STATEMENT OF SIGNIFICANCE
Leukemia-Lymphoma cell lines are important and widely used research tools. Choosing the right cell line is contingent upon its applicability as in vitro model and can be assured by authentication of the cell line, verification of derivation from the desired tissue/cell of origin, and additional characterization of the cells.
LEUKEMIA-LYMPHOMA CELL LINES
Over the last decades, there has been an explosion in understanding of the biology, pathogenesis and treatment options for various types of tumors. Part of this progress was made possible through laboratory investigations using continuous cell lines.
Due to their high relevance for human disease, easy manipulation, and relative low costs, leukemia-lymphoma (LL) cell lines (including here cell lines derived from multiple myeloma and related entities) continue to represent vital models for a large range of ongoing investigations.1
While their advantages and benefits as in vitro tools are undeniable, misidentification, contamination issues, and data over interpretation are limitations that afflict the use of LL cell lines. Furthermore, the stubbornly ineradicable myths of genetic instability of cell lines and the alleged acquisition of additional alterations in culture have contributed to almost ruin the once high reputation of LL cell lines and insinuated an unfavorable profile.
WORKING WITH THE CORRECT CELL LINE
In a series of previous articles we have tried to alert the scientific community to the wide-spread cross-contamination of tumor cell lines in general and LL cell lines in particular.2–5 The root of the problem is the same for cross-contamination and the epidemic of mycoplasma contamination which can be attributed predominantly to inadequate cell culture practice.
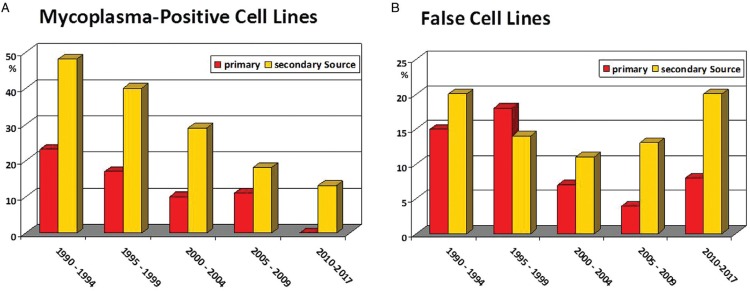
Figure 1A shows the percentage of mycoplasma-positive LL cell lines received at DSMZ, a public non-profit biological ressources center and cell lines bank, between 1990 and 2017 (data updated from ref. 5). There was steady improvement on the mycoplasma-issue by period of receipt in both groups, cell lines received from the original investigators who had established the cell line and cell lines received from secondary sources. Significantly easier mycoplasma detection techniques and the option to eradicate the contaminants with potent antibiotics led to this improvement.
Figure 1.
Incidences of mycoplasma-positive and false leukemia-lymphoma cell lines. This cell lines-based-analysis used data on overall 792 cell lines which were examined by mycoplasma detection assays (A) and on overall 878 cell lines which were subjected to DNA authentication methods (B); cell lines were received between 1990 and 2017. Shown are the percentages of mycoplasma-positive and false (cross-contaminated) cell lines for the calendar periods indicated on the X-axis. The source of cell lines is indicated in the key of each panel: primary sources (red columns) vs secondary sources (orange columns). Updated from reference.5.
Perhaps more important than treating mycoplasma-contaminated cell lines is preventing the infection through improved good cell culture practice.6 While the “mycoplasma-problem” seems to be more or less under control, this is clearly not the case for the “false cell lines-problem” where the percentages of cross-contaminated cell lines still remain stubbornly at a disturbing high level (Fig. 1B).
Real-world data show that poor adherence to legally, scientifically and socially appropriate conduct in obtaining LL cell lines appears to be associated with a very high rate of “false cell lines” being used. This is not only a problem for the laboratories working with the wrong cell lines, but also for the entire research community since many publications contain data on cell lines that are mislabeled. Spoken out clearly, the usual practice of cell cultures wandering from lab to lab without commensurate permission and without authenticity check does not help in reducing the incidence of cross-contaminated cell lines. For example, in China where clearly many LL cell lines still arrive on long and unusual routes very high levels of cross-contaminated cell lines were uncovered recently.7,8
Authentication needs to become a more central tenet of culturing LL cell lines since this is not so difficult to do. The most often used and most informative methodology is STR analysis (also known as “DNA fingerprinting”, commonly taking advantage of single tandem repeat polymorphisms) which allows for definitive and comprehensive searches in web-based databases. Creating awareness and systematic use of authenticity screening may improve the still unsatisfactory status quo. Scientists should be cognizant of the possibility to deal with false LL cell lines. An easy way to avoid working with false cell lines is to obtain cell lines from reputable sources, and not just from the lab nextdoor. It is to be hoped that improvements in awareness und correct acquisition of cell lines will translate to improvements in the quality and reproducibility of research data.
It is the mandate of public cell line banks to raise awareness about this underappreciated threat. Several journals, including HemaSphere, are now asking for proof of cell line authentication in publications, to help raise awareness and to avoid such mistakes in published data.
THE CORRECT TISSUE/CELL
Besides the proven derivation from the index patient (authentication and exclusion of cross-contamination), cell line integrity also depends on the correct clinical diagnosis of the patient (fidelity of diagnosis) and the derivation of the cell line from the malignant tissue and not from residual non-malignant bystander cells (correct tissue of origin identity or cell of origin identity).9
In order to avoid misinterpretations of findings, any misdiagnosis of the primary tumor and incorrect assignment of the cell line to a type of tissue/cell must be resolved. Various methods exist to verifiy the correct tissue/cell of origin, for example immunoprofiling of surface markers, cytogenetic karyotyping, or molecular cytogenomics.
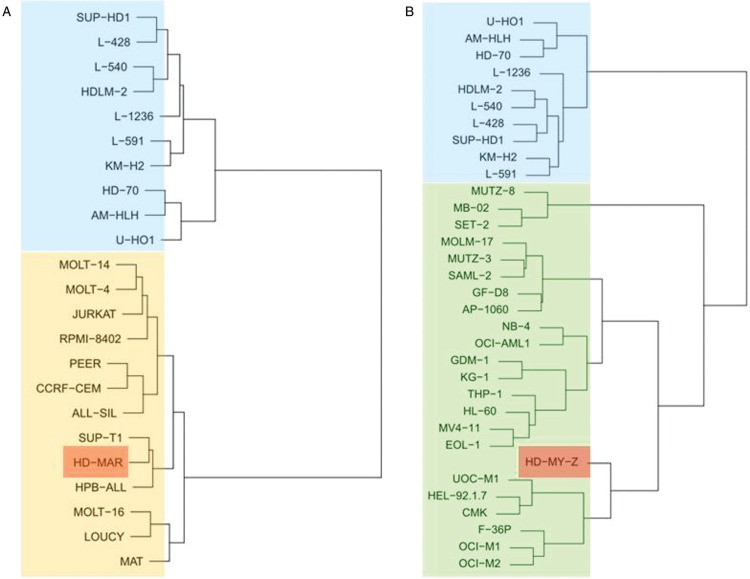
Gene expression profiling which can be used in the hierarchical clustering of LL cell lines has allowed for a more nuanced understanding of their place in the hematopoietic system and represents a pragmatic, experience-informed approach, also allowing to assess the fidelity of the diagnosis (examples are shown in Fig. 2). These data provide additional information beyond the traditional assignment of cell lines which is based mainly on the original diagnosis of the patient (clinical presenting features) and on surface marker profiles.
Figure 2.
Verification of origin identity. Dendrograms based on gene expression signatures showing unsupervised hierarchical clustering of cell line HD-MAR (red box), cell line HD-MY-Z (red box), classical Hodgkin lymphoma cell lines (blue boxes), T-cell leukemia/lymphoma cell lines (golden box) and myelomonocytic leukemia cell lines (green box). (A) Note: cell line HD-MAR which was originally published as being derived from a patient with Hodgkin lymphoma10 does not cluster with eight bona fide classical Hodgkin lymphoma cell lines, but clusters with the T-cell leukemia-lymphoma cell lines. (B) Note: cell line HD-MY-Z which had been described originally as Hodgkin lymphoma-derived11 clusters with the myelomonocytic cell lines and not with the classical Hodgkin lymphoma cell lines. Modified from reference.9.
As has been done for ovarian cancer cell lines,12 we can envision a next generation of analyses whereby cell lines are compared with primary tumor cells to document the similarities of the samples and allowing to propose the most suitable cell line model.
FURTHER CHARACTERIZATION
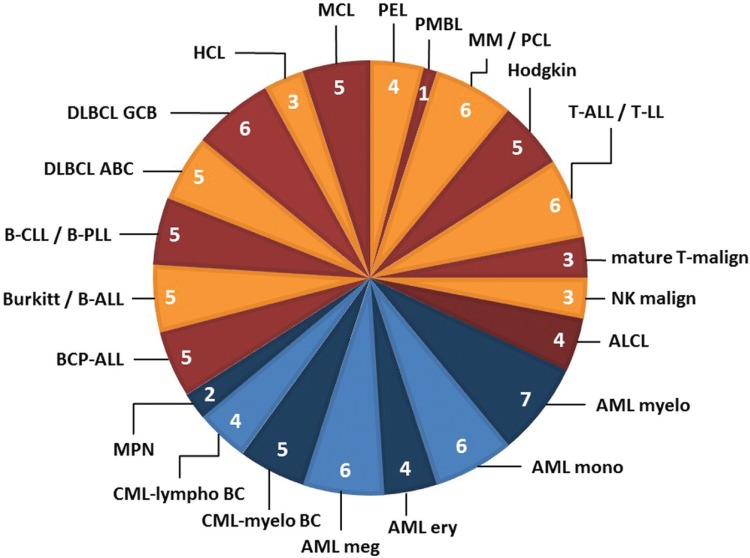
The true value of specific cell lines as models lies in their unique features and characteristics.13 Following the lead of the NCI-60 panel (which covers mainly solid tumor-derived cell lines),14 we developed a reliable set of LL cell lines that we termed “LL-100 panel”, being composed of 100 LL cell lines (Fig. 3). As there is a rationale for large-scale sequencing of LL cell lines that are cytogenetically, immunologically and clinically fully annotated and rigorously authenticated and validated, we established a formal programme for the molecular characterization of the LL-100 panel in 2017.15
Figure 3.
LL cell line panel: Stratification of cell lines by disease category. 100 LL cell lines derived from the 22 disease entities shown here were assembled in the LL-100 panel; the number of cell lines in each category is indicated. For details see reference.15 ABC = activated B-cell; ALCL = anaplastic large cell lymphoma; ALL = acute lymphoblastic leukemia; AML = acute myeloid leukemia; BC = blast crisis; BCP = B-cell precuror; Burkitt = Burkitt lymphoma; CLL = chronic lymphocytic leukemia; CML = chronic myeloid leukemia; DLBCL = diffuse large B-cell lymphoma; ery = erythrocytic; GCB = germinal center B-cell; HCL = hairy cell leukemia; Hodgkin = Hodgkin lymphoma; LL = lymphoblastic lymphoma; lympho = lymphoid; malign = malignancy; MCL = mantle cell lymphoma; meg = megakaryocytic; MM = multiple myeloma; mono = monocytic; MPN = myeloproliferative neoplasms; myelo = myelocytic; NK = natural killer; PCL = plasma cell leukemia; PEL = primary effusion lymphoma; PLL = prolymphocytic leukemia; PMBL = primary mediastinal B-cell lymphoma.
The LL-100 represents the corpus of a modern cell line panel and provides a solid background for studying genomic and transcriptomic alterations. Its huge publicly available database offers unique opportunities to query a multitude of gene alteration parameters.15 Clearly, the public availability of the most important and validated LL cell lines will be crucial to explore new molecular and biological aspects of leukemia-lymphoma.
There has been tremendous progress in the understanding of leukemia-lymphoma genomics while establishing and describing preclinical models that capture this pathobiology. The complexity of the mutational landscape highlights the need to interrogate a broad array of LL cell lines derived from different entities and different phases of the disease. Building on these recent advances, further efforts are warranted to provide a comprehensive characterization of additional LL cell lines as there are dozens and hundreds of unexplored cell lines covering the complete gamut of hematopoietic malignancies with their striking diversity.
In order for LL cell lines to continue to represent important and reliable research tools in hematological oncology, cell line integrity (fidelity of diagnosis, authentication and exclusion of cross-contamination, tissue/cell of origin identity) and extensive characterization are essential for the suitability of any given cell line to serve as a faithful in-vitro model. Finally, it will be also of utmost importance that new LL cell lines are established.
Footnotes
Citation: Drexler HG, Eberth S, Nagel S, Quentmeier H. There is a Scientific Need for the Right Leukemia-Lymphoma Cell Lines. HemaSphere, 2019;3:6. http://dx.doi.org/10.1097/HS9.0000000000000315
The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Drexler HG. Guide to Leukemia-Lymphoma Cell Lines. 2nd Edition.2010;Braunschweig, [Google Scholar]
- 2.MacLeod RA, Dirks WG, Matsuo Y, et al. Widespread intraspecies cross-contamination of human tumor cell lines arising at source. Int J Cancer. 1999;83:555–563. [DOI] [PubMed] [Google Scholar]
- 3.Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci USA. 2001;98:8012–8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drexler HG, Dirks WG, Matsuo Y, et al. False leukemia-lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416–426. [DOI] [PubMed] [Google Scholar]
- 5.Drexler HG, Dirks WG, MacLeod RA, et al. False and mycoplasma-contaminated leukemia-lymphoma cell lines: time for a reappraisal. Int J Cancer. 2017;140:1209–1214. [DOI] [PubMed] [Google Scholar]
- 6.Freshney RI. Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications. 7th Edition.2016;Wiley Blackwell, [Google Scholar]
- 7.Bian X, Yang Z, Feng H, et al. A combination of species identification and STR profiling identifies cross-contaminated cells from 482 human tumor cell lines. Sci Rep. 2017;7:9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye F, Chen C, Qin J, et al. Genetic profiling reveals an alarming rate of cross-contamination among human cell lines used in China. FASEB J. 2015;29:4268–4272. [DOI] [PubMed] [Google Scholar]
- 9.Drexler HG, Pommerenke C, Eberth S, et al. Hodgkin lymphoma cell lines: to separate the wheat from the chaff. Biol Chem. 2018;399:511–523. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Bassat H, Mitrani-Rosenbaum S, Gamliel H, et al. Establishment in continuous culture of a T-lymphoid cell line (HD-MAR) from a patient with Hodgkin's lymphoma. Int J Cancer. 1980;25:583–590. [DOI] [PubMed] [Google Scholar]
- 11.Bargou RC, Mapara MY, Zugck C, et al. Characterization of a novel Hodgkin cell line, HD-MyZ, with myelomonocytic features mimicking Hodgkin's disease in severe combined immunodeficient mice. J Exp Med. 1993;177:1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domcke S, Sinha R, Levine DA, et al. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abaan OD, Polley EC, Davis SR, et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73:4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. [DOI] [PubMed] [Google Scholar]
- 15.Quentmeier H, Pommerenke C, Dirks WG, et al. The LL-100 panel: 100 cell lines for blood cancer studies. Sci Rep. 2019;9:8218. [DOI] [PMC free article] [PubMed] [Google Scholar]