Abstract
The impact of the protein-rich in vivo environment on targeted binding of functionalized nanoparticles has been an active field of research over the past several years. Current research aims at better understanding the nature of the protein corona and how it may be possible for targeted binding to occur even in the presence of serum. Much of the current research focuses on nanoparticles targeted to particular cell receptors or features with the aim of cellular uptake. However, similar research has not been performed on nanoparticles that are targeted to non-protein disease features, such as hydroxyapatite (HA). HA is a crystalline calcium-phosphate mineral that is present in large quantities in bone, and in smaller quantities in diseased cardiovascular tissue in cases of atherosclerosis or various stenoses. Our work aims to gain a better understanding of the behavior of PEGylated, peptide-coated superparamagnetic iron oxide nanoparticles (SPIONs) in a biologically-relevant high-protein environment (50% serum). We first determined that specific binding to HA occurs at significantly higher rates than non-specific binding in the absence of serum protein. We then examined nanoparticle interactions with serum proteins, including determination of the relative quantities of protein in the hard vs. soft protein corona. Finally, we examined specific and non-specific binding of targeted SPIONs in 50% serum, and determined that targeted binding may still occur with significant (p < 0.05) selectivity. We hypothesize that this may be because the nature of the binding interactions between the peptides and the HA are, by definition, less specific than the protein-protein interactions required for nanoparticles to bind to specific cells or cell features. These results suggest that these targeted SPIONs may be further developed for use in early detection of heart diseases such as atherosclerosis and aortic stenosis.
Keywords: iron oxide nanoparticles, targeted nanoparticles, serum, protein corona, non-specific binding
Graphical Abstract
Introduction
Nanomedical materials, whether for therapeutic or diagnostic use, are often intended to target and impact a specific tissue or location in vivo. To this end, several methods of chemically functionalizing nanoparticle surfaces, thereby targeting them to specific cells or tissues, have been developed. These methods include attachment of biological or biologically-derived molecules to the nanoparticle surface, including antibodies, peptides, and aptamers intended to interact with specific proteins.1–5 For some applications (e.g. cancer treatment) these specific interactions may be intended to cause uptake of drug-carrying nanoparticles into cells via receptor-mediated endocytosis.6–9 For other applications (e.g. disease detection) it is simply necessary for the interaction between the targeting moiety and the target to cause association of the nanoparticle with the disease target.10,11 These targeting methods have shown high rates of success in vitro; however, they have often failed in vivo.12–14
One of the primary causes of this in vivo failure is the formation of the so-called protein corona that occurs almost instantaneously upon exposure of nanoparticles to protein-rich media such as blood serum.15–17 It has been established that the composition of the protein corona in terms of protein type, concentration, and conformation varies depending on the specific surface functionalization of the nanoparticles,18,19 and can define their biological fate including their biodistribution, cellular uptake, and immune response.20,21 Additionally, the presence of the corona can block targeting moieties from interacting with their targets, causing a loss of targeting efficiency.22,23 Previous results in the literature indicate that the protein corona can inhibit the targeting capability of nanoparticles by as much as 94%, even in a solution that is only 10% serum.22 Furthermore, it has been established that the protein corona consists of two layers: (1) the hard corona close to the nanoparticle surface, comprised of higher affinity proteins that may irreversibly bind to nanoparticles, and (2) the soft corona formed by loosely affiliated, lower affinity proteins that are reversibly bound and may exchange over time with proteins in the surrounding solution.24,25 It has been hypothesized that the hard protein corona, which is both closer to the nanoparticle surface and more tightly associated, may have a greater impact on nanoparticle targeting capabilities.20,26
To prevent deposition of serum proteins, nanoparticles are commonly coated in an antibiofouling layer such as poly(ethylene glycol) (PEG).27,28 PEG is thought to display good resistance against nonspecific protein adsorption in large part due to its extensive formation of hydrogen bonds with surrounding water.29 However, recent findings have suggested that protein adsorption to the nanoparticle surface occurs even in nanoparticles that have been coated in PEG.30,31 Studies show that, even at low concentrations of serum (10%), protein deposition still occurs onto PEGylated nanoparticle surfaces.32 Given that blood is comprised of approximately 50–55% protein-rich serum, this would present some obvious problems in the in vivo environment.
Alternative strategies to maintain cell-targeting abilities of nanoparticles in a protein-rich environment have emerged in recent years.13,33,34 Most of these strategies aim to address the problem of the protein corona in order to still allow cellular uptake of nanoparticles, thus facilitating drug delivery for applications including cancer treatment. Cellular uptake of nanoparticles often requires specific protein-protein or peptide-protein interactions to occur.3,35,36 These interactions are highly susceptible to blockage by the protein corona. However, the effect of the protein corona, and various strategies for circumnavigating this effect, have not been so thoroughly investigated for other applications or other targeting strategies that do not require cellular uptake. Targeting of non-protein disease features, for example, has not been examined at all.
One such non-protein disease feature is hydroxyapatite (HA). HA is a non-organic calcium phosphate crystal and a major component of bone. It is also a prominent disease feature in a variety of cardiovascular diseases, including aortic stenosis and atherosclerosis.37,38 In both aortic stenosis as well as atherosclerosis, the deposition of HA in vascular or valve tissues –where there is no such mineral in non-diseased tissue – is well-documented, even in early disease stages.37 Thus, detection of HA in cardiovascular tissue could serve as an early marker for disease detection, and nanoparticles targeted to HA could aid in the early detection and assessment of cardiovascular disease.
To test the ability of nanoparticles targeted to a non-protein disease feature to maintain specific targeting capabilities even in the presence of serum proteins, we conjugated two different HA-binding peptides to superparamagnetic iron oxide nanoparticles (SPIONs). SPIONs were selected as the core of the nanoparticle because of their well-documented usage as magnetic resonance imaging (MRI) contrast agents. Because of their superparamagnetic nature, SPIONs are able to act to enhance contrast in T2-weighted MR images.39,40 SPIONs have been extensively researched over the past decades as MRI contrast agents for a variety of biomedical applications.39,41,42
SPIONs conjugated to a Hydroxyapatite Binding Peptide (HABP), identified via ribosome display and previously characterized,43 were tested in parallel with SPIONs conjugated to OPN, a highly-phosphorylated peptide derived from the bone-binding protein osteopontin.44 We assessed the specific and non-specific binding of these functionalized SPIONs in sodium phosphate buffer before characterizing the interaction of the nanoparticles with serum proteins in a 50% serum environment, to more accurately imitate the protein-rich in vivo environment than similar studies using 5–10% serum. We additionally assessed some characteristics of the protein corona of the particles such as the quantity of protein in the hard vs. soft corona to develop a better understanding of factors that may influence the ability of nanoparticles in general to bind to their targets in the presence of serum proteins.
Our findings demonstrated that, while our peptide-coated SPIONs show substantial interactions with serum proteins and the immediate development of a protein corona, the majority of the observed interactions with serum proteins are loose associations (i.e., part of a soft corona). This has allowed these peptide-linked SPIONs to maintain successful binding to their target even in the presence of serum. We hypothesize that this is due to the primarily charge-based interaction between the targeting peptides and HA,43,44 which may help to avoid some of the typical problems observed with relying on specific protein-protein interactions to allow for targeted binding in high-protein environments. This work has implications in the selection of targets for future work on targeted nanoparticles, in addition to offering proof-of-concept that these HA-targeted SPIONs may have utility in the detection of heart disease due to their superparamagnetic nature and the well-documented use of SPIONs for MRI contrast.
Materials and Methods
Materials
Iron tri(acetylacetonate), 1,2-tetradecanediol, oleic acid, oleylamine, dibenzyl ether, citric acid, diethyl ether, 2-methoxyethylamine, N-(3- Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-Hydroxysuccinimide (NHS), 1,2-dichlorobenzene, and N,N’-dimethylformamide are all purchased from Sigma-Aldrich (St. Louis, Missouri). The LIVE/DEAD Assay kit was purchased from ThermoFisher Scientific (Rockford, Illinois). Human Umbilical Cord Vascular Endothelial Cells (HUVECs) were acquired from Lonza (Portsmouth, New Hampshire). Where it was purchased, HA powder was also purchased from Sigma-Aldrich (St. Louis, Missouri). All functionalized PEG reagents are obtained from Nanocs (Boston, Massachusetts). Slide-A-Lyzer dialysis cassettes are acquired from Thermo Scientific (Rockford, Illinois). Fetal Bovine serum is obtained from Hyclone (Logan, Utah). Media used to culture cells in these experiments is also obtained from Lonza (Portsmouth, New Hampshire).
Iron oxide particle synthesis and surface modification
Oleic acid-coated iron oxide nanoparticles are synthesized via high-temperature thermal decomposition as previously described.45 Briefly, the following reagents are mixed and stirred magnetically under nitrogen flow: 2mmol iron(III) tri(acetylacetonate), 10mmol 1,2-tetradecanediol, 6mmol oleic acid, 6mmol oleylamine, and benzyl ether. The resulting nanoparticles are precipitated in excess ethanol and pulled down using a magnet. The precipitated particles are washed with ethanol, dissolved in hexane, and centrifuged to remove aggregates. The SPIONs, which at this point are oleic acid-coated, are dried in a vacuum oven.
A ligand exchange is performed to replace oleic acid with citric acid. Oleic acid-coated nanoparticles and citric acid are dissolved in a 1:1 mixture of dichlorobenzene and N,N-dimethylformamide. The mixture is heated to 100˚C and left stirring for 24 hours. Citric acid-coated nanoparticles (CA-SPIONs) are precipitated in diethyl ether and washed with acetone and diethyl ether before drying in a vacuum oven.
CA-SPIONs are further coated with a layer of bifunctionalized poly(ethylene glycol) (PEG) using a protocol similar to those previously described.45,46 CA-SPIONs are dissolved in pH 9 sodium phosphate buffer. N-hydroxysuccinimide ester (NHS) and 1-ethyl-3-(3-dimethylaminoproply) carbodiimide (EDC) are agitated for 24 hours at room temperature with amine-terminated PEG. PEG was added at a 1:4 molar ratio of NH2-PEG3.4k-iodoacetyl to NH2-PEG2k. The resulting PEGylated SPIONs are purified by dialysis. SPIONs not functionalized to bind to HA are PEGylated with a 1:4 molar ratio of NH2-mPEG3.4k to NH2-mPEG2k.
Two different HA-binding peptides are purchased from Peptide 2.0 (Chantilly, Virginia). HA Binding Peptide (HABP – SVSVGMKPSPRPGGGKC) and an osteopontin-derived peptide (OPN - DDVDD(pT)DD(pS)HQ(pS)DEC) are linked to SPIONs via an iodoacetyl-thiol interaction to produce either HABP-SPIONs or OPN-SPIONs. Peptides are added to iodoacetyl-PEG-SPION solutions in appropriate concentrations to target the addition of 25 peptide residues per SPION. Peptide addition reactions are allowed to run overnight at 4˚C, and conjugated particles are purified from excess peptide by dialysis against deionized water at 4˚C.
A schematic of the overall SPION functionalization process can be seen in Figure 1 below.
Figure 1:
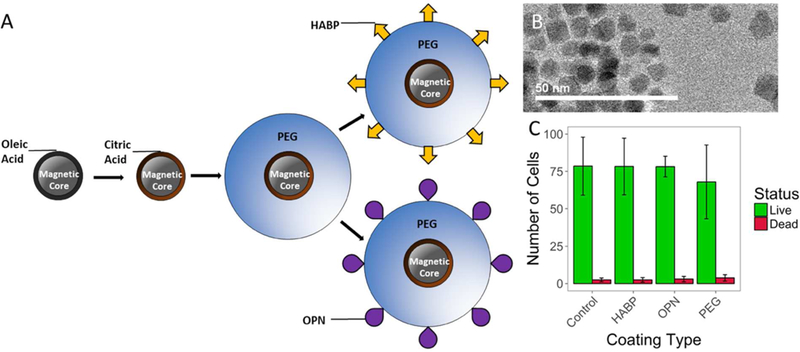
(A) Schematic of particle fabrication and functionalization. A ligand exchange is performed on magnetic SPION cores coated in oleic acid to produce SPIONs coated in citric acid (CA-SPIONs), and citric acid coated SPIONs are PEGylated using EDC/NHS chemistry. Targeting peptides HABP and OPN are added via an iodoacetyl/thiol interaction between iodoacetyl-PEG and peptides with cysteine residues.TEM images of ~10nm SPION cores. SPION core sizes were quantified in ImageJ, which yielded an average core size of 9.39 ± 0.98nm (mean ± standard deviation). (C) Results from a LIVE/DEAD assay indicating that HABP-, OPN-, and PEG-SPIONs do not show higher rates of cell death after incubation than a control sample incubated without nanoparticles.
LIVE/DEAD Assay
A LIVE/DEAD assay is performed to assess potential cytotoxicity of functionalized SPIONs. Briefly, HUVECs are cultured as recommended on tissue-coated plastic. When cells are visibly confluent, enough SPION solution to reach a concentration of 100 parts per million (ppm) is diluted in standard cell media and added to the HUVECs. SPIONs are incubated with cells at 37°C for 2 hours. Cells are rinsed with sterile PBS and stained as proscribed in the LIVE/DEAD kit protocol, with optimal concentrations of 1µM Calcein AM and 2µM Ethidium homodimer-1 having been previously determined in other HUVEC samples. Cells are imaged on an Olympus IX81 microscope equipped with the appropriate fluorescent filters. Live and dead cells are quantified using ImageJ, and subsequent statistical analysis is performed in R.
Dynamic Light Scattering, Zeta Potential, BCA Assay Characterization, and TEM
The size and zeta potential of HA-SPIONs are measured via dynamic light scattering (DLS) in pH 9 sodium phosphate buffer. All measurements are made with a 90Plus Dynamic Light Scattering Analyzer (Brookhaven Instruments) at a 90˚ detection angle. All samples are diluted 10x before measurement, as it has been previously indicated that high SPION concentrations may impact DLS readings.47 DLS measures the impact of nanoparticle movement on light scattered through a solution of small nanoparticles, and from these measurements, the size of the particles causing light to scatter can be inferred. DLS results are output from the instrument in the form of a histogram or a correlation function (data not shown).48–51The bicinchoninic acid assay (BCA assay) is used to quantify success of peptide addition to SPIONs. For the BCA, each peptide (HABP or OPN) is used to construct a calibration curve rather than the much-larger bovine serum albumin provided, in order to obtain a more accurate measurement of the quantity of peptide present.
The shape and size of the SPIONs are examined on a JEOL 2100 (200 kV) transmission electron microscope (Tokyo, Japan). The samples are prepared by placing a few drops of nanoparticle suspension onto a standard carbon-coated copper grid and then drying at ambient conditions. The TEM images are processed in ImageJ (NIH) to determine particle size and size distribution.
MR Measurements
The R2 relaxivity of the SPIONs at 3 Tesla was determined by imaging tubes with DI water plus concentrations from 0 to 100ppm of HABP-, OPN-, or PEG-SPIONs with a clinical MR imager (GE Signa HDx).
T2 measurements were obtained with a previously-described multi-echo three-dimensional Carr-Purcell-Meiboom-Gill sequence with modifications to alleviate stimulated and indirect echoes, thereby enabling accurate T2 quantitation.52–54 Data was acquired using this sequence at 16 equispaced echo times with TE ranging from 7.7 to 123 ms. Other sequence parameters were TR = 1.2sec, 1mm slice thickness, 68 slice phase encodes, and 96 × 96 matrix over a 7 cm field-of-view. A mono-exponential signal decay model (!(#$) = ‘( )*+,/+., where s the measured signal and ‘( the proton density) was fitted to the signal measured in regions-of-interest placed in the center of each tube using a non-negative least squares routine implemented in MATLAB (version R2016b, Natick, MA), thereby providing an estimate of the relaxation rate for each SPION at each concentration. R2 relaxivity was subsequenctly determined from the linear fit of relaxation rate vs. SPION concentration for each type of SPION using Excel.
In Vitro Binding Tests in Sodium Phosphate Buffer
HA-SPIONs are shaken at room temperature (RT) for 30 minutes with HA in Costar Spin-X centrifuge tube filters over a range of HA quantities (0–200mg). After incubation, all samples are centrifuged at 3000 rpm for 3 minutes. The absorbance of the filtrate is measured at 510nm to determine the relative percentage of particles bound to the HA. Particles bound to HA do not successfully pass through the filter, while unbound particles pass through easily. These experiments are repeated over a range of pH values (7.1–7.8).
The specificity of HA-SPION binding to HA is tested by comparing results from HA binding tests with results from experiments testing off-target binding to cholesterol, collagen, fibrin, fibrinogen, and human vascular endothelial cells (HUVECs). A collagen-coated six-well plate is used to test off-target HA-SPION binding to collagen, fibrin, and fibrinogen. For both the fibrin and fibrinogen gels, fibrinogen (Bovine plasma, Sigma-Aldrich) in pH 7.1 sodium phosphate buffer is incubated at RT for 10 minutes to allow binding to the collagen surface coating. Thrombin in PBS (Bovine plasma, Sigma-Aldrich) is used to crosslink fibrinogen into a fibrin gel for 1 hour. For the HA-coated plates, a protocol similar to that developed by de Bruyn et al. is adapted;55 in short, two working solutions are prepared (12mM CaCl2 and 7.5mM Na2HPO4), and are added in appropriate ratios to stoichiometrically yield 25mg of HA (Ca10(PO4)6(OH)2) upon full conversion of the salt components to a crystalline form. These solutions are mixed and allowed to dry on top of the fibrin gels, producing a thin HA coating that effectively adheres to the plate. All wells are washed twice with buffer and allowed to dry under vacuum.
When the wells have dried, a 10x diluted SPION solution is added to each well and incubated for 30 minutes at RT. After incubation, samples are pulled from each well and filtered through a 0.2µm filter. The absorbance of each sample is measured at 510 nm and compared to the absorbance of prepared samples of known SPION concentration or quantity to assess the number of SPIONs present after binding has been allowed to occur.
To test HA-SPION binding to cholesterol, one well of a tissue culture-treated six-well plate is filled with cholesterol in ethanol solution, and the ethanol is allowed to completely evaporate. After incubation, samples are pulled from each well and filtered through a 0.2µm filter. The absorbance of each sample is measured at 510 nm and compared to the absorbance of prepared samples of known SPION concentration or quantity to assess the number of SPIONs present after binding has been allowed to occur.
To test binding against HUVECs, the cells are cultured at recommended conditions in 6-well plates. After a week in culture, cells are pre-incubated at 4˚C for 10 minutes to halt endosomal trafficking. 50µL of an HA-SPION solution is added to each well of the 6-well plate, and plates are incubated at 4ºC for 30 minutes. Absorbance at 510nm is measured for samples of the media taken after incubation to determine the percentage of HA-SPIONs that remain in solution and do not adhere to HUVECs.
SPION binding to Serum Proteins
HABP-SPIONs, OPN-SPIONs, and PEG-SPIONs are each mixed 1:9 with a 50% fetal bovine serum (Hyclone) solution. The solution is transferred to a cuvette and allowed to sit at room temperature for 24 hours. DLS measurements are taken at specific time points (0 and 30 minutes, 1, 2, 4, 8, 12, and 24 hours) to observe any alterations in effective diameter of the SPIONs. These results are compared with samples that have been incubated similarly with DI water. These experiments are repeated without a 24-hour incubation at various pH levels (6–9).
To assess the amount of protein that is bound to the SPIONs, HABP-SPIONs, OPN-SPIONs, and PEG-SPIONs are each mixed 1:9 with 50% serum solution and incubated at room temperature for two hours. None of the prepared solutions show visible signs of aggregation. All particles are washed by centrifugation 3x to remove unbound serum proteins (17900g for 30 min at 4˚C) and resuspended in 200∝L of DI water. DI water was chosen as a diluent and resuspension agent in these studies because the presence of salt can screen the repulsion between nanoparticles and induce aggregation;56 for these studies, it was important to limit aggregation as much as possible during resuspension, in order to obtain an accurate final assessment of the final number of proteins per particle after all wash steps. Samples are removed after each washing step for subsequent measurement using the BCA assay, where they are compared to samples incubated in DI water as a control. Aliquots of samples incubated with either 50% serum or DI water and washed 3x are mixed 4:1 with Laemmli sample buffer and boiled in water for 5 minutes. These samples are loaded on a 4–20% Tris-glycine gel in Laemmli running buffer and resolved at 150V for 50 minutes at room temperature. After electrophoresis, the gel is fixed with 10% v/v acetic acid and 30% v/v ethanol for at least 30 minutes and stained with silver nitrate stain (Pierce) according to the manufacturer’s protocol. The stained gel is scanned using a VersaDoc Scanner from Bio Rad and the scanned image is imported into ImageJ and cropped.
In Vitro Binding Tests in Serum
Serum absorbs light at ~510nm, so an orthogonal method is used to measure SPION binding to specific surfaces in serum, although the protocol remains identical aside from the method of measurement. To coat plates in cholesterol, wells of a tissue culture-treated six-well plate are filled with 25mg cholesterol in ethanol solution, and the ethanol is allowed to completely evaporate. To coat plates in collagen, fibrin, fibrinogen, and HA, a collagen-coated six-well plate is used. For both the fibrin and fibrinogen gels, 25mg fibrinogen (Bovine plasma, Sigma-Aldrich) in DI water is incubated at room temperature for 10 minutes to allow binding to the collagen surface coating. Thrombin in PBS (Bovine plasma, Sigma-Aldrich) is used to crosslink the fibrinogen into a fibrin gel for 1 hour. To further add a layer of crystalline HA, the same protocol as previously described is used. All wells are washed twice with buffer and allowed to dry under vacuum.
Each coated well is incubated in a 50% fetal bovine serum solution for 30 minutes, while functionalized SPIONs (HABP-SPIONs, OPN-SPIONs, PEG-SPIONs, and CA-SPIONs) are simultaneously incubated at room temperature for 30 minutes. Serum is removed from the wells, and the SPIONs are diluted 1:9 in a 50% serum solution and incubated on the coated wells for 30 minutes at room temperature. After incubation, the SPION solutions are removed and wells are washed with DI water. SPIONs that remain bound are stained with a Prussian Blue stain for 35 minutes. Wells are rinsed again, allowed to dry under vacuum, and imaged on an Axiovert S100 microscope. To quantify the number of particles remaining, these images are compared to images of these same, identically-produced surfaces with known quantities of nanoparticles deposited on the surfaces. For example, fibrin surfaces with known quantities of PEG-SPIONs deposited onto the surface were used to act as a “calibration curve” for experimental samples of PEG-SPIONs deposited onto fibrin surfaces. From this information for all unique combinations of functionalized SPIONs and surfaces, the number of particles (as a function of the “image darkness” due to the stain) can be estimated. Image greyscale analysis is performed in ImageJ.
Results and Discussion
We successfully fabricated nanoparticles engineered to have both stealth and targeting properties. SPIONs with a core size of ~10nm were used, as confirmed by TEM (Figure 1B). This core size was selected to reflect the size of SPIONs that are typically developed for MR applications in the literature.57–59 SPIONs are often grouped into three categories based on their size: (1) standard SPIONs (5–180nm), (2) ultrasmall SPIONs (10–50nm), and (3) very small SPIONs (<10nm).60 The 10nm core size was chosen because the most commonly used MRI agents are ultrasmall SPIONs; subsequent to all coating steps described below, our SPIONs with a core size of ~10nm remain categorized as “ultrasmall.”58
SPIONs produced by high-temperature thermal decomposition are coated in oleic acid (Figure 1A), which is highly hydrophobic. A ligand exchange was performed with citric acid to produce hydrophilic SPION cores (CA-SPIONs) that can be further functionalized by well-characterized methods utilizing the large number of available carboxylic acid residues present on the modified SPION surface. CA-SPIONs are further functionalized with PEG. PEG is purported to aid in making these “stealth” nanoparticles by preventing protein deposition and therefore recognition by the immune system.27–29 However, these claims have been refuted or confounded by many recent studies showing that proteins present in blood or serum bind to PEGylated nanoparticles.30,31 Nevertheless, PEG was used in this study in part to create a more neutral surface that could be compared with the more highly charged (anionic) peptide-coated particles. PEGylated SPIONs coated with a mono-functionalized PEG (a 4:1 ratio of mPEG2k – NH2 and mPEG3.4k – NH2) were used as a control in studies related to serum protein adsorption. SPIONs to which targeting moieties were added were coated with hetero-bifunctionalized PEG (a 4:1 ratio of mPEG2k – NH2 and iodoacetyl – PEG3.4k – NH2) to allow for later attachment of peptides via covalent iodoacetyl-thiol interactions. Two peptides were selected for testing, as they were expected to have widely different properties. Ribosome-derived HABP is a synthetic peptide that, in addition to binding to HA, may show some binding to metallic surfaces.61 OPN is a highly-phosphorylated peptide that has been derived from osteopontin, a bone-binding sialoprotein that plays roles in biomineralization, bone remodeling, and various immune functions.62–64 Although both of these peptides bind to HA, their differing properties in terms of amino acid composition and origin (i.e. synthetic vs. biologically-derived) may mean that they have different properties in terms of interactions with serum proteins.
As expected, TEM images of HABP- and OPN-SPIONs did not show any significant difference in size from the uncoated SPIONs (data not shown). PEG is known to be highly radiation sensitive, and will thus evaporate rapidly in the TEM beam. TEM measurements of the core were captured to ascertain the specific core size, which is challenging using dynamic light scattering (DLS); SPION cores must be modified with citric acid to be soluble in water, and therefore the cores alone are not suitably measured with DLS, where particles must be in solution. However, while TEM is able to accurately image particle cores, the process of preparing samples for TEM can be destructive, and imaging of both a polymer layer and a protein coating is not often observed in SPION literature as a method for obtaining useful particle size information in solution.65–68 For these reasons, DLS, zeta potential measurements, and the BCA assay were chosen to assess the success of SPION functionalization (see below).
A LIVE/DEAD assay was performed to examine the cytotoxicity of various functionalized SPIONs (Figure 1C). HUVECs were chosen for these studies because SPIONs entering the body via injection are most likely to encounter endothelial cells. After incubation with various functionalized SPIONs, samples that had been incubated with HABP-, OPN-, and PEG-SPIONs did not show significantly higher rates of cell death than control samples (p > 0.05). These studies confirm what has been previously shown in the literature: that polymer-coated and functionalized SPIONs are not cytotoxic, and are generally safe for biological use at physiologically-relevant concentrations.46,69,70
Successful coating of SPIONs with citric acid, PEG, and desired peptides was confirmed by DLS results, zeta potential measurements, and the BCA assay (Table 1). CA-SPIONs are expected to be the smallest in size, with size increasing at every step of functionalization. Additionally, CA-SPIONs are expected to be highly negatively charged due to the presence of multiple carboxylic acid residues around the core. PEGylated SPIONs are expected to have a zeta potential closer to neutral, which was confirmed (Table 1). Successful conjugation of either peptide (ribosome-display derived HABP or osteopontin-derived OPN) was confirmed via the BCA assay. BCA assay results yield a peptide concentration value for a given solution of peptide-coated SPIONs; this peptide concentration, along with the measured quantity of SPIONs in the solution, is used to calculate the approximate number of peptides per SPION. There should be no BCA assay signal present at all in samples to which peptides have not been bound covalently; this was observed in these studies. Because the conjugation reaction was performed to target 25 peptides per SPION, we are able to calculate from these results an approximate reaction efficiency and approximate number of peptides conjugated per SPION. BCA assay results after peptide addition indicated a 72% reaction efficiency for the addition of HABP to SPIONs (~18 peptides/SPION) and a 92% reaction efficiency for the addition of OPN to SPIONs (~23 peptides/SPION).
Table 1:
DLS and zeta potential measurements of SPIONs at various stages of fabrication indicate success of functionalization. As expected, particle size increases with the addition of each surface modification. Additionally, zeta potential changes reflect the change in surface groups at each stage. Data presented as mean of 3 independent runs of particle fabrication.
| Fabrication Stage |
Effective Diameter (nm) |
Zeta Potential (mV) |
|---|---|---|
| CA-SPIONs | 13.2 | −35.1 |
| PEG-SPIONs | 19.2 | −11.4 |
| HABP-SPIONs | 20.5 | −30.8 |
| OPN-SPIONs | 25.1 | −31.3 |
It can be observed that the effective diameter of CA-SPIONs as measured by DLS is larger than the effective diameter of OA-SPIONs as measured by TEM. This was somewhat expected, as the CA coating of the particle is hydrophilic and therefore likely to associate with water molecules, making the “wet” particles observed via DLS larger in size than the “dry” particles obsereved via TEM. Similar results were observed by Liao, et al.71,72 An additional factor that may contribute to this observed difference is that DLS measures effective diameter, which includes associated ions as well as water molecules; as these measurements were carried out in pH 9 phosphate buffer, it is likely that there are also ions associated with the nanoparticles.
The T2 relaxivities of HABP-, OPN-, and PEG-SPIONs were evaluated via MR to ensure that they generated measurable signal (Figure 2). Since SPIONs are a negative contrast agent, it is expected that samples will appear darker at increasing concentrations as measured in parts per million (ppm). This was observed across all samples. All functionalized SPIONs generated an MR signal in the range of previous results measuring the MR signal of functionalized SPIONs, although the intensity of the signal did vary with different surface functionalizations.46,73 This, too, is unsurprising, given previous results.46,73 These studies confirm that HABP-, OPN-, and PEG-SPIONs as generated in these studies can act as MR contrast agents.
Figure 2:
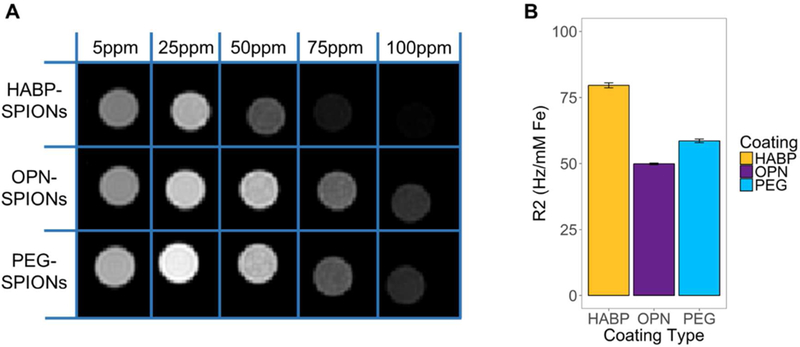
(A) T2-weighted MRI (3 Tesla) of saline solutions containing HABP-, OPN -, and PEG-SPIONs at different concentrations (ppm) exhibits decreasing signal intensity with increasing concentration for all 3 types of SPION (i.e., the color of each tube gets darker with increasing ppm). (B) The loss in signal intensity with increasing concentration is a consequence of the high R2 relaxivities of HABP-, OPN-, and PEG-SPIONs determined from the relaxation rate of each sample measured from MR images like those in (A) but acquired at multiple echo times. Error bars shown are standard error in the calculated slope.
Functionalized SPION binding to HA was evaluated in sodium phosphate buffer via an in vitro assay developed for this purpose. HA-targeted SPIONs, coated with either HABP or OPN, bind to HA in centrifugal filter tubes in a dose-dependent manner as expected – e.g. the solution in the tube farthest to the left (0mg of HA) is much darker in color than the solution in the tube farthest to the right (200mg HA), indicating that fewer nanoparticles remain unbound and in solution with increasing quantities of HA (Figure 3, A and B). HABP-SPIONs appear to bind in significantly (p < 0.05) higher quantities than OPN-SPIONs to comparable amounts of HA at lower quantities of HA. Notably, however, both HA-targeted SPIONs bind to <200mg of HA, which is a physiologically relevant quantity in cardiovascular disease.37,74
Figure 3:
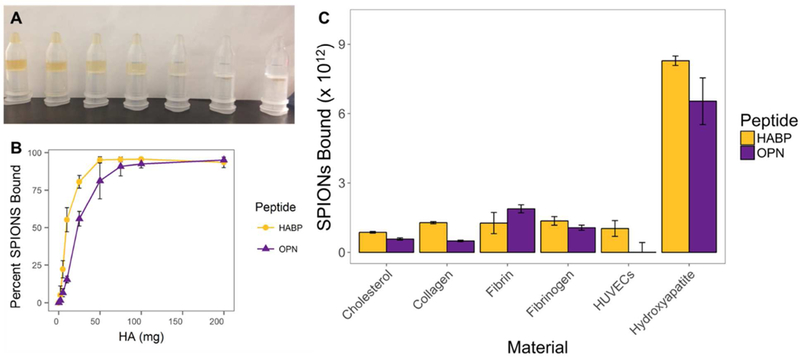
(A) Binding of HABP SPIONs to HA. From left to right, tubes contain 0, 5, 10, 25, 50, 100, and 200mg of HA. After filtration of the HA, the concentration of SPIONs remaining in the tubes is visibly different depending on the amount of HA in each tube – higher quantities of HA result in fewer particles remaining unbound. (B) Quantification of SPION binding from samples bound in the same setup as that illustrated in (A). Generally, 50mg of HA was sufficient to fully bind 9.5 × 1013 HABP-SPIONs, and 7.7 × 1013 OPN-SPIONs. (C) Quantification of HABP- and OPN-SPION binding to various biologically-relevant surfaces. HA-targeted SPIONs bind significantly more to HA than to other surfaces tested, although some degree of off-target binding did occur. Error bars are standard deviation; n=3.
For the studies in Figure 3B, powdered HA was purchased from Sigma. It was observed that this purchased form of HA did not lead to issues with the centrifugal filtration used for these experiments. Equal flow of liquid through the filter upon centrifugation was observed in all samples (Figure 3A); only the quantity of nanoparticles, assessed via an absorbance assay, differed. On the other hand, HA as formed by evaporation of mixed calcium and phosphate solutions did lead to issues with filtration in the centrifugal filter tubes, but not in the syringe filters (data not shown). Therefore, for Figure 3C, the evaporation method was preferred, as this method is more likely to produce the mix of crystalline HA and amorphous, insoluble calcium phosphate that is more reflective of what may be found biologically.75,76 It has been suggested that both HABP and OPN binding to HA is charge-mediated.43,44,77 Therefore, HABP- and OPN-SPION binding to HA was assessed in sodium phosphate buffer at different pH levels (Supporting Information, Figure S1). The pH levels of 7.1, 7.4, and 7.8 were selected because they are physiologically relevant; pH levels in this range can be detected close to plaques in atherosclerotic disease.78 The different pH levels tested did not significantly impact HABP- or OPN-SPION binding to HA in terms of binding quantity; however, higher pH levels did appear to lead to more variability in binding strength of SPIONs to HA.
In addition, SPIONs suspended in sodium-phosphate buffer do not bind to other biologically-relevant off-target molecules (Figure 3C). These experiments were performed by coating the surface of cell culture plates in biologically-relevant substances (cholesterol, collagen, fibrinogen, fibrin, or HUVECS) and allowing binding to occur. For these studies, all surfaces (including HA) were fabricated such that there were ~25mg (as close as possible to this target) of each material present for binding tests. For HUVECs, these studies were performed once HUVECs reached confluence. The number of SPIONs was determined by comparing the absorbance of the filtrate to the absorbance of solutions with known quantities of nanoparticles. This method was chosen to account for any potential differences in the starting concentrations of the various nanoparticle solutions used. Although both HABP- and OPN-SPIONS do demonstrate some binding to off-target surfaces, low levels of non-specific interactions are to be expected. Binding to HA in this experimental setup occurs significantly more than binding to off-target surfaces (p 0.05); binding occurs at least 5.23x the rate that binding to off-target surfaces occurs for HABP-SPIONs, and 4.28x for OPN-SPIONs. These results demonstrate that, as expected, both HABP- and OPN-SPIONs bind specifically to HA in vitro in the absence of serum.
However, like most if not all other nanoparticles, these functionalized nanoparticles also demonstrate binding to serum proteins. To assess this, PEG-SPIONs as well as HABP-SPIONs and OPN-SPIONs were incubated with a 50% fetal bovine serum solution for 24 hours, and compared with SPIONs incubated in DI water to observe differences in apparent diameter that would indicate association with serum proteins. Serum was chosen as it is commonly used in nanoparticle work to assess the formation of a protein corona and targeted binding in the presence of proteins.79–81 Thus, these studies are more readily comparable to previous work. However, many previous studies have been performed in lower concentrations of serum (e.g. 5–10%); to better assess the impact of a more biologically-relevant quantity of protein, we chose to use 50% serum.
In assessing size, a variety of particle diameters even within a single sample was expected due to slight differences in SPION shape and surface functionalization of different individual particles. TEM imaging may be unable to capture the full range and variability of particle diameters or properties present. DLS studies, on the other hand, are non-destructive, can be performed on the same set of samples multiple times, and capture a somewhat wider snapshot of particle properties. In addition, DLS studies assess the behavior of particles in solution, which is of higher biological relevance than samples that have undergone preparation for TEM. It was therefore determined that, for the purposes of these studies, effective diameter data as measured via DLS would be of more use in obtaining information about functionalized SPION behavior in solution than TEM.
For these experiments, PEG-SPIONs were tested in addition to HA-targeted SPIONs in order to evaluate whether the effects shown were due specifically to the presence of the peptides. Results indicate that HABP-, OPN-, and PEG-SPIONs all associate with serum proteins immediately upon incubation. Samples developed an apparent protein corona as soon as serum was added to the nanoparticle solution. This corona did not significantly (p > 0.05) alter in size over 24 hours in any case (Figure 4). These effective diameter measurements include both the hard corona closer to the SPIONs themselves as well as the more loosely-associated soft corona. There were observed differences in the size of the associated protein corona depending on the SPION coating. HABP-SPIONs exhibited the largest effective diameter, indicating either larger proteins are preferentially binding, more proteins are bound, or more aggregation is occurring. This may, in part, be due to the synthetic nature of the peptide, which, as its sequence was determined by ribosome display, is unlikely to have biological analogs. On the other hand, OPN-SPIONs, with their peptide sequence based on a common biological protein, have the smallest effective diameter, potentially indicating that fewer serum proteins tend to be associated with these particles, or that less aggregation occurs. Despite the fact that PEG is intended to prevent deposition of serum proteins, PEG-SPIONs also demonstrate a substantial (~3x) alteration in effective diameter. This indicates that serum proteins do adhere to or associate with PEGylated SPIONs, as has been previously reported by other groups.30,31The fact that the protein corona does not substantially alter in size over the course of 24 hours does not necessarily indicate that it remains constant. It is well accepted that the nanoparticle corona is made up of a “hard” corona that consists of tightly bound proteins that unlikely to change significantly in composition over time, and a “soft” corona that consists of proteins that are associated with low affinity that may readily adsorb and desorb over time.
Figure 4:
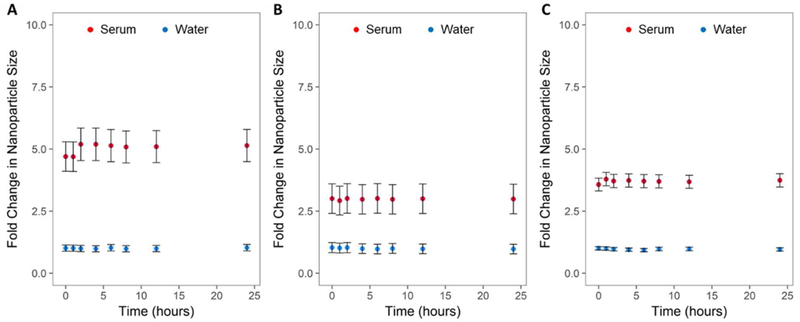
Fold change in nanoparticle size over time in a 50% serum solution. Effective nanoparticle diameter would include the nanoparticles in addition to any tightly associated and loosely associated proteins (i.e. both the hard and the soft protein corona). (A) HABP-SPIONs, (B) OPN-SPIONs, and (C) PEG-SPIONs were all tested under identical conditions. Both targeted (HABP- and OPN-SPIONs) and non-targeted (PEG-SPIONs) bind to proteins in serum, as evidenced by changes in effective particle diameter upon exposure to 50% serum. The protein corona that develops around all 3 functionalized SPIONs tested does not appear to alter significantly over the course of a 24-hour incubation. HABP-SPIONs appear to accumulate the largest protein corona, while OPN-SPIONs have the smallest. Despite the intended anti-biofouling function of PEG, PEG-SPIONs do still appear to accumulate a protein corona in 50% serum. Error bars are standard deviation; n = 3.
To better understand the protein corona of HABP-, OPN-, and PEG-SPIONs, several follow-up experiments were performed. Firstly, it was determined that formation of this protein corona around HABP-, OPN-, and PEG-SPIONs does not seem to be dependent on pH (Supporting Information, Figures S2 and S3). For these experiments, the pH of a 50% serum solution was altered over a broad range (pH 6–9), and functionalized SPIONs were added to each solution. The effective diameters and zeta potentials of each SPION sample were analyzed by DLS. Diameters were normalized to the effective diameter of that particular functionalized SPION at a pH of 7.4, i.e. the pH of unaltered 50% serum solution. If the observable (via DLS) protein corona were the result of specific and strong binding interactions, it is likely that such large alterations in pH would disrupt these interactions, and a change in the size of the protein corona or the resulting zeta potential would be observed. However, altering the pH of the 50% serum solution did not appear to significantly impact the effective diameter of SPION samples (p > 0.05), indicating that the measured or observable protein corona is likely to be a result of non-specific and therefore weaker binding interactions.
Subsequently, SPIONs were incubated for different amounts of time (0 minutes, 30 minutes, and 2 hours) with 50% serum and subjected to 3 wash steps via centrifugation (Figure 5). These times were selected for their clinical relevance; patients may be administered a contrast agent some time prior to clinical MR, but that time does not typically exceed 2 hours.82,83 Proteins still associated with the nanoparticles after 3 washes are considered to be tightly associated (i.e., part of a “hard” corona), while those that are removed with wash steps are considered to be more loosely associated (i.e., part of a “soft” corona). Samples were taken after each wash, and the BCA assay was used to determine the amount of protein that remained associated with the particles. In all cases, the amount of associated protein was significantly reduced (p < 0.05, reduction by at least 10.78x with no incubation, at least 3.26x with 30 minutes of incubation, and at least 2.82x with 2 hours of incubation) after just one wash, with additional subsequent reductions observed across all experiments (Figure 5).
Figure 5:
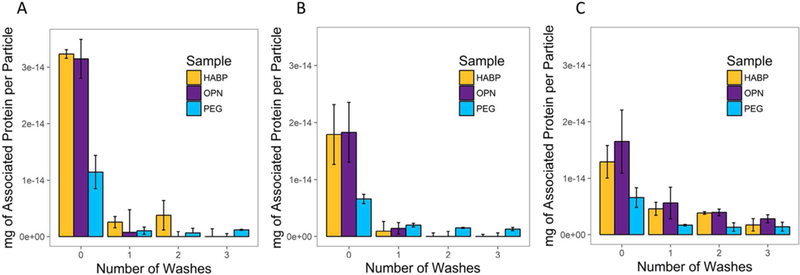
Soft and hard protein corona measured as mg of associated protein per particle after incubation in a 50% serum solution for (A) 0 minutes, (B) 30 minutes, and (C) 2 hours, followed by 3 wash steps via centrifugation and resuspension in DI water. Protein associated with the particles with no wash steps is considered to be part of the soft corona; protein associated with the particles after 3 washes is considered to be part of the hard corona. Error bars are standard deviation; n=3.
Notably, these results corroborate some of the findings from Figure 4; specifically, that the association of serum proteins with all SPIONs occurs immediately upon mixing of the nanoparticles with serum. Additionally, these results support previous literature, demonstrating that PEGylated particles do show reduced protein association in comparison to SPIONs with attached peptide prior to any performed washes. However, after 3 washes, PEGylated SPION samples that have been incubated in 50% serum for 0 or 30 minutes show increased protein in comparison to HABP- and OPN-SPIONs (for no incubation in comparison to OPN-SPIONs, and 30 minutes incubation with both HABP- and OPN-SPIONs, this difference is significant, with p < 0.05). This indicates that, while there may be less of a “soft” protein corona associated with PEGylated particles, there is an increased protein presence in the “hard” protein corona; barely any protein remains associated with HABP- or OPN-SPIONs after 3 washes for samples that have been incubated for 0 or 30 minutes.
As might be expected, samples that had been incubated for 2 hours showed an increased level of protein in the hard corona in comparison to samples that were incubated for 0 and 30 minutes (Figure 5C). There were statistically significant (p < 0.05) differences in the amount of protein per particle left after 3 washes for HABP- as well as OPN-SPIONs that had been incubated for 0 and 30 minutes in comparison to those that had been incubated for 2 hours. However, the amount of protein in the hard corona for PEGylated samples does not change significantly (p > 0.05). This may suggest an ultimately decreased targeting efficiency in peptide-coated SPIONs upon longer incubation with serum. For applications in which continued targeting capabilities after 2 hours are important (e.g. drug delivery), it may be necessary or beneficial to assess the hard vs. soft protein corona after longer incubations in a 50% serum solution.
The protein corona of HABP-, OPN-, and PEG-SPIONs after 3 washes was qualitatively examined by SDS-PAGE gel electrophoresis followed by silver nitrate staining (Figure 6). These results were compared to samples that had not been incubated with serum (Figure 6A). The darkness of the banding of the PEGylated SPION samples (Figure 5B, C, and D, rightmost well) in comparison to all other samples reflects, in part, the increased number of PEGylated SPIONs that were successfully pelleted with each was step. Qualitatively, PEGylated SPIONs formed a pellet more readily than HABP- and OPN-SPIONs, and were more difficult to resuspend in water with each wash. This is likely the result of some degree of hydrophobic interactions between PEG chains, which are known to have some hydrophobic character, and is best reflected in Figure 5.84 The results in Figure 4 are normalized for the number of particles remaining in solution; this is likely why the SDS-PAGE results appear to show a much higher quantity of proteins adsorbed to PEGylated samples, while that trend is not reflected in the actual quantification in Figure 5C.
Figure 6:
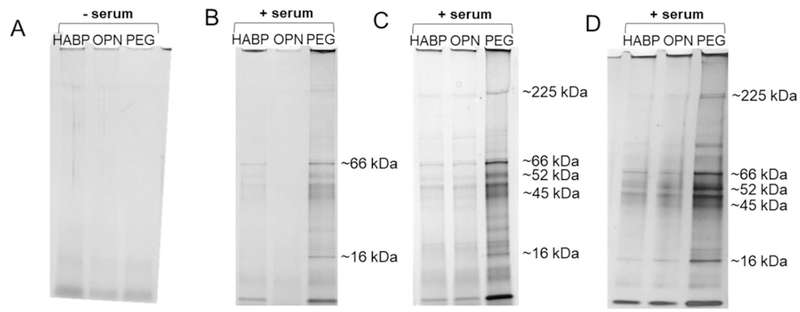
SDS-PAGE gels stained with silver nitrate. (A) Samples (HABP-, OPN-, and PEG-SPIONs, respectively) that were not incubated with serum. Wells in (B), (C), and (D) are samples (HABP-, OPN-, and PEG-SPIONs in each well, respectively) that were incubated with serum, washed 3x with water by centrifugation, and then mixed with an SDS-based running buffer and boiled to denature any protein remaining attached to the particles. Differences between wells reflect differences in which proteins may have preferentially adsorbed to different surface functionalizations; differences between gels reflect potential differences in the hard protein corona with longer incubations times. Samples were incubated with serum for: (B) 0 minutes, (C) 30 minutes, and (D) 2 hours prior to beginning the wash steps.
Beyond this, it is possible to qualitatively observe some differences between preferentially adsorbed proteins to HABP-, OPN-, and PEGylated SPIONs, both within a single gel, and between gels. Replicate gels were run, and showed similar results (data not shown). The differences within a gel are to be expected; it is likely that different proteins will preferentially adsorb to different surface functionalizations. Many previous studies have confirmed that different surface functionalizations yield an ultimately different protein corona composition.85,86 The observed differences between gels make sense, given that previous results in the literature have found that, with incubation, loosely associated proteins close to the nanoparticle may be replaced by different proteins that more preferentially associate with a given surface functionalization.26 These observations may also help to elucidate one of the trends observed in Figure 5; that is, differences may develop in the hard corona over time as this preferential association occurs, leading to an increased number of proteins ultimately left after the wash steps in Figure 5C in comparison to Figure 5A. The gels additionally appear to show that there is a higher amount of protein across comparable wells with longer incubation times as observed in Figure 5; i.e. the HABP-SPION well gets darker from 6A to 6B, and from 6B to 6C.
Approximate molecular weights of some of the clearer bands have been indicated in Figure 6, based on a ladder run as a control (not shown). Previous research of the common components in the protein corona of other SPIONs evaluated in FBS has generated lists of proteins commonly found in these hard corona.85 The types of protein associated with each particle were not further assessed, as these experiments were performed in 50% fetal bovine serum, and specific protein analysis may not be comparable or relevant for medical applications in humans. Additionally, it has been shown that factors such as shear stress (which would be experienced by SPIONs in circulation) contribute to observed differences in the protein corona formed on PEGylated particles.87 Because of this, and the matrix chosen, it was determined that assessing the exact composition of the corona would not yield physiologically relevant results for these particles. Further studies would be needed to assess targeted binding both in human serum as well as under flow conditions; yet even these studies would not fully recapitulate the in vivo environment.
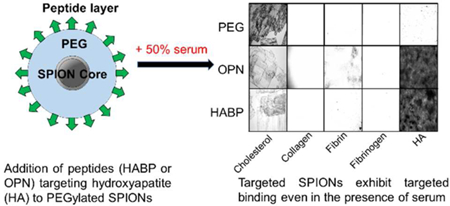
Finally, binding of HABP-, OPN-, and PEG-SPIONs both to HA as well as off-target surfaces was evaluated in 50% serum (Figure 7). For these studies, standard optical microscopy was used rather than SEM or TEM in order to get a better sense of the bulk properties of the SPION solutions as they interacted with each surface, rather than obtaining images of the precise interaction of individual particles with the surfaces. It was anticipated that this approach would yield a more accurate quantification of the bulk trends by enabling the imaging and quantification of a much larger proportion of each surface. Additionally, the preparation process required by SEM or TEM would likely destroy the somewhat delicate gel-like nature of several of the sample surfaces (in particular, fibrin and HA), thus preventing observation of the binding in situ.
Figure 7:
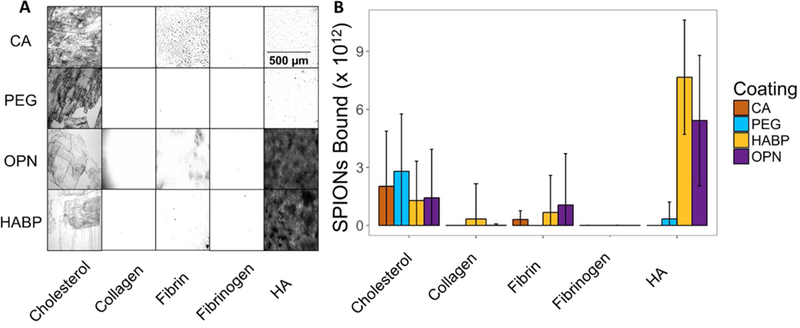
(A) Images taken at 20x of Prussian-Blue stained SPIONs bound to different surfaces to observe specific vs. non-specific binding. (B) Quantification of multiple runs of binding assays (n = 3) demonstrate that both HABP- and OPN-SPIONs bind at significantly higher rates to HA than other biologically relevant surfaces, while PEG-SPIONs and CA-SPIONs do not bind significantly to HA. PEG-SPIONs and CA-SPIONs exhibit significant off-target binding to cholesterol, potentially due to the hydrophobic nature of the cholesterol surface. All surfaces were fabricated such that ~25mg of each material is present for testing against each type of nanoparticle.
Despite the fact that HABP- and OPN-SPIONs exhibit binding to serum proteins, they still demonstrate targeted binding to HA. Binding to HA occurred at a rate that was 5.97x greater than binding to any off-target surface for HABP-SPIONs, and 4.00x greater for OPN-SPIONs. Off-target surfaces that were evaluated included: cholesterol, collagen, fibrin, and fibrinogen. HUVECs were not tested in the experiments due to concerns that a rapid increase in environmental serum concentrations would impact membrane transport of the cells due to a rapid change in environmental osmolarity.88,89 For these studies, PEG-SPIONs as well as CA-SPIONs were evaluated as negative controls to ensure that any binding trends observed in the targeted SPIONs were due to the presence of the peptide. Because CA-SPIONs, like HABP-and OPN-SPIONs, carry a negative surface charge, these results would indicate that it is not merely the presence of a negative surface charge that causes the targeted binding of HABP-and OPN-SPIONs to HA, although charge may certainly play a role.
Because HA needed to be coated on a fibrin layer to successfully stick to the plates, and because both HABP- and OPN-SPIONs did exhibit some off-target binding to fibrin, it is likely that some of the particles in the HA wells were bound to fibrin rather than HA. However, the observed binding results for fibrin occur when the particles are interacting directly with an unaltered fibrin surface, not with a fibrin surface that has been blocked by a crystalline HA layer. Therefore, the vast majority of observed binding to HA was likely to the HA itself, with a small contribution from the underlying fibrin layer.
The pre-incubation of HA with serum as described should allow for interactions between HA and serum proteins - such as the formation of an associated corona - to occur. The formation of a layer of serum proteins on HA may be interesting to quantify or characterize as part of further work, but was beyond the scope of these studies. Unlike SPIONs, which are injected into the body at a given time and perform their function (MRI contrast) within a specified amount of time after injection, HA is already in the body. Thus, the timing of the formation of a corona around HA is somewhat irrelevant, and of less interest than the time-dependent formation of the corona around the SPIONs.
One notable observation of the results in Figure 7B in comparison to earlier binding studies (Figure 3C) is that the variability of targeted and non-targeted SPION binding in serum appears to be much greater than the variability in sodium phosphate buffer. For example, in serum, between 0 – 3.43 × 1012 HABP-SPIONs bind to fibrin; in sodium phosphate buffer, that range is 0.62 × 1012 – 1.33 × 1012. In serum, between 0 – 4.27 × 1012 OPN-SPIONs bind to fibrin; in sodium phosphate buffer, that range is 1.22 × 1012 – 1.46 × 1012.This is perhaps unsurprising. However, even with this variability, binding of HABP- and OPN-SPIONs to HA was still significantly greater than off-target binding (p < 0.05). With the fairly high error, it can be observed that both HABP- as well as OPN-SPION binding to HA occurs at rates that are comparable to those in Figure 3C, despite the orthogonal method of measurement used. The apparent off-target binding of PEG-SPIONs and CA-SPIONs to cholesterol was unexpected, but is evident in microscopic images as well as the quantitative data. It is hypothesized that this binding may be due to the hydrophobic and crystalline nature of the deposited cholesterol. Similar results have been reported previously for cyclodextrin-coated nanoparticles.90,91 For diseases such as atherosclerosis, it may be worth further examining the interaction between PEGylated or CA-coated nanoparticles and cholesterol in the future.
Conclusions
In conclusion, the serum protein interactions of two peptide-coated SPIONs are evaluated and compared with the serum protein interactions of PEGylated particles. HABP- and OPN- SPIONs, while both found to interact with serums proteins, have less tightly-attached protein than PEGylated SPIONs with short (<2 hour) exposure times to serum. These particles are also still able to bind to their target surface, hydroxyapatite, potentially in part due to the somewhat non-specific nature of the target interaction when compared with protein-protein interactions. These results indicate that the selection of a non-protein disease target, where possible, may aid in minimizing the impact of the protein corona on nanoparticle binding specificity in a protein-rich environment. Further experiments will continue to assess the viability of these promising peptide-coated SPIONs as contrast agents for the detection of cardiovascular disease.
Supplementary Material
Acknowledgments
This work is supported by a Biomolecular Pharmacology Training grant from the NIH/NIGMS (5T32HL007969-13), a training grant from the NIH in Inflammatory Disorders (5T32AI089673-05), and a training grant from the NIH in Cardiovascular Biology (5T32GM008541-17). This work was performed in part at the Center for Nanoscale Systems (CNS), a member of the National Nanotechnology Coordinated Infrastructure Network (NNCI), which is supported by the National Science Foundation under NSF award no. 1541959. CNS is part of Harvard University. C.L.M. would like to acknowledge contributions to this research from Jo Ann Buczek-Thomas, Olivia Hale, Jordan Nustad, Nikita Patil, Ethan Beswick, and Adam Bossett. We additionally thank the Undergraduate Research Opportunities Program (UROP) at Boston University for their support.
References
- (1).Gao X; Cui Y; Levenson RM; Chung LWK; Nie S In Vivo Cancer Targeting and Imaging with Semiconductor Quantum Dots. Nat. Biotechnol 2004, 22 (8), 969. [DOI] [PubMed] [Google Scholar]
- (2).Sardan M; Eren ED; Ozdemir A; Tekinay AB; Guler MO Bioactive Peptide Functionalized Superparamagnetic Iron Oxide Nanoparticles (SPIONs) for Targeted Imaging with MRI. In 2015 5th International Workshop on Magnetic Particle Imaging (IWMPI); 2015; pp 1–1.
- (3).Choi CHJ; Alabi CA; Webster P; Davis ME Mechanism of Active Targeting in Solid Tumors with Transferrin-Containing Gold Nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2010, 107 (3), 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mu K; Zhang S; Ai T; Jiang J; Yao Y; Jiang L; Zhou Q; Xiang H; Zhu Y; Yang X; Zhu W Monoclonal Antibody-Conjugated Superparamagnetic Iron Oxide Nanoparticles for Imaging of Epidermal Growth Factor Receptor-Targeted Cells and Gliomas. Mol. Imaging 2015, 14. [DOI] [PubMed] [Google Scholar]
- (5).Farokhzad OC; Cheng J; Teply BA; Sherifi I; Jon S; Kantoff PW; Richie JP; Langer R Targeted Nanoparticle-Aptamer Bioconjugates for Cancer Chemotherapy in Vivo. Proc. Natl. Acad. Sci 2006, 103 (16), 6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kohler N; Sun C; Wang J; Zhang M Methotrexate-Modified Superparamagnetic Nanoparticles and Their Intracellular Uptake into Human Cancer Cells. Langmuir 2005, 21 (19), 8858–8864. [DOI] [PubMed] [Google Scholar]
- (7).Peer D; Karp JM; Hong S; Farokhzad OC; Margalit R; Langer R Nanocarriers as an Emerging Platform for Cancer Therapy. Nat. Nanotechnol 2007, 2 (12), 751. [DOI] [PubMed] [Google Scholar]
- (8).Davis ME; Chen Z (Georgia); Shin DM Nanoparticle Therapeutics: An Emerging Treatment Modality for Cancer. Nat. Rev. Drug Discov 2008, 7 (9), 771. [DOI] [PubMed] [Google Scholar]
- (9).Malam Y; Loizidou M; Seifalian AM Liposomes and Nanoparticles: Nanosized Vehicles for Drug Delivery in Cancer. Trends Pharmacol. Sci 2009, 30 (11), 592–599. [DOI] [PubMed] [Google Scholar]
- (10).Tassa C; Shaw SY; Weissleder R Dextran-Coated Iron Oxide Nanoparticles: A Versatile Platform for Targeted Molecular Imaging, Molecular Diagnostics, and Therapy. Acc. Chem. Res 2011, 44 (10), 842–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Georganopoulou DG; Chang L; Nam J-M; Thaxton CS; Mufson EJ; Klein WL; Mirkin CA Nanoparticle-Based Detection in Cerebral Spinal Fluid of a Soluble Pathogenic Biomarker for Alzheimer’s Disease. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (7), 2273–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Mahon E; Salvati A; Baldelli Bombelli F; Lynch I; Dawson KA Designing the Nanoparticle–Biomolecule Interface for “Targeting and Therapeutic Delivery.” J. Controlled Release 2012, 161 (2), 164–174. [DOI] [PubMed] [Google Scholar]
- (13).Mirshafiee V; Kim R; Park S; Mahmoudi M; Kraft ML Impact of Protein Pre-Coating on the Protein Corona Composition and Nanoparticle Cellular Uptake. Biomaterials 2016, 75, 295–304. [DOI] [PubMed] [Google Scholar]
- (14).Bertrand N; Leroux J-C The Journey of a Drug-Carrier in the Body: An Anatomo-Physiological Perspective. J. Control. Release Off. J. Control. Release Soc 2012, 161 (2), 152–163. [DOI] [PubMed] [Google Scholar]
- (15).Lynch I; Dawson KA Protein-Nanoparticle Interactions. Nano Today 2008, 3 (1), 40–47. [Google Scholar]
- (16).Mahmoudi M; Lynch I; Ejtehadi MR; Monopoli MP; Bombelli FB; Laurent S Protein−Nanoparticle Interactions: Opportunities and Challenges. Chem. Rev 2011, 111 (9), 5610–5637. [DOI] [PubMed] [Google Scholar]
- (17).Cedervall T; Lynch I; Lindman S; Berggård T; Thulin E; Nilsson H; Dawson KA; Linse S Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc. Natl. Acad. Sci. U. S. A 2007, 104 (7), 2050–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Townson JL; Lin Y-S; Agola JO; Carnes EC; Leong HS; Lewis JD; Haynes CL; Brinker CJ Re-Examining the Size/Charge Paradigm: Differing in Vivo Characteristics of Size- and Charge-Matched Mesoporous Silica Nanoparticles. J. Am. Chem. Soc 2013, 135 (43), 16030–16033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shah NB; Vercellotti GM; White JG; Fegan A; Wagner CR; Bischof JC Blood–Nanoparticle Interactions and in Vivo Biodistribution: Impact of Surface PEG and Ligand Properties. Mol. Pharm 2012, 9 (8), 2146–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Monopoli MP; Walczyk D; Campbell A; Elia G; Lynch I; Bombelli FB; Dawson A Physical-Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc 2011, 133 (8), 2525–2534. [DOI] [PubMed] [Google Scholar]
- (21).Lesniak A; Salvati A; Santos-Martinez MJ; Radomski MW; Dawson KA; Åberg C Nanoparticle Adhesion to the Cell Membrane and Its Effect on Nanoparticle Uptake Efficiency. J. Am. Chem. Soc 2013, 135 (4), 1438–1444. [DOI] [PubMed] [Google Scholar]
- (22).Mirshafiee V; Mahmoudi M; Lou K; Cheng J; Kraft ML Protein Corona Significantly Reduces Active Targeting Yield. Chem. Commun 2013, 49 (25), 2557–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Salvati A; Pitek AS; Monopoli MP; Prapainop K; Bombelli FB; Hristov DR; Kelly PM; Åberg C; Mahon E; Dawson KA Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat. Nanotechnol 2013, 8 (2), 137. [DOI] [PubMed] [Google Scholar]
- (24).Mahmoudi M; Bertrand N; Zope H; Farokhzad OC Emerging Understanding of the Protein Corona at the Nano-Bio Interfaces. Nano Today 2016, 11 (6), 817–832. [Google Scholar]
- (25).Nguyen VH; Lee B-J Protein Corona: A New Approach for Nanomedicine Design. Int. Nanomedicine 2017, 12, 3137–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Casals E; Pfaller T; Duschl A; Oostingh GJ; Puntes V Time Evolution of the Nanoparticle Protein Corona. ACS Nano 2010, 4 (7), 3623–3632. [DOI] [PubMed] [Google Scholar]
- (27).Gref R; Lück M; Quellec P; Marchand M; Dellacherie E; Harnisch S; Blunk T; Müller RH ‘Stealth’ Corona-Core Nanoparticles Surface Modified by Polyethylene Glycol (PEG): Influences of the Corona (PEG Chain Length and Surface Density) and of the Core Composition on Phagocytic Uptake and Plasma Protein Adsorption. Colloids Surf. B Biointerfaces 2000, 18 (3), 301–313. [DOI] [PubMed] [Google Scholar]
- (28).Yu M; Huang S; Yu KJ; Clyne AM Dextran and Polymer Polyethylene Glycol (PEG) Coating Reduce Both 5 and 30 Nm Iron Oxide Nanoparticle Cytotoxicity in 2D and 3D Cell Culture. Int. J. Mol. Sci 2012, 13 (5), 5554–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Wang RLC; Kreuzer HJ; Grunze M Molecular Conformation and Solvation of Oligo(Ethylene Glycol)-Terminated Self-Assembled Monolayers and Their Resistance to Protein Adsorption. J. Phys. Chem. B 1997, 101 (47), 9767–9773. [Google Scholar]
- (30).Kim HR; Andrieux K; Delomenie C; Chacun H; Appel M; Desmaële D; Taran F; Georgin D; Couvreur P; Taverna M Analysis of Plasma Protein Adsorption onto PEGylated Nanoparticles by Complementary Methods: 2-DE, CE and Protein Lab-on-Chip® System. ELECTROPHORESIS 2007, 28 (13), 2252–2261. [DOI] [PubMed] [Google Scholar]
- (31).Pozzi D; Colapicchioni V; Caracciolo G; Piovesana S; Capriotti AL; Palchetti S; De Grossi S; Riccioli A; Amenitsch H; Laganà A Effect of Polyethyleneglycol (PEG) Chain Length on the Bio-Nano-Interactions between PEGylated Lipid Nanoparticles and Biological Fluids: From Nanostructure to Uptake in Cancer Cells. Nanoscale 2014, 6 (5), 2782–2792. [DOI] [PubMed] [Google Scholar]
- (32).Walkey CD; Olsen JB; Guo H; Emili A; Chan WCW Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J. Am. Chem. Soc 2012, 134 (4), 2139–2147. [DOI] [PubMed] [Google Scholar]
- (33).Safavi-Sohi R; Maghari S; Raoufi M; Jalali SA; Hajipour MJ; Ghassempour A; Mahmoudi M Bypassing Protein Corona Issue on Active Targeting: Zwitterionic Coatings Dictate Specific Interactions of Targeting Moieties and Cell Receptors. ACS Appl. Mater. Interfaces 2016, 8 (35), 22808–22818. [DOI] [PubMed] [Google Scholar]
- (34).Zanganeh S; Spitler R; Erfanzadeh M; Alkilany AM; Mahmoudi M Protein Corona: Opportunities and Challenges. Int. J. Biochem. Cell Biol 2016, 75 (Supplement C), 143– 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sun C; Sze R; Zhang M Folic Acid-PEG Conjugated Superparamagnetic Nanoparticles for Targeted Cellular Uptake and Detection by MRI. J. Biomed. Mater. Res. A 2006, 78A (3), 550–557. [DOI] [PubMed] [Google Scholar]
- (36).Giljohann DA; Seferos DS; Patel PC; Millstone JE; Rosi NL; Mirkin CA Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett 2007, 7 (12), 3818–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Koos R; Mahnken AH; Sinha AM; Wildberger JE; Hoffmann R; Kühl HP Aortic Valve Calcification as a Marker for Aortic Stenosis Severity: Assessment on 16-MDCT. Am. J. Roentgenol 2004, 183 (6), 1813–1818. [DOI] [PubMed] [Google Scholar]
- (38).Lee JS; Morrisett JD; Tung C-H Detection of Hydroxyapatite in Calcified Cardiovascular Tissues. Atherosclerosis 2012, 224 (2), 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Stephen ZR; Kievit FM; Zhang M Magnetite Nanoparticles for Medical MR Imaging. Mater. Today Kidlington Engl 2011, 14 (7–8), 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gossuin Y; Muller RN; Gillis P Relaxation Induced by Ferritin: A Better Understanding for an Improved MRI Iron Quantification. NMR Biomed 17 (7), 427–432. [DOI] [PubMed] [Google Scholar]
- (41).Kehagias DT; Gouliamos AD; Smyrniotis V; Vlahos LJ Diagnostic Efficacy and Safety of MRI of the Liver with Superparamagnetic Iron Oxide Particles (SH U 555 A). J. Magn. Reson. Imaging 14 (5), 595–601. [DOI] [PubMed] [Google Scholar]
- (42).Revia RA; Zhang M Magnetite Nanoparticles for Cancer Diagnosis, Treatment, and Treatment Monitoring: Recent Advances. Mater. Today 2016, 19 (3), 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Roy MD; Stanley SK; Amis EJ; Becker ML Identification of a Highly Specific Hydroxyapatite-Binding Peptide Using Phage Display. Adv. Mater 2008, 20 (10), 1830–1836. [Google Scholar]
- (44).Wang L; Guan X; Tang R; Hoyer JR; Wierzbicki A; De Yoreo JJ; Nancollas GH Phosphorylation of Osteopontin Is Required for Inhibition of Calcium Oxalate Crystallization. J. Phys. Chem. B 2008, 112 (30), 9151–9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Park Y; Whitaker RD; Nap RJ; Paulsen JL; Mathiyazhagan V; Doerrer LH; Song Y-Q; Hürlimann MD; Szleifer I; Wong JY Stability of Superparamagnetic Iron Oxide Nanoparticles at Different PH Values: Experimental and Theoretical Analysis. Langmuir 2012, 28 (15), 6246–6255. [DOI] [PubMed] [Google Scholar]
- (46).Park YC; Smith JB; Pham T; Whitaker RD; Sucato CA; Hamilton JA; Bartolak-Suki E; Wong JY Effect of PEG Molecular Weight on Stability, T2 Contrast, Cytotoxicity, and Cellular Uptake of Superparamagnetic Iron Oxide Nanoparticles (SPIONs). Colloids Surf. B Biointerfaces 2014, 119, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Szydlowski J; Van Hook WA Concentration and Temperature Dependence of Dynamic Light Scattering for Some Polystyrene Solutions in Toluene, Cyclohexane, Methylcyclohexane and Deuteromethylcyclohexane. Macromolecules 1998, 31 (10), 3266–3274. [Google Scholar]
- (48).NanoBrook 90Plus PALS Particle Size Analyzer | Brookhaven https://www.brookhaveninstruments.com/nanobrook-90plus (accessed Jun 25, 2018).
- (49).Sun S; Murray CB; Weller D; Folks L; Moser A Monodisperse FePt Nanoparticles and Ferromagnetic FePt Nanocrystal Superlattices. Science 2000, 287 (5460), 1989–1992. [DOI] [PubMed] [Google Scholar]
- (50).Tiraferri A; Sethi R Enhanced Transport of Zerovalent Iron Nanoparticles in Saturated Porous Media by Guar Gum. J. Nanoparticle Res 2009, 11 (3), 635. [Google Scholar]
- (51).Sonvico F; Mornet S; Vasseur S; Dubernet C; Jaillard D; Degrouard J; Hoebeke J; Duguet E; Colombo P; Couvreur P Folate-Conjugated Iron Oxide Nanoparticles for Solid Tumor Targeting as Potential Specific Magnetic Hyperthermia Mediators: Synthesis, Physicochemical Characterization, and in Vitro Experiments. Bioconjug. Chem 2005, 16 (5), 1181–1188. [DOI] [PubMed] [Google Scholar]
- (52).Mitsouras D; Owens CD; Conte MS; Ersoy H; Creager MA; Rybicki FJ; Mulkern RV In Vivo Differentiation of Two Vessel Wall Layers in Lower Extremity Peripheral Vein Bypass Grafts: Application of High Resolution Inner-Volume Black Blood 3D FSE. Magn. Reson. Med. Off. J. Soc. Magn. Reson. Med. Soc. Magn. Reson. Med 2009, 62 (3), 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Mitsouras D; Mulkern RV; Owens CD; Conte MS; Ersoy H; Luu TM; Whitmore AG; Creager MA; Rybicki FJ High-Resolution Peripheral Vein Bypass Graft Wall Studies Using High Sampling Efficiency Inner Volume 3D FSE. Magn. Reson. Med 2008, 59 (3), 650–654. [DOI] [PubMed] [Google Scholar]
- (54).Mitsouras D; Mulkern RV; Rybicki FJ Strategies for Inner Volume 3D Fast Spin Echo Magnetic Resonance Imaging Using Nonselective Refocusing Radio Frequency Pulsesa). Med. Phys 2006, 33 (1), 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).de Bruyn JR; Goiko M; Mozaffari M; Bator D; Dauphinee RL; Liao Y; Flemming RL; Bramble MS; Hunter GK; Goldberg HA Dynamic Light Scattering Study of Inhibition of Nucleation and Growth of Hydroxyapatite Crystals by Osteopontin. PLoS ONE 2013, 8 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Abbas S; Kumar S; Aswal VK; Kohlbrecher J Aggregation in Charged Nanoparticles Solutions Induced by Different Interactions. AIP Conf. Proc 2016, 1731 (1), 040011. [Google Scholar]
- (57).Jin R; Lin B; Li D; Ai H Superparamagnetic Iron Oxide Nanoparticles for MR Imaging and Therapy: Design Considerations and Clinical Applications. Curr. Opin. Pharmacol 2014, 18, 18–27. [DOI] [PubMed] [Google Scholar]
- (58).Wahajuddin; Arora S Superparamagnetic Iron Oxide Nanoparticles: Magnetic Nanoplatforms as Drug Carriers. Int. J. Nanomedicine 2012, 7, 3445–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Di Marco M; Sadun C; Port M; Guilbert I; Couvreur P; Dubernet C Physicochemical Characterization of Ultrasmall Superparamagnetic Iron Oxide Particles (USPIO) for Biomedical Application as MRI Contrast Agents. Int. J. Nanomedicine 2007, 2 (4), 609–622. [PMC free article] [PubMed] [Google Scholar]
- (60).Corot C; Robert P; Idée J-M; Port M Recent Advances in Iron Oxide Nanocrystal Technology for Medical Imaging. Adv. Drug Deliv. Rev 2006, 58 (14), 1471–1504. [DOI] [PubMed] [Google Scholar]
- (61).Estephan E; Dao J; Saab M-B; Panayotov I; Martin M; Larroque C; Gergely C; Cuisinier FJG; Levallois B SVSVGMKPSPRP: A Broad Range Adhesion Peptide. Biomed. Tech. (Berl) 2012, 57 (6), 481–489. [DOI] [PubMed] [Google Scholar]
- (62).Sodek J; Ganss B; McKee MD Osteopontin. Crit. Rev. Oral Biol. Med. Off. Publ. Am. Assoc. Oral Biol 2000, 11 (3), 279–303. [DOI] [PubMed] [Google Scholar]
- (63).Choi ST; Kim JH; Kang E-J; Lee S-W; Park M-C; Park Y-B; Lee S-K Osteopontin Might Be Involved in Bone Remodelling Rather than in Inflammation in Ankylosing Spondylitis. Rheumatol. Oxf. Engl 2008, 47 (12), 1775–1779. [DOI] [PubMed] [Google Scholar]
- (64).Wang KX; Denhardt DT Osteopontin: Role in Immune Regulation and Stress Responses. Cytokine Growth Factor Rev 2008, 19 (5–6), 333–345. [DOI] [PubMed] [Google Scholar]
- (65).Xie J; Chen K; Lee H-Y; Xu C; Hsu AR; Peng S; Chen X; Sun S Ultrasmall c(RGDyK)-Coated Fe3O4 Nanoparticles and Their Specific Targeting to Integrin Αvβ3-Rich Tumor Cells. J. Am. Chem. Soc 2008, 130 (24), 7542–7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Kim BH; Lee N; Kim H; An K; Park YI; Choi Y; Shin K; Lee Y; Kwon SG; Na HB; Park JG; Ahn TY; Kim YW; Moon WK; Choi SH; Hyeon T Large-Scale Synthesis of Uniform and Extremely Small-Sized Iron Oxide Nanoparticles for High-Resolution T1 Magnetic Resonance Imaging Contrast Agents. J. Am. Chem. Soc 2011, 133 (32), 12624–12631. [DOI] [PubMed] [Google Scholar]
- (67}.Shevtsov MA; Nikolaev BP; Yakovleva LY; Marchenko YY; Dobrodumov AV; Mikhrina AL; Martynova MG; Bystrova OA; Yakovenko IV; Ischenko AM Superparamagnetic Iron Oxide Nanoparticles Conjugated with Epidermal Growth Factor (SPION–EGF) for Targeting Brain Tumors. Int. J. Nanomedicine 2014, 9, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Shaik AP; Shaik AS; Majwal AA; Faraj AA Blocking Interleukin-4 Receptor α Using Polyethylene Glycol Functionalized Superparamagnetic Iron Oxide Nanocarriers to Inhibit Breast Cancer Cell Proliferation. Cancer Res. Treat. Cancer Res. Treat 2016, 49 (2), 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Gupta AK; Gupta M Cytotoxicity Suppression and Cellular Uptake Enhancement of Surface Modified Magnetic Nanoparticles. Biomaterials 2005, 26 (13), 1565–1573. [DOI] [PubMed] [Google Scholar]
- (70).Gupta AK; Wells S Surface-Modified Superparamagnetic Nanoparticles for Drug Delivery: Preparation, Characterization, and Cytotoxicity Studies. IEEE Trans. NanoBioscience 2004, 3 (1), 66–73. [DOI] [PubMed] [Google Scholar]
- (71).Liao G; Chen J; Zeng W; Yu C; Yi C; Xu Z Facile Preparation of Uniform Nanocomposite Spheres with Loading Silver Nanoparticles on Polystyrene-methyl Acrylic Acid Spheres for Catalytic Reduction of 4-Nitrophenol https://pubs.acs.org/doi/abs/10.1021/acs.jpcc.6b09356 (accessed Aug 15, 2018).
- (72).Liao G; Li Q; Zhao W; Pang Q; Gao H; Xu Z In-Situ Construction of Novel Silver Nanoparticle Decorated Polymeric Spheres as Highly Active and Stable Catalysts for Reduction of Methylene Blue Dye. Appl. Catal. Gen 2018, 549, 102–111. [Google Scholar]
- (73).LaConte LEW; Nitin N; Zurkiya O; Caruntu D; O’Connor CJ; Hu X; Bao G Coating Thickness of Magnetic Iron Oxide Nanoparticles Affects R2 Relaxivity. J. Magn. Reson. Imaging 2007, 26 (6), 1634–1641. [DOI] [PubMed] [Google Scholar]
- (74).Guo W; Morrisett JD; DeBakey ME; Lawrie GM; Hamilton JA Quantification In Situ of Crystalline Cholesterol and Calcium Phosphate Hydroxyapatite in Human Atherosclerotic Plaques by Solid-State Magic Angle Spinning NMR. Arterioscler. Thromb. Vasc. Biol 2000, 20 (6), 1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Kim KM; Trump BF Amorphous Calcium Precipitations in Human Aortic Valve. Calcif. Tissue Res 1975, 18 (1), 155–160. [DOI] [PubMed] [Google Scholar]
- (76).Towler DA Molecular and Cellular Aspects of Calcific Aortic Valve Disease. Circ. Res 2013, 113 (2), 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Weiger MC; Park JJ; Roy MD; Stafford CM; Karim A; Becker ML Quantification of the Binding Affinity of a Specific Hydroxyapatite Binding Peptide. Biomaterials 2010, 31 (11), 2955–2963. [DOI] [PubMed] [Google Scholar]
- (78).Naghavi M; John R; Naguib S; Siadaty MS; Grasu R; Kurian KC; van Winkle WB; Soller B; Litovsky S; Madjid M; Willerson JT; Casscells W PH Heterogeneity of Human and Rabbit Atherosclerotic Plaques; a New Insight into Detection of Vulnerable Plaque. Atherosclerosis 2002, 164 (1), 27–35. [DOI] [PubMed] [Google Scholar]
- (79).Gräfe C; Weidner A; Lühe M.v. d.; Bergemann C; Schacher FH; Clement JH; Dutz S Intentional Formation of a Protein Corona on Nanoparticles: Serum Concentration Affects Protein Corona Mass, Surface Charge, and Nanoparticle–Cell Interaction. Int. J. Biochem. Cell Biol 2016, 75, 196–202. [DOI] [PubMed] [Google Scholar]
- (80).Zhu Y; Li W; Li Q; Li Y; Li Y; Zhang X; Huang Q Effects of Serum Proteins on Intracellular Uptake and Cytotoxicity of Carbon Nanoparticles. Carbon 2009, 47 (5), 1351–1358. [Google Scholar]
- (81).Sikora A; Shard AG; Minelli C Size and ζ-Potential Measurement of Silica Nanoparticles in Serum Using Tunable Resistive Pulse Sensing. Langmuir 2016, 32 (9), 2216–2224. [DOI] [PubMed] [Google Scholar]
- (82).Brix G; Semmler W; Port R; Schad LR; Layer G; Lorenz WJ Pharmacokinetic Parameters in CNS Gd-DTPA Enhanced MR Imaging. J. Comput. Assist. Tomogr 1991, 15 (4), 621–628. [DOI] [PubMed] [Google Scholar]
- (83).Essig M; Shiroishi MS; Nguyen TB; Saake M; Provenzale JM; Enterline D; Anzalone N; Dörfler A; Rovira À; Wintermark M; Law M Perfusion MRI: The Five Most Frequently Asked Technical Questions. AJR Am. J. Roentgenol 2013, 200 (1), 24– 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Israelachvili J The Different Faces of Poly(Ethylene Glycol). Proc. Natl. Acad. Sci. U. S. A 1997, 94 (16), 8378–8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Sakulkhu U; Mahmoudi M; Maurizi L; Salaklang J; Hofmann H Protein Corona Composition of Superparamagnetic Iron Oxide Nanoparticles with Various Physico-Chemical Properties and Coatings. Sci. Rep 2014, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Vogt C; Pernemalm M; Kohonen P; Laurent S; Hultenby K; Vahter M; Lehtiö J; Toprak MS; Fadeel B Proteomics Analysis Reveals Distinct Corona Composition on Magnetic Nanoparticles with Different Surface Coatings: Implications for Interactions with Primary Human Macrophages. PLoS ONE 2015, 10 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Palchetti S; Colapicchioni V; Digiacomo L; Caracciolo G; Pozzi D; Capriotti AL; La Barbera G; Laganà A The Protein Corona of Circulating PEGylated Liposomes. Biochim. Biophys. Acta BBA - Biomembr 2016, 1858 (2), 189–196. [DOI] [PubMed] [Google Scholar]
- (88).Elmoazzen HY; Elliott JAW; McGann LE Osmotic Transport across Cell Membranes in Nondilute Solutions: A New Nondilute Solute Transport Equation. Biophys. J 2009, 96 (7), 2559–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Isaacks RE; Bender AS; Kim CY; Prieto NM; Norenberg MD Osmotic Regulation Of<Emphasis Type=“Italic”>myo</Emphasis>-Inositol Uptake in Primary Astrocyte Cultures. Neurochem. Res 1994, 19 (3), 331–338. [DOI] [PubMed] [Google Scholar]
- (90).Li H; El-Dakdouki MH; Zhu DC; Abela GS; Huang X Synthesis of β-Cyclodextrin Conjugated Superparamagnetic Iron Oxide Nanoparticles for Selective Binding and Detection of Cholesterol Crystals. Chem. Commun 2012, 48 (28), 3385–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Varan G; Öncül S; Ercan A; Benito JM; Ortiz Mellet C; Bilensoy E Cholesterol-Targeted Anticancer and Apoptotic Effects of Anionic and Polycationic Amphiphilic Cyclodextrin Nanoparticles. J. Pharm. Sci 2016, 105 (10), 3172–3182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


