Abstract
In this study, we describe a method to reliably characterize intrahepatic leukocyte populations using flow cytometry and next‐generation RNA sequencing on fresh human liver biopsies. Over the last decades, immune responses of viral hepatitis patients, and of other liver diseases, have been incompletely characterized. Most studies include peripheral blood samples only, foregoing the possibility to investigate the site of inflammation directly. Here, we show that with an optimized protocol that combines cell sorting and RNA sequencing, we can perform a side by side comparison of both intrahepatic and peripheral immune cells. Using core liver biopsies from chronic hepatitis B virus patients, we show that the expression levels of IFN‐stimulated genes and leukocyte‐specific genes are markedly different in the liver compartment as compared to the peripheral blood. These observations emphasize the need to sample the liver directly. The variation of gene expression profiles in these chronic hepatitis B patients was considerable, despite the uniform treatment with nucleotide analogs and absence of liver inflammation in these patients. Finally, we show that this method can provide a detailed characterization of previously undetected liver‐specific effects of novel candidate therapeutic compounds.
Keywords: Cells: Natural Killer, Cells: T Lymphocytes, Process: Host‐Pathogen Interactions, Tissue/System: Lymphoid, Tissue/System: Digestive, Process: Lymphoid Cell Mediated Immunity, Process: Gene Regulation, Techniques: Multiparameter FACS
The use of fluorescence‐activated cell sorting and next generation RNA sequencing to maximize the information yield of studies investigating tissue‐resident leukocytes in human liver.
Abbreviations
- FSC
forward scatter
- HBV
hepatitis B virus
- ISG
IFN‐stimulated genes
- RIN
RNA integrity number
- SSC
sideward scatter
1. INTRODUCTION
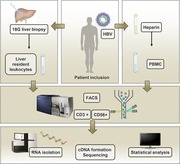
Our knowledge on immune processes in diseased tissues is still incomplete. The main reason for this is that sampling of many diseased organs is not routinely performed for research purposes, and because blood is the most accessible compartment, the more invasive procedure to obtain tissue is less frequently conducted. This is very clear in case of infections of the liver with hepatitis B virus (HBV) or hepatitis C virus: most studies have examined peripheral blood. Importantly, the few studies that examined the liver demonstrated that the phenotype and function of intrahepatic immune cells differ considerably to that of their peripheral counterparts.1, 2, 3, 4, 5 In recent years, an increasing number of viral hepatitis researchers are describing efforts to fill this gap of knowledge, and are in agreement that a better understanding of liver immunology is crucial for the development of new curative antiviral treatment strategies for chronic hepatitis B.1, 5, 6, 7 Unfortunately, progress in the field of liver immunology is hampered by practical and technical difficulties.6 Core liver biopsies are rarely collected for research purposes only, because the associated complication rate is only accepted if the biopsy assists clinical decision making.8 Also, with the expanding use of noninvasive tests of liver disease, such as fibroscan,9, 10 rest material from liver biopsies is less frequently available. Finally, not all clinical units have a research laboratory able to receive and process liver tissue within hours, which is essential because liver cells are prone to die from apoptosis.11, 12 Because the number of liver biopsies available is limited, it is essential to make optimal use of all material for research purposes. In the current study, we therefore examined the possibility to combine cell sorting of lymphocytes from liver biopsies by flow cytometry with RNA sequencing to obtain gene expression profiles of liver‐resident leukocytes. To do this, we used liver biopsies of 6 patients with chronic hepatitis B who were on continuous nucleotide analog therapy (Supplementary Table 1). A number of groups have reported on the evaluation of liver material collected by fine needle aspiration using a 22–25 Gauge needle for flow cytometry or bulk gene expression profiling.13, 14 In this study, we aimed to combine these approaches and purify T cell and NK cells from liver biopsies, followed by next‐generation sequencing of the RNA isolated from these cells, to increase the information gained from biopsies during future immunological studies.
2. MATERIALS AND METHODS
2.1. Collection of liver biopsies and blood samples
We collected core liver biopsies (18 Gauge) of 6 patients chronically infected with HBV and treated with nucleotide analog therapy. Biopsies were collected in 10 ml PBS + 1% FCS in a 15 ml tube, and kept on ice. The liver biopsy tissue was homogenized by carefully mashing the biopsy using the plunger of a 1 ml syringe in a 48 well flat bottom plate. PBS + 1% FCS was added until a 200 μl cell suspension was formed. Erythrocyte lysis was not performed and only fresh material was used (no cryopreservation). All biopsies were taken between 9–11 AM and processed for FACS analysis and sorting within the hour. Rapid processing and analysis was crucial to minimize cell death. Patients were monitored for at least 2 h following biopsy collection and no complications related to the liver biopsies were recorded. Heparinized blood was collected from all patients immediately prior to the biopsy procedure. PBMC were isolated by ficoll separation (Ficoll‐Paque™ plus, GE Healthcare Life Sciences, Amersham, United Kingdom), counted and 106 cells stained using the same antibody panel as the liver biopsies.
2.2. FACS sorting procedure
Intrahepatic and peripheral leukocytes were sorted using the FACS Aria II (BD, Franklin Lakes, New Jersey, United States) equipped with a 130 μm nozzle. Blood and liver samples were immunophenotyped and various leukocyte populations were subsequently sorted for gene expression analysis by RNA sequencing, using the gating strategy depicted in Fig. 1A and B. We selected CD235a‐negative, CD45‐positive cells (nonerythrocyte, all leukocytes) cells, followed by further gating on forward scatter (FSC) and sideward scatter (SSC), positivity for CD56 or CD3 (Fig. 1), and exclusion of dead cells (5–10% in the liver biopsies) and doublets (using FSC‐W/SSC‐W and FSC‐H/SSC‐H). This sorting strategy resulted in T and NK cells with purities of at least 95%. The following populations were purified (Fig. 1C): CD45+CD3+ (T cells) and CD45+CD56+ (total NK cells) using CD3:PeCy7 (clone UCHT1, eBioscience, Waltham, Massachusetts, United States), CD8:APC‐H7 (clone SK1, BD Biosciences), Live/Dead:AmCyan (Miltenyi Biotec), CD45:PE‐eFluor610 (eBioscience, HI30), and CD235a:FITC (clone HIR2, eBioscience).
Figure 1.
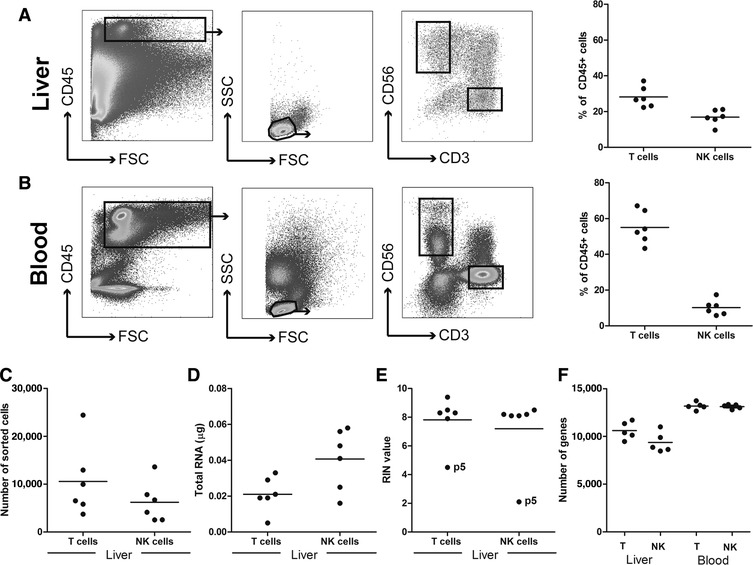
Description of the gating strategy and quality control steps for isolated mRNA derived from liver immune cell populations. (A) FACS plots showing the gating strategies for liver T cells (CD45+CD3+CD56‐ lymphocytes), and NK cells (CD45+CD3‐CD56+ lymphocytes) of 6 chronic HBV patients treated with nucleotide analogs. The panel on the right‐hand side shows the T cell and NK cell frequencies of total intrahepatic CD45+ cells. (B) The gating strategy for peripheral blood T cells and NK cells, including the T cell and NK cell frequencies of the total blood CD45+ population (right panel). (C) The absolute number of intrahepatic T and NK cells sorted by flow cytometry. (D) The total amount of RNA obtained from the sorted cells. (E) The RNA integrity number (RIN value) of the RNA samples. (F) The total number of unique genes identified in T cells and NK cells from the 5 liver biopsies passing quality control, in comparison to blood‐derived T and NK cells
2.3. RNA isolation
RNA was isolated using Arcturus® PicoPure® RNA Isolation Kit according to the manufacturer instructions: Briefly, sorted cells were centrifuged (3000 × g, 9 min), the supernatant was discarded, and the pellet incubated for 30 min with extraction buffer. Ethanol (70%) was pipetted onto the supernatant and the cell extract and ethanol mixture was then pipetted onto the prepared purification column and centrifuged twice (2 min at 100 × g, 30 s at 16,000 × g). DNA was removed using by DNAse treatment. RNA integrity was measured using Eukaryote Total RNA Pico assay (Agilent Technologies Inc, Santa Clara, California, United States). Technical information on the RNA sequencing of purified leukocyte populations, the alignment of short reads to the genome and, the analysis of expression of genes and pathways can be found in the Extended Methods on page in the Supplementary Information.
2.4. Clinical trial
This study was part of the international trial investigating the safety, tolerability, and efficacy of vesatolimod (GS‐9620) in patients with chronic hepatitis B on nucleotide analog therapy (Clinical Trial Number: GS‐US‐283‐1059; NCT 02166047).15 The liver biopsies (18 Gauge) used for the current study were performed at baseline and 24 h after the last eighth dose of vesatolimod. Vesatolimod (GS‐9620) is an oral agonist of TLR7. It is currently under investigation for the treatment of both HBeAg‐positive and negative chronic HBV. Our center was the only site to perform liver immune cell isolation (in tandem with PBMC isolation), using an 18 Gauge needle, before and after vesatolimod treatment, and on continuous nucleotide analog therapy. Six noncirrhotic virally suppressed chronic hepatitis B patients on nucleotide analog therapy (age: 29–63 years; 4 males and 2 females) were treated once a week for 7 weeks with 1 mg GS‐9620 (n = 3), 2 mg GS‐9620 (n = 1), 4 mg GS‐9620 (n = 1), or placebo (n = 1). Blood was collected from the patients prior (week 0) and at 4, 8, 12, 24, and 48 weeks after start of GS‐9620 treatment.
2.5. Ethical statement
The study was performed in concordance with the Declaration of Helsinki as adopted by the 64th WMA General Assembly, Fortaleza, Brazil, October 2013. The institutional ethical review board of the Erasmus Medical Center approved the protocols, and written informed consent was obtained from all individuals.
3. RESULTS AND DISCUSSION
As expected, the frequency of NK cells was higher in the liver as compared to the blood (Fig. 1A and B), while the opposite was observed for T cells. Surprisingly, a significant percentage of the CD45+ liver immune cells were not identified as T cells or NK cells (Fig. 1A) and may reflect the higher fraction of CD3+CD56+ NKT cells (16.5% vs. 2.9%) and the relative enrichment of B cells and myeloid cells in the liver. All samples that passed the quality control were analyzed further. In this project, bulk CD3+ T cells were sorted to map general gene expression of T cells in liver and blood, but because the frequencies of CD4+/CD8+ T cells usually differ in liver and blood, sorting T cell subset separately would likely be more informative. The numbers of genes detected were higher in the peripheral blood‐derived immune cells, as compared to liver‐derived T cells and NK cells (Fig. 1F), which is likely due to higher input of sorted T and NK cells (average of 1,609,073 vs. 11,242 T cells and 389,820 vs. 7289 NK cells in blood vs. liver, respectively). Ninety‐five percent of the genes detected in liver‐resident NK cells and T cells overlapped with their peripheral counterpart (data not shown). The transcriptional profiles of T and NK cells in the liver biopsies, and in the blood, were evaluated for expression of IFN‐stimulated genes (ISG; Fig. 2A and B, Supplementary Table 2), T cell genes (Supplementary Table 3) and NK cell genes (Fig. 2C, Supplementary Table 4). It should be noted here that for a relevant assessment of clinical parameters of the patients included, a larger cohort of patients would be necessary. As previously described at the protein level in both cellular compartments, the immune profiles in blood and liver of chronic hepatitis B patients exhibit considerable variation2, 16: only small gene clusters (<20 genes) were modulated in the same direction in 2 individual patients (Fig. 2). We observed less variation in the blood compared to the liver, which emphasizes the need for sampling of the liver, preferably using multiple measurements. In sequential biopsies from a single individual, the frequency of liver‐resident lymphocyte populations, including T cells and NK cells, did not vary >2% between biopsies, confirming the reproducibility of the protocol used in our study. All RNA samples derived from T cells and NK cells had good RNA integrity scores, with RIN (RNA integrity number) values of 7 or higher (Fig. 1D and E). In the current study, a similar pattern of ISG induction could be observed in T cells and NK cells in liver for each patient, suggesting that these intrahepatic lymphocyte populations are exposed to the same signals (such as type I IFNs), and possibly regulate the transcription of ISG in a similar way. The expression of T cell genes in CD3+ cells in the liver (Fig. 2C) was different from the blood, as was the expression of NK cell genes in NK cells. In addition to type I IFNs and lymphocyte specific genes, we detected several genes involved in T or NK cell‐mediated immune responses (Table 1). Of these genes, a few were identified to be highly expressed in the liver T cells (CXCR6, TNF, CD160, CCR5, and IFNG; blood/liver ratio 0.03, 0.04, 0.06, 0.06, and 0.07, respectively) and NK cells (CXCR6, CCL3L3, and TNFSF14; blood/liver ratio 0.015, 0.09, and 0.13, respectively). This observation corresponds to previous reports17 that liver‐resident NK cells, but not peripheral blood leukocytes, express CXCR6, as well as high levels of Eomes,18 but low levels of T‐bet (blood/liver ratio EOMES 0.2, TBX21 2.3). Interestingly, we detected high expression of genes coding for TNF receptor ligands (Table 1), like TNF and FASLG (liver T cells), as well as TNFSF14 (liver NK cells). These genes were among the most liver‐enriched genes in these HBV patients, which may be related to earlier reports of their role in initiation of fibrogenesis.19
Figure 2.
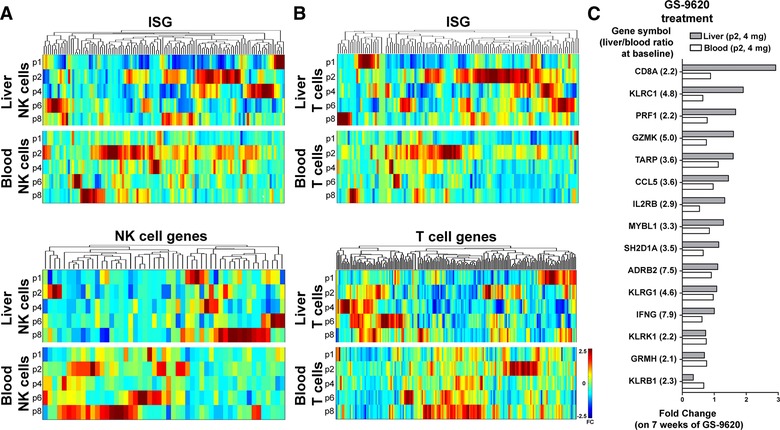
Relative expression of ISG and cell type specific genes in liver and blood of virally suppressed patients with chronic hepatitis B. (A) The relative baseline expression level of ISG (upper panel) and NK cell genes (lower panel) in liver and peripheral blood NK cells of 5 nucleotide analog‐treated chronic HBV patients. The colors represent a range of fold difference from −2.5 (blue) to 2.5 (red). (B) The relative baseline expression of ISG in T cells of 5 nucleotide analog‐treated chronic HBV patients is shown in the upper panel and of T cell genes in the lower panel. (C) The modulation of ISG expression after 7 weeks of treatment with GS‐9620 (Vesatolimod), evaluated longitudinally in T cells isolated from liver biopsies in 1 virally suppressed chronic HBV patient (patient 2, treated with 4 mg Vesatolimod weekly). Vesatolimod (GS‐9620) is a TLR7 agonist under investigation for treatment of chronic HBV. The 15 genes whose expressions are most specific to the liver at baseline (before Vesatolimod treatment) are shown, as identified by calculating the liver/blood ratio of expression levels in both compartments. The FC before and after treatment in liver (grey bars) and blood (white bars) is shown for each individual gene, revealing liver‐specific treatment effects
FC; fold change
Table 1.
The top liver‐enriched genes in T cells and NK cells derived from blood and liver during virally suppressed HBV
| T cells | NK cells | ||||||
|---|---|---|---|---|---|---|---|
| Rank | Gene | P‐value | Ratio blood/liver | Rank | Gene | P‐value | Ratio blood/liver |
| 1 | CXCR6 | 0.0006 | 0.02869 | 1 | CXCR6 | 0.00001 | 0.01512 |
| 2 | TNF | 0.00001 | 0.04143 | 2 | SPRY2 | 0.00006 | 0.02319 |
| 3 | CD160 | 0.0007 | 0.0593 | 3 | EXPH5 | 0.01919 | 0.08083 |
| 4 | CCR5 | 0.00002 | 0.05955 | 4 | CCL3L3 | 0.00219 | 0.08839 |
| 5 | SLC4A10 | 0.00801 | 0.05977 | 5 | LDB2 | 0.00011 | 0.09216 |
| 6 | IFNG | 0.00024 | 0.07045 | 6 | SLC4A10 | 0.00615 | 0.09528 |
| 7 | B3GALT2 | 0.00211 | 0.07435 | 7 | DET1 | 0.02772 | 0.10272 |
| 8 | FASLG | 0.00008 | 0.0766 | 8 | IGIP | 0.00241 | 0.10989 |
| 9 | MS4A1 | 0.00014 | 0.09095 | 9 | MBOAT1 | 0.00188 | 0.12441 |
| 10 | CMTM1 | 0.00208 | 0.10766 | 10 | TNFSF14 | 0.00479 | 0.13107 |
Finally, using pathway analysis, we aimed to identify the degree of pathway activation in our samples, and compare the blood and liver compartment (Table 2). In liver T cells, the NK‐cell‐mediated cytotoxicity, hepatitis B and Th1 and Th2 differentiation pathway are relatively active, which was expected given the more direct exposure to the virally infected liver tissue and increased T cell differentiation state in most tissues. The oxidative phosphorylation genes were active in T cells and NK cells in the peripheral blood, which may be due to its superior oxygenation compared to the portal blood entering the liver sinusoids. In both T cells and NK cells, cell cycle genes (and DNA Mismatch repair genes) have low expression in the blood suggesting that these cells proliferate slightly faster in the liver. Interestingly, in NK cells in the blood, the pathways related to direct NK cell effector functions, like Lysosome and NK cell mediated cytotoxicity, were more active, possibly reflective of the immune suppressive environment of the liver.
Table 2.
Relative activation of signaling pathways in T cells and NK cells from blood and liver
| Pathway name | Genes in pathway (total) | Blood CD3 FC | Liver CD3 FC | Blood NK FC | Liver NK FC |
|---|---|---|---|---|---|
| Glycolysis / gluconeogenesis | 68 (100%)a | 2.8 (64%) | 2.5 (58%) | −2.7 (64%) | −2.2 (51%) |
| Cell cycle | 124 (100%) | 0.0 (84%) | 0.9 (77%) | −1.3 (89%) | 1.7 (73%) |
| Lysosome | 123 (100%) | −4.2 (89%) | −4.0 (82%) | 7.3 (88%) | 1.8 (71%) |
| NK cell‐mediated cytotoxicity | 133 (100%) | −11.7 (71%) | −3.7 (70%) | 3.6 (77%) | 2.6 (69%) |
| Oxidative phosphorylation | 133 (100%) | 7.2 (68%) | −5.0 (67%) | 4.5 (68%) | −4.3 (62%) |
| Hepatitis B | 144 (100%) | −0.2 (77%) | 1.7 (73%) | 1.6 (79%) | −1.4 (67%) |
| Th1 and Th2 cell differentiation | 92 (100%) | −5.9 (86%) | 1.5 (82%) | 1.6 (85%) | −1.0 (72%) |
| Mismatch repair | 23 (100%) | 2.3 (96%) | −0.5 (88%) | 0.8 (98%) | −1.6 (83%) |
The percentage of overlap with the original list of genes in that pathway to estimate the degree of representation of each pathway in every sample.
FC; fold change.
In this study, we had the opportunity to obtain a second 18 Gauge core liver biopsy from the same patients at a later time point to map the modulation by immunotherapeutic intervention on the phenotype and gene expression profile of specific lymphocyte populations in addition to baseline gene expression. Analysis of the paired samples showed the modulation of gene expression in T cells from a chronic HBV patient before and after 7 weeks of treatment with 4 mg GS‐9620 (Vesatolimod), an oral TLR7 agonist under investigation for the treatment of chronic HBV and may (re‐)activate host immune responses to the virus20 (Clinical Trial Number: GS‐US‐283‐1059; NCT 02166047). To detect possible liver‐specific effects of the compound, we evaluated the modulation of genes highly enriched in the liver (high liver/blood ratio). In the example depicted in Fig. 2C, on the gene expression modulation of liver T cells of patient 2, treated with the highest dose of GS‐9620 (4 mg), we found CD8A, KLRC1, PRF1, GRMK, and IL2RB (Fig. 2C) to be up‐regulated by GS‐9620 treatment in this patient's liver, but not in the blood.
In summary, we describe here a method to investigate intrahepatic leukocyte populations using FACS and next generation RNA sequencing on fresh human liver biopsies directly ex vivo. Fewer liver biopsies are available for research, and more sophisticated methodologies are needed to provide an insight into the pathogenesis of immune‐mediated liver disease, including viral hepatitis, non‐alcoholic steatohepatitis (NASH)/Non‐alcoholic fatty liver disease (NAFLD), and primary sclerosing cholangitis (PSC). The expression of ISG and leukocyte‐specific genes are markedly different in the liver compartment as compared to the peripheral blood, emphasizing the need to sample the liver directly. The variation of gene expression profiles in these nucleotide analog‐treated chronic hepatitis B patients was considerable, despite undetectable viral loads and the absence of liver inflammation in all included subjects. We show that this method can provide a detailed characterization of the effect of new immune therapeutic compounds on liver‐residing leukocyte populations.
Supporting information
Supplementary Information
AUTHORSHIP
L.L.B. and R.K. contributed to clinical follow up of the included patients; L.L.B., M.H.M., A.G., S.F., and G.W.O. contributed to acquisition of data; L.L.B., J.H., L.L., and A.B. contributed to the analysis and interpretation of data; J.H. and L.L. contributed to the statistical analysis; L.L.B. and A.B. contributed to writing of the manuscript; A.B. supervised the study and acquired funding; A.G., S.F., R.K., G.W.O., M.H.M., and A.B. contributed to critical revision of the manuscript.
ACKNOWLEDGEMENTS
This study was funded by Gilead Sciences. Data have not been previously presented at national or international meetings. We thank Heleen van Santen and Melek Polat for help with collecting patient material and follow up of the included patients. We thank Jasmijn Selten for assistance with figure layout and the Erasmus MC Cancer Computational Biology Center for giving access to the IT‐infrastructure and software that was used for the analysis in this study.
DISCLOSURES
A.B. was advisory board member for Gilead Sciences and has received research support from Gilead Sciences, Fujirebio, BMS, and Roche. L.L.B., G.W.O., J.H., M.H.M., and R.K. have no conflicts of interest. A.G., L.L., and S.F. are employees of Gilead Sciences.
Boeijen LL, van Oord GW, Hou J, et al. Gene expression profiling of human tissue‐resident immune cells: Comparing blood and liver. J Leukoc Biol. 2019;105:603–608. 10.1002/JLB.6AB0718-278R
REFERENCES
- 1. Lebosse F, Testoni B, Fresquet J, et al. Intrahepatic innate immune response pathways are downregulated in untreated chronic hepatitis B. J Hepatol. 2017;66:897–909. [DOI] [PubMed] [Google Scholar]
- 2. Tjwa ET, Zoutendijk R, van Oord GW, et al. Similar frequencies, phenotype and activation status of intrahepatic NK cells in chronic HBV patients after long‐term treatment with tenofovir disoproxil fumarate (TDF). Antiviral Res. 2016;132:70–75. [DOI] [PubMed] [Google Scholar]
- 3. Spaan M, Claassen MA, Hou J, Janssen HL, de Knegt RJ, Boonstra A. The intrahepatic T cell compartment does not normalize years after therapy‐induced hepatitis C virus eradication. J Infect Dis. 2015;212:386–390. [DOI] [PubMed] [Google Scholar]
- 4. Pallett LJ, Davies J, Colbeck EJ, et al. IL‐2(high) tissue‐resident T cells in the human liver: sentinels for hepatotropic infection. J Exp Med. 2017;214:1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boeijen LL, Hoogeveen RC, Boonstra A, Lauer GM. Hepatitis B virus infection and the immune response: the big questions. Best Pract Res Clin Gastroenterol. 2017;31:265–272. [DOI] [PubMed] [Google Scholar]
- 6. Gill US, Pallett LJ, Kennedy PTF, Maini MK. Liver sampling: a vital window into HBV pathogenesis on the path to functional cure. Gut. 2018;67:767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singh AK, Rooge SB, Varshney A, et al. Global micro RNA expression profiling in the liver biopsies of hepatitis B virus infected patients suggests specific miRNA signatures for viral persistence and hepatocellular injury. Hepatology. 2018;67:1695–1709. [DOI] [PubMed] [Google Scholar]
- 8. Piccinino F, Sagnelli E, Pasquale G, Giusti G. Complications following percutaneous liver biopsy. A multicentre retrospective study on 68276 biopsies. J Hepatol. 1986;2:165–173. [DOI] [PubMed] [Google Scholar]
- 9. Goyal R, Mallick SR, Mahanta M, et al. Fibroscan can avoid liver biopsy in Indian patients with chronic hepatitis B. J Gastroenterol Hepatol. 2013;28:1738–1745. [DOI] [PubMed] [Google Scholar]
- 10. Lipp MJ, D'Souza LS, Clain DJ, Jr Bodenheimer HC, , Min AD. Trends in the indication and method of liver biopsy for hepatitis B and C. Dig Dis Sci. 2010;55:2971–2976. [DOI] [PubMed] [Google Scholar]
- 11. Castera L. Noninvasive methods to assess liver disease in patients with hepatitis B or C. Gastroenterology. 2012;142:1293–1302.e4. [DOI] [PubMed] [Google Scholar]
- 12. Papatheodoridis GV, Manolakopoulos S, Liaw YF, Lok A. Follow‐up and indications for liver biopsy in HBeAg‐negative chronic hepatitis B virus infection with persistently normal ALT: a systematic review. J Hepatol. 2012;57:196–202. [DOI] [PubMed] [Google Scholar]
- 13. Demirkiran A, Baan CC, Kok A, Metselaar HJ, Tilanus HW, van der Laan LJ. Intrahepatic detection of FOXP3 gene expression after liver transplantation using minimally invasive aspiration biopsy. Transplantation. 2007;83:819–823. [DOI] [PubMed] [Google Scholar]
- 14. Sprengers D, van der Molen RG, Kusters JG, et al. Flow cytometry of fine‐needle‐aspiration biopsies: a new method to monitor the intrahepatic immunological environment in chronic viral hepatitis. J Viral Hepat. 2005;12:507–512. [DOI] [PubMed] [Google Scholar]
- 15. Janssen HLA, Brunetto MR, Kim YJ, et al. Safety, efficacy and pharmacodynamics of vesatolimod (GS‐9620) in virally suppressed patients with chronic hepatitis B. J Hepatol. 2018;68:431–440. [DOI] [PubMed] [Google Scholar]
- 16. Boeijen LL, Montanari NR, de Groen RA, et al. Mucosal‐associated invariant T cells are more activated in chronic hepatitis B, but not depleted in blood: reversal by antiviral therapy. J Infect Dis. 2017;216:969–976. [DOI] [PubMed] [Google Scholar]
- 17. Stegmann KA, Robertson F, Hansi N, et al. CXCR6 marks a novel subset of T‐bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep. 2016;6:26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cuff AO, Robertson FP, Stegmann KA, et al. Eomeshi NK cells in human liver are long‐lived and do not recirculate but can be replenished from the circulation. J Immunol. 2016;197:4283–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang YM, Seki E. TNFalpha in liver fibrosis. Curr Pathobiol Rep. 2015;3:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boni C, Vecchi A, Rossi M, et al. TLR7 agonist increases responses of hepatitis B virus‐specific T cells and natural killer cells in patients with chronic hepatitis B treated with nucleos(t)ide analogues. Gastroenterology. 2018;154:1764–1777.e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


