Abstract
Purpose:
With the updated World Health Organization (WHO) 2016 neuropathological diagnostic criteria, radiographic prognostic associations in lower-grade gliomas (LGG, WHO grade II and III) are undergoing re-evaluation.
Methods:
We identified 316 LGG patients (151 grade II and 165 grade III) for a combined cohort from three independent databases. We analyzed the preoperative axial FLAIR, axial T2-weighted and post-gadolinium volumetric T1-weighted MR images. The molecular data collected included the status of IDH1/2, TP53, TERT promoter and ATRX mutations, in addition to 1p/19q co-deletions. In a subset of cases (n=133), we assessed the “T2-FLAIR mismatch” sign.
Results:
Gliomas were assigned to one of the three molecular groups: Group O (IDH-mutant, 1p/19q co-deleted oligodendrogliomas, n=95), Group A (IDH-mutant, ATRX inactivated astrocytomas, n=175) and Group G (IDH wild-type, GBM-like, n=46). A contrast-enhancing tumor was seen in 98 patients (31%), most frequently in Group G (n=28/45, 57%), when compared to Group A (n= 49/175, 28%) and Group O (n= 24/95, 25.3%) tumors (p=0.008 and p=0.0011, respectively). Consistent with previous reports, T2-FLAIR mismatch was preferentially found in Group A tumors (73.1%, 60 of 82), although its presence was not associated with survival, after controlling for molecular group. False positive mismatch sign was noted in 28.5% (12/42) Group O tumors, but none of the tumors in Group G. A combination of all three factors: age under 40 years at first diagnosis, a tumor size larger than 6 cm and T2-FLAIR mismatch was highly specific for IDH mutant astrocytoma (Group A).
Conclusion:
We identify radiographic correlates of molecular groups in lower-grade gliomas, which join clinical demographic features in defining the characteristic presentation of these tumors. Radiographic correlates of prognosis in LGG require re-evaluation within molecular group.
Keywords: Glioma, IDH mutation, contrast enhancement, T2-FLAIR mismatch, radiographic correlates
Introduction
Imaging plays an essential role in the management of lower-grade gliomas (LGG). Indeed, magnetic resonance imaging (MRI) represents the first-line modality in the diagnosis of these tumors, and in the assessment of response to treatment. Recent advances in MRI techniques have allowed for an improved, non-invasive, pre-therapeutic characterization of LGG, expanding our knowledge beyond the historical view of LGG as non-enhancing areas of increased signal on T2-weighted imaging [1–3]. Moreover, the legacy classification of adult diffuse LGG as a homogenous group of neoplasms is shifting as evidence has emerged that emphasizes the distinct origin of tumors with differing molecular subtypes, and the subsequent impact on response to treatment and patient outcomes [4–9]. Codified in the recent 2016 update to the World Health Organization (WHO) Classification of Tumors of the Central Nervous System [10], this has resulted in an ongoing re-evaluation of the radiographic assessment of LGG patients [11, 12].
In the era preceding WHO 2016, well-established clinical and radiographic features, such as contrast enhancement, mass effect and necrosis were used to correlate MRI features to pathological grade in gliomas and, consequently, to predict outcome [13–16]. In addition, contrast enhancement was routinely used to predict higher grade or progressive malignant transformation and was associated with poor overall survival. Radiologic features such as tumor size and midline infiltration were synthesized into the widely used Pignatti criteria for prognostic assessment[17]. However, the significant overlap of imaging characteristics between different histological entities, limited the utility of MRI as a definitive predictor of grade – some higher-grade lesions were non-enhancing, some lower-grade tumors had enhancing components.
With the new WHO 2016 criteria enforcing a molecular homogeneity within these previously mixed histologic cohorts, recent studies have identified specific radiographic features that are characteristic of certain molecular groups, most notably the “T2-FLAIR mismatch” sign, which is reported to be highly specific for IDH-mutant, TP53/ATRX inactivated astrocytic gliomas [18, 19]. We hypothesized that a combined “radio-genomic” approach composed of clinical and pre-operative radiographic features could prove useful in the diagnosis and survival prediction of LGG patients. Here, we aim to establish a radiographic score that facilitates identifying patients within certain molecular classifications.
Methods
Patient selection and clinical data
Our retrospective study was reviewed and approved by the human subjects’ institutional review board of the Dana-Farber Cancer Institute/Harvard Cancer Center and complied with HIPAA guidelines, protocol number 2011P002334. The study utilized three separate databases to develop a combined cohort of 316 patients who underwent surgical resection of a WHO grade II/III glioma with available imaging and molecular data. Patient data was acquired from The Cancer Genome Atlas (TCGA) (n=162), a National Cancer Institute-supported publicly available dataset derived from de-identified patients, the Department of Neurosurgery at Massachusetts General Hospital (n=87) and the Department of Neurosurgery at University Hospital Dresden (n=69). Included cases met the following criteria: (1) Tumors were WHO grade II or WHO grade III, (2) preoperative imaging data with fluid-attenuated inversion recovery (FLAIR), T2-weighted and post-gadolinium T1 contrast sequences was available, and (3) tumors had known IDH mutation status. The primary reason for patient ineligibility (n=127) was due to lack of preoperative MR scans acquired from the Cancer Imaging Archive (TCIA).
Molecular analysis
The molecular data collected from the TCGA cohort included the status of IDH1/2, TP53 and ATRX mutations, in addition to 1p/19q co-deletions. In the remaining cohorts, IDH mutations were assessed using either Next Generation Sequencing (MGH Cohort)[20] or Sanger sequencing (Dresden cohort)[21] or Immunohistochemistry (MGH and Dresden)[22]. ATRX inactivation was evaluated using Immunohistochemistry[23]. TERT promoter (TERTp) mutations were assessed either by amplification using Sanger sequencing performed with ABI Prism 3730 DNA Analyzer or using the fluorescence PCR technique, as previously described [24, 25]. 1p/19q co-deletions were detected by fluorescence-in situ hybridization analysis on samples of paraffin-embedded tumor tissues [26, 27]
Radiographic Annotation
MR images from the TCGA cohort were obtained from The Cancer Imaging Archive (TCIA), a National Cancer Institute-supported imaging network that provides radiographic data corresponding to the de-identified patients from the TCGA. The MR images from the remaining two datasets were locally performed and included preoperative axial FLAIR (3-5 mm sections, 1 mm interslice gaps), axial T2-weighted images (5 mm sections, 1 mm interslice gaps), coronal T1 (5 mm sections, 1 mm interslice gaps) as well as post-gadolinium T1-weighted MR images. All images were analyzed in accordance with the Response Assessment in Neuro-Oncology (RANO) guideline [12].
Using Osirix, a DICOM image processing application, we scored the preoperative axial FLAIR, axial T2-weighted and post-gadolinium volumetric T1-weighted MR images. Tumor size was measured as the largest orthogonal cross product of the tumor on the axial T2/FLAIR scans. Similarly, enhancing volume was measured as the largest orthogonal cross product of the enhancing tumor on axial T1-post gadolinium scans. For the purposes of the study, tumor enhancement was defined as present in cases with enhanced cross product greater than or equal to 10% of the overall tumor size demonstrated on the T2/FLAIR images (examples in Supplemental Figure 1).
T2-FLAIR mismatch sign
Preoperative MR scans from 133 patients were assessed for mismatched T2-weighted (“hyper-intense”) and the FLAIR (“hypo-intense”) signals, using a previously reported approach [19]. The assessment was performed by two independent clinically experienced reviewers (D.D., a neuroradiologist and T.A.J., a neurosurgeon). The reviewers were blinded to molecular status and diagnosis. In cases of disagreement (n=4), a third experienced reviewer (J.J.M.) was involved in the assessment. Inter-reviewer agreement was evaluated using the Kappa statistic (κ = 0–0.40, poor; κ = 0.41–0.60, moderate; κ = 0.61–0.80, good; κ = 0.81–1.00, excellent).
Statistical Analysis
The Kaplan-Meier method was used to estimate progression free survival (PFS) and overall survival (OS). Differences in survival were assessed using the log-rank test. PFS was defined as the interval between the day of first surgery to MRI-confirmed tumor progression, death or end of follow-up. OS was defined as the interval from the day of first surgery until death or the end of follow-up. The Mann-Whitney U and Fisher’s exact tests were used to test for association of radiographic variables and the three molecular subgroups. A multivariate Cox proportional hazards model was constructed to assess the impact of radiographic features on patient outcome. A p value (two-sided) of less than 0.05 was considered significant. All analyses were conducted using the SPSS software package (Version 21.0 SPSS Inc., Chicago, IL, USA).
Results
Molecular classification
The 2016 World Health Organization (WHO) classification of central nervous system tumors was used to categorize tumors [28]. Based on the presence or absence of TERTp mutations, IDH mutations, ATRX inactivation and lp/19q co-deletion, the vast majority of adult diffuse gliomas can be clustered into three distinct molecular subgroups [9]: Group O, Group A and Group G. The molecular oligodendroglial “Group O” is defined by the presence of IDH and TERTp mutations and 1p/19q co-deletions. The molecular astrocytic “Group A” is characterized by IDH mutation with concomitant TP53/ATRX inactivation, or the absence of lp/19q co-deletions and/or TERTp mutations. Group G (“glioblastoma-like”) can then be defined by the absence of a mutation in an IDH gene. In our cohort, a total of 316 WHO grade II/III gliomas (151 WHO grade II and 165 WHO grade III) were assigned to one of the three molecular groups: Group O (n=95), Group A (n=175) and Group G (n=46). Table 1 provides a summary of the clinical characteristics of the three molecular subgroups.
Table 1:
Clinical characteristics of the study patients according to the molecular groups
| Characteristic | Total (n=316) | Group O (n=95) | Group A (n=175) | Group G (n=46) |
|---|---|---|---|---|
| WHO grade | ||||
| Grade II | 151 (47.8%) | 59 (62.1%) | 80 (45.7%) | 12 (26.1%) |
| Grade III | 165 (52.2%) | 36 (37.9%) | 95 (54.3%) | 34 (73.9%) |
| Age at diagnosis (years) | ||||
| Median | 40 | 43 | 37 | 54 |
| Range | 18-86 | 20-86 | 18-74 | 23-72 |
| Tumor size | ||||
| x-dimension (cm) | ||||
| Median | 4 | 3.73 | 4.24 | 3.53 |
| Range | 1-9.5 | 1-9.5 | 1.3-8.4 | 1-7.4 |
| y-dimension (cm) | ||||
| Median | 5.9 | 5.3 | 6.2 | 5.2 |
| Range | 1.38-12.3 | 2.3-11.6 | 1.9-12.3 | 1.38-10.5 |
| Total area (cm2) | ||||
| Median | 24.25 | 19.6 | 276.46 | 18.22 |
| Range | 1.87-87.87 | 2.9-87.87 | 3.23-84 | 1.87-49.56 |
| Largest tumor diameter | ||||
| < 6 cm | 163 (51.6%) | 53 (55.8%) | 82 (46.9%) | 28 (60.9%) |
| ≥ 6 cm | 153 (48.4%) | 42 (44.2%) | 93 (53.1%) | 18 (39.1%) |
| Crossing midline | ||||
| Yes | 56 (17.7%) | 23 (24.2%) | 28 (16%) | 5 (10.9%) |
| No | 260 (82.3%) | 72 (75.8%) | 147 (84%) | 41 (89.1%) |
The median age for all of the patients at the time of diagnosis was 44 years (range 19 - 86 years). The median age of patients in Group A was 36.7 years (range 19-74), which is significantly younger than patients in Group G (54 years, range 23-72) and Group O (43.1 years, range 20-86) (p<0.0001). The median PFS and OS of all patients were 5.06 years (95% CI 4.2 – 5.8) and 11.3 years (95% CI 9.2 – 13.4 years), respectively. During the follow-up time, 144 patients (45.5%) experienced progressive disease and 72 patients (22.8%) died. As expected, patients in Groups A and O had a significantly longer PFS (5.3 years, 95% CI 4.6 -6.1 years for Group A and 6.1 years, 95% CI 4.3 – 8.0 years for Group O, respectively) than their counterparts in Group G (1.3 years, 95% CI 0.9 – 1.7 years, p < 0.0001) (Figure 1).
Figure 1:
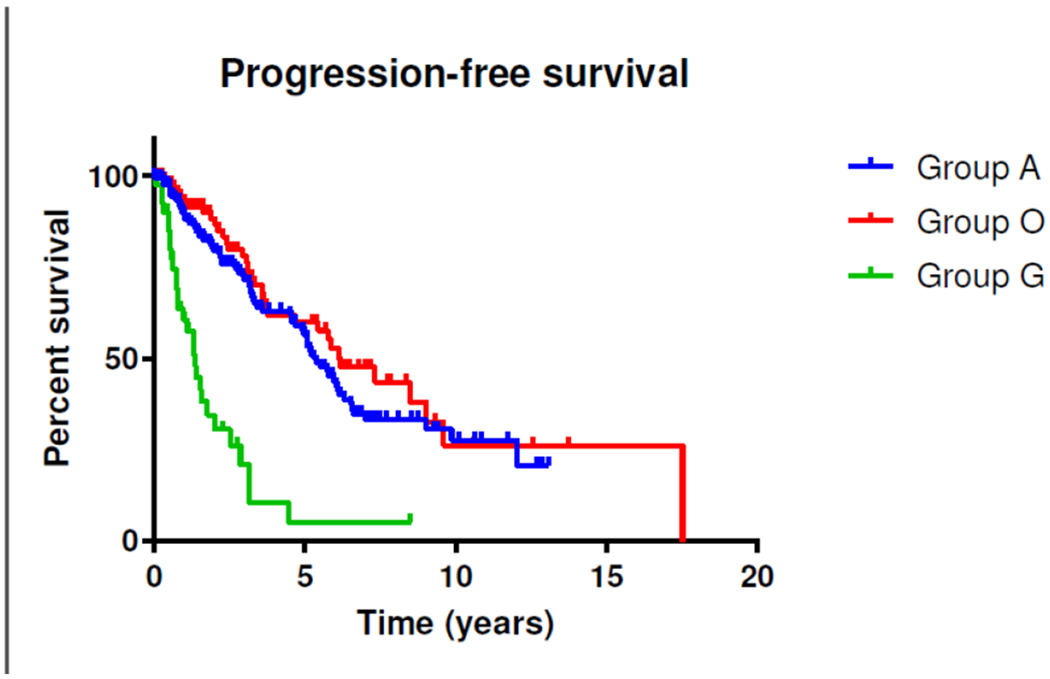
Kaplan-Meier estimates of progression-free survival in LGG in relation to the molecular groups. Patients in Groups A and O had a significantly longer PFS (5.3 years, 95% CI 4.6 -6.1 years for Group A and 6.1 years, 95% CI 4.3 – 8.0 years for Group O, respectively) than their counterparts in Group G (1.3 years, 95% CI 0.9 – 1.7 years, p < 0.0001).
Tumor Size
Analyses were performed to determine if tumor size varied by molecular group. Gliomas in Group A had the largest tumor diameter, with a median size of 26.4 mm2 (range 3.2-84 mm2). In accordance with the Pignatti criteria [17], gliomas in Group A were more likely to have at least one diameter equal to or larger than 6 cm (Fisher’s exact p=0.044 and p=0.045 compared to Group G and Group O, respectively). In aggregate however, Group A gliomas were comparable in size to those in Group O (median size 19.6 mm2, range 2.9 – 87.8); a difference in tumor diameter which was not significant (p=0.2). Interestingly, LGG in Group G were significantly smaller at first presentation than those in Group A (median 18.2 mm2, range 1.8-49.5, p = 0.002). We speculate that because IDH mutant tumors have a slower growth trajectory, they are able to achieve a larger size prior to becoming clinically symptomatic and then being diagnosed for the first time [29]. Furthermore, while patients with IDH-mutant WHO grade II and III gliomas had similar age at first diagnosis (median 40.1 years, 95% CI 18 – 86 years), IDH mutant WHO grade III gliomas had a significantly larger size at diagnosis than IDH mutant WHO grade II tumors (p< 0.01) (Table 1).
Moreover, a multivariate Cox regression analysis revealed molecular Group G status and larger tumor size as factors that are significantly associated with worse PFS (Table 2).
Table 2:
Multivariate Cox regression analysis associated with progression-free survival.
| Hazard ration (HR) | 95% CI | p-value | |
|---|---|---|---|
| Molecular group (G vs. A/O) | 5.2 | 3.2-7.1 | 0.000 |
| Tumor size (>6cm vs. < 6cm) | 1.6 | 1.3-2.2 | 0.007 |
Further tested variables which were not independent factors were: patients’ age at first diagnosis, contrast enhancement in the MRI, tumor crossing the midline and WHO grade II vs. III.
Contrast enhancement
Out of 316 patients, 98 patients (31%) showed a contrast-enhancing tumor on their preoperative MR scans (see Methods). The inter-reviewer agreements were excellent (κ = 0.93, 95% CI, 0.89 – 0.95). Based on the molecular analysis, enhancing gliomas were found to be significantly more common in Group G (n=28/45, 57%), when compared to Group A (n= 49/175, 28%) and Group O (n= 24/95, 25.3%) tumors (p=0.008 and p=0.0011, respectively). Strikingly, we did not observe a significant difference in PFS when comparing the enhancing and non-enhancing gliomas (Figure 2).
Figure 2:
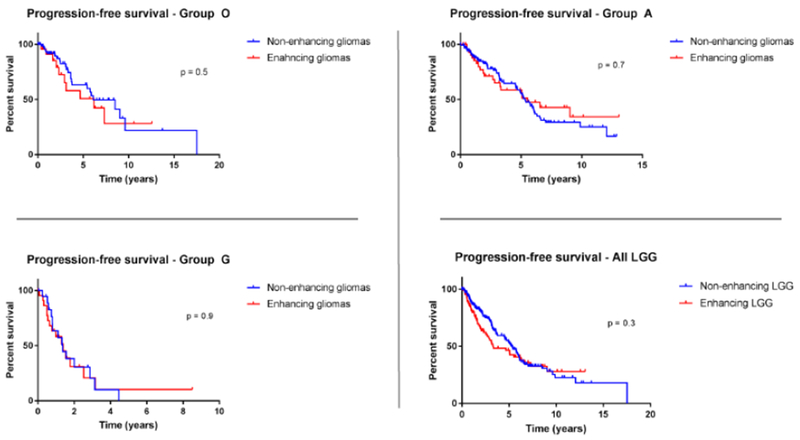
Kaplan-Meier estimates of progression-free survival in all three LGG molecular groups (A/O/G) in relation to contrast enhancement. Out of 316 patients, 98 patients (31%) showed a contrast-enhancing tumor on their preoperative MR scans. No significant differences in PFS were observed when comparing the enhancing and non-enhancing gliomas.
Notably, although we observed a significantly higher rate of gadolinium enhancement in IDH-mutant WHO grade III gliomas (50/131, 38.2%) compared to their WHO II counterparts (23/139, 16.5%, p< 0.0001), this did not clearly influence prognosis as there was no significant difference in PFS between IDH-mutant WHO grade II and grade III gliomas (median PFS of 5.5 years (95% CI 4.6-6.4) for WHO grade II and median 5.9 years (95% CI 4.8-7.1) for WHO grade III, p = 0.65.)
Likewise, the OS of patients with enhancing gliomas was nearly identical to those with non-enhancing gliomas [11.3 years (95% CI 8.0 -14.3) vs. 11.3 years (95% CI 8.3 – 14.3), p = 0.2]. We hypothesized that this lack of association reflects the molecular heterogeneity of the overall cohort, and therefore examined the molecular subgroups separately. We did not observe an association between contrast enhancement and prognosis after stratification within molecular group. Accordingly, although there were 22 deaths in the Group G cohort, only 12 of them demonstrated enhancing features on their MRI scans, suggesting that the poor prognosis commonly associated with IDH wild-type LGG is not necessarily correlated with contrast-enhancement.
T2-FLAIR Mismatch analysis
In a recently published study, Broen et al. confirmed a strong association between the T2-FLAIR mismatch sign and classification as an IDH-mutant astrocytoma[19]. While this study exclusively included non-enhancing gliomas, we analyzed the mismatch signal in our multi-institutional cohort, which included contrast-enhancing gliomas as well. The results of the analysis are shown in Table 3. Consistent with the prior reports, T2-FLAIR mismatch was preferentially found in Group A tumors, 68.7% and 76% of WHO grades II and III, respectively. Additionally, we observed a mismatch sign in 28% of WHO grade II and 29.5% of WHO grade III oligodendrogliomas (Group O), but none of the tumors in Group G. Furthermore, we evaluated the inter-reviewer consensus with the kappa statistic. The inter-reviewer agreement for T2-FLAIR mismatch was largely consistent (κ = 0.86, 95% CI, 0.79 – 0.91). We did not observe a difference in PFS of patients in Groups A and O with and without a T2-FLAIR mismatch sign, (Figure 3) suggesting that T2-FLAIR mismatch does not identify cases with different prognosis, within the A and O groups.
Table 3:
Analysis of the T2-FLAIR mismatch signal in 133 cases from two cohorts (MGH and Dresden)
| Molecular group | WHO grade | T2-FLAIR mismatch sign (n=133) |
|---|---|---|
| Group A (n=82) | Grade II | 22/32 (68.7%) |
| Grade III | 38/50 (76%) | |
| Group O (n=42) | Grade II | 7/25 (28%) |
| Grade III | 5/17 (29.5%) | |
| Group G (n=9) | Grades II/III | 0/9 |
Figure 3:
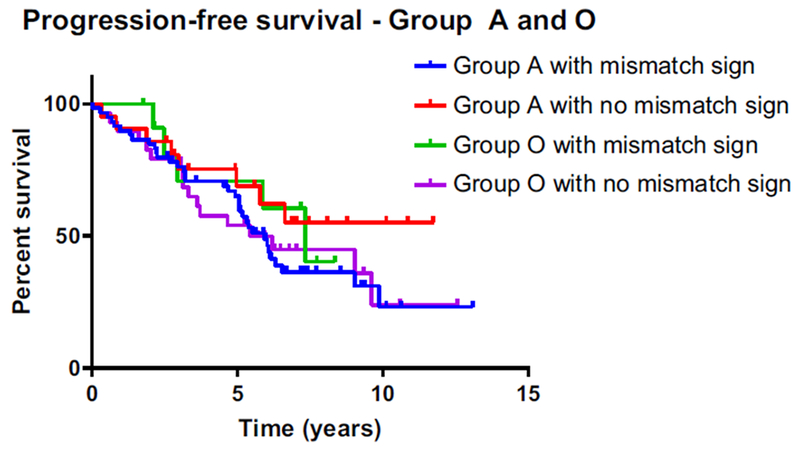
Kaplan-Meier estimates of progression-free survival in the molecular groups A and O in relation to the T2-FLAIR mismatch sign. The PFS of patients in Groups A and O with and without a T2-FLAIR mismatch sign were similar.
Group A probability criteria
We considered that radiographic correlates could be combined with clinical demographic features to robustly identify typical presentations of molecular categories of glioma. In particular, a radiographic method of confidently identifying patients with a Group A glioma preoperatively would have significant clinical utility, as these patients tend to benefit most from aggressive upfront surgical resection [30, 31]. Considering the potential combination of features, we noted that patients younger than 40 years of age with a tumor larger than 6cm in diameter that displays T2-FLAIR mismatch were highly likely to have an IDH mutant astrocytoma (Group A), with a specificity of 96% and a positive predictive value of 88% when all three criteria were fulfilled. However, it should be noted that many group A tumors did not fulfill all of these criteria, as there was only 27% sensitivity (Table 4).
Table 4:
Group A probability criteria: Patients who were younger than 40 years of age at first diagnosis, with a tumor larger than 6cm in diameter that displays T2-FLAIR mismatch were most likely (96% specificity and likelihood ratio of 5.7) to have an IDH mutant astrocytoma (Group A).
| Statistic | Value | 95% CI |
|---|---|---|
| Sensitivity | 27.1% | 17.99% to 37.79% |
| Specificity | 94.3% | 84.34% to 98.82% |
| Positive Likelihood Ratio | 4.7 | 1.51 to 15.15 |
| Negative Likelihood Ratio | 0.77 | 0.67 to 0.89 |
| Positive Predictive Value | 88.4% | 70.76% to 96.05% |
| Negative Predictive Value | 44.6% | 41.09% to 48.26% |
| Disease Prevalence | 61.6% | 52.4% to 69.74% |
Discussion
With the updated WHO 2016 diagnostic criteria, modernization of the radiographic clinical heuristics for decision-making is undergoing re-evaluation, as the relationship between imaging characteristics and the underlying molecular features remains to be fully elucidated. Utilizing a large cohort of WHO grade II and grade III glioma, our study provides evidence that the conventional MR features of low-grade gliomas do not seamlessly correspond with the new molecular WHO 2016 classification of brain tumors. The evaluation of radiographic metrics in the WHO 2016 era is complex, since many prior metrics were initially developed using mixed molecular cohorts, in populations enriched for IDH wild-type high-grade gliomas [32].
Contrast enhancement is one of the most common radiographic features used in clinical practice for prediction of malignant behavior. In addition, it has historically thought to serve as a strong negative prognostic factor [14–16]. In our cohort, although enhancement was present within each molecular subgroup in our study, the IDH wild-type Group G, a biologically more aggressive group, exhibited the highest prevalence of contrast enhancement compared with gliomas in Groups A and O (IDH-mutant). Interestingly, despite this finding, we did not observe significant outcome differences when comparing enhancing and non-enhancing tumors within each molecular subgroup, suggesting that the prognostic significance of contrast enhancement may lose its impact after controlling for molecular subclass. Thus, contrary to the prevailing assumption, our analysis suggests that contrast enhancement may prove to be a less useful marker of prognosis for LGG. This finding partly confirms the results of a previous study that showed no significant survival differences between patients with and without enhancing IDH-mutant gliomas but reported longer survival in patients without gadolinium enhancement in the IDH wild-type glioblastomas group [33]. In contrast, we did not detect an improved survival in patients in Group G harboring LGG with no contrast enhancement when compared with their counterparts with enhancement. The difference between our findings and those of Hemple et al. could be explained by the fact that our Group G included exclusively WHO grades II/III gliomas and no glioblastomas. Moreover, we cannot completely exclude the possibility that some of the patients included- in Group G may have actually had other enhancing IDH wild-type subtypes of LGGs, such as gangliogliomas or pilocytic astrocytomas.
In contrast to the clinical similarities that have been previously described [34, 35], we did note distinct radiographic differences between IDH-mutant WHO grade II and grade III gliomas. IDH mutant grade III gliomas were significantly larger at diagnosis and were more likely to exhibit enhancement when compared with their WHO grade II counterparts. On one hand, this finding shows substantial differences in radiologic appearance between grades, corresponding with the histological grading of these tumors. On the other hand, this higher rate of gadolinium enhancement in IDH-mutant WHO grade III gliomas does not correspond with a more aggressive clinical course or a higher recurrence rate. Indeed, the PFS of patients with WHO grade II and III gliomas were almost identical (Supplementary Figure 2). This finding further aligns with our observation that the presence of contrast enhancement in IDH-mutant gliomas is not an obligate surrogate for poor outcome.
Overall, these observations further highlight the need for improved imaging surrogate biomarkers of prognosis. In keeping with this intention, we expanded upon the recently reported studies that utilized the T2-FLAIR mismatch sign as a diagnostic marker in IDH-mutant astrocytic LGGs [18, 19]. We detected the presence of the mismatch sign in a considerably higher rate of gliomas than previously reported [18, 19], potentially explained by inclusion of IDH-mutant WHO grade III and more contrast-enhancing gliomas in our series. Moreover, we were able to detect the T2-FLAIR mismatch sign in a substantial subset of WHO grades II and III oligodendrogliomas, which has not been yet reported. While the mismatch sign was validated in non-enhancing WHO grade II gliomas in the study by Broen et al., our dataset included contrast enhancing tumors. Though useful for predicting which tumors are IDH-mutant, the presence of the mismatch sign in either oligodendrogliomas or astrocytomas was not associated with a difference in PFS, limiting its utility as a prognostic tool. Our data nevertheless further validate the specificity of the T2-FLAIR mismatch sign and suggest that it can be applied as a diagnostic tool in both IDH-mutant WHO grade II and III gliomas. Since diffusion-weighted imaging (DWI) is considered a valuable MRI tool in the clinical setting, Wu et al. have recently demonstrated that ADC values obtained from DWI correlate with IDH mutation status and overall survival in adult diffuse gliomas [36]. The authors concluded that preoperative ADC estimates may corroborate with molecular subtypes as a prognostic marker and potentially enhance risk stratification, especially within IDH-mutant gliomas [36].
Finally, we established clinical-radiographic criteria that are specific to patients with IDH-mutant astrocytoma, by combining three parameters: age younger than 40 years, large tumor size defined as greatest diameter larger than 6 cm, and the presence of T2-FLAIR mismatch sign. These clinical and radiographic data are straightforward to determine and may find utility in clinical scenarios due to its high specificity. In addition, the predictive positive value of our score rule (88.6%) can be applied to identify patients with IDH-mutant astrocytoma in the preoperative setting. With 96% specificity (likelihood ratio of 5.7) for predicting an IDH-mutant astrocytoma (Group A), such criteria could be useful for prioritizing patients who will maximally benefit from surgical resection adjuncts, such as intraoperative MRI scanning, since an individual tumor’s molecular classification is not typically available at the time of the initial operative procedure [30, 31].
Taken together, our findings demonstrate that the relationship between imaging features and molecular classification is complex, providing evidence that the long-standing usage of contrast enhancement as a marker of prognosis in LGG needs to be re-evaluated. As molecular characterization now plays an instrumental role in predicting tumor behavior, careful clinical and radiologic assessment within specific molecular subgroups may result in the emergence of a prognostic radio-genomic signature in the future.
Supplementary Material
Acknowledgments
Funding: This work is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Projektnummer: 401837860 (to Dr. T. Juratli), the U.S. NIH K12CA090354 (to Dr. J. Miller) and the Burroughs Wellcome Fund Career Award (to Dr. D. Cahill).
References
- 1.Dhermain FG, Hau P, Lanfermann H, Jacobs AH, van den Bent MJ (2010) Advanced MRI and PET imaging for assessment of treatment response in patients with gliomas. Lancet Neurol 9: 906–920 doi: 10.1016/S1474-4422(10)70181-2 [DOI] [PubMed] [Google Scholar]
- 2.Andronesi OC, Loebel F, Bogner W, Marjanska M, Vander Heiden MG, Iafrate AJ, Dietrich J, Batchelor T, Gerstner ER, Kaelin W, Chi AS, Rosen B, Cahill DP (2015) Treatment response assessment in IDH-mutant glioma patients by non-invasive 3D functional Spectroscopic Mapping of 2-Hydroxyglutarate. Clin Cancer Res doi: 10.1158/1078-0432.CCR-15-0656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, Salamon N, Chou AP, Yong WH, Soto H, Wilson N, Driggers E, Jang HG, Su SM, Schenkein DP, Lai A, Cloughesy TF, Kornblum HI, Wu H, Fantin VR, Liau LM (2012) Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol 107: 197–205 doi: 10.1007/s11060-011-0737-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, Fink K, Souhami L, Laperriere N, Curran W, Mehta M (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 31: 337–343 doi: 10.1200/JCO.2012.43.2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, Buckner JC, Fink KL, Souhami L, Laperriere NJ, Huse JT, Mehta MP, Curran WJ (2014) Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol 32: 783–790 doi: 10.1200/JCO.2013.49.3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckner JC, Shaw EG, Pugh SL, Chakravarti A, Gilbert MR, Barger GR, Coons S, Ricci P, Bullard D, Brown PD, Stelzer K, Brachman D, Suh JH, Schultz CJ, Bahary JP, Fisher BJ, Kim H, Murtha AD, Bell EH, Won M, Mehta MP, Curran WJ (2016) Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. N Engl J Med 374: 1344–1355 doi: 10.1056/NEJMoa1500925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller JJ, Shih HA, Andronesi OC, Cahill DP (2017) Isocitrate dehydrogenase-mutant glioma: Evolving clinical and therapeutic implications. Cancer 123: 4535–4546 doi: 10.1002/cncr.31039 [DOI] [PubMed] [Google Scholar]
- 8.Juratli TA, Lautenschläger T, Geiger KD, Pinzer T, Krause M, Schackert G, Krex D (2015) Radio-chemotherapy improves survival in IDH-mutant, 1p/19q non-codeleted secondary high-grade astrocytoma patients. J Neurooncol doi: 10.1007/s11060-015-1822-1 [DOI] [PubMed] [Google Scholar]
- 9.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, Sarkar G, Caron AA, Kollmeyer TM, Praska CE, Chada AR, Halder C, Hansen HM, McCoy LS, Bracci PM, Marshall R, Zheng S, Reis GF, Pico AR, O’Neill BP, Buckner JC, Giannini C, Huse JT, Perry A, Tihan T, Berger MS, Chang SM, Prados MD, Wiemels J, Wiencke JK, Wrensch MR, Jenkins RB (2015) Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med 372: 2499–2508 doi: 10.1056/NEJMoa1407279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131: 803–820 doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 11.Lai A, Kharbanda S, Pope WB, Tran A, Solis OE, Peale F, Forrest WF, Pujara K, Carrillo JA, Pandita A, Ellingson BM, Bowers CW, Soriano RH, Schmidt NO, Mohan S, Yong WH, Seshagiri S, Modrusan Z, Jiang Z, Aldape KD, Mischel PS, Liau LM, Escovedo CJ, Chen W, Nghiemphu PL, James CD, Prados MD, Westphal M, Lamszus K, Cloughesy T, Phillips HS (2011) Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 29: 4482–4490 doi:JCO.2010.33.8715 [pii] 10.1200/JCO.2010.33.8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Bent MJ, Wefel JS, Schiff D, Taphoorn MJ, Jaeckle K, Junck L, Armstrong T, Choucair A, Waldman AD, Gorlia T, Chamberlain M, Baumert BG, Vogelbaum MA, Macdonald DR, Reardon DA, Wen PY, Chang SM, Jacobs AH (2011) Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol 12: 583–593 doi: 10.1016/S1470-2045(11)70057-2 [DOI] [PubMed] [Google Scholar]
- 13.Barker FG, Chang SM, Huhn SL, Davis RL, Gutin PH, McDermott MW, Wilson CB, Prados MD (1997) Age and the risk of anaplasia in magnetic resonance-nonenhancing supratentorial cerebral tumors. Cancer 80: 936–941 [PubMed] [Google Scholar]
- 14.Ginsberg LE, Fuller GN, Hashmi M, Leeds NE, Schomer DF (1998) The significance of lack of MR contrast enhancement of supratentorial brain tumors in adults: histopathological evaluation of a series. Surg Neurol 49: 436–440 [DOI] [PubMed] [Google Scholar]
- 15.Pallud J, Capelle L, Taillandier L, Fontaine D, Mandonnet E, Guillevin R, Bauchet L, Peruzzi P, Laigle-Donadey F, Kujas M, Guyotat J, Baron MH, Mokhtari K, Duffau H (2009) Prognostic significance of imaging contrast enhancement for WHO grade II gliomas. Neuro Oncol 11: 176–182 doi: 10.1215/15228517-2008-066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pallud J, Mandonnet E, Duffau H, Kujas M, Guillevin R, Galanaud D, Taillandier L, Capelle L (2006) Prognostic value of initial magnetic resonance imaging growth rates for World Health Organization grade II gliomas. Ann Neurol 60: 380–383 doi: 10.1002/ana.20946 [DOI] [PubMed] [Google Scholar]
- 17.Pignatti F, van den Bent M, Curran D, Debruyne C, Sylvester R, Therasse P, Afra D, Cornu P, Bolla M, Vecht C, Karim AB, Group EOfRaToCBTC, Group EOfRaToCRC (2002) Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol 20: 2076–2084 [DOI] [PubMed] [Google Scholar]
- 18.Patel SH, Poisson LM, Brat DJ, Zhou Y, Cooper L, Snuderl M, Thomas C, Franceschi AM, Griffith B, Flanders AE, Golfinos JG, Chi AS, Jain R (2017) T2-FLAIR Mismatch, an Imaging Biomarker for IDH and 1p/19q Status in Lower-grade Gliomas: A TCGA/TCIA Project. Clin Cancer Res 23: 6078–6085 doi: 10.1158/1078-0432.CCR-17-0560 [DOI] [PubMed] [Google Scholar]
- 19.Broen MPG, Smits M, Wijnenga MMJ, Dubbink HJ, Anten MHME, Schijns OEMG, Beckervordersandforth J, Postma AA, van den Bent MJ (2018) The T2-FLAIR Mismatch Sign as an Imaging Marker for Non-Enhancing IDH-mutant, 1p/19q-intact Lower Grade Glioma: A Validation Study. Neuro Oncol doi: 10.1093/neuonc/noy048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, Chen J, Robinson HE, Shim HS, Chmielecki J, Pao W, Engelman JA, Iafrate AJ, Le LP (2014) Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 20: 1479–1484 doi: 10.1038/nm.3729 [DOI] [PubMed] [Google Scholar]
- 21.Juratli TA, Kirsch M, Robel K, Soucek S, Geiger K, von Kummer R, Schackert G, Krex D (2012) IDH mutations as an early and consistent marker in low-grade astrocytomas WHO grade II and their consecutive secondary high-grade gliomas. J Neurooncol 108: 403–410 doi: 10.1007/s11060-012-0844-1 [DOI] [PubMed] [Google Scholar]
- 22.Capper D, Sahm F, Hartmann C, Meyermann R, von Deimling A, Schittenhelm J (2010) Application of mutant IDH1 antibody to differentiate diffuse glioma from nonneoplastic central nervous system lesions and therapy-induced changes. Am J Surg Pathol 34: 1199–1204 doi:00000478-201008000-00016 [pii] 10.1097/PAS.0b013e3181e7740d [DOI] [PubMed] [Google Scholar]
- 23.Heckman LD, Chahrour MH, Zoghbi HY (2014) Rett-causing mutations reveal two domains critical for MeCP2 function and for toxicity in MECP2 duplication syndrome mice. Elife 3 doi: 10.7554/eLife.02676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shankar GM, Francis JM, Rinne ML, Ramkissoon SH, Huang FW, Venteicher AS, Akama-Garren EH, Kang YJ, Lelic N, Kim JC, Brown LE, Charbonneau SK, Golby AJ, Sekhar Pedamallu C, Hoang MP, Sullivan RJ, Cherniack AD, Garraway LA, Stemmer-Rachamimov A, Reardon DA, Wen PY, Brastianos PK, Curry WT, Barker FG, Hahn WC, Nahed BV, Ligon KL, Louis DN, Cahill DP, Meyerson M (2015) Rapid Intraoperative Molecular Characterization of Glioma. JAMA Oncol 1: 662–667 doi: 10.1001/jamaoncol.2015.0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juratli TA, Thiede C, Koerner MVA, Tummala SS, Daubner D, Shankar GM, Williams EA, Martinez-Lage M, Soucek S, Robel K, Penson T, Krause M, Appold S, Meinhardt M, Pinzer T, Miller JJ, Krex D, Ely HA, Silverman IM, Christiansen J, Schackert G, Wakimoto H, Kirsch M, Brastianos PK, Cahill DP (2017) Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget 8: 109228–109237 doi: 10.18632/oncotarget.22650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woehrer A, Sander P, Haberler C, Kern S, Maier H, Preusser M, Hartmann C, Kros JM, Hainfellner JA, Societies RCotECoN (2011) FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice - a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS). Clin Neuropathol 30: 47–55 [DOI] [PubMed] [Google Scholar]
- 27.Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, Hosek SM, Kimmel D, O’Fallon J, Yates A, Feuerstein BG, Burger PC, Scheithauer BW, Jenkins RB (1999) Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 18: 4144–4152 doi: 10.1038/sj.onc.1202759 [DOI] [PubMed] [Google Scholar]
- 28.Louis DNOH, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, Perry A, Reifenberger G, Von Deimling A (2016) WHO Classification of Tumours of the Central Nervous System. [DOI] [PubMed]
- 29.Wefel JS, Noll KR, Rao G, Cahill DP (2016) Neurocognitive function varies by IDH1 genetic mutation status in patients with malignant glioma prior to surgical resection. Neuro Oncol doi: 10.1093/neuonc/now165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M, Shonka N, Gilbert MR, Sawaya R, Prabhu SS, Weinberg J, Lang FF, Aldape KD, Sulman EP, Rao G, McCutcheon IE, Cahill DP (2014) IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol 16: 81–91 doi: 10.1093/neuonc/not159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijnenga MMJ, French PJ, Dubbink HJ, Dinjens WNM, Atmodimedjo PN, Kros JM, Smits M, Gahrmann R, Rutten GJ, Verheul JB, Fleischeuer R, Dirven CMF, Vincent AJPE, van den Bent MJ (2018) The impact of surgery in molecularly defined low-grade glioma: an integrated clinical, radiological, and molecular analysis. Neuro Oncol 20: 103–112 doi: 10.1093/neuonc/nox176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, Dunn WD, Scarpace L, Mikkelsen T, Jain R, Wintermark M, Jilwan M, Raghavan P, Huang E, Clifford RJ, Mongkolwat P, Kleper V, Freymann J, Kirby J, Zinn PO, Moreno CS, Jaffe C, Colen R, Rubin DL, Saltz J, Flanders A, Brat DJ (2013) MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267: 560–569 doi: 10.1148/radiol.13120118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hempel JM, Brendle C, Bender B, Bier G, Skardelly M, Gepfner-Tuma I, Eckert F, Ernemann U, Schittenhelm J (2018) Contrast enhancement predicting survival in integrated molecular subtypes of diffuse glioma: an observational cohort study. J Neurooncol doi: 10.1007/s11060-018-2872-y [DOI] [PubMed] [Google Scholar]
- 34.Reuss DE, Mamatjan Y, Schrimpf D, Capper D, Hovestadt V, Kratz A, Sahm F, Koelsche C, Korshunov A, Olar A, Hartmann C, Reijneveld JC, Wesseling P, Unterberg A, Platten M, Wick W, Herold-Mende C, Aldape K, von Deimling A (2015) IDH mutant diffuse and anaplastic astrocytomas have similar age at presentation and little difference in survival: a grading problem for WHO. Acta Neuropathol 129: 867–873 doi: 10.1007/s00401-015-1438-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olar A, Wani KM, Alfaro-Munoz KD, Heathcock LE, van Thuijl HF, Gilbert MR, Armstrong TS, Sulman EP, Cahill DP, Vera-Bolanos E, Yuan Y, Reijneveld JC, Ylstra B, Wesseling P, Aldape KD (2015) IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol 129: 585–596 doi: 10.1007/s00401-015-1398-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu CC, Jain R, Radmanesh A, Poisson LM, Guo WY, Zagzag D, Snuderl M, Placantonakis DG, Golfinos J, Chi AS (2018) Predicting Genotype and Survival in Glioma Using Standard Clinical MR Imaging Apparent Diffusion Coefficient Images: A Pilot Study from The Cancer Genome Atlas. AJNR Am J Neuroradiol doi: 10.3174/ajnr.A5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


