Abstract
Transposable elements colonize genomes and with time may end up being incorporated into functional regions. SINE Alu elements, which appeared in the primate lineage, are ubiquitous in the human genome and more than a thousand overlap annotated coding exons. Although almost all Alu-derived coding exons appear to be in alternative transcripts, they have been incorporated into the main coding transcript in at least 11 genes. The extent to which Alu regions are incorporated into functional proteins is unclear, but we detected reliable peptide evidence to support the translation to protein of 33 Alu-derived exons. All but one of the Alu elements for which we detected peptides were frame-preserving and there was proportionally seven times more peptide evidence for Alu elements as for other primate exons. Despite this strong evidence for translation to protein we found no evidence of selection, either from cross species alignments or human population variation data, among these Alu-derived exons. Overall, our results confirm that SINE Alu elements have contributed to the expansion of the human proteome, and this contribution appears to be stronger than might be expected over such a relatively short evolutionary timeframe. Despite this, the biological relevance of these modifications remains open to question.
INTRODUCTION
Transposable elements are mobile DNA sequences that are able to copy themselves into new genomic locations (1). Approximately half the human genome is made up of active and inactive transposable element segments (2–4) but the actual proportion of mobile element-derived sequences in the human genome may be considerably higher since many inactive mobile elements have diverged beyond the detection of normal search algorithms (5).
Transposable elements can be divided into four major and many smaller classes (2). DNA transposons encode the transposase protein, which they need to cut and paste themselves into new genomic regions (6). There are three types of retrotransposons that use RNA intermediates to copy themselves throughout the genome (7). Long terminal repeat (LTR) retrotransposons are derived from endogenous retroviruses with LTRs, most of which are no longer active in the human genome (8). Non-LTR retrotransposons are made up of long interspersed nuclear elements (LINEs), which, like the LTRs, encode a reverse transcriptase, and short interspersed nuclear elements (SINEs), which do not encode any ORF and rely on the LINEs to carry out the copying process (7).
Active transposons in the human genome are relatively infrequent and are vastly outnumbered by a ‘graveyard’ of fossil transposon copies (3). Active retrotransposons exist among the non-LTR retrotransposons, including LINE-1, SINE Alu and SINE-VNTR-Alu (SVA) elements (3). These three families, which together make up more than a quarter of the human genome, have appeared and proliferated over the past 80 million years (9). However, most copies of these retrotransposons are no longer active due to decay by truncations and mutations. For example, although there are more than 500 000 copies of the LINE-1 retrotransposon in the human genome (10), fewer than 100 copies are still intact and capable of transposition (11,12).
Accumulation of transposable elements has been shown to have a deleterious effect on fitness (13) and their presence has been associated with many diseases (14,15). However, with time transposable element sequences can also add to the functionality of genomic features through a process of co-option in which the transposable element sequence, or part of it, is recruited to perform some function. The incorporation of transposable elements (exaptation) has been shown to contribute to the evolution of regulatory motifs (16), promoters (17) and lncRNA (18) among others, and transposable elements have been co-opted into ancient protein-coding genes, either in their main isoform (19–21) or as alternative splice variants (22).
The SINE Alu family of retrotransposons are primate-specific elements (23) that derived from the small cytoplasmic 7SL RNA and are ∼300 nt long. The majority map to non-functional regions of introns or intergenic sequences (24). Alu elements can be divided into three large sub-families. The oldest, the AluJ sub-family, arose 65 million years ago and has become entirely extinct through deleterious sequence changes (25). The AluS family evolved 30 million years ago and almost all elements are fossils, though some sub-families have been found to contain active members (25). Almost all active Alu elements are from the youngest subfamily, AluY (26), though not all AluY elements are active. Like other transposable elements, Alu elements are potentially deleterious (27,28).
Unlike most transposable elements, Alu elements have a pair of dinucleotides that can form a weak 3′ splice site and facilitate their conversion into exons (29). In addition, 5′ splice sites (30) and polyadenylation sites (31) can be generated from a minimal number of base substitutions. Sorek et al (32) found that while SINE Alu elements are incorporated into exons, they are found predominantly in alternative exons rather than constitutive exons. These alternative exons are included in transcripts at lower frequencies than alternatively spliced exons derived from other sources, and they found that the vast majority would lead to a frameshift or a premature termination codon. However, since exons generated from Alu elements are almost always alternatively spliced, the main isoform is intact, allowing the Alu exons to acquire functionality over time (29).
It is not clear to what extent exaptation of primate-specific Alu elements contributes to cellular proteins. Gotea and Makałowski (20) concluded that functional proteins were unlikely to contain regions derived from young transposable elements like LINE-1 and Alu. However support for the incorporation of Alu elements in coding genes has come from microarrays (33) and proteomics data (34). Lin et al (34) found peptide evidence for 85 Alu-derived exons, which led them to suggest that Alu elements may be a substantial source of novel coding exons and may represent species-specific differences between humans and other primates. However, the peptides that supported these 85 Alu-derived exons came from the PRIDE proteomics database (35). While the PRIDE database is an important repository of experimental data, it is uncurated and the false discovery rate cannot easily be controlled in such a huge database (36). Because of this, many novel sequences identified solely via PRIDE are likely to be false positives (37,38). The Lin et al. study (34) only managed to validate two of the Alu-derived exons when they searched the FDR-controlled Peptide Atlas database (39).
Here we investigate to what extent SINE Alu elements are incorporated into coding genes in the human reference set and attempt to determine what proportion of the Alu elements that overlap coding exons are likely to code for functional proteins.
MATERIALS AND METHODS
Human reference set
The human reference gene set used in this study was v28 of the GENCODE manual annotation (40), which is equivalent to Ensembl 92 (41). The GENCODE v28 gene set is annotated with 97 713 protein-coding transcripts.
APPRIS
The APPRIS database (42) annotates splice isoforms with structural and functional information and cross-species conservation. It also selects a single protein sequence unique isoform as the principal isoform for that gene (43). We have shown that most genes have a main isoform at the cellular level (44) and that the principal isoforms selected by APPRIS are a highly reliable predictor of this main cellular isoform (44). Transcripts from the GENCODE v28 reference set were tagged as principal or alternative by the APPRIS database. The distinction can also be made at the level of exons. We tagged exons whose translation would be included in the principal isoform as principal exons and the remainder, exons that belong exclusively to alternative splice variants, were tagged as alternative exons.
RepeatMasker
RepeatMasker regions [Smit AFA, Hubley R and Green P, http://repeatmasker.org] were obtained from the UCSC genome browser at http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/hg38.fa.out.gz and mapped to transcripts from the GENCODE v28 reference set. For the SINE Alu analysis if a transposon mapped to both principal and alternative isoforms, we counted just the principal isoform. Where a transposon or repeat mapped to more than one gene (generally where the transposon was present in a coding gene and in a read-through gene), we only counted the transposon once.
Selection tests
Using human population variation data (45) we estimated a global dN/dS value with the dNdScv R package (46) for sets of exons overlapping simple repeats, low complexity regions, and transposable elements (all defined by RepeatMasker). dNdScv reports the ratio of the non-synonymous to synonymous substitution rates (dN/dS). Although dNdScv was originally designed for cancer genomic studies, it can and has been used to quantify selection in population variation data (46).
A dN/dS lower than 1 implies purifying selection. Under purifying selection, dN/dS values are expected to be lower for common alleles than for rare alleles. Values of dN/dS close to one for both rare and common alleles are compatible with neutral evolution, but can also mean there is not enough statistical power to infer negative or positive selection, or also that there is a perfect balance between negative and positive selection.
To estimate dN/dS ratios cross-species we obtained primate CDS alignments from the 100 vertebrate alignments generated with MultiZ (47) for each Alu-containing exon or exon fraction with evidence of protein expression. Alignments were visually inspected for frame-shifts and STOP codons and species carrying any of these were discarded from dN/dS calculations. To gain statistical power, the alignments of the coding portions of the 36 Alu elements with peptide evidence were concatenated into a single alignment. Based on this alignment a phylogenetic tree was inferred with Phyml 3.0 (48), selecting the best fit model with SMS (49). Then we used codeml from the PAML package (50) to optimize branch lengths, estimate dN/dS ratios and calculate likelihoods. The likelihood of a M0 model with a free dN/dS ratio parameter was compared to the null hypothesis in which dN/dS was fixed at 1 (neutral evolution). P-values were calculated using a Likelihood Ratio Test (LRT) with one degree of freedom. We tested three different alignments/trees: one containing all simians (Green monkey, Marmoset, Orangutan, Human, Chimp, Gorilla, Gibbon, Squirrel monkey, Baboon, Rhesus and Crab eating macaque), one containing apes (Orangutan, Human, Chimp, Gorilla and Gibbon), and one with just Chimp and Human. In addition, we conducted a similar analysis but fitting M0 selection models separately for each individual exon and then gathering all the individual likelihoods together (sum of Log-likelihoods). A LRT with degrees of freedom equal to the number of exons tested was conducted to compare the neutral evolution and selection models.
We also carried out an analysis of selective pressure within primates using PhyloCSF (51), which uses likelihood ratios calculated from multi-species alignments and pre-computed substitution frequencies to determine whether a given nucleotide sequence is likely to represent a functional, conserved protein-coding sequence. Scores were calculated for the simian subset of the 100-vertebrates MultiZ alignment and the primate subset (simian plus Bushbaby) using the PhyloCSF ‘mle’ option. A P-value was calculated for each region by estimating the probability a non-coding region of the same length would get the same or higher PhyloCSF score, using the non-coding model previously described for PhyloCSF-psi (51), with a Holm–Bonferroni correction applied for the number of regions tested (36).
Gene family analysis
We performed a phylostratification analysis following a previously described pipeline (52) based on the gene family phylogenetic reconstructions of Ensembl Compara (53). Compara v95 is constructed out of genes from 152 species, providing 43,716 annotated gene family trees. Only species with enough coverage (>5×) were considered for the analysis. Compara assigns the speciation or duplication events represented by each internal tree node to the phylogenetic level in which these events were detected (53).
To estimate the gene family age and the individual gene age for all protein coding genes annotated in GENCODE v28 human coding genes were classified in the following classes or phylostrata: Fungi/Metazoa, Bilateria, Chordata, Vertebrata, Euteleostomi, Sarcopterygii, Tetrapoda, Amniota, Mammalia, Theria, Eutheria, Boreoeutheria, Primates, Simiiformes, Catarrhini, Hominoidea, Hominidae, HomoPanGorilla and Homo sapiens.
Gene family age was defined as the age class at the root of the family tree (the oldest common ancestor with a member of the gene family), while gene age is the phylostratum in which the most recent genomic event took place. Gene age for duplicated genes represents the phylostratum of the last duplication, whereas gene age always agrees with family gene age for genes without a detectable duplication origin in their gene trees. Duplication events with a consistency score (54) below 0.3 were tagged as unclear and nodes with a score of 0 were dismissed from the analysis.
Primate-derived exons
To determine whether an exon arose in the primate clade we defined as alternative all those exons that did not overlap with any exon integrated in a principal isoform in APPRIS. We removed sequences shorter than 45 bases, as these exons are likely to be too short to identify homology in the TBLASTN search (55). There were 12 540 exons in the GENCODE v28 gene set that met these criteria. The translated sequences of these exons were used as query to search against six different mammalian non-primate genomes, cat, dog, mouse, sheep, polar bear and pig, retrieved from Ensembl v95 (41), equivalent to GENCODE v28. In the TBLASTN search we turned off low complexity filtering, defined gap opening and extension penalties of 13 and 1, respectively, and set a maximum E-value threshold of 0.1. All exons that had significant homology hit in one of these species were discarded. We also used APPRIS annotations to filter out non-primate exons. Any alternative exon that formed part of a transcript with a conservation score of more than 1.5 (conservation in human plus chimp) was also discarded from the primate exon list. We defined 7566 primate-derived alternative exons. A total of 777 of these overlapped an Alu element so were discarded. The final list of exons that we were not able to map to any of the six non-primate mammalian species totaled 6789 exons.
Proteomics analysis
The proteomics analysis was carried out using the January 2019 human build of PeptideAtlas (39). We mapped peptides validated by PeptideAtlas to the 1224 Alu elements in the human proteome and to the 6789 alternative primate-derived exons. The advantage of using the PeptideAtlas database is that identifications from large-scale MS experiments are first subject to a pre-processing step that reduces the numbers of false positive matches. For this analysis we also rejected non-tryptic peptides, peptides that mapped to more than one gene and peptides shorter than seven amino acids.
The remaining peptides mapping to SINE Alu regions or primate-derived alternative exons were validated by manual inspection of the spectra. Expert curation of peptide spectrum matches is an essential step when validating peptides that identify novel coding regions. Only those peptide-spectrum matches that passed manual inspection were deemed sufficiently reliable to confirm the translation of the inserted Alu elements or primate-derived exons.
Transcript evidence
Pext (proportion expressed across transcripts) scores are normalized transcript level measures of RNAseq expression. They are generated as part of the GNOMAD project from the large-scale RNAseq analyses carried out by the GTex consortium (56). Pext scores have been shown to distinguish highly conserved exons from exons with poor conservation. Here the Pext scores were used to measure the inclusion rates of Alu-derived exons and primate-derived exons with peptide evidence.
cDNA support for Alu-derived exons and primate-derived exons with peptide evidence came from the European Nucleotide Archive (57) and NCBI RefSeq (58). Exons were counted as supported by a cDNA when the cDNA mapped to the 5′ and 3′ boundaries of the exon. cDNAs that included the exon as part of a retained intron were not counted as supporting the exon.
RESULTS
According to RepeatMasker remnants of transposon-based elements (not including regions predicted as Simple Repeats and Low Complexity) make up just over half of the bases in the human reference genome (50.66%). More than 20% of the fragments predicted as transposable element-derived in the human genome are SINE Alu elements, though LINE/L1 elements are the most common by number of bases because LINE/L1 elements are longer than Alu elements. By bases LINE/L1 elements make up 17.3% of the genome compared to the 10.4% of the genome that is contributed by Alu elements (Supplementary Figure S1A).
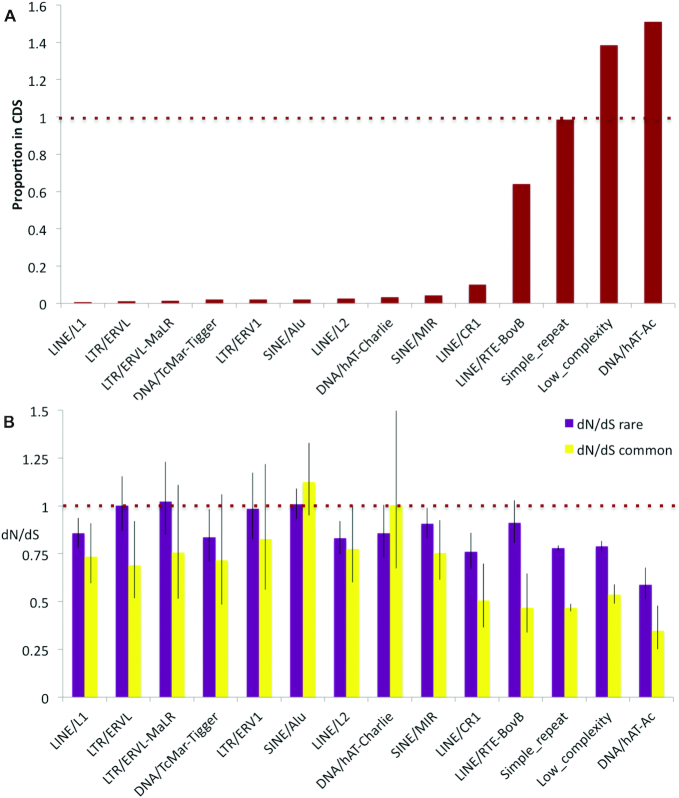
Transposon-based elements were predicted to overlap CDS in 9% of GENCODE v28 transcripts (40). Almost 25% of the transposable elements that overlap coding exons are SINE Alu elements. Alu elements overlapped a total of 1224 distinct coding exons. The next most common transposable element classes were SINE MIR (789) and LINE/L1 (684). Almost all common transposon classes were found in much lower proportions within coding sequences (CDS) than within the whole genome (Supplementary Figure S1B); Alu elements total just 0.23% of the bases in the human coding reference set and LINE/L1 elements 0.12%. This is what would be expected if the presence of transposable elements were selected against in coding exons. However, some transposable element families are exceptions to the rule. The proportion of LINE/RTE-BovB elements are almost as high in CDS regions as they are in the whole genome, and DNA/hAT-Ac elements are actually more prevalent in CDS than in the genome as a whole (Figure 1A).
Figure 1.
The relative proportion of elements overlapping coding exons and their dN/dS. (A) The ratio of the percentage of transposable element bases in coding exons to the percentage of transposable element bases across the whole genome. Values close to one suggest that the presence of elements in coding sequences have not been selected against. SINE Alu elements have a ratio that is much lower than 1. Predicted simple repeats and low complexity regions included as a comparison. (B) The dN/dS for transposable element families overlapping human coding exons for both rare and common allele frequencies. Values below one and lower dN/dS with common allele frequencies than with rare allele frequencies indicate purifying selection, while values close to one suggest that the elements are generally under neutral selection. SINE Alu elements have dN/dS values close to 1.
Taken at face value these proportions might suggest DNA/hAT-Ac transposable elements are not selected against in CDS regions. However, these are ancient transposable elements (2,59). While DNA/hAT-Ac elements preserved in CDS regions are still detectable by RepeatMasker, those outside CDS regions will not have been subject to purifying selection and may no longer be recognizable as deriving from transposable elements. This suggests that many of the ancient DNA/hAT-Ac elements have been co-opted and are evolving under purifying selection. The same is probably true for many LINE/RTE-BovB elements.
Selection
In order to determine whether transposable elements that overlap annotated coding exons have acquired functional importance as proteins, we measured selection using the ratio of the rates of non-synonymous and synonymous changes (dN/dS). We estimated a global dN/dS value for exons overlapping each of the most common categories of RepeatMasker regions using dNdScv (46). The results (Figure 1B) suggest that in general DNA/hAT-Ac, and LINE/RTE-BovB transposable elements (along with LINE1/CR1 elements, simple repeats and low complexity regions) are under purifying selection, as might be expected from their partitioning between genome and proteome (Figure 1A), whereas exons overlapping most other elements (including SINE Alu elements) are not, in general, under selection and are therefore less likely to have functional importance.
SINE Alu elements locate preferentially to alternative exons
The APPRIS database (42) divides transcripts into those that give rise to the principal protein isoform and those that if translated would produce alternative isoforms (see ‘Materials and Methods’ section for more details). Exons that overlapped all RepeatMasker transposon classes were separated into those found in the APPRIS-defined principal transcripts, and those found solely in alternative transcripts.
Alternative exons make up just over 10% of the exons in the reference genome, so if transposable elements were randomly distributed, we would expect to find 1 in 10 transposable elements in alternative elements and the other 90% should overlap with principal exons. This is true for exon-overlapping simple repeats (87.8%) and some older transposable elements are also found at the expected frequency in principal exons, including DNA/hAT-ac (88.8%), SINE/SS-Deu-L2 (83.3%), SINE/tRNA (78.9%) and LINE/RTE-BovB (85.4%) elements (Supplementary Figure S2). By contrast, just 9.2% of SINE Alu elements were found in principal exons.
It should be noted that APPRIS determines principal isoforms based on conserved structural and functional features and cross-species conservation. Since Alu elements arose in the primate lineage and do not form part of conserved functional or structural domains, we would expect few Alu element-derived exons to be classified as principal by APPRIS. In any case, APPRIS predictions are backed up by transcript level studies showing that internal exons overlapping Alu elements are predominantly alternatively spliced (32).
SINE Alu elements in the human reference genome
A total of 1074 distinct coding genes in GENCODE v28 have coding exons that overlap SINE Alu elements. There are 1224 Alu elements that overlap coding exons, but several genes harbour more than one element. For example ZNF506 contains four distinct Alu overlaps in alternative 3′ exons and 23 genes overlap three different Alu elements.
Genes with coding regions that overlap Alu elements are significantly enriched in zinc finger motifs relative to the whole genome. A total of 93 genes are annotated with C2H2 zinc finger domains (Fisher's test, P-value of 9.4 e-16) according to SMART (60). Only one other protein domain is significantly enriched in this set, KRAB domains (P-value of 4.8 e-24). KRAB domains are generally found in tandem with C2H2 zinc finger domains. Many of these genes are from the cluster of KRAB-ZNF genes at the centromere on chromosome 19. Just over half of these genes overlap a range of different Alu elements, including all six members of the ZNF431 clade (61).
SINE Alu elements are more often found in the final coding exon: almost half of the coding exons that overlap Alu elements are 3′ CDS (591). Sixty per cent of the Alu elements that overlap zinc finger genes are found in the final exon. This elevated number has two possible explanations. It may be because Alu elements are likely to produce fewer deleterious effects when inserting into a 3′ exon, or it may be caused by out of frame insertions that generate premature stop codons. The fact that Alu insertions can easily form polyadenylation signals (33) would clearly facilitate the establishment of 3′ exons.
Alu elements that insert into internal CDS may generate frameshifts in downstream exons. In fact 50.2% of annotated Alu elements that overlap internal or first CDS are predicted to lead to frameshifts. This is somewhat fewer than expected by chance and in contrast to what was found by Sorek et al. (32). This lower number may be evidence in favour of these being truly functional exons, but it could also be caused by systematic bias given the composition of Alu sequences.
Almost all SINE Alu elements that overlap coding exons are inactive
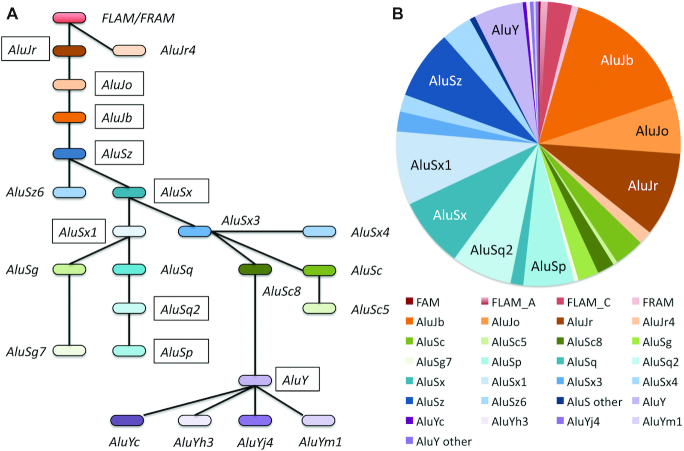
More than 50% of the Alu elements that overlap exons are from the AluS family (55.2%) against just 7.4% of the youngest Alu family (AluY family). The AluY sub-family itself is partly active (28), but only three copies of elements from sub-families known to be active (28) are annotated in (alternative) coding exons in the reference genome. The proportions of Alu sub-families overlapping coding exons are shown in Figure 2.
Figure 2.
SINE Alu sub-families that overlap coding exons. (A) The SINE Alu family tree based on the family tree in RepeatMasker. The most common sub-families are marked with a black box. (B) The proportion of each Alu sub-family that overlaps coding exons. Members of the FRAM/FLAM, AluJ, AluS and AluY families by their proportion in coding exons in the reference genome. The most common sub-families are labeled in the chart.
Over 37% of the Alu elements that overlap coding exons are from the older FRAM/FLAM or AluJ families, compared to just 29.4% across the whole genome (Supplementary Figure S3). The difference is significant in a Chi squared test (<0.0001). This may be partly because older Alu elements are often no longer detectable outside of conserved regions such as coding exons.
The NPIPB sub-family
Genes with Alu-derived exons annotated in the reference genome have a similar age distribution to the rest of the human reference set, except that there are proportionally more genes that have arisen in the primate lineage (Supplementary Figure S4). Though difference is significant (Chi-squared test, P-value of 0.00014), it is entirely due to the 10 duplications in the NPIPB sub-family, which itself arose in the primate clade (62).
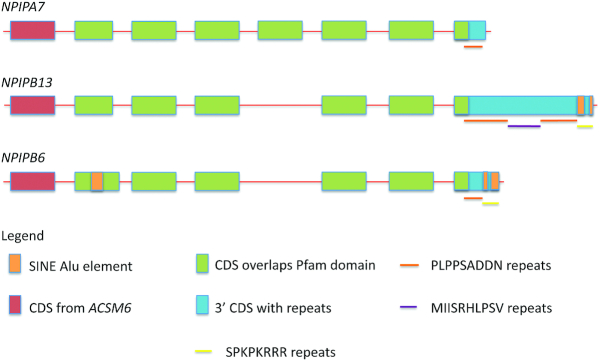
The 15 members of the nuclear pore complex-interacting protein family are primate specific and found in segmental duplications on chromosome 16 (62). The nuclear pore complex-interacting proteins (NPIPs) are made up of one or two membrane-interacting regions, a central coiled-coil domain and a variable number of C-terminal repeats. Three subfamilies can be distinguished by the length and composition of the repeats; the NPIPA subfamily does not contain any SINE Alu elements, but RepeatMasker defines two distinct SINE Alu elements for each member of the two NPIPB sub-families, NPIPB3/4/5/11/13 and NPIPB6/7/8/9/15. In fact one of the three distinct types of repeats that make up the final exon in this family seems to have derived from Alu elements (Figure 3). The NPIPB6/7/8/9/15 sub-family also has an Alu-derived insertion in the second coding exon.
Figure 3.
A schematic representation of the three NPIP subfamilies. The relationship between coding exons, SINE Alu elements, Pfam domains and repeats in the three NPIP sub-families. One member of each family is taken as the representative. Exons are not to scale. Each family member has an initial coding exon duplicated from an acyl-COA synthetase medium chain family member (there is also an alternative 5′ coding exon annotated for most family members), five or six internal exons that define the Pfam domain that is unique to NPIP family members, and a variable-sized 3′ CDS that is essentially composed of repeats. The Pfam domain overlaps one set of repeats. A second set of repeats, found at the 3′ end of the final CDS in the NPIPB subfamilies, appears to be composed entirely of SINE Alu element fragments.
Phylogenetic reconstruction suggests that the NPIPB sub-families derived from the ancestral NPIPA in stepwise manner and that the evolution of NPIPB sub-families within the great apes clade coincided with the insertion of Alu elements in the coding region and a number of further retrotransposon events within the 5′ and 3′ UTRs of the NPIPB sub-family members.
Since the duplications are so recent, the genes are very similar. It is not easy to distinguish whether all annotated genes are coding, or whether only some are coding and others are pseudogenes. However, at least one member of the NPIPB6/7/8/9/15 sub-family has clear evidence of protein expression in testis. All the peptide evidence in PeptideAtlas mapped to a single gene (NPIPB6), so NPIPB6 was used to represent the whole sub-family.
Alu elements in principal isoforms
Alu elements were predicted to be present in the principal exons of 103 coding genes. We carried out a detailed manual analysis of these genes to determine whether the Alu element had been incorporated into the main transcript or an alternative variant and whether or not the Alu elements were part of bona fide coding genes (63). Details of the manual annotation can be found in the Supplementary Results section.
We found that the Alu element forms part of the main coding isoform of 10 genes and all the members of the NPIPB sub-family (Table 1).
Table 1.
Genes in which the Alu element is part of the main coding isoform
| Gene | Gene family age | Function |
|---|---|---|
| BEND2 | Euteleostomi | Unknown function. Expressed in testis. Alu element inserts a whole exon into the highly divergent N-terminal. |
| HSD17B7 | Fungi-Metazoa | 3-keto-steroid reductase, part of the estrogen synthesis pathway. Adds eight amino acids to the N-terminal. |
| NLRP1 | Euteleostomi | Part of the NLRP1 inflammasome (64). The Alu region corresponds to an inserted exon that adds 27 amino acids. |
| NPIPB6 | Simiiformes | Unknown function. Expressed in testis. Represents a primate-derived sub-family with three Alu inserts. All three extend exons. |
| TTF1 | Chordata | Transcript termination factor in ribosome biogenesis. The Alu element adds 23 amino acids to the C-terminal. |
| USP19 | Fungi-Metazoa | A multi-functional deubiquitinating enzyme. The Alu element extends exon 2 by 46 amino acids. |
| ZNF101 | Bilateria | Unknown function. The Alu element inserts 49 base pairs and a stop codon into the 3′ exon of the CDS. |
| ZNF394 | Euteleostomi | A transcriptional repressor in MAP kinase signaling (65). The element adds 8 amino acids to the C-terminal. |
| ZNF433 | Bilateria | Activation of beta-catenin/TCF signaling. The Alu region changes a single amino acid at the C-terminal. |
| ZNF669 | Bilateria | Unknown function. Adds 22 amino acids to the stop codon. |
| ZNF91 | Bilateria | SVA transposable element repressor (66). The Alu element displaces two zinc finger motifs while adding 33 amino acids. |
Gene family age is the age of the oldest common ancestor of the gene family.
Five of the 11 genes in which Alu elements have modified the main coding sequence code forzinc finger proteins and all but ZNF394 are primate-specific duplications of the same zinc finger family (61). The most interesting case is ZNF91. Here the Alu element, which only appears in the great apes, adds 33 amino acids to the C-terminal while displacing eight zinc-binding residues from the ancestral protein. A further change in the human lineage led to the upstream insertion of seven zinc finger binding motifs. The gain of these zinc fingers has enabled ZNF91 to become a repressor of SVA transposable elements (66). It is not clear whether the Alu insertion also contributes to this role.
Eight of the Alu elements, including those in all five zinc finger genes, would extend the C-terminal of the resulting protein. It is known that zinc finger proteins are highly plastic at their C-terminals (67). All the elements, except those in BEND2 and NLRP1, have integrated into the principal isoform by ‘hijacking’ existing coding exons rather than creating new coding exons.
Peptide evidence for SINE Alu functionality
It is possible that other Alu-derived exons, besides those present in principal isoforms, have evidence for functionality. We attempted to confirm the translation to protein of the SINE Alu elements in the human proteome. We searched the PeptideAtlas database for validated peptides that mapped to the 1224 unique Alu-derived exons and manually verified the peptide-spectrum matches (PSMs) for these peptides (see ‘Materials and Methods’ section).
The peptide evidence validated the translation of 33 of Alu-derived exons from 29 different genes (SLC3A2 and NPIPB6 both contain three Alu-derived exons and all three were validated by spectra from PeptideAtlas). The 29 genes with translated Alu-derived exons are shown in Supplementary Table S1.
There are validated peptides for 8 of the 13 of the Alu elements in principal isoforms, including all three SINE Alu regions in NPIPB6. The elements in ZNF91 and USP19 are supported by two non-overlapping peptides. Although we do not find peptides that map to the Alu elements present in zinc finger proteins ZNF101, ZNF394, and ZNF669, there are peptides that uniquely identify the exons that the Alu elements are part of, so we can assume that all these Alu elements are translated as well.
The remaining 25 Alu elements with validated translation are all in alternative isoforms, though some of the variants have so much peptide and RNAseq evidence that they could be considered at least as strong alternative isoforms. The alternative C-terminal in CD55 is supported by three non-overlapping peptides and the inserted Alu region in NEK4 is supported by four peptides. The peptide data for these two Alu regions suggests that the Alu exons have at least as much support as the ancestral isoforms.
Twenty two of the Alu elements for which we found valid peptides are inserts in the ancestral transcripts, and all but one insert was frame preserving (the indel in DLGAP5 adds four amino acids and a stop codon from the last coding exon of the principal variant as a result of a frameshift). Nine of the remaining Alu-derived exons (and DLGAP5) would affect the C-terminal of the proteins while two extend the N-terminal.
All SINE Alu elements for which we found verified peptide evidence modified existing CDS. In all cases the ancestral gene family predated the Alu element insertions, though we cannot be sure whether SINE Alu insertion occurred before or after gene duplication for genes ZNF101, ZNF195 and ZNF669.
We crosschecked the 85 genes identified in the Lin et al. (34) paper against evidence from the PeptideAtlas database. We validated just five of the peptides detected by Lin et al. for SINE Alu elements.
How do Alu-derived exons compare to other primate-derived exons?
In order to determine whether the peptide evidence we found for 33 SINE Alu elements was similar to what might be expected for primate derived alternative exons, we repeated the PeptideAtlas analysis with exons that arose in the primate clade as comparison. We only looked at primate exons tagged by APPRIS as alternative because exons within principal isoforms would be expected to form part of the expressed proteins (we found peptides for 8 of the 13 SINE Alu overlaps in verified principal exons).
We curated a set of 6789 primate-derived alternative exons (see methods section for details). In comparison the curated set of alternative SINE Alu-derived exons totalled 777 exons. SINE Alu elements make up 10.4% of the bases in the human genome and just over 10% of annotated primate exons are Alu-derived, which suggests that Alu elements are not any more likely to be annotated as coding exons than other non-coding region.
We mapped peptides from the PeptideAtlas database to the exons (as described in the ‘Materials and Methods’ section). After manual curation we found reliable peptide identifications for just 25 primate-derived alternative exons, 0.37%. As a comparison, we found peptide evidence for 22 of the 777 SINE Alu-derived alternative exons (2.83%), proportionally more than seven times as much and significantly more than would be expected for standard primate-derived exons (P-value of <0.0001 in Chi-squared tests). This shows that a significantly higher proportion of SINE Alu elements are incorporated into expressed proteins than would be expected.
Transcript evidence
We analysed transcript evidence in the form of cDNA support and Pext scores (normalized exon inclusion rates) for the 47 alternative exons with peptide evidence. There was more supporting transcript evidence for the translated Alu-derived exons than for the translated primate-derived exons. cDNA evidence supported the expression of 19 of the 22 alternative Alu-derived exons against just 14 of the 25 primate-derived exons, while 8 of the 22 alternative Alu-derived exons had Pext scores >0.5, against none of the primate-derived alternative exons. The differences between the two sets of exons are significant: Fisher's tests showed a P-value of 0.0293 for the differences in cDNA support and 0.001 for the Pext scores.
Several of the Alu-derived exons had higher tissue-specific expression patterns. For example the Alu-derived exon in DLGAP5 had an average Pext score of just 0.1, but was completely included in endocervix, while the inclusion of the 3′ Alu-derived exon in CMC2 was noticeably higher in brain than in other tissues.
SINE Alu inserts and domain composition conservation
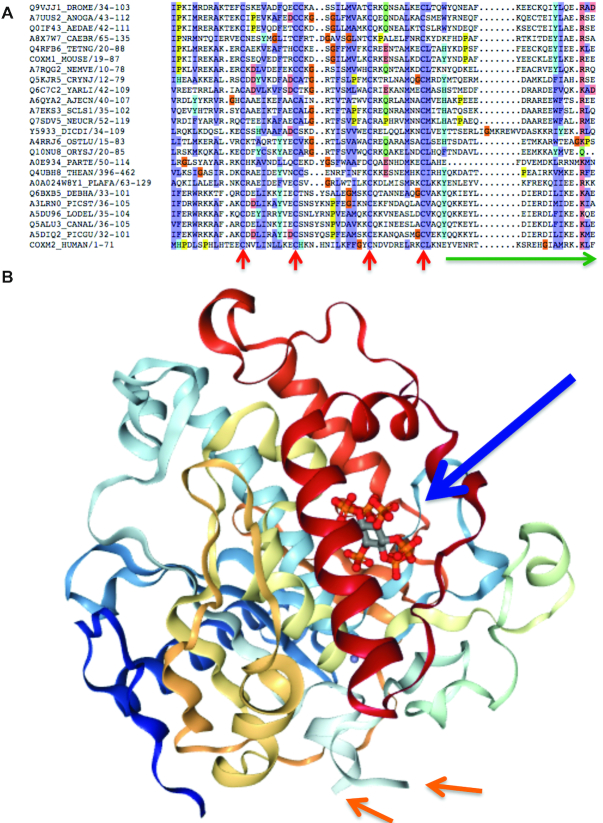
Events that cause changes in Pfam (68) domain composition tend not to be detected in proteomics experiments (69). This is presumably because, like frame-changing indels, this would normally lead to gross functional changes in the protein and be selected against. Even though all detected SINE Alu element inserts were frame preserving, six of the events for which we found peptides would break Pfam functional domains.
While this is somewhat surprising, five of the six domain-disrupting events may not actually have much effect on the functional domain. For example, the insertion in the domain in TKT is relatively short, occurs in a loop region, and the Pfam seed alignment (68) includes sequences with similar sized inserts at the same position. In CMC2 the Alu exon removes the C-terminal portion of the Pfam domain, but the C-terminal swap does not affect the beta-hairpin that this protein forms, nor the conserved cysteines. The C-terminal of the Cmc1 domain that is broken by the SINE Alu insertion is not conserved in the Pfam seed alignment (Figure 4A). The A_deamin domain in RNA-editing deaminase 1 from gene ADARB1 has two conserved N- and C-terminal sections and a central linker section without conservation. Sequences from Danio Rerio, chicken and Xenopus are among those that also have insertions in this central ‘linker’ region and the central linker region is just where the ADARB1 SINE Alu exon inserts. The insertion can be visualized mapped onto the crystallized structure (Figure 4B)—it inserts into an already disordered region away from the catalytic site, in contrast to what is reported by Lin et al.
Figure 4.
The effect of the SINE Alu insertions on Pfam domains. (A) The seed alignment for Pfam domain Cmc1. Conserved cysteines are marked with red arrows, the region of Pfam domain equivalent to the region replaced by the SINE Alu element insert is shown by the green arrow. It would not affect the four conserved cysteines. (B) The structure of the ADARB1 catalytic domain (PDB (70) structure: 1ZY7). The catalytic region and the phytic acid co-factor are shown with the large arrow. The SINE Alu element would be inserted into the disordered region, the start and end of which is marked by the smaller arrows and would therefore not interact directly with the catalytic domain of ABARB1.
SINE Alu element translation and selection
The substantial evidence for the expression and translation of a small set of Alu-derived exons suggested that this subset of Alu elements might have gained functional roles in the cell. We investigated whether there was data to support this hypothesis. We defined ‘functional role’ for the purpose of this analysis as having evidence of protein-like purifying selection (71). Although SINE Alu elements as a whole are not under selective pressure (Figure 1B), it is possible that the subset of Alu elements with evidence of translation is under measurable selective constraints.
Using PAML we estimated dN/dS from concatenated primate alignments (50) of the coding portion of the 33 elements with peptide evidence and for the Alu elements that overlap expressed coding exons in ZNF101, ZNF394 and ZNF669 that we can assume are also expressed as proteins. The estimated dN/dS values were not significantly different from one for the alignments of all simians, of apes, or of human and chimp (see Supplementary Table S2). An alternative analysis fitting the selection models separately for each individual exon and then multiplying the resulting likelihoods did not reject the null hypothesis of neutral evolution either. Furthermore, we found stop gains and frame-shifts in 24 of the 36 Alu-derived exons across primates, suggesting that these Alu elements have not established important functional roles across the primate clade. In order to test for significance, we looked at stop gains and frame-shifts in the primate clade for 36 exons of similar size selected at random from the 32 genes with Alu-derived exons that we analysed. Just four of these exons had frame-shifts or stop gains in the primate clade.
Analysis of the same 36 elements using PhyloCSF (51), a measure of evolutionary coding potential, produced similar conclusions. The average PhyloCSF score for the coding portion of these Alu elements using alignments of the primate and simian clades is negative, suggesting that these regions have not been under protein-coding constraint in aggregate. However, there is one case for which we found weak evidence for coding selection. The 8-codon Alu-derived region in ZNF394 has a PhyloCSF score of 29.4, which is higher than would be expected for a region of that length that was not under protein-coding selection (uncorrected P = 0.003, multiple-hypothesis corrected P = 0.12). Further support comes from the fact that there are no indels and the stop codon immediately following it is perfectly conserved (CMC2 is the only other C-terminal addition that conserves its stop codon throughout primates). The alignment of the ZNF394 region can be seen in Supplementary Figure S5.
From the point of view of human population variation there is not enough data to assess selection on this small set of exons. However, just eight of the 35 variants with a MAF greater than 0.1% are synonymous, while six (17.1%) are high impact (four stop gains and two frameshifts). By way of comparison just 3 of the 271 variants with an MAF above 0.1% in non Alu-derived exons from the relevant principal transcripts were high impact variants (1.1%). The two proportions of high impact variants are significantly different (Fisher's exact test P-value of 0.0002). The high impact variants in the Alu-derived exons occurred in both principal (two) and alternative (four) Alu-derived exons. Although the data is scarce, the frequency of high impact variants further supports the hypothesis that these Alu-derived exons have not yet gained relevant functions.
DISCUSSION
SINE Alu elements make up more than 10% of the human genome; in total the genome has been colonized by close to 1.2 million SINE Alu fragments. The vast majority map to intergenic and intronic regions and just 1224 Alu fragments (0.1%) overlap annotated coding exons. The reduced proportion of SINE Alu elements in exons suggests that there is selective pressure against their inclusion in coding regions.
Even where Alu fragments overlap coding exons, they do not appear to be functionally important. Coding regions that derive from SINE Alu elements are not under selective pressure and almost all annotated Alu-derived exons are found in alternative coding transcripts. Little is known about the cellular roles of any of these Alu-derived exons, though the Alu-derived exon in LIN28B has been shown to be necessary for oncogene activation (72). Alu-derived coding exons are highly enriched in zinc finger proteins (67).
Although Alu elements as a whole are not under selective pressure, we find that Alu-derived exons have become part of the principal splice variant in at least 11 coding genes. In all but two genes the Alu elements have ‘colonized’ the principal isoform by merging with existing coding exons. This is perhaps not surprising since merging with functioning coding exons is likely to be a shortcut to becoming established as part of the main transcript.
Large-scale proteomics experiments tend not to detect evidence for alternative splice variants (69), nor genes that have evolved de novo in the primate lineage (63), so we would expect to find little evidence of translation for Alu-derived exons. Despite this there is clear evidence for the translation of 33 Alu-derived exons and peptide and transcript evidence suggests that many of these alternative exons are strongly expressed. All but one of the 22 insertion events we detected were in-frame, significantly more than would be expected by chance. The proportion of SINE Alu-derived exons detected in large-scale proteomics experiments was also significantly higher than expected; more than seven times higher than that of other primate-derived exons. This may be related to the splice signals present in Alu elements (29,30). Transcription evidence supported the strength of expression of these Alu-derived exons: both inclusion rates and cDNA support were significantly stronger for the Alu-derived exons with peptide evidence than they were for the other primate-derived exons with peptide evidence. A small subset of the 1224 Alu-derived exons has clearly added to the human proteome.
All the evidence suggests that these SINE Alu elements have added to the human proteome via gene modification rather than de novo gene generation. In 26 of the 29 genes with peptide evidence, the SINE Alu elements added to an established (often ancient) protein-coding gene, while in the remaining three genes the SINE Alu event may have been concurrent with, or just after, a gene duplication. We find no evidence for the conversion of any SINE Alu element into a de novo human coding gene.
Despite the lack of evidence for selection in SINE Alu-derived coding exons at the population level, we expected to find some evidence of evolutionary pressure for those Alu-derived exons with evidence of translation. However, we found none. There was no evidence for any selection from cross-species alignments within the primate clade or even among great apes. While there were too few variants in common alleles to be able to draw any conclusions about purifying or positive selection from human population variation, the sizable frequency of high impact variations among the common variants supports the possibility that even those Alu-derived exons with peptide evidence have yet to gain biologically important roles. Overall it seems that although SINE Alu elements contribute to the human proteome, they add little to the range of protein functions.
Supplementary Material
Notes
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
National Human Genome Research Institute of the National Institutes of Health [2 U41 HG007234 to M.K., I.J., L.M., J.M.M., F.P., M.L.T., R01 HG004037 to I.J., M.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1. McClintock B. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 1956; 21:197–216. [DOI] [PubMed] [Google Scholar]
- 2. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al.. Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 3. Mills R.E., Bennett E.A., Iskow R.C., Devine S.E.. Which transposable elements are active in the human genome?. Trends Genet. 2007; 23:183–191. [DOI] [PubMed] [Google Scholar]
- 4. Tang W., Mun S., Joshi A., Han K., Liang P.. Mobile elements contribute to the uniqueness of human genome with 15,000 human-specific insertions and 14 Mbp sequence increase. DNA Res. 2018; 25:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Koning A.P., Gu W., Castoe T.A., Batzer M.A., Pollock D.D.. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2001; 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feschotte C., Pritham E.J.. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007; 41:331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Havecker E.R., Gao X., Voytas D.F.. The diversity of LTR retrotransposons. Genome Biol. 2004; 5:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Konkel M.K., Walker J.A., Batzer M.A.. LINEs and SINEs of primate evolution. Evol. Anthropol. 2010; 19:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levin H.L., Moran J.V.. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011; 12:615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H. Jr.. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V.. LINE-1 retrotransposition activity in human genomes. Cell. 2010; 141:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pasyukova E.G., Nuzhdin S.V., Morozova T.V., Mackay T.F.. Accumulation of transposable elements in the genome of Drosophila melanogaster is associated with a decrease in fitness. J. Hered. 2004; 95:284–290. [DOI] [PubMed] [Google Scholar]
- 14. Reilly M.T., Faulkner G.J., Dubnau J., Ponomarev I., Gage F.H.. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 2013; 33:17577–17586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burns K.H. Transposable elements in cancer. Nat. Rev. Cancer. 2017; 17:415–424. [DOI] [PubMed] [Google Scholar]
- 16. Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008; 9:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cohen C.J, Lock W.M., Mager D.L.. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009; 448:105–114. [DOI] [PubMed] [Google Scholar]
- 18. Johnson R., Guigó R.. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA. 2014; 20:959–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bejerano G., Lowe C.B., Ahituv N., King B., Siepel A., Salama S.R., Rubin E.M., Kent W.J., Haussler D.. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006; 441:87–90. [DOI] [PubMed] [Google Scholar]
- 20. Gotea V., Makałowski W.. Do transposable elements really contribute to proteomes?. Trends Genet. 2006; 22:260–267. [DOI] [PubMed] [Google Scholar]
- 21. Tellier M., Chalmers R.. Human SETMAR is a DNA sequence-specific histone-methylase with a broad effect on the transcriptome. Nucleic Acids Res. 2019; 47:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abascal F., Tress M.L., Valencia A.. Alternative splicing and co-option of transposable elements: the case of TMPO/LAP2α and ZNF451 in mammals. Bioinformatics. 2015; 31:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kriegs J.O., Churakov G., Jurka J., Brosius J., Schmitz J.. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007; 23:158–161. [DOI] [PubMed] [Google Scholar]
- 24. Krull M, Brosius J., Schmitz J.. Alu-SINE exonization: en route to protein-coding function. Mol. Biol. Evol. 2005; 22:1702–1711. [DOI] [PubMed] [Google Scholar]
- 25. Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E.. Active Alu retrotransposons in the human genome. Genome Res. 2008; 18:1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Konkel M.K., Walker J.A., Hotard A.B., Ranck M.C., Fontenot C.C., Storer J., Stewart C., Marth G.T. 1000 Genomes Consortium 1000 Genomes Consortium Batzer M.A.. Sequence Analysis and Characterization of Active Human Alu Subfamilies Based on the 1000 Genomes Pilot Project. Genome Biol. Evol. 2015; 7:2608–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Payer L.M., Steranka J.P., Yang W.R., Kryatova M., Medabalimi S.I., Ardeljan D., Liu C., Boeke J.D., Avramopoulos D., Burns K.H.. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3984–E3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larsen P.A., Lutz M.W., Hunnicutt K.E., Mihovilovic M., Saunders A.M., Yoder A.D., Roses A.D.. The Alu neurodegeneration hypothesis: a primate-specific mechanism for neuronal transcription noise, mitochondrial dysfunction, and manifestation of neurodegenerative disease. Alzheimers Dement. 2017; 13:828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lev-Maor G., Sorek R., Shomron N., Ast G.. The birth of an alternatively spliced exon: 3′ splice-selection in Alu exons. Science. 2003; 300:1288–1291. [DOI] [PubMed] [Google Scholar]
- 30. Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G.. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004; 14:221–231. [DOI] [PubMed] [Google Scholar]
- 31. Lavi E., Carmel L.. Alu exaptation enriches the human transcriptome by introducing new gene ends. RNA Biol. 2018; 15:715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sorek R., Ast G., Graur D.. Alu-containing exons are alternatively spliced. Genome Res. 2002; 12:1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin L., Shen S., Tye A., Cai J.J., Jiang P., Davidson B.L., Xing Y.. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008; 4:e1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin L., Jiang P., Park J.W., Wang J., Lu Z.X., Lam M.P., Ping P., Xing Y.. The contribution of Alu exons to the human proteome. Genome Biol. 2016; 17:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al.. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ezkurdia I., Calvo E., Del Pozo A., Vázquez J., Valencia A., Tress M.L.. The potential clinical impact of the release of two drafts of the human proteome. Expert. Rev. Proteomics. 2015; 12:579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gascoigne D.K., Cheetham S.W., Cattenoz P.B., Clark M.B., Amaral P.P., Taft R.J., Wilhelm D., Dinger M.E., Mattick J.S.. Pinstripe: a suite of programs for integrating transcriptomic and proteomic datasets identifies novel proteins and improves differentiation of protein-coding and non-coding genes. Bioinformatics. 2012; 28:3042–3050. [DOI] [PubMed] [Google Scholar]
- 38. Guerzoni D., McLysaght A.. De novo genes arise at a slow but steady rate along the primate lineage and have been subject to incomplete lineage sorting. Genome Biol. Evol. 2016; 8:1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kusebauch U., Deutsch E.W., Campbell D.S., Sun Z., Farrah T., Moritz R.L.. Using PeptideAtlas, SRMAtlas, and PASSEL: comprehensive resources for discovery and targeted proteomics. Curr. Protoc. Bioinformatics. 2014; 46:13.25.1–13.25.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J. et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. et al.. Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodriguez J.M., Rodriguez-Rivas J., Di Domenico T., Vázquez J., Valencia A., Tress M.L.. APPRIS 2017: principal isoforms for multiple gene sets. Nucleic Acids Res. 2018; 46:D213–D217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rodriguez J.M., Carro A., Valencia A., Tress M.L.. APPRIS WebServer and WebServices. Nucleic Acids Res. 2015; 43:W455–W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ezkurdia I., Rodriguez J.M., Carrillo-de Santa Pau E., Vázquez J., Valencia A., Tress M.L.. Most highly expressed protein-coding genes have a single dominant isoform. J. Proteome Res. 2015; 14:1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. 1000 Genomes Project Consortium A global reference forhuman genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martincorena I., Raine K.M., Gerstung M., Dawson K.J., Haase K., Van Loo P., Davies H., Stratton M.R., Campbell P.J.. Universal patterns of selection in cancer and somatic tissues. Cell. 2018; 17:1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D. et al.. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004; 14:708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lefort V., Longueville J-E., Gascuel O.. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017; 34:2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O.. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010; 59:307–321. [DOI] [PubMed] [Google Scholar]
- 50. Ziheng Y. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007; 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- 51. Lin M.F., Jungreis I., Kellis M.. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011; 27:i275–i282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ezkurdia I., Juan D., Rodriguez J.M., Frankish A., Diekhans M., Harrow J., Vazquez J., Valencia A., Tress M.L.. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 2014; 23:5866–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Herrero J., Muffato M., Beal K., Fitzgerald S., Gordon L., Pignatelli M., Vilella A.J., Searle S.M., Amode R., Brent S. et al.. Ensembl comparative genomics resources. Database. 2016; 2016:baw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vilella A.J., Severin J., Ureta-Vidal A., Heng L., Durbin R., Birney E.. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009; 19:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 56. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Silvester N., Alako B., Amid C., Cerdeño-Tarrága A., Clarke L., Cleland I., Harrison P.W., Jayathilaka S., Kay S., Keane T. et al.. The European Nucleotide Archive in 2017. Nucleic Acids Res. 2018; 46:D36–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arensburger P., Hice R.H., Zhou L., Smith R.C., Tom A.C., Wright J.A., Knapp J., O’Brochta D.A., Craig N.L., Atkinson P.W.. Phylogenetic and functional characterization of the hAT transposon superfamily. Genetics. 2011; 188:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Letunic I., Doerks T., Bork P.. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015; 43:D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hamilton A.T., Huntley S., Tran-Gyamfi M., Baggott D.M., Gordon L., Stubbs L.. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006; 16:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johnson M.E., Viggiano L., Bailey J.A., Abdul-Rauf M., Goodwin G., Rocchi M., Eichler E.E.. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001; 413:514–519. [DOI] [PubMed] [Google Scholar]
- 63. Abascal F., Juan D., Jungreis I., Kellis M., Martinez L., Rigau M., Rodriguez J.M., Vazquez J., Tress M.L.. Loose ends: almost one in five human genes still have unresolved coding status. Nucleic Acids Res. 2018; 46:7070–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Finger J.N., Lich J.D., Dare L.C., Cook M.N., Brown K.K., Duraiswami C., Bertin J.J., Bertin J., Gough P.J.. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J Biol Chem. 2012; 287:25030–25037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang C., Wang Y., Li D., Li Y., Luo J., Yuan W, Ou Y., Zhu C., Zhang Y., Wang Z. et al.. Inhibition of transcriptional activities of AP-1 and c-Jun by a new zinc finger protein ZNF394. Biochem. Biophys. Res. Commun. 2004; 320:1298–1305. [DOI] [PubMed] [Google Scholar]
- 66. Jacobs F.M., Greenberg D., Nguyen N., Haeussler M., Ewing A.D., Katzman S., Paten B., Salama S.R., Haussler D.. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014; 516:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Emerson R.O., Thomas J.H.. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009; 5:e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshim M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Abascal F., Ezkurdia I., Rodriguez-Rivas J., Rodriguez J.M., del Pozo A., Vázquez J., Valencia A., Tress M.L.. Alternatively spliced homologous exons have ancient origins and are highly expressed at the protein level. PLoS Comput. Biol. 2015; 11:e1004325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Burley S.K., Berman H.M., Christie C., Duarte J.M., Feng Z., Westbrook J., Young J., Zardecki C.. RCSB Protein Data Bank: Sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education. Protein Sci. 2017; 27:316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ophir R., Itoh T., Graur D., Gojobori T.. A simple method for estimating the intensity of purifying selection in protein-coding genes. Mol. Biol. Evol. 1999; 16:49–53. [DOI] [PubMed] [Google Scholar]
- 72. Jang H.S., Shah N.M., Du A.Y., Dailey Z.Z., Pehrsson E.C., Godoy P.M., Zhang D., Li D., Xing X., Kim S. et al.. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019; 51:611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hamilton A.T., Huntley S., Tran-Gyamfi M., Baggott D.M., Gordon L., Stubbs L.. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006; 16:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.