T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive malignancy of the lymphocytes that is driven by the cooperation of various mutations. Constitutive activation of JAK-STAT signaling is observed in one-third of T-ALL patients and is caused by activating mutations in the interleukin 7 receptor alpha chain (IL7R), in the Janus kinases JAK1 or JAK3, or in the Signal transducer and activator of transcription 5B (STAT5B).1–3 STAT5B mutations are most frequently the N642H variant and are associated with an unfavorable prognosis and higher risk of relapse.1,2,4,5 We set out to test the efficacy of JAK1/JAK3 kinase inhibitors ruxolitinib, tofacitinib, upadacitinib, baricitinib, decernotinib, and peficitinib in the murine Ba/F3 cell model transformed by different mutated components of JAK/STAT signaling. We show that cells carrying mutations in any component of IL7 receptor signaling are sensitive to JAK-inhibition, including mutations in STAT5B.
JAK2 inhibition by ruxolitinib is a well-established treatment mechanism for polycythemia vera and myelofibrosis.6,7 The central role for the JAK1 and JAK3 kinases in IL7R/JAK/STAT driven T-ALL provides a rationale for JAK inhibition in this disease. The use of ruxolitinib, which has selectivity towards JAK1 and JAK2 and tofacitinib, targeting JAK1 and JAK3, in T-ALL is supported by preclinical evidence, showing sensitivity of IL7R, JAK1, and JAK3 mutated cases to these inhibitors in vitro8 and in vivo in patient-derived xenograft models.9 Indeed, mutant IL7R and mutant JAK3 typically require JAK1 activity and are sensitive to JAK1 inhibition.10,11 PhaseI/II clinical trials for acute lymphoblastic leukemia with ruxolitinib are currently ongoing.
IL7R signaling is important for normal development, maturation and homeostasis of the B- and T-cell compartment. The IL7 receptor is a heterodimer that consists of the IL7Ralpha chain and the common gamma receptor protein (IL2Rγ). Upon binding of interleukin 7, the ligand, to the IL7R, a conformational change occurs that brings both chains closer together, exposing tyrosine kinases JAK1 and JAK3 for phosphorylation and activation. They in turn phosphorylate downstream effectors including STAT5A and STAT5B. After phosphorylation of tyrosine 694, the STATs dimerize and translocate to the nucleus to activate transcription of STAT5 target genes,3 including known oncogenes (eg, Myc) and anti-apoptotic factors such as BCL2. Mutations in IL7R or JAK1/3 increase phosphorylation activity of the JAKs, while the STAT5B(N642H) mutant displays stronger intra-dimer interaction, increased and prolonged phosphorylation of tyrosine Y694 as well as increased DNA binding capacities and transcriptional activity compared to wild type STAT5B.4,5
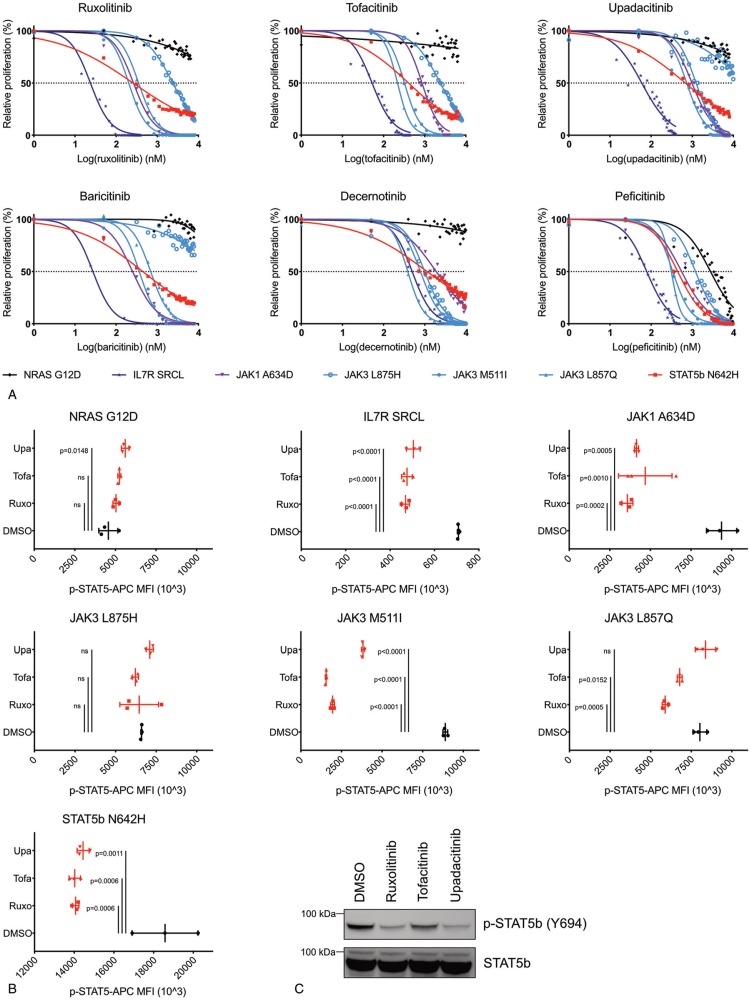
We used BaF/3 cells that were transformed to cytokine-independent growth by expression of IL7R(insSRCL), JAK1(A634D), JAK3(M511I), JAK3(L857Q), JAK3(L875H), STAT5B(N642H), or NRAS(G12D). We treated these cells with ruxolitinib, tofacitinib, upadacitinib, baricitinib, decernotinib, and peficitinib in a concentration range from 1 nM to 10 μM for 24 hours. We used ATPlite luminescence as a read-out for proliferation and established dose-response curves and calculated IC50 values (Fig. 1A, Table 1). Cells expressing mutant JAK1 or JAK3 were sensitive to JAK-inhibition with IC50 values between 203 nM and 2395 nM. IC50 values for different mutants were determined by both specificity and potency of the inhibitor, as well as intrinsic sensitivity to inhibition of the given mutant. Importantly, all tested new inhibitors showed efficacy in inhibiting proliferation of JAK mutants comparable to well established JAK inhibitor ruxolitinib. The JAK3(L875H) mutant was the least sensitive mutant and showed high resistance towards upadacitinib and baricitinib. (Fig. 1A) Both inhibitors selectively inhibit JAK1, confirming the JAK1 independent character of mutations in the kinase domain of JAK3. The IL7R(insSRCL) transformed Ba/F3 cells showed the highest sensitivity to all inhibitors, except to decernotinib, which is a more JAK3 selective inhibitor. Surprisingly, the STAT5B(N642H) expressing Ba/F3 cells that were grown in the absence of cytokines and do not contain JAK kinase mutations, were also sensitive to JAK-inhibition with IC50 values within range of the other cells and even higher sensitivity than JAK3(L875H).
Figure 1.
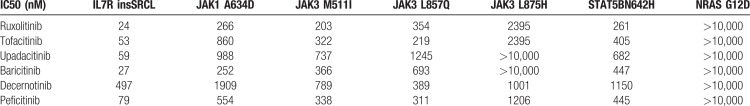
Ba/F3 cells transformed by any mutated member of IL7-JAK-STAT signaling are sensitive to JAK-inhibition. (A) Dose-response curves for ruxolitinib, tofacitinib, upadacitinib, baricitinib, decernotinib, and peficitinib treatment on Ba/F3 cells transformed by IL7R(insSRCL), JAK1(A634D), JAK3(M511I), JAK3(L857Q), JAK3(L875H), STAT5B(N642H), and NRAS(G12D) after 24 hours incubation. Data represent the mean of 3 experiments. (B) Mean fluorescence intensity values for p-STAT5B Y694 of Ba/F3 cells transformed by IL7R(insSRCL), JAK1(A634D), JAK3(M511I), JAK3(L857Q), JAK3(L875H), STAT5B(N642H), and NRAS(G12D) after treatment with upadacitinib, tofacitinib, ruxolitinib 2 μM or DMSO for 3 hours. Symbols represent biological replicates, mean and standard deviation (SD) are shown. Significance was calculated using one-way analysis of variance (ANOVA) with Bonferroni multiple comparisons correction. ns = not significant (p ≥ 0.05). Upa: upadacitinib, Tofa: tofacitinib, Ruxo: ruxolitinib. (C) Western blot analysis of phospho-STAT5 Y694 and STAT5 in STAT5B(N642H) transformed Ba/F3 cells after treatment with baricitinib, decernotinib, peficitinib, ruxolitinib, tofacitinib, and upadacitinib 2 μM or DMSO for 90 minutes.
Table 1.
IC50 values (nM) for all inhibitors in Figure 1 in Ba/F3 cells overexpressing IL7R(insSRCL), JAK1(A634D), JAK3(M511I), JAK3(L857Q), JAK3(L875H), STAT5B(N642H), or NRAS(G12D).
To determine if the sensitivity to JAK-inhibition was an on-target effect, we performed flow cytometry analysis of phospho-STAT5B Y694 after treatment of the different Ba/F3 variants with 2 μM ruxolitinib, tofacitinib, upadacitinib, or DMSO for 3 hours (Fig. 1B). These data showed that growth inhibition correlated with loss of phospho-STAT5 signal for all sensitive JAK-STAT mutants. The JAK3(L875H) mutant was not very sensitive to these inhibitors and showed also no response in the phospho-STAT5 measurements. The STAT5B(N642H) cells were sensitive to ruxolitinib, tofacitinib, upadacitinib, and showed clear decreases in STAT5 phosphorylation upon treatment with these inhibitors. This was observed by phospho-flow measurement after 3 hours of treatment (Fig. 1B) or Western blot analysis of STAT5 Y694 phosphorylation after 90 minutes of treatment (Fig. 1C). Moreover, we observed reduced expression of STAT5 target genes in the STAT5B(N642H) expressing cells upon ruxolitinib treatment, as measured by quantitative RT-PCR (Fig. 2A). These results confirm a rapid loss of phosphorylation of STAT5 and loss of its transcriptional activity upon treatment of the cells with JAK inhibitors. Thus, despite the absence of activating mutations in the JAK kinases, the STAT5B(N642H) mutant showed high levels of phosphorylation that could be repressed with the use of JAK-inhibitors (Fig. 1).
Figure 2.
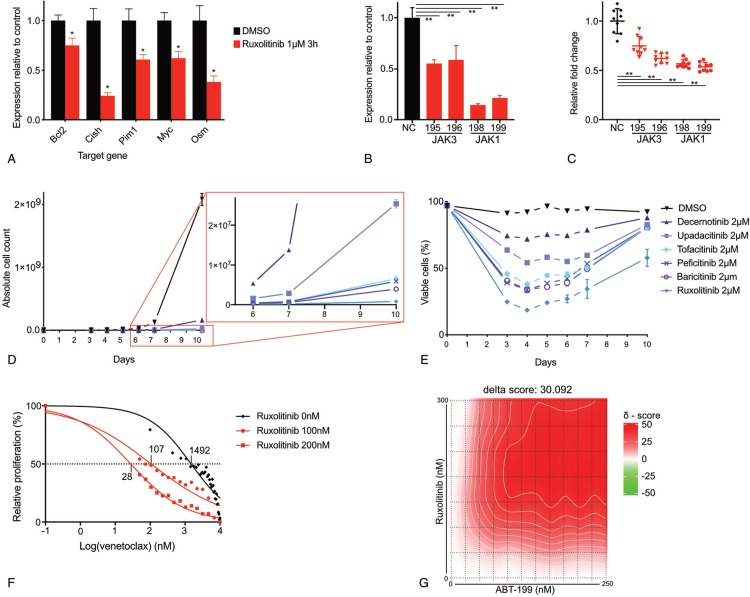
JAK-inhibition in Ba/F3 STAT5B(N642H) reduces expression of STAT5 target genes and causes a proliferation deficit that is synergistic with Bcl2-inhibition. (A) RT-qPCR of Ba/F3 STAT5B(N642H) for STAT5 target genes Bcl2, Cish, Pim1, Myc, and Osm after treatment with 1 μM ruxolitinib or DMSO for 3 hours. Data show the mean of 3 experiments +/− SD. ∗ p < 0.05. in unpaired two-tailed t test with equal variance. (B) RT-qPCR of Ba/F3 STAT5B(N642H) for JAK1 and JAK3 24 hours after electroporation with negative control siRNA (NC), siRNA targeting JAK3 (195, 196) or siRNA targeting JAK1 (198, 199). Data represent the mean of 3 experiments +/− SD. ∗∗ p < 0.0001, p values were calculated using one-way analysis of variance (ANOVA) with Bonferroni multiple comparisons correction. Data represent the mean of 3 experiments +/− SD. (C) Relative fold change of proliferation of Ba/F3 STAT5B(N642H) 72 hours after electroporation with negative control siRNA (NC), siRNA targeting JAK3 (195, 196) or siRNA targeting JAK1 (198, 199). ∗∗ p < 0.0001, significance was calculated using one-way analysis of variance (ANOVA) with the Bonferroni multiple comparisons correction. Data are presented as mean of 3 experiments +/− SD. (D-E) Ba/F3 STAT5B(N642H) were cultured for 10 days in the presence of DMSO, ruxolitinib, tofacitinib, upadacitinib, baricitinib, decernotinib, or peficitinib and analyzed by flowcytometry for concentration (D) and viablility (E). Data represent the mean of 3 experiments +/− SD. (F) Dose-response curves for venetoclax on Ba/F3 cells transformed by STAT5B(N642H) after 24 h incubation in the presence of 0 nM, 100 nM or 200 nM ruxolitinib. The mean of 3 experiments is shown. (G) Synergy matrix plot showing δ-scores for Ba/F3 STAT5B(N642H) treated with ruxolitinib and venetoclax. δ-score = the average d-score for the whole range of concentrations shown in the synergy matrix.
These data indicate that the STAT5B(N642H) mutant itself induces activation of the JAK kinases and that this is required for its phosphorylation on Y694 and its transcriptional activity. To further confirm this, we electroporated Ba/F3 STAT5B(N642H) cells with siRNA targeting Jak1, Jak3 or negative control and confirmed knockdown by RT-PCR (Fig. 2B). Proliferation of the STAT5B(N642H) transformed cells was significantly reduced by Jak1 or Jak3 knock-down (Fig. 2C), supporting the hypothesis that JAK kinases are essential for proliferation of STAT5B(N642H) mutated cells. While this work was in progress, Pham and colleagues12 came to similar conclusions in a mouse T-cell lymphoma/leukemia model driven by STAT5B(N642H). They documented strong phosphorylation of STAT5B(N642H) and sensitivity of the leukemia cells to JAK inhibition.
To investigate whether JAK-inhibition could sustainably inhibit proliferation of STAT5B(N642H) overexpressing cells, we cultured the cells at a starting concentration of 100.000/mL for 10 days in culture medium supplemented with 2 μM concentration of inhibitor or equal amount of DMSO as control (Fig. 2D). When compared to DMSO, proliferation was clearly reduced for every inhibitor. However, in contrast to what has previously been described for JAK1 and JAK3 mutants, complete eradication of the cultured STAT5B(N642H) overexpressing cells could not be achieved. This corresponds with the inability to reduce relative proliferation to zero in the dose-response curves (Fig. 1A). Upon treatment, a fast decrease in viability occurs. For tofacitinib, upadacitinib, baricitinib, decernotinib, and peficitinib this is accompanied by a strong reduction in rate of proliferation. Treatment with ruxolitinib causes cell death during the first 4 days of treatment, after which viability starts to increase for all inhibitors (Fig. 2E). Despite incomplete response, proliferation rates remained strongly reduced for the entire duration of the growth experiment (Fig. 2D).
It can be expected that Ba/F3 STAT5B(N642H) cells would rely on the expression of STAT5B target genes such as BCL2 for survival. Indeed, these cells were sensitive to BCL2-inhibition by venetoclax (IC50 1492 nM) and addition of 100 nM or 200 nM ruxolitinib showed a drastic decrease of the IC50 value of venetoclax to 107 nM and 28 nM respectively (Fig. 2F). This synergistic activity between ruxolitinib and venetoclax was indicated by high synergy scores calculated according to the zero interaction potential method.13 We observed a clear overall synergistic effect (δ-score 30.09), with a maximum ZIP synergy score of 43.23, which means that there was >40% additional inhibition compared to the additive effect of the two compounds14 (Fig. 2G).
From this work, we conclude that the recently developed JAK-inhibitors with higher specificity towards JAK1 and JAK3 when compared to ruxolitinib, all show activity against hyperactivation of JAK-STAT signaling in T-ALL with similar or higher efficacy compared to ruxolitinib. Interestingly, despite its position downstream of the JAK kinases, we find that the STAT5B(N642H) mutant also depends on the JAK1/JAK3 kinase activity. Therefore, expression of STAT5 target genes and cell growth was reduced by JAK-inhibition in cells that express mutant STAT5B and this effect was synergistic with the BCL2-inhibitor venetoclax. In vivo, the sensitivity to JAK-inhibition is affected by the mutational landscape and clonal architecture of the leukemia for any given patient.4,15 However, our data suggest that T-ALL patients who carry the STAT5B(N642H) mutation may still benefit from treatment with a JAK-inhibitor especially in combination with a BCL2 inhibitor.
Footnotes
Citation: Govaerts I, Jacobs K, Vandepoel R, Cools J. JAK/STAT Pathway Mutations in T-ALL, Including the STAT5B N642H Mutation, are Sensitive to JAK1/JAK3 Inhibitors. HemaSphere, 2019;3:6. http://dx.doi.org/10.1097/HS9.0000000000000313
This study was funded by KU Leuven (C14/18/104) and the Foundation against Cancer (JC). IG was supported by a fellowship ‘Emmanuel van der Schueren’ of Kom op tegen Kanker (Stand up to Cancer), the Flemish cancer society and by FWO-Vlaanderen.
The authors declare no conflicts of interest.
References
- 1.Vicente C, Schwab C, Broux M, et al. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica. 2015;100:1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2012;32:2601–2613. [DOI] [PubMed] [Google Scholar]
- 4.Kontro M, Kuusanmäki H, Eldfors S, et al. Novel activating STAT5B mutations as putative drivers of T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:1738–1742. [DOI] [PubMed] [Google Scholar]
- 5.Bandapalli OR, Schuessele S, Kunz JB, et al. The activating STAT5B N642H mutation is a common abnormality in pediatric T-cell acutelymphoblastic leukemia and confers a higher risk of relapse. Haematologica. 2014;99:e188–e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vannucchi AM, Kantarjian HM, Kiladjian JJ, et al. A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica. 2015;100:1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vannucchi AM, Kiladjian JJ, Griesshammer M, et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N Engl J Med. 2015;372:426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maude SL, Dolai S, Delgado-Martin C, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood. 2015;125:1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Losdyck E, Hornakova T, Springuel L, et al. Distinct acute lymphoblastic leukemia (ALL)-associated Janus Kinase 3 (JAK3) mutants exhibit different cytokine-receptor requirements and JAK inhibitor specificities. J Biol Chem. 2015;290:29022–29034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degryse S, De Bock CE, Cox L, et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood. 2014;125:1107–1115. [DOI] [PubMed] [Google Scholar]
- 12.Pham HTT, Maurer B, Prchal-Murphy M, et al. STAT5B N642H is a driver mutation for T cell neoplasia. J Clin Invest. 2018;128:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav B, Wennerberg K, Aittokallio T, et al. Searching for drug synergy in complex dose–response landscapes using an interaction potency model. Comput Struct Biotechnol J. 2015;13:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ianevski A, He L, Aittokallio T, et al. SynergyFinder: a web application for analyzing drug combination dose–response matrix data. Stegle O, ed. Bioinformatics. 2017;33:2413–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson EI, Pützer S, Yadav B, et al. Discovery of novel drug sensitivities in T-PLL by high-throughput ex vivo drug testing and mutation profiling. Leukemia. 2018;32:774–787. [DOI] [PubMed] [Google Scholar]