Abstract
Gastric cancer (GC) is one of the major diseases that threaten human health. Although the development of novel drugs has significantly improved the efficacy of GC chemotherapy, the 5-year survival rate of patients with GC remains unsatisfactory. In the present study, the role and mechanism of the long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in GC proliferation was investigated. Clinical specimens and cancer cells were analyzed by western blotting or immunofluorescence. Reverse transcription-quantitative polymerase chain reaction analysis of 57 paired GC and non-tumorous tissues revealed elevated expression of MALAT1 in GC tissues compared with controls. In addition, increased MALAT1 was associated with elevated levels of microtubule-associated protein 1 light chain 3β (LC3B) and antigen Ki67, which are autophagy and proliferation markers, respectively. MTT and colony formation assay results demonstrated that MALAT1 promoted GC cell proliferation. To the best of our knowledge, the present study was the first to demonstrate that upregulated MALAT1 was associated with increased autophagy activation in GC tissues. Furthermore, this study reported that MALAT1 increased cell proliferation and enhanced autophagy activation in GC cells. In addition, the results revealed that MALAT1 inhibited microRNA (miR)-204 expression in GC cells. The present study also demonstrated that miR-204 repressed autophagy through the downregulation of LC3B and transient receptor potential melastatin 3 expression in GC cells. These results indicated that MALAT1 activated autophagy and promoted cell proliferation by downregulating miR-204 expression in GC.
Keywords: gastric cancer, MALAT1, autophagy, proliferation, miR-204
Introduction
Gastric cancer (GC) is the fifth most frequent cancer (1,033,701 new cases in 2018) and the third leading cause of cancer-associated mortality 782,685 deaths in 2018) worldwide (1). Incidence rates are the highest in East Asia, particularly in Japan and China (1). Despite comprehensive use of surgical treatment, chemotherapy, radiotherapy and molecular-targeted treatment, the 5-year survival rate of patients with GC remains <20% in China (2–4). Further investigation is therefore required to fully elucidate the molecular mechanisms underlying the progression of GC and to identify novel therapeutic targets.
Autophagy is a highly conserved catabolic process in which cytoplasmic material and organelles are delivered to lysosomes for degradation. Autophagy is a multi-step and complex process that is regulated by ~30 autophagy-associated proteins and multiple signaling pathways, including AMPK, PERK (5,6). Dysregulation of autophagy has been detected in numerous human diseases, including cancer (7). In addition, increasing evidence indicates that autophagy serves a crucial role in tumor cell proliferation and differentiation, and that it affects the efficacy of anti-tumor drugs (8,9). Although a number of studies have investigated autophagy in GC (10–15), the precise role of autophagy in the development of GC remains to be completely identified.
Long non-coding RNAs (lncRNAs) serve crucial roles in various biological processes, including immune responses, angiogenesis, cell proliferation, differentiation, apoptosis and autophagy (16–18). Previous studies have also reported the abnormal expression and roles of lncRNAs in cancer (19–21). Investigation of the tumor-associated molecular mechanisms of lncRNAs may therefore contribute to the prevention and treatment of cancer. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA discovered in 2003 that is closely associated with the clinical outcome and progression of lung cancer (22). MALAT1 expression has been reported to be significantly upregulated in lung cancer, hepatocellular carcinoma, bladder cancer and other types of malignancy (23–25). A recent study reported that MALAT1 may be used as a diagnostic marker of GC metastasis (26). However, the precise mechanism of MALAT1 in the development of GC remains not fully understood.
MicroRNA (miR)-204 is a well-studied tumor suppressor, which is commonly downregulated in breast and prostate cancer, renal cell carcinoma and GC (27–32). Numerous studies have indicated that miR-204 can repress the development of GC (31,33,34). Furthermore, loss of miR-204 leads to upregulated expression of transient receptor potential melastatin 3 (TRPM3), which stimulates oncogenic autophagy by regulating microtubule-associated protein 1 light chain 3α (MAP1LC3A, also known as LC3A) and LC3B and promotes cancer growth (33,35). However, in cholangiocarcinoma and lung cancer, miR-204 is negatively regulated by MALAT1 (36,37).
The present study aimed to clarify the association between MALAT1, miR-204, and GC progression, and to investigate the underlying molecular mechanism of GC.
Materials and methods
Clinical samples
A total of 57 human GC tissues and corresponding non-cancerous adjacent tissues (5 cm from the edge of tumor) were collected from 18 women and 39 men who received surgery at Jiangyin Hospital Affiliated to Nantong University (Jiangyin, China) between September 2017 and June 2018. The mean age of patients was 56.3 years (age range, 37–72 years). All patient materials were obtained with written informed consent in accordance with the requirements of the Clinical Research Ethics Committees of Jiangyin Hospital Affiliated to Nantong University; The experimental protocols were approved by the Research Ethics Committee in Jiangyin Hospital Affiliated to Nantong University (approval no. JY1608AC97). All methods used in this study were in accordance with the approved guidelines.
Cell lines and transfection
The human normal gastric epithelial cell line GES-1 and the GC cell line MKN45 were purchased from the American Type Culture Collection. The gastric adenocarcinoma-derived circulating tumor cell lines CTC141 and CTC105 (38) were kindly donated by the Laboratory of Stem Cell Biology of Sichuan University (Chengdu, China). All cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Sigma-Aldrich; Merck KGaA) and placed at 37°C in a humidified incubator containing 5% CO2. Cells were passaged every 2–3 days.
For the transfection, the small interfering (si)RNA were obtained from GenSript. Sequences for the siRNAs were designed as follows: Negative control siRNA (si-NC), 5′-CUUGCCUGGACCAGCUUAAdTdT-3′; si-MALAT1−1, 5′-CAGCCCGAGACTTCTGTAAdTdT-3′; and si-MALAT2, 5′-AGCCCGAGACTTCTGTAAAdTdT-3′. The pcDNA vector and MALAT1 overexpression plasmid (pcDNA-MALAT1) were obtained from Nanjing KGI Biological Technology Development Co., Ltd. miR-204 mimic (cat. no. miR10022693-1-5) and mimic control were purchased from Guangzhou RiboBio Co., Ltd. Transcription efficiency was evaluated by quantitative PCR.
CTC 141 and CTC105 cells were transiently transfected with siRNA (100 nM/1×105 cells), plasmid (1 µg/1×105 cells) or mimic (100 nM/1×105 cells) using Lipofectamine® 3000 (12 µl/1×105 cells; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Following 48 h transfection, cells were subjected to experiments. The transcription efficiency was evaluated by quantitative real-time PCR.
MTT assay
CTC 141 and CTC105 cell proliferation was measured with MTT assay (Sigma-Aldrich). Cells were seeded into 96-well plates (3×103 cells/well) for 24, 48 or 72 h and were treated for 3 h with 2 mM 3-methyladenine (3-MA; Selleck, cat. no. S2767), used as a cell autophagy inhibitor, before assessing cell proliferation. MTT (20 µl) was then added to each well for 4 h at 37°C. Cell viability was assessed by detecting absorbance at 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.). All tests were performed in quadruplicate.
Colony formation assay
Transfected CTC105 cells were plated in a 6-well plate (200 cells/well) and cultured for 14 days under standard culture conditions. Cell culture medium was replaced every 2–3 days. Cells were fixed with methanol for 20 min, and stained with 0.5% crystal violet for 5 min at room temperature. Stained colonies (cells number >30) were imaged using a Nikon camera (Nikon, Tokyo, Japan). Each experiment was performed at least three times.
Immunofluorescence staining
Paraffin-embedded GC tissues were collected and fixed with 10% formalin overnight at room temperature. Section (0.5 µm thick) were dehydrated by increasing ethanol gradient (50, 70, 85, 95 and 100%) at room temperature for 2 h, deparaffinized with dimethylbenzene at room temperature for 15 min and rehydrated by decreasing gradient of ethanol (100, 95, 85 and 75%) for 5 min at room temperature. Tissue sections were incubated in 0.5 M sodium citrate buffer (JRDUN Biotechnology, Co., Ltd.) at 37°C for 15 min and heated in a microwave at 92–98°C for 20 min for antigen retrieval. Tissue sections were permeabilized with 0.1% Triton X-100 (Sangon Biotech Co. Ltd.) in PBS for 25 min at room temperature, and blocked with 2% bovine serum albumin (Sigma-Aldrich) in PBS for 1 h at room temperature. Tissue sections were then incubated with anti-LC3B (Cell Signaling Technology, Inc.; cat. no. 83506; 1:200) and anti-Ki67 antibodies (Cell Signaling Technology, Inc.; cat. no. 9449; 1:300) at 4°C overnight. Tissue sections were then incubated with fluorescein isothiocyanate-labeled secondary anti-mouse antibody (Cell Signaling Technology Europe, B.V.; cat. no. 4410; 1:500) for 2 h at room temperature. Cell nuclei were stained with DAPI (1 µg/ml; Sigma-Aldrich; Merck KGaA) for 5 min at room temperature. Stained sections were observed under a CX41RF fluorescence microscope (Olympus Corporation) with ×200 magnification.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol® reagent (Takara Biotechnology Co., Ltd.) was used to extract total RNA from gastric tissues and cells. cDNAs were generated using the PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. SYBR® Premix Ex Taq (Takara Biotechnology Co., Ltd.) was used for qPCR according to the manufacturer's instructions. The thermocycling conditions were as follows: 95°C for 10 sec, followed by 40 cycles of 95°C for 5 sec, 60°C for 15 sec, and 72°C for 30 sec, the final extension was 72°C for 5 min. Primer sequences used for qPCR were as follows: MALAT1 forward, 5′-AGCGGAAGAACGAATGTAAC-3′ and reverse, 5′-GAACAGAAGGAAGAGCCAAG-3′; LC3B forward, 5′-GATGTCCGACTTATTCGAGAGC-3′ and reverse, 5′-TTGAGCTGTAAGCGCCTTCTA-3′; TRPM3 forward, 5′-ATACCCAGCACCAAAGACC-3′ and reverse 5′-TCTGAAGCACGGAGATACTG-3′; and GAPDH forward, 5′-TGAACGGGAAGCTCACTGG-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′. The relative expressions levels were normalized to the endogenous control GAPDH and calculated using the 2−ΔΔCq method (39).
To detect miRNA-204, reverse transcription and qPCR were performed using a Bulge-Loop™ miRNA qPCR Primer Set for hsa-miR-204 (Guangzhou RiboBio Co., Ltd.) and U6 snRNA (Guangzhou RiboBio Co., Ltd.) according to the manufacturer's instructions and as previously described (40). U6 served as an internal control.
Western blotting
Total cellular proteins from CTC105 and CTC141 cells were extracted using radioimmunoprecipitation assay buffer (Auragene). Protein concentration was determined with bicinchoninic acid assay (Thermo Fisher Scientific, Inc.). Proteins (20 µg) were separated by 12% SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Membranes were blocked with 5% skimmed milk at room temperature for 1 h and incubated with primary antibodies against p62 (cat. no. 39749; 1:2,000; Cell Signaling Technology, Inc.), LC3B (cat. no. 3868; 1:1,000; Cell Signaling Technology, Inc.), Ki67 (cat. no. 13110; 1:5,000; Cell Signaling Technology, Inc.), β-actin (cat. no. 4970; 1:1,000; Cell Signaling Technology, Inc.) and TRPM3 (cat. no. ab56171; 1:1,000; Abcam) at room temperature for 2 h. Bands were detected using enhanced chemiluminescence substrate (Applygen Technologies, Inc.) according the manufacturer's protocol. Protein quantification was performed by ImageJ software (National Institutes of Health).
Statistical analysis
Data are presented as the mean ± standard deviation. SPSS 17.0 statistical software (SPSS, Inc.) was used for statistical analyses. Comparison between groups was performed with paired Student's t-test or analysis of variance followed by Holm-Sidak's or Dunnett's multiple comparisons test (GraphPad Software, Inc.). The expression correlation was analyzed using Pearson's correlation test. P<0.05 was considered to indicate a statistically significant difference.
Results
MALAT1 is associated with autophagy activation in GC tissues
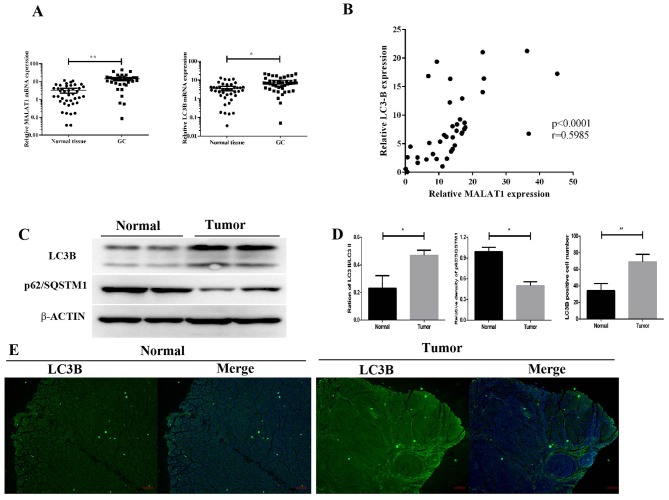
To examine the role of MALAT1 in GC and to identify potential molecular events, RT-qPCR was used to detect MALAT1 expression levels in 57 GC and paired non-tumorous tissues. The results demonstrated that MALAT1 expression was significantly increased in GC tissues compared with paired non-tumorous tissues (P<0.01; Fig. 1A). As a structural protein of autophagosome membranes (41), LC3B mRNA level was upregulated in GC tumors (P<0.05; Fig. 1A). Furthermore, the mRNA level of LC3B was positively correlated with MALAT1. As a marker of autophagy, the ratio of LC3-II/LC3-I was also increased in GC tissues compared with the control group (Fig. 1C and D). The results of the western blotting analysis demonstrated that p62 expression was lower in GC tissues compared with the control group (Fig. 1C and D). In addition, fluorescence microscopy revealed that the number of FITC-LC3 puncta was higher in GC tissues compared with controls (Fig. 1D and E). These results demonstrated that upregulated MALAT1 levels were associated with increased expression of autophagy markers in GC tissues.
Figure 1.
Upregulated MALAT1 is associated with increased autophagy activation in GC tissues. (A) MALAT1 expression levels in 57 paired GC tissues measured by reverse transcription-quantitative polymerase chain reaction. (B) Correlation analysis between MALAT1 and LC3B mRNA expression in GC tissues compared with paired non-tumorous tissues. (C) Western blotting of LC3B and p62 protein levels in GC and paired non-tumorous tissues. (D) LC3B, p62 protein level and LC3B immunofluorescence quantification in clinic tissues. (E) Immunofluorescence detection of endogenous LC3B in GC and paired non-tumorous tissues. *P<0.05 and **P<0.01. GC, gastric cancer; MALAT1, metastasis-associated lung adenocarcinoma transcript 1.
MALAT1 enhances GC cell proliferation
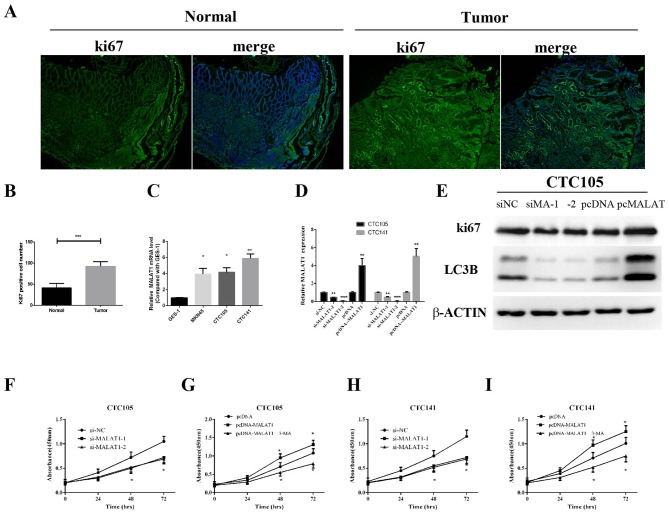
To examine whether MALAT1 may be associated with GC cell proliferation, Ki67 expression levels were assessed in clinical specimens by immunofluorescence analysis. Ki67 expression levels in GC tissues were demonstrated to be higher compared with the control group (Fig. 2A and B), which indicated increased proliferation in GC samples.
Figure 2.
MALAT1 promotes GC cell proliferation. (A and B) Immunofluorescence detection of Ki-67 in GC and non-tumorous tissues. (C) MALAT1 expression levels in the normal human gastric epithelial cell line GES-1 and human GC cell lines MKN45, CTC105 and CTC141 were measured by RT-qPCR. (D and E) CTC105 and CTC141 cell lines were transfected with the MALAT1 overexpression vector, MALAT1 siRNAs or the indicated controls and examined by (D) RT-qPCR and (E) western blotting. (F-I) CTC141 and CTC105 cells were transfected with (F and H) MALAT1 siRNAs, (H and I) the MALAT1 overexpression vector or the indicated controls, and MTT assays were performed at 0, 24, 48 and 72 h, with or without 3-MA (2 mM, 3 h). *P<0.05, **P<0.01 and ***P<0.001 vs. the corresponding control. MA, MALAT1; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si, small interfering.
MALAT1 expression levels in GC cells lines were evaluated, and RT-qPCR results demonstrated that MALAT1 expression was significantly upregulated in CTC105 and CTC141 cells compared with normal gastric epithelial GES-1 cells (Fig. 2C). To evaluate the function of MALAT1 in GC-derived circulating cells, MALAT1 overexpression plasmid and siRNAs targeting MALAT1 were used. MALAT1 mRNA expression increase in cells transfected with the MALAT1 overexpression plasmid and decrease in si-MALAT1- and si-MALAT2-transfected cells were confirmed (Fig. 2D). Similar results were obtained by western blotting in CTC105 cells (Fig. 2E).
MALAT1 downregulation significantly decreased CTC105 and CTC141 cell proliferation compared with controls (Fig. 2F and H), whereas upregulated MALAT1 enhanced CTC105 and CTC141 cell proliferation, and the increased proliferation was inhibited by the autophagy inhibitor, 3-methyladenine (3-MA) (Fig. 2G and I). These results demonstrated that MALAT1 may increase GC cell proliferation through autophagy.
MALAT1 regulates miR-204 in GC
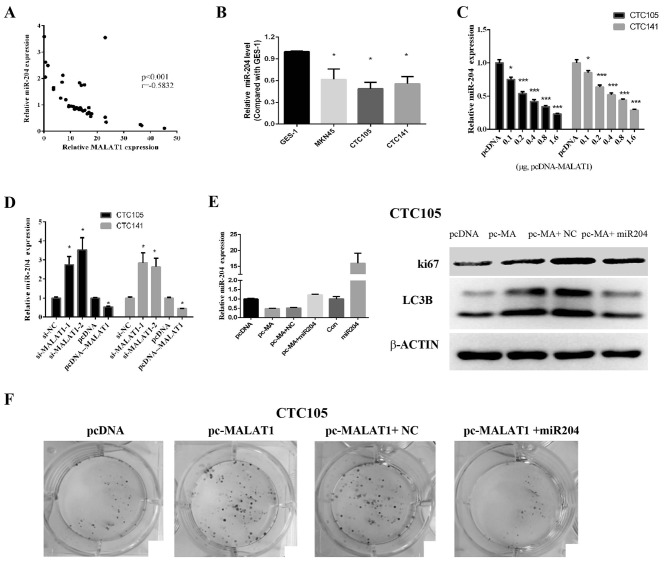
A previous study reported that MALAT1 exerted its effects by binding and inhibiting miR-204 (36). The results from the present study demonstrated that miR-204 and MALAT1 expression levels were negatively correlated in GC tissues (Fig. 3A), which suggested that MALAT1 may inhibit miR-204 expression in GC. In addition, miR-204 expression levels in CTC105 and CTC141 cells were significantly lower compared with those in GES-1 cells (Fig. 3B), and the levels or miR-204 decreased following MALAT1 overexpression (Fig. 3C). Furthermore, following MALAT1 downregulation in CTC105 and CTC141 cells, miR-204 expression level was increased, whereas miR-204 expression level was decreased in CTC105 and CTC141 cells overexpressing MALAT1 (Fig. 3D).
Figure 3.
MALAT1 regulates miR-204 levels in GC cells. (A) MALAT1 expression was negatively correlated with miR-204 expression in GC tissues. (B) RT-qPCR analysis of miR-204 expression in GC cells compared with GES-1 cells. (C and D) miR-204 levels were detected in GC cell lines transfected with (C) MALAT1 overexpression vector, (D) siRNAs, or the appropriate controls as indicated. (E) After the miR-204 expression was detected by RT-qPCR, western blot analysis of LC3B and Ki67 was performed in CTC105 cells transfected with the MALAT1 overexpression vector or miR-204 mimic as indicated. (F) Colony formation assay of CTC105 cells transfected with the MALAT1 overexpression vector or miR-204 mimic as indicated. *P<0.05 and ***P<0.001 vs. the corresponding control. MA, MALAT1; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; miR-204, microRNA-204; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Furthermore, MALAT1 overexpression increased LC3B and Ki67 expression in CTC105 cells, which was abolished following miR-204 mimic transfection (Fig. 3E). In addition, colony formation assays indicated that MALAT1 promoted GC cell proliferation and that miR-204 rescued the effect of MALAT1 on cancer cells (Fig. 3F). These results suggested that MALAT1 may induce miR-204 downregulation and that MALAT1-induced cell proliferation and autophagy marker upregulation was blocked by miR-204. These results suggested that MALAT1 may work by negatively regulating miR-204.
miR-204 inhibits MALAT1-induced regulation of LC3B and TRPM3 in GC
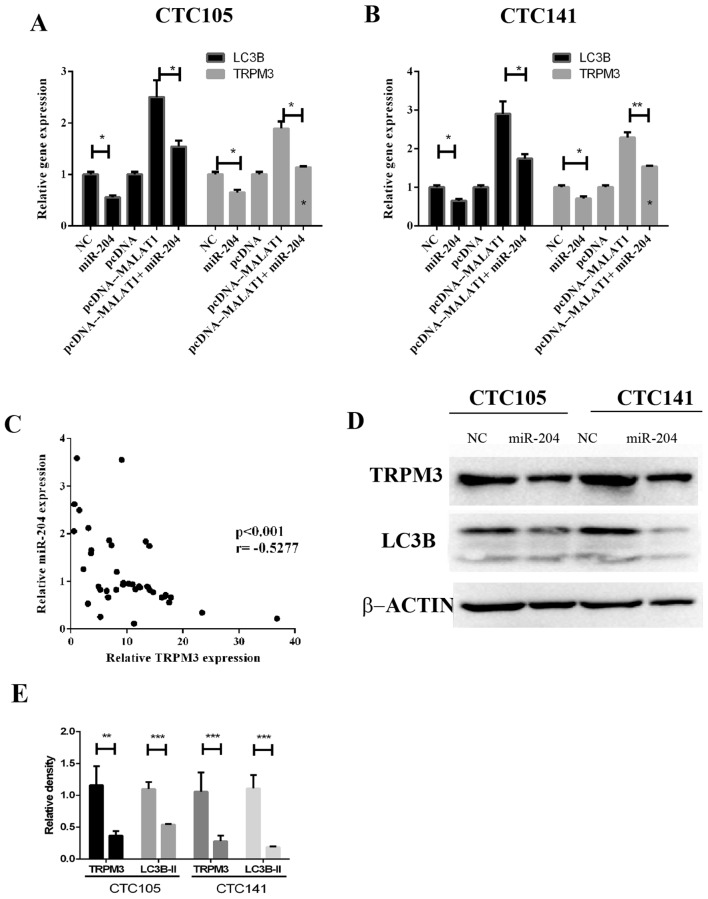
The regulatory effects of MALAT1 and miR-204 on TRPM3, which is an autophagy activator, were evaluated. The results demonstrated that MALAT1 increased LC3B and TRPM3 mRNA expression levels, whereas miR-204 co-expression eliminated these effects in CTC105 (Fig. 4A) and CTC141 (Fig. 4B) cell lines. In addition, TRPM3 and miR-204 expression levels were negatively correlated GC tissues (Fig. 4C). Western blotting demonstrated that LC3B and TRPM3 protein levels were significantly decreased in CTC105 and CTC141 cells transfected with the miR-204 mimic (Fig. 4D and E).
Figure 4.
miR-204 reverses MALAT1-induced regulation of LC3B and TRPM3 in GC. (A and B) Reverse transcription-quantitative polymerase chain reaction analysis of LC3B and TRPM3 mRNAs in (A) CTC105 and (B) CTC141 cells transfected as indicated. (C) Correlation analysis of miR-204 and TRPM3 expression in GC tissues. (D) LC3B and TRPM3 western blotting in GC cells transfected with miR-204. (E) LC3B and TRMP3 level quantification. *P<0.05, **P<0.01 and ***P<0.001. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; MA, MALAT1; miR-204, microRNA-204; NC, negative control; TRPM3, transient receptor potential melastatin 3.
Discussion
Numerous studies have demonstrated that lncRNAs serve crucial roles in the development of various types of cancer (16–21). However, the function of lncRNAs in GC remains unclear. Identification of cancer-associated lncRNAs and their targets is therefore critical for understanding their roles in tumorigenesis and for the development of novel targets for GC therapy. The present study investigated the role of lncRNA MALAT1 in GC and its potential underlying mechanisms.
MALAT1 has recently been proposed as a marker of GC (26). Previous studies have reported that MALAT1 promotes GC cell proliferation and metastasis (42,43). In addition, MALAT1 serves a crucial role in the regulation of autophagy, and its overexpression is associated with chemoresistance (44). Although these studies indicate that MALAT1 is involved in GC progression and autophagy-associated chemoresistance, the correlations between MALAT1, autophagy and proliferation in vivo remain unclear.
In the present study, the correlation between MALAT1 expression levels and autophagy activation in GC cell lines and tissues and the association between MALAT1 expression and GC proliferation were examined. The results demonstrated that MALAT1 upregulation was associated with increased autophagy in GC tissues. MALAT1 expression levels were significantly increased in GC tissues, and LC3B expression was proportional to the level of MALAT1. In addition, expression levels of LC3B were also significantly higher in GC tissues compared with non-tumor tissues. In the pcDNA negative control group, LC3B was downregulated compared with the siNC group, which may be due to endotoxin effect following plasmid extraction that may have affected cell autophagy (45). However, this did not affect our conclusion.
However, p62 expression was lower in GC tissues compared with the control group, p62, which is a scaffolding/adaptor protein, is involved in numerous physiological processes, including inflammation, mitosis and autophagy and is a crucial factor during tumorigenesis (46,47). These findings demonstrated that upregulated MALAT1 was associated with increased autophagy activation in GC. The results observed in GC cell lines were consistent with these findings. In addition, the present study demonstrated that MALAT1 overexpression increased GC cell proliferation.
As a target of MALAT1, miR-204 prevents tumor development (33,36,37,48,49) and regulates TRPM3-induced oncogenic autophagy (50,51). The results of the present study revealed that the miR-204 mimic reversed the effects of MALAT1 overexpression on LC3B and TRPM3 in GC cells, which indicated that MALAT1 may exert its roles in GC by negatively regulating miR-204. Furthermore, the results suggested that miR-204 may regulate the expression of the autophagy markers LC3B and TRPM3 in GC cells. The results of the present study demonstrated that MALAT1 overexpression resulted in decreased miR-204 levels, and that MALAT1 and miR-204 expression levels were negatively correlated in GC tissues, which further indicated that MALAT1 may negatively regulate miR-204 in GC cells. Although the MALAT1/miR-204/LC3B axis has been reported to regulate autophagy in myocardial ischemia-reperfusion injury (52), the effect of the microenvironment on cell autophagy through this axis remains unclear. Preliminary data from a mouse model of GC indicated that this axis was involved in Helicobacter pylori-induced GC and regulated the infection by causing autophagy (data not shown). Identifying how the MALAT1/miR-204/LC3B axis regulates the H. pylori-induced infection and tumorigenesis through autophagy will be the aim of our further study.
The present study explored the molecular mechanism of MALAT1 in the occurrence and development of GC. The results may provide potential novel targets for the development of molecular targeted therapy of GC.
Acknowledgements
The authors would like to thank Dr Xianming Mo (Laboratory of Stem Cell Biology of Sichuan University) for providing the cancer cell lines CTC105 and CTC141.
Funding
The present study was supported by the National Natural Science Foundation of China (grant. no. 81602636), the Wuxi Health and Family Planning Commission Youth Research Foundation (grant no. Q201720) and the Li Jie-shou Gut Barrier Foundation (grant no. LJS-201708).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
GS, ZZ and WZ analyzed and interpreted the patient data. GS, ZZ and GH performed the cellular and molecular experiments, LZ, WL, CX and XZ analyzed the data. WZ and CX were major contributors in writing the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of Jiangyin Hospital Affiliated to Nantong University (approval no. JY1608AC97). All patients provided written informed consent for the present study.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Russo AE, Strong VE. Gastric cancer etiology and management in Asia and the West. Annu Rev Med. 2019;70:353–367. doi: 10.1146/annurev-med-081117-043436. [DOI] [PubMed] [Google Scholar]
- 3.Irino T, Takeuchi H, Terashima M, Wakai T, Kitagawa Y. Gastric cancer in Asia: Unique features and management. Am Soc Clin Oncol Educ Book. 2017;37:279–291. doi: 10.1200/EDBK_175228. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Zhang K. Surgical treatment for metastatic gastric cancer. Zhonghua Wei Chang Wai Ke Za Zhi. 2017;20:731–734. (In Chinese) [PubMed] [Google Scholar]
- 5.Li S, Le W. An insight review of autophagy biology and neurodegenerative diseases: Machinery, mechanisms and regulation. Sci China Life Sci. 2017;60:1457–1459. doi: 10.1007/s11427-017-9214-7. [DOI] [PubMed] [Google Scholar]
- 6.Yin Z, Pascual C, Klionsky DJ. Autophagy: Machinery and regulation. Microb Cell. 2016;3:588–596. doi: 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fulda S. Targeting autophagy for the treatment of cancer. Biol Chem. 2018;399:673–677. doi: 10.1515/hsz-2018-0105. [DOI] [PubMed] [Google Scholar]
- 8.Tian Y, Xu H, Farooq AA, Nie B, Chen X, Su S, Yuan R, Qiao G, Li C, Li X, et al. Maslinic acid induces autophagy by down-regulating HSPA8 in pancreatic cancer cells. Phytother Res. 2018;32:1320–1331. doi: 10.1002/ptr.6064. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Feng L, Zhang H, Zhang J, Zhang Y, Li S, Qin L, Yang Z, Xiong J. Effects of miR-144 on the sensitivity of human anaplastic thyroid carcinoma cells to cisplatin by autophagy regulation. Cancer Biol Ther. 2018;19:484–496. doi: 10.1080/15384047.2018.1433502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song J, Zhou Y, Gong Y, Liu H, Tang L. Rottlerin promotes autophagy and apoptosis in gastric cancer cell lines. Mol Med Rep. 2018;18:2905–2913. doi: 10.3892/mmr.2018.9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D, Gao M, Zhao S. Autophagy as a novel strategy for treatment of gastric cancer: A hypothesis. Med Sci Monit. 2013;19:794–796. doi: 10.12659/MSM.889486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li LQ, Pan D, Zhang SW, Xie DY, Zheng XL, Chen H. Autophagy regulates chemoresistance of gastric cancer stem cells via the Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22:3402–3407. doi: 10.26355/eurrev_201806_15162. [DOI] [PubMed] [Google Scholar]
- 13.Bai XY, Liu YG, Song W, Li YY, Hou DS, Luo HM, Liu P. Anticancer activity of tetrandrine by inducing pro-death apoptosis and autophagy in human gastric cancer cells. J Pharm Pharmacol. 2018;70:1048–1058. doi: 10.1111/jphp.12935. [DOI] [PubMed] [Google Scholar]
- 14.Fan H, Jiang M, Li B, He Y, Huang C, Luo D, Xu H, Yang L, Zhou J. MicroRNA-let-7a regulates cell autophagy by targeting Rictor in gastric cancer cell lines MGC-803 and SGC-7901. Oncol Rep. 2018;39:1207–1214. doi: 10.3892/or.2018.6194. [DOI] [PubMed] [Google Scholar]
- 15.Li B, Wang W, Li Z, Chen Z, Zhi X, Xu J, Li Q, Wang L, Huang X, Wang L, et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017;410:212–227. doi: 10.1016/j.canlet.2017.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res. 2018;129:151–155. doi: 10.1016/j.phrs.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhu M, Sun Y, Li W, Wang Y, Yu W. Upregulation of lncRNA CASC2 suppresses cell proliferation and metastasis of breast cancer via inactivation of the TGF-β signaling pathway. Oncol Res. 2019;27:379–387. doi: 10.3727/096504018X15199531937158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ, Hsu CW, Chen WS, Wang JH. Linc00659, a long noncoding RNA, acts as novel oncogene in regulating cancer cell growth in colorectal cancer. Mol Cancer. 2018;17:72. doi: 10.1186/s12943-018-0821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Wu K, Liu K, Miao R. Effects of MALAT1 on proliferation and apo- ptosis of human non-small cell lung cancer A549 cells in vitro and tumor xenograft growth in vivo by modulating autophagy. Cancer Biomark. 2018;22:63–72. doi: 10.3233/CBM-170917. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Cui Y, Liu LF, Ren WB, Li QQ, Zhou X, Li YL, Li Y, Bai XY, Zu XB. High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin Genitourin Cancer. 2017;15:570–576. doi: 10.1016/j.clgc.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Wang CJ, Shi SB, Tian J, Xu J, Niu ZX. lncRNA MALAT1, HOTTIP and PVT1 as predictors for predicting the efficacy of GEM based chemotherapy in first-line treatment of pancreatic cancer patients. Oncotarget. 2017;8:95108–95115. doi: 10.18632/oncotarget.19345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, Ren M, Chen L, Yuan D, Zhang Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209–56218. doi: 10.18632/oncotarget.10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuai F, Wang B, Dong S. MicroRNA-204 inhibits the growth and motility of colorectal cancer cells by downregulation of CXCL8. Oncol Res. 2018;26:1295–1305. doi: 10.3727/096504018X15172747209020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Shu L, Zhang Z, Cai Y. MicroRNA-204 inhibits cell migration and invasion in human cervical cancer by regulating transcription factor 12. Oncol Lett. 2018;15:161–166. doi: 10.3892/ol.2017.7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu Y, Ren L, Xie B, Liang Z, Chen J. MiR-204 enhances mitochondrial apoptosis in doxorubicin-treated prostate cancer cells by targeting SIRT1/p53 pathway. Oncotarget. 2017;8:97313–97322. doi: 10.18632/oncotarget.21960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen SQ, Huang LS, Xiao XL, Zhu XF, Xiong DD, Cao XM, Wei KL, Chen G, Feng ZB. MiR-204 regulates the biological behavior of breast cancer MCF-7 cells by directly targeting FOXA1. Oncol Rep. 2017;38:368–376. doi: 10.3892/or.2017.5644. [DOI] [PubMed] [Google Scholar]
- 31.Canu V, Sacconi A, Lorenzon L, Biagioni F, Lo Sardo F, Diodoro MG, Muti P, Garofalo A, Strano S, D'Errico A, et al. MiR-204 down-regulation elicited perturbation of a gene target signature common to human cholangiocarcinoma and gastric cancer. Oncotarget. 2017;8:29540–29557. doi: 10.18632/oncotarget.15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T, Pan H, Li R. The dual regulatory role of miR-204 in cancer. Tumour Biol. 2016;37:11667–11677. doi: 10.1007/s13277-016-5144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrestha S, Yang CD, Hong HC, Chou CH, Tai CS, Chiew MY, Chen WL, Weng SL, Chen CC, Chang YA, et al. Integrated microRNA-mRNA analysis reveals miR-204 inhibits cell proliferation in gastric cancer by targeting CKS1B, CXCL1 and GPRC5A. Int J Mol Sci. 2017;19(pii):E87. doi: 10.3390/ijms19010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Liu XS, Liu HY, Lu YY, Li Y. Reduced expression of serum miR-204 predicts poor prognosis of gastric cancer. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15027702. [DOI] [PubMed] [Google Scholar]
- 35.Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Wang J, Chen Y, Li S, Jin M, Wang H, Chen Z, Yu W. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res. 2016;6:1099–1107. [PMC free article] [PubMed] [Google Scholar]
- 37.Tan X, Huang Z, Li X. Long non-coding RNA MALAT1 interacts with miR-204 to modulate human hilar cholangiocarcinoma proliferation, migration, and invasion by targeting CXCR4. J Cell Biochem. 2017;118:3643–3653. doi: 10.1002/jcb.25862. [DOI] [PubMed] [Google Scholar]
- 38.Chen T, Yang K, Yu J, Meng W, Yuan D, Bi F, Liu F, Liu J, Dai B, Chen X, et al. Identification and expansion of cancer stem cells in tumor tissues and peripheral blood derived from gastric adenocarcinoma patients. Cell Res. 2012;22:248–258. doi: 10.1038/cr.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao W, Chang C, Cui Y, Zhao X, Yang J, Shen L, Zhou J, Hou Z, Zhang Z, Ye C, et al. Steroid receptor coactivator-3 regulates glucose metabolism in bladder cancer cells through coactivation of hypoxia inducible factor 1α. J Biol Chem. 2014;289:11219–11229. doi: 10.1074/jbc.M113.535989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin JJ, Liang B, Zhan XR. MicroRNA-204 inhibits cell proliferation in T-cell acute lymphoblastic leukemia by down-regulating SOX4. Int J Clin Exp Pathol. 2015;8:9189–9195. [PMC free article] [PubMed] [Google Scholar]
- 41.Bolanos JM, Moran AM, da Silva CM, Dávila MP, Muñoz PM, Aparicio IM, Tapia JA, Ferrusola CO, Peña FJ. During cooled storage the extender influences processed autophagy marker light chain 3 (LC3B) of stallion spermatozoa. Anim Reprod Sci. 2014;145:40–46. doi: 10.1016/j.anireprosci.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557–564. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 43.Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan S, Yu Q, Li YY, Kang Y, Li H, et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget. 2016;7:12693–12703. doi: 10.18632/oncotarget.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung KW, Kim KM, Choi YJ, An HJ, Lee B, Kim DH, Lee EK, Im E, Lee J, Im DS, et al. The critical role played by endotoxin-induced liver autophagy in the maintenance of lipid metabolism during sepsis. Autophagy. 2017;13:1113–1129. doi: 10.1080/15548627.2017.1319040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moscat J, Diaz-Meco MT. p62: A versatile multitasker takes on cancer. Trends Biochem Sci. 2012;37:230–236. doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z, Long J, Du R, Ge C, Guo K, Xu Y. MiR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumour Biol. 2016;37:8327–8335. doi: 10.1007/s13277-015-4627-0. [DOI] [PubMed] [Google Scholar]
- 49.Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J, Tan D, Liu S. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumour Biol. 2017;39:1010428317718135. doi: 10.1177/1010428317718135. [DOI] [PubMed] [Google Scholar]
- 50.Hall DP, Cost NG, Hegde S, Kellner E, Mikhaylova O, Stratton Y, Ehmer B, Abplanalp WA, Pandey R, Biesiada J, et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell. 2014;26:738–753. doi: 10.1016/j.ccell.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cost NG, Czyzyk-Krzeska MF. Regulation of autophagy by two products of one gene: TRPM3 and miR-204. Mol Cell Oncol. 2015;2:e1002712. doi: 10.1080/23723556.2014.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S, Yu W, Luo X, Chen J, Deng F. MALAT1/miR-204/LC3-II: A potential regulated axis of autophagy in myocardial ischemia-reperfusion injury. Int J Cardiol. 2019;277:222. doi: 10.1016/j.ijcard.2018.08.075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.