Abstract
We present cleared tissue Axially Swept Light-Sheet Microscopy (ctASLM), which enables isotropic, subcellular resolution, high optical sectioning capability, and large field of view imaging over a broad range of immersion media. ctASLM can image live, expanded, and both aqueous and organic chemically cleared tissue preparations. Depending on the optical configuration, ctASLM provides up to 260 nm axial resolution, an improvement over confocal and other reported cleared tissue light-sheet microscopes by a factor 3–10. We image millimeter-scale tissues with subcellular 3D resolution, which enabled us to automatically detect with computer vision multicellular tissue architectures, individual cells, synaptic spines, and rare cell-cell interactions.
Human tissues are composed of multiple polarized cell types organized in well-defined three-dimensional architectures. Interestingly, it has been shown that rare subsets of cells play a crucial role in disease progression,1 and interdisciplinary efforts now aim to generate comprehensive atlases of human cells in diverse tissue types. To date, this has largely relied on massively parallel sequencing and machine learning-based analyses to identify unique sub-populations of cells. Combined with advanced imaging, such efforts could not only shed light on the diversity of cell types, but the biological context in which each population operates. However, imaging large tissues with subcellular resolution remains challenging due to the heterogeneous refractive index and composition of tissues, which results in complex aberrations and an increased scattering coefficient, both of which decrease spatial resolution and limit imaging depth.2
To circumvent this challenge, a large variety of optical clearing techniques have been developed that aim to homogenize the optical properties of the tissue using aqueous or organic solvents.3 Today, these clearing techniques routinely render biological specimens sufficiently transparent, such that – if the imaging technology were to exist – entire organisms could be imaged with subcellular resolution and with molecular specificity. As the specimens are three-dimensional (3D), an ideal imaging system should possess isotropic resolution, in order to accurately measure 3D cellular morphology, biomolecular localization, and signaling activity.4 Nevertheless, imaging chemically cleared specimens with diffraction-limited or super-resolution presents technical challenges. For example, each clearing mechanism provides advantages and disadvantages, and requires unique immersion media with refractive indices that range between 1.33 and 1.559. Thus, the imaging system must optimally operate throughout this refractive index range without suffering from deleterious aberrations, which decreases both resolution and sensitivity.
Light-sheet fluorescence microscopy (LSFM), because of its parallelized image acquisition, inherent optical sectioning, and ability to image large biological structures quickly and with high optical resolution, serves as an ideal candidate for cleared tissue imaging (Supplementary Table 1).5 Nevertheless, to the best of our knowledge, there is not yet a light-sheet microscope for cleared tissue specimens that possesses submicron, isotropic resolution, and is also compatible with the full range of clearing methods.6–9 Although Lattice light-sheet microscopy readily achieves high spatial resolution, it relies on specialized high numerical aperture lenses that are only compatible with aqueous solvents (refractive index = 1.333). Here, we address these limitations, and present a scalable imaging platform that provides sub-cellular anatomical detail in any spatial dimension across millimeter cubes of tissue. The system, which we refer to as cleared tissue Axially Swept Light-Sheet Microscopy (ctASLM), builds upon previous work where the sample was mechanically scanned through a thin light-sheet.10, 11 Instead of slow sample scans, ctASLM scans the illumination in its propagation direction using high-speed aberration-free remote focusing (Supplementary Note 1, Supplementary Figure 1), refractive index-independent illumination and detection optics, residual aberration correction, and syncronous camera readout to deliver isotropic high-resolution, large field of view, and high-contrast imaging (Figure 1a).12, 13 Compared to other light-sheet variants that lack aberration free refocusing (such as mesoSPIM) and camera synchronization (such as tiling light-sheet microscopy), ctASLM is better suited for using thin light-sheets over large field of views (Supplementary Note 1).14, 15 Consequently, we present two implementations of ctASLM using matched multi-immersion objectives of NA 0.4 or 0.7, which provide isotropic subcellular imaging at two resolution levels (~600 and ~300 nm, respectively, Supplementary Figure 2, Supplementary Note 2). By tiling multiple volumes16 ctASLM permits routine visualization of sub-cellular features throughout millimeters of tissue, regardless of the clearing method (Supplementary Figure 3, Supplementary Movie 1).
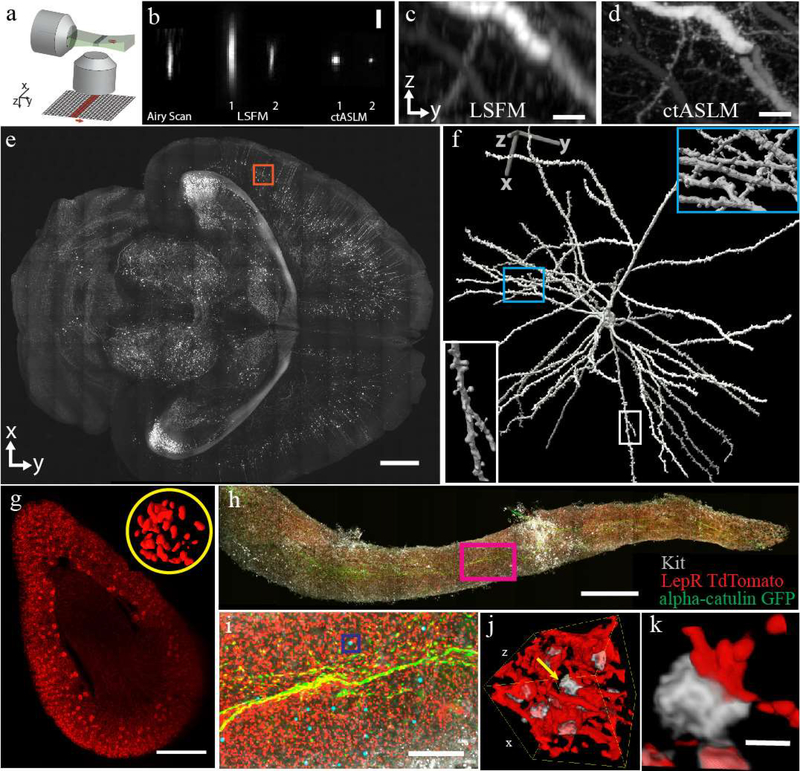
Figure 1 -. ctASLM enables isotropic, sub-micron imaging over large field of views.
(a) Working principle of axially swept light-sheet microscopy: a thin light-sheet is scanned in its propagation direction. The rolling shutter readout of an sCMOS camera, adjusted to the size of the beam waist, is tightly synchronized to the light sheet scan. (b) Experimentally measured PSF of Airy scan confocal microscopy, conventional light-sheet fluorescence microscopy (LSFM), and ctASLM. LSFM and ctASLM PSFs are shown for NA 0.4 (indicated with “1”) and 0.7 objectives (indicated with “2”). (c-d) Maximum intensity projections of CLARITY cleared cortical Thy1-eYFP neurons as imaged by LSFM and ctASLM, respectively. (e) An XY view of a section 2.5mm from the top surface of a Thy1-eGFP PEGASOS cleared brain. (f) Zoom-in of the red box with a rendered neuron. Insets provide magnified views of synaptic spines. (g) Volume rendering of a mouse kidney labeled with Flk1-GFP. Inset shows a 3D rendering of endothelial cells in one selected glomerulus. (h) Maximum intensity projection of a mouse tibia bone marrow plug. (i) Zoom-in of magenta box shows alpha-catulin-GFP+ cells (green, Alexa Fluor 488), Lepr+ niche cells (red, tdTomato) and cKit+ cells (grey, Alexa Fluor 647). Alpha-catulin-GFP+/cKit+ hematopoietic stem cells (HSC) are labeled with cyan spheres. (j&k) are the volume-rendering of boxed region in (h), showing progenitor cells in grey and niche cells in red. All biological data acquired with NA 0.4 objectives. Scale bars are: (b) 3 μm, (c,d) 20 μm, (e) 1 mm, (g) 500 μm, (h) 1 mm, (i) 200 μm, (j) 40×40×40 μm3, (k) 5 μm.
To evaluate optical performance, we imaged sub-diffraction beads with ctASLM, conventional light-sheet microscopy and an Airy Scan confocal microscope (Figure 1b), each equipped with long working distance multi-immersion objectives (detailed Full-Width Half-Maximum values are given in Table 1 in the online methods). Importantly, ctASLM improves axial resolution sixfold over conventional LSFM and three- to seven-fold over Airy scan confocal microscopy. Depending on the refractive index of the immersion media, ctASLM achieves an isotropic, raw resolution of 800–900 nm with the NA 0.4 objectives and 400–500 nm with the NA 0.7 objectives, respectively, over the same field of view encompassed by conventional light-sheet microscopy (Figure 1b, Supplementary Figures 4 and 5, Supplementary Tables 2 and 3, and Supplementary Note 3).17, 18 Iterative deconvolution improves the resolution of ctASLM by ~30%, leading to 300 nm resolution with the NA 0.7 objective (Table 1 online methods, Supplementary Figures 6 and 7), which we further confirmed inside a biological sample (Supplementary Figure 8). To the best of our knowledge, ctASLM’s axial resolving power of 260–290 nm in solvent based media is the highest reported z-resolution in diffraction limited light-sheet microscopy (i.e. without requiring super-resolution mechanisms such as stimulated emission depletion, nonlinear structured illumination, or localization approaches). Owing to ctASLM’s excellent optical sectioning and high-quality raw data, deconvolution is however not always needed (Supplementary Figure 9). In comparison to a conventional light-sheet microscope (Figure 1c), individual synaptic spines could be clearly resolved in any spatial dimension (Figure 1d), and neurons could be traced in 3D using unprocessed data.
Table 1.
Experimental values for point spread functions. Note that the magnification and resolving power slightly changes for the multi-immersion lenses used for ctASLM for refractive indices (η) between 1.333 and 1.56, respectively. All numbers presented are the mean ± the standard deviation of the Full-Width Half-Maximum, and the sample size is 5 for each measurement. --- not measured.
| Optical Performance | |||||||
|---|---|---|---|---|---|---|---|
| Microscope | NA | Field of View (μm2) | Lateral Resolution (μm) | Axial Resolution (μm) | Field of View (μm2) | Lateral Resolution (μm) | Axial Resolution (μm) |
| (η = 1.333) | (η = 1.56) | ||||||
| ctASLMI | 0.4 | 870 × 870 | 0.95 ± 0.030 (raw, n= 10) | 0.95 ± 0.115 (raw, n= 10) | 737×737 | 0.83 ± 0.035 (raw, n= 10) | 0.94 ± 0.075 (raw, n= 10) |
| 870 × 870 | 0.77 ± 0.033 (deconv., n= 10) | 0.75 ± 0.087 (deconv., n= 10) | 737×737 | 0.62 ± 0.032 (deconv., n= 10) | 0.72 ± 0.064 (deconv., n= 10) | ||
| ctASLM2 | 0.7 | 327 × 327 | 0.48 ± 0.012 (raw, n= 10) | 0.48 ± 0.029 (raw, n= 10) | 310×310 | 0.45 ± 0.016 (raw, n= 6) | 0.38 ± 0.032 (raw, n= 6) |
| 327 × 327 | 0.36 ± 0.012 (deconv., n= 10) | 0.37 ± 0.018 (deconv., n= 10) | 310×310 | 0.31 ± 0.018 (deconv., n= 6) | 0.26 ± 0.018 (deconv., n= 6) | ||
| LSFM1 | 0.4 | 870 × 870 | 1.04 ± 0.04 (raw, n= 5) | 6.7 ± 0.4 (raw, n=5) | 737×737 | --- | --- |
| LSFM2 | 0.7 | 327 × 327 | 0.5 ± 0.02 (raw, n= 5) | 2.66 ± 0.04 (raw, n= 5) | 310×310 | --- | --- |
| Airy Scan | 0.8 | --- | --- | --- | --- | 0.32 ± 0.03 (deconv., n= 5) | 3.1 ± 0.4 (deconv., n= 5) |
Because of its large field of view and long working distance, we explored the ability of ctASLM equipped with the NA 0.4 objectives to image large tissues. Figure 1e shows a PEGASOS cleared, Thy1-GFP brain, (Supplementary Movie 2) and Figure 1f shows a volume rendering of a Neuron at a depth of 2.5 mm. Even at this depth, we were able to resolve fine neuronal structures and individual spines (Supplementary Movies 3 and 4, and Supplementary Figures 10 and 11). Using benzyl alcohol and benzyl benzoate clearing, we imaged a neonatal kidney labeled with the vasculature marker Flk1-GFP, and maintained sufficiently high spatial resolution to distinguish individual cells throughout the 3.4×2.6×2.5 mm3 volume (Figure 1g, Supplementary Movie 5). We also imaged the hematopoietic stem cell (HSC) niche in a bone marrow plug (Figure 1h–k). HSCs are responsible for the continued production of blood and immune cells throughout life, and HSCs reside within a perisinusoidal niche in the bone marrow where Leptin Receptor+ (LepR+) stromal cells and endothelial cells synthesize the factors required for HSC maintenance.19 However, careful analysis of the interactions between these cells has not been done, as HSCs are rare (0.003% of all bone marrow cells) and imaging large specimens has been prohibitively time-consuming.19 ctASLM imaging allowed us to identify all α-catulin+ c-kit+ HSCs, markers that identify all HSCs in young adult bone marrow and which give very high levels of purity (Figure 1i). The isotropic resolution revealed extensive interactions of LepR+ cell projections and HSCs (Figure 1j–k), which were distorted when imaged with a confocal microscope (Supplementary Movie 6). Further we imaged the entire murine olfactory bulb (Supplementary Movie 7), which included ossified structures that would otherwise be incompatible with aqueous clearing methods and demonstrate the importance of being able to image in solvents that range in refractive index from 1.333 to 1.559.
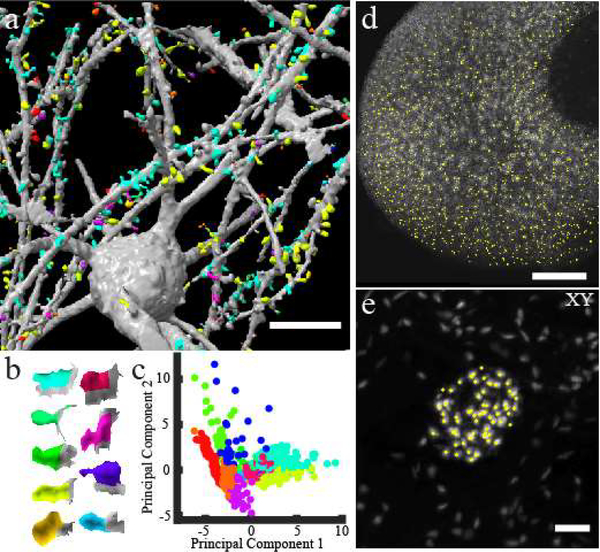
Next, we tested whether the quality of ctASLM data is sufficient for the application of automated and unbiased approaches to image quantification. Using open source software, we could readily detect dendritic spines and cluster them based on their 3D morphology (Figure 2a, and Supplementary Movies 8 and 9).20 These clusters span a wide range of morphologies (Figure 2b), form a structured space following principal component analysis (Figure 2c), and are separable via interpretable measures, such as the ratio of spine neck area to spine surface area (Supplementary Figure 12). For the kidney, damage to glomeruli is associated with a spectrum of clinical outcomes, including proteinuria and reduced filtration, which are the hallmarks of chronic kidney diseases. To evaluate tissue composition in an automated fashion, we developed a multiscale watershed algorithm to identify glomeruli (Figure 2d). Further, owing to the high axial resolution, we can detect endothelial cells within an individual glomerulus (Figure 2e, Supplementary Figure 13, Supplementary Movie 10). Formerly, such an analysis was performed manually on hundreds of serial-sectioned tissue preparations, as identification of individual cells in confocal datasets was particularly challenging due to its diminished z-resolution.
Figure 2 -. ctASLM allows detection of cellular and subcellular features in large tissues.
(a) Machine learning based supervised detection and unsupervised clustering of spines in a Clarity cleared mouse brain. (b) The shape of the median spine in each cluster. (c) Principal component analysis of the spines. (d) automatic detection of glomeruli in the mouse kidney data set shown in Fig 2 (e). (e) Lateral maximum intensity projection of one glomerulus and detection of individual endothelial cells (yellow circles). Data acquired with NA 0.4 objectives. Scale bar: (a) 20 μm, (d) 200 μm, (e) 50 μm
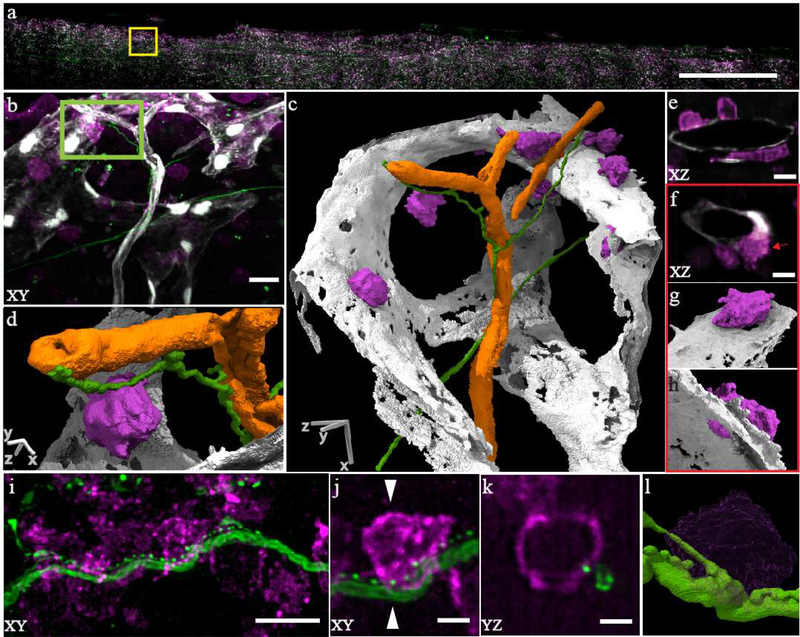
Lastly, we imaged HSCs, nerve fibers and vasculature in mouse bone marrow with the NA 0.7 objectives and 300 nm-scale resolution (Figure 3a–d, Supplementary Figure 14. Many c-kit+ HSCs were associated with fenestrations in the sinusoidal blood vessels, consistent with observations that HSCs migrate through these fenestrations to enter and exit circulation (Figure 3e–h).19 Importantly, detailed features of the HSCs could not be resolved with a confocal microscope (Supplementary Figure 15), but were readily resolved by ctASLM due to its improved resolution. Intriguingly, we observed punctate staining along nerve fibers that resembled synapses, raising the possibility that nerve fibers directly interact with a subset of c-kit+ cells (Figure 3i–l, Supplementary Movie 11).
Figure 3 -. ctASLM allows visualization of subcellular interactions amongst cells in large tissues.
(a) Maximum intensity projection of a mouse bone marrow specimen. (b) Zoom-in of yellow box shows c-kit+ stem/progenitor cells (magenta, Alexa 647), nerve fibers (green, Alexa 488) and blood-vessels (gray, tdTomato). (c) 3D segmented and rendered view of (b). Orange depicts arterioles. (d) Zoomed in view (segmented & rendered) view of the green box in (b) showing c-kit+ cells in contact with a nerve fiber. (e) XZ cross-section of cKit+ cells residing on the sinusoids. (f) c-kit+ cells are positioned along fenestrations in the walls of the sinusoids. (g&h) 3D rendering of cell highlighted in (f). (i) Nerve fibers with punctate staining adjacent to cKit+ positive cells. (j) Zoom in view shows a possible interaction. (k) YZ view along a plane indicated by white triangles in (j) showing an interaction between a nerve fiber and a c-kit+ cell. (l) Volume rendering of (j). Data acquired with NA 0.7 objectives. Scale bar: (a) 500 μm (b&c) 10 μm (d) 2.5 μm (e&f) 5 μm (i) 10 μm (j&k) 2.5 μm
In summary, ctASLM simultaneously provides subcellular detail and tissue scale anatomy. As tissues are inherently 3D, the isotropic resolution of ctASLM is an important attribute to describe 3D morphologies and molecular concentrations in an unbiased way. Indeed, even higher resolutions can be achieved by combining ctASLM with Expansion Microscopy (Supplementary Figures 16–18, and Supplementary Movies 12 and 13), albeit with one order of magnitude larger field of view than Lattice Light-Sheet Microscopy. Of note, ctASLM imposes a higher out-of-focus excitation load on the sample than a conventional light-sheet system and illuminates a smaller portion at a time, both of which can increase photobleaching. However, even at the highest resolution level (NA 0.7), we were not limited by photobleaching and were able to image a sample multiple times with high signal to noise ratio. Some steering of the light-sheet was observed deep in the PEGASOS brain and likely higher order aberrations are present in some of our CLARITY cleared samples (Supplementary Figure 19). Automated alignment routines, as well as adaptive optics could help to achieve diffraction limited performance in less than optimally cleared samples. With the given resolution level, we readily show that we can combine ctASLM with computer vision to detect and classify biological features throughout millimeters of tissue. Consequently, we believe that ctASLM will expedite human cell atlas efforts, providing much-needed insight into how tissue function manifests in health and disease, from the heterogenous cellular populations that compose it.
Methods
Animal Specimens
All animal protocols were approved by local Institutional Animal Care and Use Committees (IACUC) as directed by the National Instutes of Health, and strictly followed. These protocols include AC-AAAR0417 (to R.T., Columbia University), 2017–102370 (to J.M., UT Southwestern Medical Center), 101917 (to D.M., UT Southwestern Medical Center), 101715 (to I.B., UT Southwestern Medical Center), and 102632 (to S.M., UT Southwestern Medical Center).
Microscope Control
The data acquisition computer for ctASLM-v1 was a Dell Precision 5810 Tower equipped with an Intel Xeon E5–1650W v3 processor operating at 3.5 GHz with 6 cores and 12 threads, 128 GB of 2133 MHz DDR4 RAM, and an integrated Intel AHCI chipset controlling 4× 512 GB SSDs in a RAID0 configuration. The data acquisition computer for ctASLM v2 was a Colfax ProEdge SXT9800 Workstation equipped with two Intel Xeon Silver 4112 processors operating at 2.6 GHz with 8 cores and 16 threads, 96 GB of 2.667 GHz DDR4 RAM, a Intel DC P3100 1024 GB M.2 NVMe drive, and a Micron 5200 ECO 7680 GB hard-drive for file storage. All software was developed using a 64-bit version of LabView 2016 equipped with the LabView Run-Time Engine, Vision Development Module, Vision Run-Time Module and all appropriate device drivers, including NI-RIO Drivers (National Instruments). Software communicated with the camera (Flash 4.0, Hamamatsu) via the DCAM-API for the Active Silicon Firebird frame-grabber and delivered a series of deterministic TTL triggers with a field programmable gate array (PCIe 7852R, National Instruments). These triggers included control of the resonant mirror galvanometers, voice coils, stage positioning, laser modulation and blanking, camera fire and external trigger. Some of the core functions and routines in the microscope control software are licensed under a Material Transfer Agreement from Howard Hughes Medical Institute, Janelia Research Campus. The control software code for ctASLM can be requested from the corresponding authors and will be distributed under MTA with HHMI Janelia Research Campus.
ctASLM-v1 Microscope Layout
A schematic layout of the ctASLM-v1 microscope is shown in Supplementary Figure 20 and a part list is provided in Supplementary Table 4. The illumination train consisted of four Coherent Obis lasers (LX 405–100C, LX 488–50C, LS 561–50, LX 637–140C) that were combined with dichroic beam splitters (LM01–427-25, LM01–503-25, LM01–613-25, Semrock), focused through a 30 μm pinhole (P30D, ThorLabs), with a 50 mm achromatic doublet (AC254–50-A, ThorLabs), recollimated with a 200 mm achromatic doublet, (AC254–200-A-ML, ThorLabs), and directed through a 3× Galilean beam expander (GEB03-A). Thus, the initial beams were expanded by a factor of 12× before being focused onto a resonant mirror galvanometer (CRS 4 kHz, Cambridge Technology) with a cylindrical lens (ACY254–50-A, ThorLabs). The mirror galvanometer was used to wobble the light sheet and was driven with a 12V power supply (A12MT400, Acopian), and the 1D focus of the cylindrical lens was relayed with a 200 mm achromatic doublet (AC508–200-A, ThorLabs), passed through a 50:50 polarizing beam splitter and a quarter waveplate, and imaged onto a mirror with a 4× microscope objective (XL Fluor 4×/340, NA 0.28, Olympus Life Sciences). The mirror was mounted on a voice coil with 10 mm of travel, sub-50 nm positioning repeatability, and a sub-3 millisecond response time (LFA-2010, Equipment Solutions). The reflected light was recollected by the same 4× objective, the polarization state was rotated by the second passage through the quarter waveplate (AQWP3, Boldervision) reflected with the cube beam splitter (10FC16PB.7, Newport) and relayed to the illumination objective (Cleared Tissue Objective, NA 0.4 Advanced Scientific Imaging) with 200 mm and 75 mm achromatic doublets (AC508–200-A and AC508–75-A, ThorLabs). To correct for spherical aberrations, 3 mm N-BK7 glass piece (37–005, Edmund Optics) was placed between the 4× objective and the voice coil actuated mirror (Supplementary Figure 21).
The detection arm consisted of an identical microscope objective (Cleared Tissue Objective, Advanced Scientific Imaging), a tube lens (ITL200-A, ThorLabs), a 4-position filter wheel (LB10-WHS4 and a Lambda 10–3, Sutter Instruments), and a sCMOS camera (Flash 4.0, Hamamatsu Corporation). For emission filters, we used 445/20, 525/30, 605/15, and 647 long-pass filters (FF01–444/20–25, FF01–525/30–25, FF01–505/15–25, and BLP01–647R-25, Semrock), for blue, green, red, and far-red, respectively. Both the illumination and detection objectives were immersed in a large chamber that was used to house the specimen. To collect a Z-stack, a voice coil direct drive was used to scan the specimen in a sample-scanning mode (V-522.1AA, C-413.2GA, and C-413.1IO, Physik Instrumente). Sub-volumes were acquired by moving the position of the specimen and the voice coil stage with a 3-axis motorized stage (3DMS and MP-285A, Sutter Instruments). To image a 1mm3 volume using ctASLM takes 17.6 minutes, using 100 ms exposure time, an isotropic step size of 0.425 μm and 18% overlap between stacks for faithful stitching.
ctASLM-v2 Microscope Layout
ctASLM-v2 was built essentially identical to ctASLM-v1, with a few minor differences noted here. A schematic drawing of the setup is provided in Supplementary Figure 22 and a parts list is provided in Supplementary Table 5. The illumination train consisted of three Coherent Obis lasers (LS 561–100C, LX 647–120C, and 488–150C) housed in an Obis LX/LS Laser Box that permits user-selectable input impedances and improved thermal management. All three lasers were combined with dichroic beamsplitters (LM01–503-25, LM01–613-25, Semrock), their polarization rotated with an achromatic half wave plate (10RP52–1B, Newport), focused with a 50 mm achromatic doublet (AC254–50-A, ThorLabs) through a 30 μmpinhole (P30D, ThorLabs), recollimated with a 100 mm achromatic doublet (AC254–100-A, ThorLabs), and expanded an additional 5x with a beam expander (BEO5M, ThorLabs). A cylindrical lens (ACY254–50-A, ThorLabs) was used to create a 1-dimensional laser line focus on a 4 kHz resonant galvo (6SC04KA040–01Y, Cambridge Technology), which was used to wobble the light sheet and was driven with a 12V power supply (A12MT400, Acopian), and the 1D focus of the cylindrical lens was relayed with a 100 mm achromatic doublet (AC254–100-A, ThorLabs), passed through a polarizing polarizing beam splitter (10FC16PB.7, Newport), an achromatic quarter wave plate (10RP54–1B, Newport), and imaged onto a mirror with a 40x microscope objective (CFI S Plan Fluor ELWD 40XC, NA 0.60, Nikon Instruments). The mirror was mounted on a voice coil with 10 mm of travel, sub-50 nm positioning repeatability, and a sub-3 millisecond response time (LFA-2010, Equipment Solutions). The reflected light was recollected by the same 40× objective, the polarization state was rotated by the second passage through the quarter waveplate, reflected with the polarizing beam splitter and relayed to the illumination objective (Cleared Tissue Objective, NA 0.7 Advanced Scientific Imaging) with 80 mm and 200 mm achromatic doublets (AC508–200-A and AC508–75-A, ThorLabs).
The detection arm consisted of an identical NA 0.7 microscope objective (Cleared Tissue Objective, Advanced Scientific Imaging), a 300 mm achromatic doublet (AC500–300-A, ThorLabs), a 4-position filter wheel (LB10-WHS4 and a Lambda 10–3, Sutter Instruments), and a liquid cooled (ThermoCube 400, Solid State Cooling Systems) sCMOS camera (Flash 4.0, Hamamatsu Corporation). For emission filters, we used 525/30, 605/15, and 647 long-pass filters (FF01–525/30–25, FF01–505/15–25, and BLP01–647R-25, Semrock), for green, red, and far-red, respectively. Both the illumination and detection objectives were immersed in a large chamber that was used to house the specimen. To collect a Z-stack, a 480 μmtravel piezo (Piezo Jena NanoX 400) was used to scan the specimen in a sample-scanning mode. Sub-volumes were acquired by moving the position of the specimen and the piezo stage with a 3-axis motorized stage (3DMS and MP-285A, Sutter Instruments).
Sample mounting
Samples were glued with cyanoacrylate (PT09, Pacer Technology, California) or with silicone (GE Silicone 2+) on to a 1×2.5 cm2 glass slide cut from a 2.5×7.5 cm2 microscope slide (Cat. No. 3050, Thermo Scientific). The glass slides were mounted with a custom holder to the sample scanning stage. CAD drawings of the sample holder assembly are shown in Supplementary Figure 23.
Conventional Light-Sheet Microscopy
For conventional low-NA light-sheet imaging, the same optical train was used as in ctASLM-v1, but with the presence of a variable width slit aperture conjugate to the back pupil of the cylindrical lens, and without axial scanning of the illumination beam. The illumination NA was selected such that the entire field of view (870 × 870 μm2) resided within two Rayleigh lengths of the illumination beam. This resulted in a Gaussian beam with a e−2 thickness of ~7 μm, which is in agreement with resolution measurements obtained using sub-diffraction fluorescence beads. With the ctASLM-v2 microscope (NA 0.7 objectives), an axial resolution of 2.5 microns resulted, which we believe is in large part due to the close to diffraction limited detection PSF.
Confocal Microscopy
Imaging of the bone plug was performed on a Zeiss LSM880 equipped with a long working-distance multi-immersion objective (LD LCI Plan-Apo 25× 0.8 NA) as previously described.21, 22 PEGASOS cleared mouse brain were also imaged using a Leica SP8 confocal laser scanning microscope equipped with an HCX APO L20x/0.95 immersion objective. For best performance a 56 μm pinhole size with a pixel dwell time of 1.84 μs was used. A comparison of ctASLM to the data acquired with the Leica SP8 is shown in Supplementary Figure 24.
Sample Preparation
Zebrafish
The zebrafish embryo is a transgenic line (krdl:mCherry) expressing mCherry on the vascular endothelium. A 3-day post fertilization zebrafish embryo is mounted in 2% low-melting agarose (A9045–25G, Sigma-Aldrich) in an FEP (Fluorinated ethylene propylene) tube (FEP HS .029 EXP/ .018 REC, Zeus). The working solvent of the agarose is E3 fish water. The FEP tube is sonicated in 70% Ethanol and then washed with clean distilled water prior to the mounting of the zebrafish embryo. We first pick and leave a zebrafish embryo in a few (~50 μL) E3 media on a petri dish. Next, 5 μL of 0.1% Tricaine is added to anesthetize the zebrafish embryo. About 50 μL of the mixture of the Tricaine and E3 medium is removed, and then the liquid low-melting agarose is added on the zebrafish embryo. The zebrafish embryo in the liquid agarose is aspirated directly into the clean FEP tube with a pipette. Within a few minutes, the agarose is solidified, and the zebrafish embryo is mounted in the FEP tube. For the last step, we use a needle to poke two holes on the FEP tube above and below the zebrafish embryo to increase the gaseous exchange allowing the embryo to breathe. The zebrafish embryo is then transferred into the microscope chamber and ready to be imaged.
CLARITY Brain
Thy1-eYFP brain were extracted and cleared using passive CLARITY23 with the following hydrogel monomer (HM) solution recipe: 1% (wt/vol) acrylamide, 0.05% (wt/vol) bisacrylamide, 4% paraformaldehyde, 1x Phosphate Buffered Saline, deionized water, and 0.25% thermal initiation VA-044 (Wako Chemicals, NC0632395). The transcardiac perfusion was performed with 20 ml HM solution, followed by brain extraction and overnight incubation at 4°C. The Hydrogel was polymerized by incubating the brain samples at 37°C for 3–4 hours, and the clearing was performed by shaking in clearing buffer (4% (wt/vol) SDS, 0.2 M boric acid, pH 8.5 at 37°C, ~3 weeks). After washing with the buffered solution (0.2 M boric acid buffer, pH 7.5, 0.1% Triton X-100), the brains were transferred into 50–87% glycerol solution for refractive index matching (as described previously24) and imaging.
Olfactory Bulb
Double transgenic mice were made by crossing Ai9 mice (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J, Jax Laboratory Stock No. 007909) with FosTRAP mice25 and resulting mice were termed cFosxAi9 mice. Briefly, animals were anesthetized with IP injections of ketamine/xylazine (120mg/kg ketamine and 16mg/kg xylazine). After verification of complete anesthesia via the toe pinch test, the chest cavity was exposed and a syringe needle was inserted into the left ventricle. Mice were perfused with 20mL of PBS solution followed by 20mL of a 4% PFA Solution. After perfusion, mice were decapitated and the brain, including the olfactory bulbs, were carefully dissected out and placed into 4% PFA solution overnight. These samples were then cleared using the PEGASOS method as described in Jing et al.26 All samples were kept at 42°C for this process. Briefly, samples were decolorized for 2 days in 25% Quadrol before undergoing delipidation in a series of 30%, 50%, and 75% tert-Butanol solutions. This was followed by a final delipidation step with TBP (75% tert-butanol and 25% PEGMMA solution). Finally, samples were submerged in BB-PEG (75% benzyl benzoate, 22% PEGMMA, 3% Quadrol) until imaging. These data shown here were based upon a male mouse aged 3.32 months.
Bone Marrow
For imaging of blood vessels or niche cells, double or triple transgenic mice were generated by crossing B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (Jax#007909) mice to Cdh5-creER (B6.CBA-Tg(Cdh5-cre/ERT2)1Rha) or Leptin receptor-cre and alpha-catulin-GFP mice.21, 27 To induce Cre recombinase activity, Cdh5-creER;Rosa26-tdTomato mice received an intraperitoneal injection of 2 mg of tamoxifen dissolved in corn oil every second day for 10 days (5 injections total). For imaging of blood vessels, bones were embedded into OCT embedding medium.21 Using a cryostat, femurs were then cut in half lengthwise and then re-mounted at a 90 degree angle to the original cut, and cut in half lengthwise again, resulting in a quarter bone. For all other samples, intact bone marrow plugs from freshly dissected tibias of 6−8-week-old mice were extruded from the bone using a PFA-filled syringe with a 25-gauge needle and placed directly into 4% PFA solution for 6 h at room temperature. Processing of samples were carried out as previously described in Acar et al.21 Briefly, fixed quarter bones or plugs were then washed in PBS and blocked in whole mount staining medium (PBS with 5% donkey serum, 0.5% NP-40, and 10% DMSO) overnight at room temperature. After blocking, plugs were stained for three days at room temperature with primary antibodies (chicken anti-GFP: GFP-1020, Aves Labs and goat anti-c-kit: BAF1356, R&D Systems) or (rabbit anti tyrosine hydroxylase at 1–250 (EMD Millipore cat#AB152), rabbit anti peripherin at 1–250 (EMD Millipore cat#AB1530), and goat anti cKit at 1–250 (R&D Systems cat# BAF1356)) in staining solution. Then the samples were washed multiple times in PBS at room temperature for one day and put into staining solution containing secondary antibodies (Alexa Fluor 488-AffiniPure F(ab’)2 fragment donkey anti-chicken IgY and Alexa Fluor 647-AffiniPure F(ab’)2 fragment donkey anti-goat IgG) or (Alexa Fluor 488-AffiniPure F(ab’)2 fragment donkey anti-rabbit IgG, Alexa Fluor 647-AffiniPure F(ab’)2 fragment donkey anti-goat IgG) for three days followed by a one-day wash to remove any unbound secondary antibodies. Nerve stained marrow plugs were embedded in 2% low melt agarose in water and were dehydrated in a series methanol washes, followed by refractive index matching for 1 hour at room temperature in 1:2 benzyl alcohol (BA): benzyl benzoate (BB). Quarter bones and bone marrow niche plugs embedded in 2% low melt agarose were prepared using the PEGASOS method.26 Briefly, samples were dehydrated in a series of 30%, 50%, 70%, and 100% Tert-Butanol. Each step was performed for 8 hours at room temperature. Samples were then refractive index matched overnight at room temperature in BB-PEG. BB-PEG was prepared as a 3:1 mixture of benzyl benzoate (BB) (Sigma-Aldrich B6630) and PEGMMA500 (Sigma-Aldrich 447943) supplemented with 3% w/v Quadrol (Sigma-Aldrich 122262). Samples were imaged 1 day to 2 months after initial refractive index matching without significant loss of endogenous TdTomato or Alexa Fluor signal.
Isolated Hippocampus
Thy1-GFP mouse was transcardially perfused with cold PBS first, then 4% PFA in PBS for fixation. The brain was taken out and put in 4% PFA in PBS overnight at 4°C, then 2 mm thickness hippocampal slices was cut with vibratome coronally. The hippocampal slices were then cleared with PEGASOS clearing protocol, previously described in Jing et al.26 Briefly, the slices were decolorized in 25% Quadrol for 1 day, then delipidated in series of 30%, 50%, 70% Tert-Butanol solutions, dehydrated in tB-PEG, and finally cleared in BB-PEG.
Expanded Hippocampus
For imaging expanded hippocampal samples from either Thy1-GFP mice (Supplementary Figures 16 and 18) or WT mice injected with Synaptotag AAV (Supplementary Figure 17) were used.28 The mice were transcardially perfused with cold PBS first, then 4% PFA in PBS for fixation. The brain was taken out and put in 4% PFA in PBS overnight at 4°C, then 0.1 mm thickness coronal hippocampal slices were cut with vibratome. Samples were processed according to the original protein-expansion microscopy procedure.17, 29 Briefly, slices were permeabilized using 5% BSA, 0.5% Triton X-100 in PBS for 1 h at room temperature and stained with rat anti-mCherry (Thermo Scientific M11217, 1:500) (Supplementary Figure 17) or chicken anti-GFP (Abcam ab13970, 1:500) (Supplementary Figure 18) and rabbit anti-Homer1 (Synaptic systems, 160003, 1:200) antibodies (Supplementary Figure 18) in permeabilization buffer for 2 days at 4 C. For Homer1 staining antigen was retrieved by incubation in 1 mM EDTA at 60 C for 45 mins. Slices were washed 3 times in PBS and stained with donkey anti-rat-Alexa594 (Invitrogen, 1:200) (Supplementary Figure 17) or goat anti-chicken-Alexa488 (Invitrogen, 1:200) and goat anti-rabbit-Atto647N (Sigma-Aldrich, 1:200) (Supplementary Figure 18) conjugated antibodies at room temperature for 4 h. After 3 washing steps samples were incubated overnight with a coupling reagent 0.1% acryoyl-X succinimidyl ester (AcX) in PBS.29 For results shown on Supplementary Figure 16 the samples were directly crosslinked with AcX reagent without further antibody staining against GFP. Slices were washed 3 times with PBS for 5 minutes and incubated in polymerization solution (8.6% sodium acrylate, 2.5%, 0.15% N,N – Methylenebisacrylamide, 2 M NaCl, 0.2% APS, 0.2% TEMED, 0.01% TEMPO) for 1 h at 4 C before prior to polymerization at 37 C for 2 h. Embedded tissue was transferred to isotonic proteinase K buffer (50 mM Tris-HCl pH 8.0, 2 M NaCl, 1 mM EDTA, 0.5% Triton X-100, proK 8 u/mL) and incubated for 2 h at room temperature. Complete proK digestion was achieved overnight at room temperature in proteinase K buffer (50 mM Tris-HCl pH 8.0, 1 M NaCl, 1 mM EDTA, 0.5% Triton X-100, proK 8 u/mL). Next day, samples were expanded for 1 h in double-distilled H2O with 3 water changes. Glass holders were covered with 0.1% poly-L-lysine for 1 h, washed 3 times with ddH2O and allowed to air dry for 1 h. Hippocampus region was excised from coronary slices and mounted on a poly-L-lysine coated glass holder using wax as additional support on the sides. For long-term storage and image acquisition samples were transferred to 1 mM NaOH buffered solution.
Neonatal Kidney Preparation
Post-natal day 3 kidneys from an Flk1-GFP (Jackson Laboratories, 017006) mouse were harvested and fixed for 2h at room temperature in 4% paraformaldehyde in PBS. Procedures were performed according to UTSW-IACUC-approved guidelines. Fixed kidneys were incubated with tert-Butanol/water mixtures of increasing alcohol concentrations (30%, 50%, 70%, 80%, 96%, and twice 100%) with 2% Triethanolamine. The kidneys were then left in BABB (benzyl alcohol and benzyl benzoate in a 1: 2 volume ratio) with 10% Triethanolamine until tissue cleared.6 For prolonged storage in clearing medium (> 2 days), samples were stored at 4⁰C. Given that the tissue was harvested from neonatal mice, and no genotyping was peformed, the gender of the mouse is unknown.
Stitching and 3D visualization
Image processing was performed on a Windows 10-based workstation equipped with two Intel Xeon Gold 5120 CPUs, 1 TB of RAM, an NVIDIA Quadro P6000 GPU. To stitch the sub-volumes, the Fiji-based plugin BigStitcher30 was used. Image analysis was performed with Fiji31 and MATLAB (Mathworks), and 3D renderings were produced with ChimeraX or Arivis.
Deconvolution
We tested both Richardson-Lucy deconvolution and Blind Deconvolution (Supplementary Table 6 list details for each data set shown in this manuscript). When using Richardson-Lucy algorithm to deconvolve the images, the point spread function was extracted from the image of a single bead, centered and background subtracted and 3D-deconvolution was performed with 20 iterations of the Richardson-Lucy algorithm (Microvolution). For the Blind deconvolution, we use deconvblind function in MATLAB (Mathworks) and the following procedure. Prior to the deconvolution, we rotationally average along the optical axis the experimental image of a single 200 nm bead. We measure the mean value of the background in the rotationally average PSF and subtract it from the PSF image. In order to improve this experimental PSF, we first deconvolve the raw image with the rotationally averaged and background-subtracted PSF for 5 iterations. This yields a restored PSF from the devonvblind routine (i.e. the resulting PSF will more closely approximate the real PSF in the measurement). In a second step we then use this restored PSF to perform blind deconvolution on the raw image data, which then results in the final deconvolved image. For biological data we used 5–10 iterations, whereas for the bead analysis, we limited the number of iterations to 5. The raw images and the experimental PSF were resampled 2x with bicubic algorithm with ImageJ before the deconvolution, which was particularly necessary for the ctASLM 1 data (NA 0.4), which has not much Nyquist oversampling in its data acquistion. ctASLM2 was designed with more oversampling (larger optical magnification), hence computational resampling before deconvolution is not necessary.
Data analysis
HSC Detection
HSCs were detected as previously described.21 Briefly, individual alpha-catulin-GFP+, Kit+ HSCs were identified manually using the orthoslicer function of Bitplane Imaris v9.2.1 software. HSC coordinates and size were interactively annotated using the Imaris spots function in manual mode. Kit and LepR TdTomato surfaces were generated in Imaris using iso-surface function based on an empirically chosen intensity threshold.
Glomeruli Detection
In an effort to automatically detect the abundance of glomeruli throughout the entire neonatal kidney tissue, sub-volumes (1024 × 1024 × 470 pixels) were retrieved from the stitched volume (3042 × 4018 × 2757 pixels) and loaded into MATLAB. Sub-volumes were subjected to a three-dimensional Gaussian blur of 20 pixels before individual glomeruli were detected using an auto-adaptive watershed algorithm implemented in MATLAB.32 This algorithm uses a difference of Gaussians to suppress features that are either too large or small, here defined as 100 and 10 pixels, respectively. Segmented objects touching the border of the sub-volume were removed. After identification of glomeruli, the encompassing region was again loaded into MATLAB, and the full resolution was subjected to the same watershed algorithm, but with a difference of Gaussian of 10 and 2 pixels, respectively.
Spine Detection and Clustering
We detected spines using previously described software.20 Images were first deconvolved for 10 iterations using the MATLAB function deconvblind(), which uses a maximum likelihood algorithm to infer the fluorescence distribution in the sample. Following a gamma transform of 0.4 and 3D hole filling, a triangle mesh representing the cell surface was created as an isosurface at the Otsu threshold value of the image. The mesh was next decomposed into approximately convex patches using the parameters previously described,20 except that merging via the triangle criterion was disabled and merging via the line-of-sight criterion was performed more conservatively with a parameter of 0.8. Training data for patch merging and patch classification machine-learning models were then generated from nine non-overlapping images of size 200 pixels in the lateral dimensions and approximately 150 pixels in the axial dimension. These nine images were cropped from eight larger images. Using leave-one-out cross validation on patches labeled as certainly spines or certainly not spines, we calculated that our spine detection workflow had a precision of 0.86 and a recall of 0.88.
We performed an unsupervised clustering of detected spines into 15 clusters using agglomerative hierarchical clustering. For each cluster, we calculated the median of each of the morphological feature vectors used for clustering and plotted the spine closest to that median (Figure 2b). Only the clusters with a median spine attached to the neuron body were plotted. Principal component analysis was performed on these same features. We defined the spine neck surface area, as the patch closure surface area. We previously defined20 the closure surface area as the additional surface area needed to minimally generate a closed polyhedron from the open polyhedron comprising the dendritic spine. We also defined the variation from a sphere by fitting each spine to a sphere and measuring the ratio of the standard deviation of distances from the sphere at each mesh face to the sphere radius.
Segmentation of Neurons
The mouse brain volume was tiled in sub-volumes for parallel processing, and the signal was enhanced using a volumetric Laplacian-of-Gaussian transform with an isotropic scale of 1.5 voxels. The resulting volume was then transformed into a triangular mesh using an empirically chosen threshold. In order to reduce clutter, only the connected components with volume superior to the 99 percentiles of all connected component volume are displayed in Figure 1f. The algorithm is implemented using the open-source G’MIC software (https://gmic.eu), and the mesh was imported into Meshlab33 for coloring and image capture.
Statistics and Reproducibility
For statistical analysis, we report the mean, standard deviation, and number of observations. For PSF measurements of the different microscopes and immersion media, the number of measurements ranged from 5–10 for light-sheet microscopy and three for airy scan microscopy. The values are listed in Table 1 in the online methods.
Repeatability: Generally, only one sample was used for each imaging experiment (e.g. imaging of whole mouse brain, kidney, etc.), unless otherwise noted. Two expanded brain slices were prepared and imaged with either the NA 0.4 or NA 0.7 version of ctASLM. The number of times that each imaging operation was repeated for each figure is indicated in Supplementary Table 6 and amounted to one in most cases. Segmentation of neurons was perfomed on one data set from a CLARITY brain sub-volume and on one data set from a hippocampal slice sub-volume that was cleared with PEGASOS. Synaptic spines were detected on two different sub-volumes from a CLARITY brain.
Supplementary Material
Acknowledgements
We would like to thank the Cancer Prevention Research Institute of Texas (RR160057 to R.F., and R1225 to G.D.) for their generous funding, as well as the National Institutes of Health (F32GM116370 and K99GM123221 to M.K.D., and R01GM067230 to G.D., R01AG055577 and R01NS056224 to I.B., R01DK118032 and R01DK099478 to D.M., R01DC015784 and R21NS104826 to J.M. and R33CA235254 to R.F.). We are grateful to the Live cell imaging facility at UT Southwestern for access to the Zeiss Airy Scan confocal microscope (which is supported by 1S10OD021684-01 to Katherine Luby-Phelps). We would like to thank David Saucier and Dr. James Amatruda for providing the zebrafish specimens, and Dr. Etai Sapoznik for his assistance with CAD design.
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
The datasets acquired for this study are available from the corresponding authors upon reasonable request.
Code Availability
The instrument control software can be requested for academic use from the corresponding authors and will be delivered under material transfer agreements with Howard Hughes Medical Institute and UT Southwestern Medical Center.
References
- 1.Villani A-C et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K et al. Rapid adaptive optical recovery of optimal resolution over large volumes. Nature Methods 11, 625–628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson DS & Lichtman JW Clarifying Tissue Clearing. Cell 162, 246–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welf Erik S. et al. Quantitative Multiscale Cell Imaging in Controlled 3D Microenvironments. Developmental Cell 36, 462–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huisken J Optical Sectioning Deep Inside Live Embryos by Selective Plane Illumination Microscopy. Science 305, 1007–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Dodt H-U et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nature Methods 4, 331–336 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Stefaniuk M et al. Light-sheet microscopy imaging of a whole cleared rat brain with Thy1-GFP transgene. Scientific Reports 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pende M et al. High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nature Communications 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomer R et al. SPED Light Sheet Microscopy: Fast Mapping of Biological System Structure and Function. Cell 163, 1796–1806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buytaert JA & Dirckx JJ Tomographic imaging of macroscopic biomedical objects in high resolution and three dimensions using orthogonal-plane fluorescence optical sectioning. Appl Opt 48, 941–948 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Santi PA et al. Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. Biotechniques 46, 287–294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean Kevin M., Roudot P, Welf Erik S., Danuser G & Fiolka R Deconvolution-free Subcellular Imaging with Axially Swept Light Sheet Microscopy. Biophysical Journal 108, 2807–2815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botcherby EJ et al. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proceedings of the National Academy of Sciences 109, 2919–2924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao L Extend the field of view of selective plan illumination microscopy by tiling the excitation light sheet. Optics Express 23 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Voigt FF et al. The mesoSPIM initiative: open-source light-sheet mesoscopes for imaging in cleared tissue. bioRxiv, 577122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hörl D et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nature Methods (2019). [DOI] [PubMed] [Google Scholar]
- 17.Gao R et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Q, Martin BL, Matus DQ & Gao L Imaging multicellular specimens with real-time optimized tiling light-sheet selective plane illumination microscopy. Nature Communications 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crane GM, Jeffery E & Morrison SJ Adult haematopoietic stem cell niches. Nature Reviews Immunology 17, 573–590 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Driscoll MK, Welf ES, Dean KM, Fiolka R & Danuser G Cell morphological motif detector for high-resolution 3D microscopy images. bioRxiv, 376608 (2018). [Google Scholar]
- 21.Acar M et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comazzetto S et al. Restricted Hematopoietic Progenitors and Erythropoiesis Require SCF from Leptin Receptor+ Niche Cells in the Bone Marrow. Cell Stem Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomer R, Ye L, Hsueh B & Deisseroth K Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nature Protocols 9, 1682–1697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migliori B et al. Light sheet theta microscopy for rapid high-resolution imaging of large biological samples. BMC Biol 16, 57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guenthner Casey J., Miyamichi K, Yang Helen H., Heller HC & Luo L Permanent Genetic Access to Transiently Active Neurons via TRAP: Targeted Recombination in Active Populations. Neuron 78, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing D et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Research 28, 803–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorensen I, Adams RH & Gossler A DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113, 5680–5688 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Xu W & Sudhof TC A Neural Circuit for Memory Specificity and Generalization. Science 339, 1290–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tillberg PW et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology 34, 987–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preibisch S, Saalfeld S & Tomancak P Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Applegate KT et al. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. Journal of Structural Biology 176, 168–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cignoni P et al. MeshLab: an Open-Source Mesh Processing Tool. Eurographics Italian Chapter Conference 2008, 129–136 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.