generation and MAPK pathway activation. Oncol Rep. 2017;37:3369–3376. doi: 10.3892/or.2017.5577. [DOI] [PubMed] [Google Scholar]
- 17.Shen L, O'Shea JM, Kaadige MR, Cunha S, Wilde BR, Cohen AL, Welm AL, Ayer DE. Metabolic reprogramming in triple-negative breast cancer through Myc suppression of TXNIP. Proc Natl Acad Sci USA. 2015;112:5425–5430. doi: 10.1073/pnas.1501555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han SH, Jeon JH, Ju HR, Jung U, Kim KY, Yoo HS, Lee YH, Song KS, Hwang HM, Na YS. VDUP1 upregulated by TGF-beta1 and 1,25-dihydorxyvitamin D3 inhibits tumor cell growth by blocking cell-cycle progression. Oncogene. 2003;22:4035–4046. doi: 10.1038/sj.onc.1206610. [DOI] [PubMed] [Google Scholar]
- 19.Morrison JA, Pike LA, Sams SB, Sharma V, Zhou Q, Severson JJ, Tan AC, Wood WM, Haugen BR. Thioredoxin interacting protein (TXNIP) is a novel tumor suppressor in thyroid cancer. Mol Cancer. 2014;13:62. doi: 10.1186/1476-4598-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikarashi M, Takahashi Y, Ishii Y, Nagata T, Asai S, Ishikawa K. Vitamin D3 up-regulated protein 1 (VDUP1) expression in gastrointestinal cancer and its relation to stage of disease. Anticancer Res. 2002;22:4045–4048. [PubMed] [Google Scholar]
- 21.Park JW, Lee SH, Woo GH, Kwon HJ, Kim DY. Downregulation of TXNIP leads to high proliferative activity and estrogen-dependent cell growth in breast cancer. Biochem Biophys Res Commun. 2018;498:566–572. doi: 10.1016/j.bbrc.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Nie W, Huang W, Zhang W, Xu J, Song W, Wang Y, Zhu A, Luo J, Huang G, Wang Y, Guan X. TXNIP interaction with the Her-1/2 pathway contributes to overall survival in breast cancer. Oncotarget. 2015;6:3003–3012. doi: 10.18632/oncotarget.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W, Wang TD, Meng YY, Yuan C, Li HM, Yu YP, et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt β-catenin signaling pathway. Mol Cancer. 2019;18:15. doi: 10.1186/s12943-019-0942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong B, Zhou J, Ma K, Zhang J, Xie H, Zhang K, Li L, Cai L, Zhang N, Zhang Z, Gong K. TRIB3 promotes the proliferation and invasion of renal cell carcinoma cells via activating MAPK signaling pathway. Int J Biol Sci. 2019;15:587–597. doi: 10.7150/ijbs.29737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Syed V. TGF-β Signaling in Cancer. J Cell Biochem. 2016;117:1279–1287. doi: 10.1002/jcb.25496. [DOI] [PubMed] [Google Scholar]
- 26.Sitaram RT, Mallikarjuna P, Landstrom M, Ljungberg B. Transforming growth factor-β promotes aggressiveness and invasion of clear cell renal cell carcinoma. Oncotarget. 2016;7:35917–35931. doi: 10.18632/oncotarget.9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang HM, Noh KH, Chang TK, Park D, Cho HS, 1, Lim JH, Jung CR. Ubiquitination of MAP1LC3B by pVHL is associated with autophagy and cell death in renal cell carcinoma. Cell Death Dis. 2019;10:279. doi: 10.1038/s41419-019-1520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Fan Y, Huang S, Wang G, Han R, Lei F, Luo A, Jing X, Zhao L, Gu S, Zhao X. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci. 2018;109:3865–3873. doi: 10.1111/cas.13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang ZL, Deng Q, Chong T, Wang ZM. Autophagy suppresses the proliferation of renal carcinoma cell. Eur Rev Med Pharmacol Sci. 2018;22:343–350. doi: 10.26355/eurrev_201801_14178. [DOI] [PubMed] [Google Scholar]
- 30.Shang D, Liu Y, Ito N, Kamoto T, Ogawa O. Defective Jak-Stat activation in renal cell carcinoma is associated with interferon-alpha resistance. Cancer Sci. 2007;98:1259–1264. doi: 10.1111/j.1349-7006.2007.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.