Abstract
Fungal highly reducing polyketide synthases (HRPKSs) are highly programmed multidomain enzymes that synthesize reduced polyketide structures. Recent reports indicated salicylaldehydes are synthesized by HRPKS biosynthetic gene clusters, which are unexpected based on known enzymology of HRPKSs. Using genome mining of a Trichoderma virens HRPKS gene cluster that encodes a number of redox enzymes, we uncover the strategy used by HRPKS pathways in the biosynthesis of aromatic products such as salicylaldehyde 4, which can be oxidatively modified to the epoxycyclohexanol natural product trichoxide 1. We show selective β-hydroxyl groups in the linear HRPKS product are individually reoxidized to β-ketones by short-chain dehydrogenase/reductase (SDR) enzymes, which enabled intramolecular aldol condensation and aromatization. Our work expands the chemical space of natural products accessible through HRPKS pathways.
Graphical Abstract

Fungal highly reducing polyketide synthases (HRPKSs) are a large family of multidomain enzymes that synthesize structurally diverse natural products.1,2 With iterative and permutative use of a single set of catalytic domains, fungal HRPKSs can synthesize polyketides of defined lengths with precise combinations of β−carbon reduction, dehydration and α-carbon methylation. The resulting polyketide product can then be subjected to tailoring modifications to yield bioactive natural products, such as the cholesterol lowering lovastatin3 and mycotoxin fumonisin.4 Given most ascomycetes sequenced to date contain many HRPKSs of unknown functions, it is important to expand knowledge of HRPKS functions and completely map the chemical space of their products.
Current classification of fungal PKSs is based on the degrees of reduction of the product carbon backbones.1 While HRPKSs synthesize highly reduced products, nonreducing PKSs (NRPKSs) and partially reducing PKSs (PRPKSs) synthesize aromatic products that contain phenol groups such as 2,4-dihydroxybenezne and salicylate, respectively.5 To date, this chemical classification of fungal PKSs has remained intact. Therefore, it was surprising when an HRPKS-containing biosynthetic gene cluster (BGC) from Neurospora crassa was shown to be responsible for the biosynthesis of the salicylaldehyde sordarial (Figure 1).6 Similarly, an HRPKS from Magnaporthe oryzae was genetically verified to be required for the biosynthesis of the devastating rice mycotoxin pyriculol.7 A number of redox active enzymes in these gene clusters are proposed to morph a reduced polyketide product into the phenol-containing compound, although the biochemical logic for such transformation is not resolved.6 Salicylaldehydes have also been proposed to be precursors to a family of epoxycyclohexenol containing natural products, such as eupenoxide8 and phomoxide (Figure 1).9 The epoxycyclohexene core is proposed to be derived from oxidative dearomatization of the phenyl ring in salicylaldehydes, followed by additional redox modifications.10 To date, the biosynthesis of these compounds have remained unexplored.
Figure 1.
Fungal natural products that contain or are derived from the salicylaldehyde core. Trichoxide 1 is discovered in this work through genome mining and heterologous reconstitution.
Biosynthesis of salicylaldehydes by HRPKS-containing BGCs therefore represents a significant departure from current fungal PKS classification. In this work, we demonstrate how an HRPKS and associated tailoring enzymes can accomplish the above through genome mining and biosynthetic characterization of a new epoxycyclohexenol compound, trichoxide 1, from the biocontrol fungus Trichoderma virens.
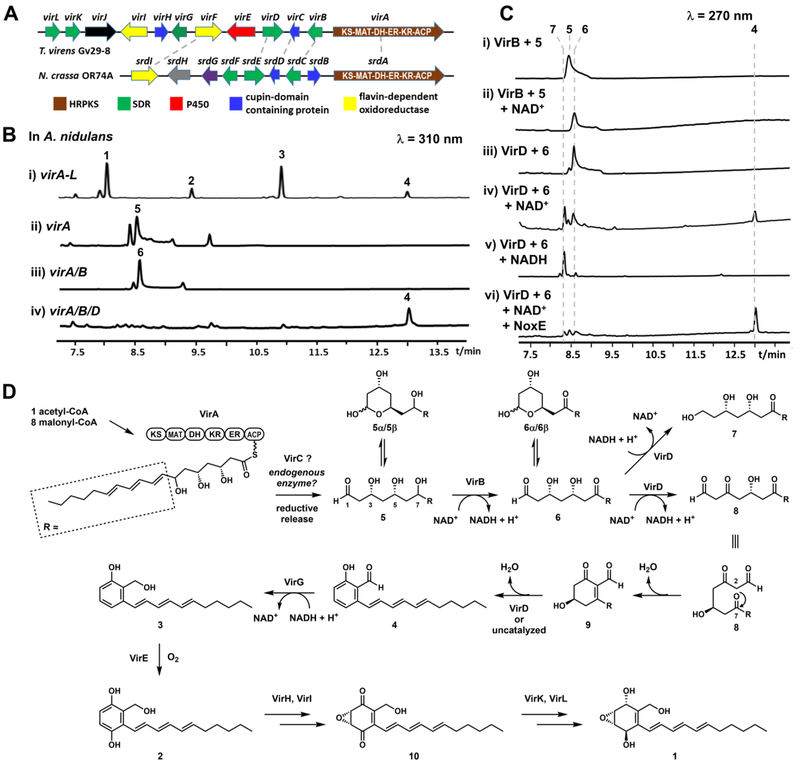
Using the srd gene cluster that synthesizes sordarial as a lead,6 we searched for related gene clusters from sequenced fungal genomes. In particular, we mined clusters that contain additional redox enzymes that may lead to the formation of epoxycyclohexenol products. We identified one such HRPKS-containing cluster well-conserved among many Trichoderma species (Figure S6), that encodes homologs to most of the srd enzymes (Figure 2A). The vir cluster from T. virens encodes the HRPKS (virA), five SDR family proteins (virB, virD, virG, virK and virL), two cupin-domain containing proteins (virC and virH), two flavin-dependent oxidoreductases (virF and virI), a cytochrome P450 (virE), and a glutamyl-tRNA synthetase like enzyme (virJ) (Figure 2A and Table S1). When all twelve genes (virA-L) were introduced into an engineered Aspergillus nidulans host using three vectors,11 analysis of the extract showed that four new metabolites (1–4) were accumulated (Figure 2B, i). The major product 1 (1.2 mg/L) was shown to be an epoxycyclohexenol based on 1D and 2D NMR data (Figure S9–15 and Table S4) and was named trichoxide. Virensol A 2 (0.3 mg/L) and virensol B 3 (0.4 mg/L) were characterized to be substituted salicylic alcohol derivatives (Figure S16–25 and Table S5, S6), while 4 (0.9 mg/L) was identified to be the known salicylaldehyde 5-deoxyaurocitrin (Figure S26–27 and Table S7).12 1 exhibited antifungal activities against Saccharomyces cerevisiae and Candida albicans with minimum inhibitory concentrations (MICs) of 11 and 18 μg/mL, respectively (Table S10). 4 has MICs 0.16 and 0.63 μg/mL against S. cerevisiae and C. albicans, respectively, and it also has MICs 0.63 and 1.25 μg/mL against Staphylococcus aureus and Bacillus subtilis, respectively (Table S10).
Figure 2.
Biosynthesis of trichoxide 1 by the vir gene cluster from T. virens Gv29–8. (A) Gene cluster comparison between vir and srd clusters shows the vir cluster contains more redox enzymes. Enzyme/domain abbreviations: KS: ketosynthase; MAT: malonyl-CoA:ACP transacylase; DH: dehydratase; ER: enoylreductase; KR: ketoreductase; ACP: acyl-carrier protein; P450: cytochrome P450 monooxygenase; (B) Metabolite analysis of heterologous reconstitution of the vir cluster in A. nidulans; (C) Enzymatic assays of SDR-like enzymes. (D) Proposed biosynthetic pathway of 1.
Based on the structures of 1–4, we reasoned that 1 is the most advanced product of the vir cluster, preceded by 2, 3 and 4. When virA was removed from the heterologous host, the production of metabolites 1–4 was abolished, confirming the essential role of HRPKS. To investigate the product of VirA, we directly expressed this enzyme in A. nidulans, which led to the production of a new compound virensol C 5 (molecular weight (MW) 310; 1.5 mg/L) (Figure 2B, ii). Structural elucidation showed 5 is (8E,10E,12E)-3,5,7-trihydroxyoctadeca-8,10,12-trienal, which exists mostly as a pair of hemiacetals (5α and 5β, Figure 2D) through ring-chain tautomerism in solution (Figure S28–S33, Table S8).13 NOESY correlation of the major 5β established the relative stereochemistry of 5. Therefore, VirA is a bona-fide HRPKS that synthesizes the reduced chain in 5. VirA has interesting programming rules: the first two ketides are fully reduced, followed by three ketides that undergo β-dehydration, and the last three ketides are only reduced to β-hydroxys to yield the trihydroxy portion. The production of aldehyde 5 by VirA alone in A. nidulans is surprising, since VirA does not contain a reductase (R) domain that is typically associated with reductive product release in HRPKS. Efforts to obtain pure VirA from a yeast expression host was not successful, which precluded a direct assay of its function. Interestingly, when VirC was coexpressed with VirA in A. nidulans, the cupin-domain enzyme elevated the amount of 5 (Figure S3B), which suggests this enzyme may be involved in enhancing product turnover.
To understand how 5 can be morphed into the aromatic 4, we expressed tailoring enzymes with VirA (Figure S3). Of the different binary combinations, only coexpression of VirA and VirB led to disappearance of 5 and emergence of a new metabolite virensol D 6 (MW 308; 1.7 mg/L) (Figure 2B, iii). 6 also exists as hemiacetals (6α and 6β, Figure 2D) in solution (Figure S34–S39, Table S9). The structural difference between 5 and 6 is oxidation of C7 alcohol in 5 to a ketone in 6, which pinpoints VirB as the SDR that performs the dehydrogenation. As confirmation of this function, purified VirB (Figure S4) (10 μM) completely converted 5 (200 μM) into 6 after 12 hours in the presence of NAD+ (1 mM) (Figure 2C, i).
We then extended the biosynthetic pathway through the addition of other redox enzymes. Coexpression of another SDR enzyme, VirD, together with VirAB led to disappearance of 6 and the emergence of salicylaldehyde 4, with a trace amount of 3 (Figure 2B, iv). To investigate the mechanism of VirD, the purified enzyme was assayed with 6 (Figure S4). VirD (20 μM) catalyzed the conversion of 6 (1 mM) into two new products when 1 mM NAD+ was present (Figure 2C, iv). One product was confirmed to be 4, while the other product 7 (MW= 310) displayed a mass increase of 2 mu corresponding to a reduced product of 6. The UV spectrum of 7 (Figure S5B) was the same as 6, suggesting the triene portion is not reduced. Therefore, we conclude that 7 is a triol compound in which the aldehyde of 6 was reduced (Figure 2D). The source of reducing equivalent (i.e. NADH) is generated from the oxidation of 6 that is required for formation of 4. When we repeated the VirD assay with only NADH, near complete conversion of 6 to 7 was observed, with no trace of 4 forming despite NAD+ generation (Figure 2C, v). We reasoned that this may be due to the reduction of aldehyde by VirD is much faster than oxidation of 6. Under cellular conditions in which NAD+/NADH ratio is high,14 formation of 7 is likely minimized. To mimic that condition, we included 20 μM of the NADH oxidase NoxE15 and substoichiometric (50 μM) amount of NAD+ in the reaction mixture. Under this reaction condition, we observed near complete conversion of 6 to 4 (Figure 2C, iv).
We proposed three different reactions are involved in the oxidative aromatization of 6 to 4 (Figure 2D): i) dehydrogenation of C3 in 6 to form the β-ketone aldehyde 8 and generate the nucleophilic C2 that is required for the ii) intramolecular aldol condensation between C2 and C7 to from 9; iii) dehydration and aromatization of 9 gives the salicylaldehyde 4. This is reminiscent of cyclization of C9-reduced β-ketone polyketides in bacterial aromatic polyketide biosynthesis.5 While the dehydrogenation of 6 is definitely catalyzed by VirD, the aldol condensation and dehydration may be uncatalyzed or assisted by VirD. To provide support for this set of proposed reactions, we performed a time-course experiment of VirD in the presence of NAD+ and NoxE.15 When the reaction was quenched at 10 minutes, two intermediates with MWs matching the proposed intermediates 8 and 9 were detected (Figure S5C, trace i). Phenylhydrazine was used to derivatize these intermediates,16 which lead to a set of phenyldrazones (Figure S5C, trace iii) with MW consistent with the derivatized products of 8 and 9 (Figure S5D).
Collectively, we demonstrated that only three enzymes, one HRPKS and two SDRs are required to synthesize salicylaldehyde 4. Three hydroxyl groups are introduced at the last three iterations of VirA, two of which are selectively oxidized back to β-ketone by SDRs to enable the aldol reaction. Using this strategy, the product 4 with both reduced and aromatic portions can be synthesized using a single HRPKS. This is in contrast to the well-studied duo PKSs system in synthesis of structurally similar compounds in fungi: an HRPKS synthesizes the reduced portion, and the chain is transferred to an NRPKS to synthesize the aromatic portion.17 Using a single HRPKS could be metabolically less demanding as the synthesis of a second megasynthase is energetically expensive.
We next investigated the transformation of 4 to 1. Minor reduction of 4 to 3 was observed in the virA-D expression strain, likely catalyzed by an endogenous alcohol dehydrogenase. A dedicated enzyme in the vir pathway should be required for complete reduction. When we expressed the SDR VirG with VirA-D, complete conversion of 4 to 3 was observed (Figure S7A). The function of VirG was further verified using purified enzyme (Figure S4 and S7C). To identify the enzyme responsible for hydroxylation of 3, we screened the remaining redox enzymes through coexpression analysis. Upon introducing virE which encodes the lone P450 in the gene cluster into the strain that produces 3, we observed formation of the hydroquinone 2 (Figure S7A). Functional verification was performed using microsomal fractions of S. cerevisiae RC0118 expressing VirE. Hydroxylation of 3 to 2 was confirmed in the in vitro assay, while no hydroxylation of 4 was detected (Figure S7B).
To complete the biosynthetic pathway to 1, we introduced additional genes to the virA-G expression strain. Expression of the flavin-dependent enzyme VirI and the cupin-domain containing protein VirH led to the emergence of 10 (MW 302), which has a UV spectrum (Figure S8) consistent with that of the epoxycyclohexene-1,4-dione shown in Figure 2D. However, the quantity of this compound was too low for structure identification. We propose that VirI may oxidize 2 to form the quinone, while VirH performs the epoxidation, a two-step reaction that is proposed for the same moiety in asukamycin.19 Finally, since coexpressing all the genes in the pathway afforded 1, we deduce that the two remaining SDR enzymes, VirK and VirL are responsible for reducing the ketones in 10 to the corresponding alcohols to furnish the epoxycyclohexanol structure in 1. Interestingly, the stereo configurations of 1,4-diol in 1 are different from related compounds (Figure 1), suggesting that the ketone reductions catalyzed by VirK and VirL have different stereochemical outcomes than the other SDRs.
In summary, we mined and verified the vir pathway that is responsible for biosynthesis of salicylaldehyde 4 and epoxycyclohexenol 1. Our work unraveled new biosynthetic logic in which β-hydroxyl groups in a highly reduced polyketide are selectively reoxidized to the corresponding β-ketones to enable intramolecular aldol condensation and form an aromatic product, which are typically associated with nonreducing and partially reducing PKSs. Our study expands the chemical space accessible by HRPKS-containing BGCs, and cautions against polyketide product prediction based entirely on the current PKS classifications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH 1R35GM118056 to YT. LL was supported by NSFC (21977113, 21772228). Chemical characterization studies were supported by shared instrumentation grants from the NSF (CHE-1048804) and the NIH NCRR (S10RR025631).
Footnotes
Supporting Information.
This material is available free of charge via the Internet at http://pubs.acs.org. Experimental procedures, chromatograms, and spectroscopic data.
REFERENCES
- (1).Cox RJ Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem 2007, 5, 2010–2026. [DOI] [PubMed] [Google Scholar]
- (2).Chooi YH; Tang Y Navigating the fungal polyketide chemical space: from genes to molecules. J. Org. Chem 2012, 77, 9933–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kennedy J; Auclair K; Kendrew SG; Park C; Vederas JC; Hutchinson CR Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 1999, 284, 1368–1372. [DOI] [PubMed] [Google Scholar]
- (4).Alexander NJ; Proctor RH; McCormick SP Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev 2009, 28, 198–215. [Google Scholar]
- (5).Zhou H; Li Y; Tang Y Cyclization of aromatic polyketides from bacteria and fungi. Nat. Prod. Rep 2010, 27, 839–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhao Z; Ying Y; Hung YS; Tang Y Genome mining reveals Neurospora crassa can produce the salicylaldehyde sordarial. J. Nat. Prod 2019, 82, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Jacob S; Grötsch T; Foster AJ; Schüffler A; Rieger PH; Sandjo LP; Liermann JC; Opatz T; Thines E Unravelling the biosynthesis of pyriculol in the rice blast fungus Magnaporthe oryzae. Microbiology 2017, 163, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Myobatake Y; Takemoto K; Kamisuki S; Inoue N; Takasaki A; Takeuchi T; Mizushina Y; Sugawara F, Cytotoxic alkylated hydroquinone, phenol, and cyclohexenone derivatives from Aspergillus violaceofuscus Gasperini. J. Nat. Prod 2014, 77, 1236–1240. [DOI] [PubMed] [Google Scholar]
- (9).(a) Liu Z; Jensen PR; Fenical W A cyclic carbonate and related polyketides from a marine-derived fungus of the genus Phoma. Phytochemistry 2003, 64, 571–574. [DOI] [PubMed] [Google Scholar]; (b) Mehta G; Roy S Enantioselective total synthesis of (+)-eupenoxide and (+)-phomoxide: revision of structures and assignment of absolute configuration. Org. Lett 2004, 6, 2389–2392. [DOI] [PubMed] [Google Scholar]; (c) Mehta G; Roy S; Davis RA On the stereostructures of (+)-eupenoxide and (−)-3’,4’ dihydrophomoxide: a caveat on the spectral comparisons of oxygenated cyclohexenoids. Tetrahedron Lett 2008, 49, 5156–5164. [Google Scholar]
- (10).(a) Teng LL; Song TY; Xu ZF; Liu X; Dai R; Chen YH; Li SH; Zhang KQ; Niu XM Selected mutations revealed intermediates and key precursors in the biosynthesis of polyketide-terpenoid hybrid sesquiterpenyl epoxy-cyclohexenoids. Org. Lett 2017, 19, 3923–3926. [DOI] [PubMed] [Google Scholar]; (b) Holm DK; Petersen LM; Klitgaard A; Knudsen PB; Jarczynska ZD; Nielsen KF; Gotfredsen CH; Larsen TO; Mortensen UH. Molecular and chemical characterization of the biosynthesis of the 6-MSA-derived meroterpenoid yanuthone D in Aspergillus niger. Chem. Biol 2014, 21, 519–529. [DOI] [PubMed] [Google Scholar]
- (11).Sato M; Yagishita F; Mino T; Uchiyama N; Patel A; Chooi YH; Goda Y; Xu W; Noguchi H; Yamamoto T; Hotta K; Houk KN; Tang Y; Watanabe K Involvement of Lipocalin-Like CghA in Decalin-forming stereoselective intramolecular [4 + 2] cycloaddition. ChemBioChem 2015, 16, 2294–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Berkaew P; Soonthornchareonnon N; Salasawadee K; Chanthaket R; Isaka M Aurocitrin and related polyketide metabolites from the wood-decay fungus Hypocrea sp. BCC 14122 J. Nat. Prod 2008, 71, 902–904. [DOI] [PubMed] [Google Scholar]
- (13).(a) Hurd CD; Saunders WH Jr. Ring-chain tautomerism of hydroxy aldehydes. J. Am. Chem. Soc 1952, 74, 5324–5329. [Google Scholar]; (b) Jones PR Ring-chain tautomerism. Chem. Rev 1963, 63, 461–487. [Google Scholar]
- (14).Bekers KM; Heijnen JJ; van Gulik WM Determination of the in vivo NAD:NADH ratio in Saccharomyces cerevisiae under anaerobic conditions, using alcohol dehydrogenase as sensor reaction. Yeast 2015, 32, 541–557. [DOI] [PubMed] [Google Scholar]
- (15).(a) Lopez de Felipe F; Hugenholtz J Purification and characterisation of the water forming NADH-oxidase from Lactococcuslactis. Int. Dairy J 2001, 11, 37–44. [Google Scholar]; (b) Sybille Tachon; Emilie Chambellon; Mireille Yvon Identification of a conserved sequence in flavoproteins essential for the correct conformation and activity of the NADH oxidase NoxE of Lactococcuslactis. J. Bacteriol 2011, 193, 3000–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Li Z; Fang M; LaGasse MK; Askim JR; Suslick KS Colorimetric recognition of aldehydes and ketones. Angew. Chem. Int. Ed. Engl 2017, 56, 9860–9863. [DOI] [PubMed] [Google Scholar]
- (17).Jana N; Nanda S, Resorcylic acid lactones (RALs) and their structural congeners: recent advances in their biosynthesis, chemical synthesis and biology. New J. Chem 2018, 42, 17803–17873. [Google Scholar]
- (18).Tang MC; Lin HC; Li DH; Zou Y; Li J; Xu W; Cacho RA; Hillenmeyer ME; Garg NK; Tang Y Discovery of unclustered fungal indole diterpene biosynthetic pathways through combinatorial pathway reassembly in engineered yeast. J. Am. Chem. Soc 2015, 137, 13724–13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rui Z; Petrícková K; Skanta F; Pospísil S; Yang Y; Chen CY; Tsai SF; Floss HG; Petrícek M; Yu TW Biochemical and genetic insights into asukamycin biosynthesis. J. Biol. Chem 2010, 285, 24915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.