Abstract
The MADS-box gene family encodes transcription factors with many biological functions that extensively regulate plant growth, development and reproduction. Erigeron breviscapus is a medicinal herb used widely in traditional Chinese medicine, and is believed to improve blood circulation and ameliorate platelet coagulation. In order to gain a detailed understanding of how transcription factor expression may regulate the growth of this potentially important medicinal plant, a genome-wide analysis of the MADS-box gene family of E. breviscapus is needed. In the present study, 44 MADS-box genes were identified in E. breviscapus and categorized into five subgroups (MIKC, Mα, Mβ, Mγ and Mδ) according to their phylogenetic relationships with the Arabidopsis MADS-box genes. Additionally, the functional domain, subcellular location and motif compositions of the E. breviscapus MADS-box gene products were characterized. The expression levels for each of the E. breviscapus MADS-box (EbMADS) genes were analyzed in flower, leaf, stem and root organs, and showed that the majority of EbMADS genes were expressed in flowers. Meanwhile, some MADS genes were found to express high levels in leaf, stem and root, indicating that the MADS-box genes are involved in various aspects of the physiological and developmental processes of the E. breviscapus. The results from gene expression analysis under different pollination treatments revealed that the MADS-box genes were highly expressed after non-pollinated treatment. To the best of our knowledge, this study describes the first genome-wide analysis of the E. breviscapus MADS-box gene family, and the results provide valuable information for understanding of the classification, cloning and putative functions of the MADS-box family.
Introduction
MADS-box gene family, one of the most extensively studied transcription factor families, are involved in developmental control and signal transduction in eukaryotes [1]. These genes have been identified in fungi [2], animals [3] and plants [4]. Members of the MADS-box gene family possess a conserved N-terminal DNA-binding domain, of approximately 60 amino acids, which binds to CArG boxes [5]. This MADS domain is named after several of its earliest members: yeast Mini chromosome maintenance 1 (MCM1) [2]; Arabidopsis thaliana AGAMOUS (AG) [5]; snapdragon DEFICIENS (DEF) [4] and human Serum response factor (SRF) [3]. The MADS-box transcription factors were initially identified as floral organ identity-determination genes [4], and play important roles in plant development [6, 7, 8, 9] especially reproduction [10]. Later reports have shown that members of this gene family also regulate other processes such as fruit development [11], embryogenesis [12] and vegetative organ development [13], suggesting a diverse role for this gene superfamily [14]. Parenicova et al. [15] have reported the functions of MADS-box genes in A. thaliana. For example, the MADS-box genes of SOC1 (Suppressor of Overexpression of Constants1), FLC1 (Flowering Locus C1), AGL24 (AGAMOUS-Like24), MAF1/FLM (MADS Affecting Flowering1/ Flowering Locus M) and SVP (Short Vegetative Phase) affect flowering time; the genes of AP1 (Apetala1), FUL (Fruitfull) and CAL (Cauliflower) determine the identity of floral meristem; these genes are related to floral organogenesis, such as AP1, SEP1 to SEP3 (Sepallata), AP3 (Apetala3), PI (Pistillata) and AG (Agamous). SHP1, SHP2 (Shatterproof) and FUL regulate fruit formation while TT16 (Transparent Testa16) influence seed pigmentation and endothelium development [15]. Based on characteristics such as gene structure, encoded protein secondary structure and phylogenetic relationship, the plant MADS-box gene family can be divided into two major lineages: type I and type II [16]. This diversity is generated by an ancestral gene duplication event [17]. The plant type II genes, which possess the highly conserved MADS domain, have been extensively studied over the last decade. These genes are also termed the MIKC type genes due to the four characteristic functional domains: the most conserved MADS (M) DNA binding domain [18], the less well conserved intervening (I) domain which is crucial for the formation of DNA dimers [19], the keratin (K) domain mediating protein-protein interactions [20] and the C-terminus (C) domain for regulating transcription activation [21]. Among the four domains, the K domain is very important to the evolution and functional diversify of the type II MADS-box genes in plants [22]. Conversely, the type I MADS-box gene subfamily in plants has remained largely unexplored [23]. Compared with type II genes, type I MADS-box genes have a relatively simple gene structure and lack the K domain. Furthermore, the type I MADS-box genes contain a highly conserved SRF-like MADS domain. In A. thaliana, type I genes are mainly divided into four subgroups: Mα, Mβ, Mγ and Mδ, based on the phylogenetic relations of the conserved MADS-box domain [15]. The Mδ group is closely related to MIKC* class [24].
With the development of high-throughput sequencing technology, the available whole genome sequences for individual species has expanded exponentially, allowing the systematic study of the expression of key genes and gene families comprehensively during plant growth and development. So far, genome-wide analysis of MADS-box genes have been reported in A. thaliana [25], Populus trichocarpa [26], Oryza sativa [10], Prunus mume [27], Brassica rapa [1], Malus domestica [28], Gossypium hirsutum [29], Beta vulgaris [30], Sesamum indicum [31], Vitis vinifera [32], Cucumis sativus [33], among others. In A. thaliana, 108 MADS-box genes have been identified, with functions for nearly half of them having been described [6]. In addition, there are 32, 34, 41, 57, 75, 80, 105, 160 and 207 MADS-box genes in V. vinifera, B. vulgaris, C. sativus, S. indicum, O. sativa, P. mume, P. trichocarpa, B. rapa and G. hirsutum, respectively. In plants, MADS-box genes widely participate in the development of the roots, leaves, flowers and fruits. For example, Tian et al. [28] cloned MdMADS5 gene from apple, which displayed high homology with AP1 from A. thaliana and could make the flowering time of A. thaliana advance, inflorescence shorten and cluster leaves decrease after the gene was transferred into A. thaliana. The PpMADS11, 12 and 19 genes of peach all showed expression profiles in stamen, petal and other floral organs [34], while in cucumber, CUM26 gene was found to play an important role in the development of petals and stamens [35].
Erigeron breviscapus, also known as dengzhanhua in Chinese, is a perennial herb in the Erigeron genus of the Compositae (Asteraceae) family. It has a beautiful flower which is comprised of yellow disk-like florets and multiple surrounding blue to purple ray florets [36]. The plant is endemic to southwestern China and grows in mid-altitude mountains, subalpine open slopes, grasslands and forest margins from 1000 m to 3500 m [37]. As an important Chinese traditional medicinal plant, E. breviscapus has been widely used to treat various diseases [38, 39, 40]. Recent studies on E. breviscapus have focused on characterizing the chemical components [41], pharmacological activities [42, 43] and germplasm resources [44, 45]. However, little is known about how growth and development of E. breviscapus is regulated at the molecular level. The recent generation of the E. breviscapus whole genome sequence makes a genome-wide analysis of MADS-box genes in E. breviscapus possible [36].
In this study, we identified 44 MADS-box genes from the E. breviscapus genome, analyzed their phylogenetic relationships and defined the conserved motifs. To investigate the underlying molecular mechanisms of MADS-box protein function, we performed a protein-protein interaction network analysis of the MADS-box gene products. In addition, according to the work of Yang et al. [36] and Zhang et al. [46], we analyzed the expression patterns of the E. breviscapus MADS-box genes in four tissues (flower, leaf, stem and root) and three pollination treatments (self-pollinated, cross-pollinated and non-pollinated), and verified by qRT-PCR. To the best of our knowledge, this extended analysis is the first comprehensive study of the MADS-box gene family in E. breviscapus and provides valuable information for understanding the classification, cloning expression and analysis of putative functions of this family. The study will also broaden our insight into the functional evolution of the MADS-box genes in plants.
Materials and methods
Identification of MADS-box genes in E. breviscapus
The genomic and protein sequences of E. breviscapus were downloaded from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/). The sequences of the A. thaliana MADS-box family were retrieved from the Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org/) [15].
To identify all candidate MADS-box genes in E. breviscapus, a local BLASTP search with a threshold e-value of 1e−10 was performed using A. thaliana MADS-box protein sequences as query sequences [47]. The identity and cover region (more than 50%) were used as filter criteria to eliminate improper MADS-box genes. Subsequently, to further verify the reliability of the selected sequences, the Pfam database (http://pfam.sanger.ac.uk/search) was used for domain analysis to ensure the presence of the MADS-box domain in each candidate EbMADS protein [48].
Multiple sequence alignment and phylogenetic analysis between E. breviscapus and A. thaliana
Multiple sequence alignments of MADS-box proteins in E. breviscapus and Arabidopsis were performed using ClustalW program in MEGA X 10.1 software with the default settings [49]. The aligned sequences were saved as a .meg extension by choosing Export Alignment from the Data menu and export the file. Choose Open a File/Session from the file menu and open the .meg aligned sequences file in MEGA X 10.1’s main window. A phylogenetic tree was constructed by the Test Maximum Likelihood (ML) method using the pairwise deletion option and poisson correction model. In addition, bootstrap values were calculated with 1000 replications to examine the statistical reliability of the result [49, 50]. The resulting phylogenetic tree data was exported as a Newick file to import a tree into the FigTree version 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). Polar tree layout was chosen to visualize a ring phylogenetic tree. Taxa and Clade program made the phylogenetic tree aesthetic.
To further confirm the accuracy of the phylogenetic tree, Bayesian analysis was conducted by using Markov chain Monte Carlo (MCMC)method with MrBayes version 3.12 software [51]. The parameters were set as follows: four Markov chains per analysis, run 2 million times with random tree as the starting tree and sample every 100 generations and repeat once. After discarding burn in sample, consensus tree is constructed based on the remaining samples [52, 53].
Analysis of conserved motifs and physicochemical properties
To identify shared motifs and structural divergences among the proteins encoded by the MADS-box genes, translated MADS-box protein sequences were subjected to MEME (version 4.12.0, http://meme-suite.org/tools/meme) analysis using the default parameters with the exception that the number of motifs was set to seven [54].
The ProtParam online tool (http://web.expasy.org/protparam/) was used to estimate the basic physicochemical properties of the protein, such as the isoelectric point and molecular weight of the gene product for each member of E. breviscapus MADS-box gene family. Finally, the subcellular localization of 44 MADS-box genes was predicted by four online analysis tools, such as WoLF PSORT Prediction (https://wolfpsort.hgc.jp/?tdsourcetag=s_pcqq_aiomsg), PSORT Prediction tool (https://www.genscript.com/psort.html), Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) and LocTree3 Prediction system (https://rostlab.org/services/loctree3/). The subcellular localization of the MADS-box genes was retained only results which were confirmed by more than one approach.
Analysis of the protein–protein interaction network
Protein-protein interaction (PPI) data was obtained from the online database STRING (https://string-db.org/cgi/info.pl), an open source software interface for predicting and visualizing complex networks. The data for PPI stored in the database were derived from experimental validation reports in peer-reviewed journals, including the physical interactions and enzymatic reactions found in signal transduction pathways. The PPI data were preprocessed, including removing redundancy and self-loops. Targets with a high confidence score >0.7 were selected to construct the PPI networks [7]. PPI networks are visualized in Cytoscape with the nodes representing the proteins/genes and the edges representing interactions between any two proteins/genes.
Expression analysis of Erigeron breviscapus MADS-box genes
The genome-wide transcriptome data of E. breviscapus in different tissues and three pollination treatments were obtained from the NCBI SRA databases under Bioproject Accession codes PRJNA352312 [36] and SRA24595 [46]. The raw reads that contained adapters or more than 5% unknown ‘N’ and low-quality bases as identified by CycleQ 30, were removed. After filtering, gene expression levels were normalized using edgeR with FPKM (Fragments Per Kilobase of transcript per Million mapped reads) value [55]. An FPKM filtering cutoff of 1.0 in at least one of the collected samples, was used to determine expressed transcripts. According to the GeneID of 44 EbMADS genes in E. breviscapus expressed transcripts, the expression data of these genes in four different tissues (root, stem, leaf and flower) and three pollination treatments (non-pollination, self-pollination and cross-pollination) were obtained. The expression profiles were displayed in a heatmap generated with the Heatmap Illustrator software (v 1.0.3.7) by the default data normalization parameter (Linear) and clustering method (Average Linkage) [56].
Quantitative real-time PCR (qRT-PCR) was performed to further confirm the reliability of the expression profile results via six selected genes (including EbMADS1, 4, 10, 13, 15 and 39). The genome-wide transcription data of E. breviscapus were obtained from four young tissues (root, stem, leaf and flower of wild-type E. breviscapus) and three pollination treatments (harvested at 24 h after non-(Sample T1), self- (Sample T2) and cross-pollination (Sample T3). Total RNA of all collected samples was extracted using the TRIzol Reagent (Takara, Beijing, China) following the manufacturer’s instructions. The qRT-PCR analysis was performed in a Roche detection system (Roche, Switzerland) using SYBR Green assays. 18s RNA was served as the reference gene to normalize the target gene expression and to correct the variation between samples. The gene-specific primers for the qRT-PCR analysis of six selected genes and reference gene were listed in the S1 Table. The reaction conditions were 30 s at 94 °C, 45 cycles of 20 s at 94 °C 20 s at 55 °C, and 30 s at 72 °C. The melting curves were analyzed from 60 °C to 95 °C to observe the specificity of the PCR products. The comparative 2−ΔΔCT method was employed to calculate the relative expression between samples [57]. All calculations were performed using PASW Statistics 18.0 [58].
Results
Identification of the MADS-box genes in E. breviscapus
To identify the members of the E. breviscapus MADS-box gene family, 108 Arabidopsis genes were employed as a query to search against the E. breviscapus database by the BLAST programs. In total, 44 putative MADS-box genes were identified in E. breviscapus and serially named as EbMADS1 through EbMADS44 for convenience (Table 1). Most of the genes contained both SRF-TF domain and K-box domain while some genes coded for either SRF-TF domain or K-box domain. In addition, the results showed that the MADS-box genes varied substantially in the length of the mRNA transcripts and their encoded protein sequences. The length of the 44 EbMADS mRNA products ranged from 117 to 981 bp and the length of the translated protein sequences varied from 39 to 327 amino acids (Table 1).
Table 1. The basic information of MADS-box family members in Erigeron breviscapus.
| Nomenclature | Accession number in NCBI | Length of mRNA | Group | Length of protein | Number of domains | Domains | MW (kDa) | PI | Subcellular location |
|---|---|---|---|---|---|---|---|---|---|
| EbMADS1 | AAO22986 | 588 | Mβ | 196 | 2 | SRF-TF K-box | 22.98 | 7.62 | Nuclear |
| EbMADS2 | CAX65571 | 459 | Mγ | 153 | 1 | K-box | 17.54 | 6.34 | Nuclear |
| EbMADS3 | CAH04879 | 849 | Mγ | 283 | 2 | SRF-TF K-box | 32.21 | 8.40 | Nuclear |
| EbMADS4 | CAA57445 | 708 | Mγ | 236 | 2 | SRF-TF K-box | 27.24 | 9.60 | Nuclear |
| EbMADS5 | AAO22982 | 489 | Mγ | 163 | 2 | SRF-TF K-box | 18.71 | 9.93 | Nuclear |
| EbMADS6 | CAA08802 | 642 | Mβ | 214 | 2 | SRF-TF K-box | 24.92 | 9.86 | Nuclear |
| EbMADS7 | AAV65497 | 501 | Mγ | 167 | 1 | K-box | 18.61 | 7.61 | Nuclear |
| EbMADS8 | EOX92192 | 981 | Mδ | 327 | 1 | SRF-TF | 37.47 | 5.72 | Nuclear |
| EbMADS9 | ADU15473 | 351 | Mβ | 117 | 1 | K-box | 14.13 | 9.83 | Nuclear |
| EbMADS10 | ACV86637 | 126 | Mγ | 42 | 1 | SRF-TF | 4.84 | 11.61 | Nuclear |
| EbMADS11 | BAL14660 | 711 | Mγ | 237 | 2 | SRF-TF K-box | 27.63 | 8.46 | Nuclear |
| EbMADS12 | BAK09621 | 501 | Mγ | 167 | 1 | K-box | 19.04 | 8.23 | Nuclear |
| EbMADS13 | XP_002327838 | 378 | Mα | 126 | 1 | SRF-TF | 14.12 | 5.71 | Nuclear |
| EbMADS14 | CAX65661 | 744 | Mγ | 248 | 2 | SRF-TF K-box | 28.27 | 9.90 | Nuclear |
| EbMADS15 | EOX92192 | 981 | Mδ | 327 | 1 | SRF-TF | 37.44 | 5.72 | Nuclear |
| EbMADS16 | CBI15681 | 285 | Mγ | 95 | 1 | K-box | 11.28 | 10.54 | Nuclear |
| EbMADS17 | AAO22980 | 771 | Mγ | 257 | 2 | SRF-TF K-box | 29.76 | 10.05 | Nuclear |
| EbMADS18 | XP_002278239 | 417 | Mγ | 139 | 1 | K-box | 16.03 | 4.49 | Nuclear |
| EbMADS19 | AAV65497 | 501 | Mγ | 167 | 1 | K-box | 18.61 | 7.61 | Nuclear |
| EbMADS20 | ADU56833 | 672 | Mβ | 224 | 2 | SRF-TF K-box | 25.14 | 5.52 | Nuclear |
| EbMADS21 | ADU56833 | 672 | Mβ | 224 | 2 | SRF-TF K-box | 25.14 | 5.52 | Nuclear |
| EbMADS22 | Q9ATE5 | 765 | MIKC | 255 | 2 | SRF-TF K-box | 29.68 | 7.16 | Nuclear |
| EbMADS23 | ACH61565 | 315 | Mγ | 105 | 1 | SRF-TF | 12.32 | 11.10 | Nuclear |
| EbMADS24 | AGQ04483 | 147 | Mγ | 49 | 1 | SRF-TF | 5.88 | 10.28 | Nuclear |
| EbMADS25 | AFA37965 | 705 | Mβ | 235 | 2 | SRF-TF K-box | 26.28 | 5.08 | Nuclear |
| EbMADS26 | EPS59489 | 117 | Mγ | 39 | 1 | SRF-TF | 4.57 | 11.29 | Nuclear |
| EbMADS27 | ACV74250 | 675 | Mβ | 225 | 2 | SRF-TF K-box | 25.41 | 5.23 | Nuclear |
| EbMADS28 | CCO61905 | 645 | MIKC | 215 | 2 | SRF-TF K-box | 24.37 | 6.78 | Nuclear |
| EbMADS29 | CBX45125 | 438 | Mγ | 146 | 1 | K-box | 17.22 | 8.21 | Nuclear |
| EbMADS30 | ADK94172 | 399 | Mγ | 133 | 1 | K-box | 15.22 | 6.27 | Nuclear |
| EbMADS31 | BAN19212 | 654 | Mγ | 218 | 2 | SRF-TF K-box | 25.24 | 8.20 | Nuclear |
| EbMADS32 | CAX65663 | 447 | Mγ | 149 | 2 | SRF-TF K-box | 17.28 | 10.27 | Nuclear |
| EbMADS33 | CBI28594 | 348 | Mβ | 116 | 1 | SRF-TF | 13.2 | 10.11 | Nuclear |
| EbMADS34 | XP_004170688 | 129 | Mγ | 43 | 1 | SRF-TF | 5.00 | 10.96 | Nuclear |
| EbMADS35 | AFO10123 | 516 | MIKC | 172 | 2 | SRF-TF K-box | 20.02 | 10.02 | Nuclear |
| EbMADS36 | CAA57445 | 708 | Mγ | 236 | 2 | SRF-TF K-box | 27.24 | 9.60 | Nuclear |
| EbMADS37 | AAO22986 | 588 | Mβ | 196 | 2 | SRF-TF K-box | 22.81 | 9.60 | Nuclear |
| EbMADS38 | AAO18231 | 441 | Mβ | 147 | 2 | SRF-TF K-box | 17.29 | 9.97 | Nuclear |
| EbMADS39 | CCO61905 | 696 | MIKC | 232 | 2 | SRF-TF K-box | 26.25 | 6.30 | Nuclear |
| EbMADS40 | XP_002522085 | 618 | Mα | 206 | 1 | SRF-TF | 23.21 | 10.37 | Nuclear |
| EbMADS41 | BAN19212 | 654 | Mγ | 218 | 2 | SRF-TF K-box | 25.24 | 10.37 | Nuclear |
| EbMADS42 | AAO22986 | 588 | Mβ | 196 | 1 | K-box | 22.98 | 7.62 | Nuclear |
| EbMADS43 | XP_002278584 | 783 | Mβ | 261 | 2 | SRF-TF K-box | 29.62 | 10.05 | Nuclear |
| EbMADS44 | BAN19217 | 204 | Mγ | 68 | 1 | K-box | 7.86 | 4.41 | Nuclear |
The physicochemical properties of the 44 complete MADS-box amino acid sequences from E. breviscapus were analyzed using ProtParam (Table 1). The results showed that the molecular weight of these EbMADS proteins ranged from 4.57 to 37.47 kDa. Most of the EbMADS proteins exhibited alkaline isoelectric points greater than 7.5, with the highest being 11.61 for EbMADS10, while 12 proteins had acidic isoelectric points of less than 6.5, of which EbMADS44 was the lowest at 4.41. Two proteins, EbMADS22 and EbMADS28 had relatively neutral isoelectric points that fell between 6.5 and 7.5.
Further analysis using four protein subcellular location prediction tools was performed to exactly predict the subcellular localization for the products of the EbMADS gene family (S2 Table). As the result shown, all MADS-box proteins were most likely to be located in the nucleus, indicating that although the physicochemical properties of MADS-box transcription factors differed greatly, subcellular location was very conservative (Table 1). Altogether, the results suggest that EbMADS proteins, as transcription factors, play a transcriptional regulatory role directly in the nucleus, consistent with the characteristics of the MADS-box family as transcription factors that regulate transcription of nuclear genomic DNA.
Classification and phylogenetic analysis of the EbMADS and AtMADS gene families
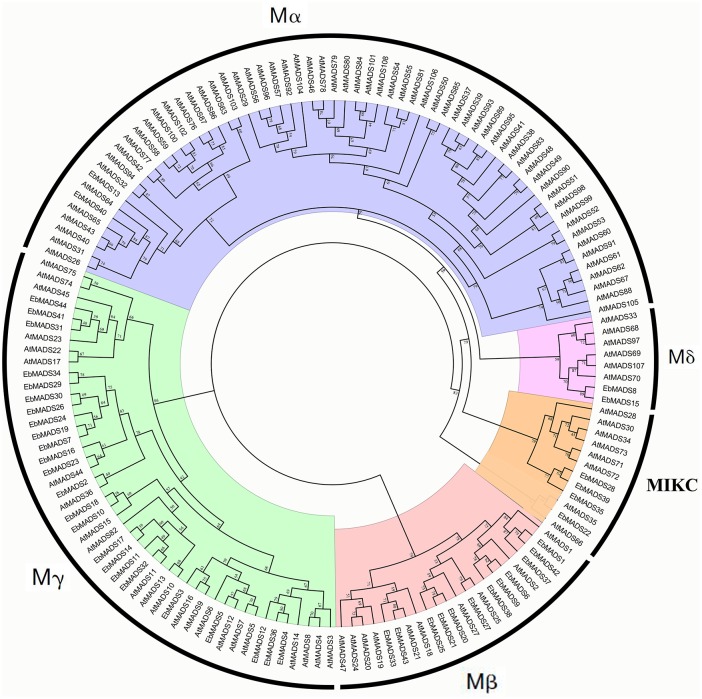
To investigate the evolutionary relationship between E. breviscapus MADS-box genes in detail, we performed multiple sequence alignments and generated a phylogenetic tree for MADS-box proteins from E. breviscapus and A. thaliana. The phylogenetic tree was constructed on the basis of the consistency of Maximum Likelihood and Bayesian phylogenetic tree. In our study, the 44 EbMADS genes were classified into functional groups according to A. thaliana MADS-box genes that had been extensively studied (Fig 1) [15]. EbMADS members could be divided into two groups: type I and type II. According to the A. thaliana phylogenetic relationships, type I proteins could be further divided into three subfamilies as follows: Mα, Mβ and Mγ whereas the type II group contained Mδ and MIKC proteins. As shown in Fig 1, among the 38 type I members in E. breviscapus, two were grouped as Mα, 12 as Mβ, 24 as Mγ. However, only two Mδ and four MIKC proteins comprised the EbMADS type II proteins, such as EbMADS8, 15, 22, 28, 35 and 39. The phylogenetic tree and the website information of Arabidopsis MADS-box Transcription Factor Gene Family (https://www.arabidopsis.org/browse/genefamily/MADSlike.jsp) further explained the relationships of the EbMADS genes classification by function and structure. For example, EbMADS13 and EbMADS40 of the Mα group, EbMADS8 and EbMADS15 of the Mδ group, EbMADS20, 21, 25, 33 and 43 of Mβ group and EbMADS2, 3, 7, 10, 16, 18, 19, 23, 24, 26, 29, 30 and 34 of the Mγ group were all AGL subgroup indicating these affected flowering time. The group of Mγ and SOC1 including EbMADS31, 41 and 44, Mβ and SVP group embodying EbMADS27 and the group of MIKC and MAF containing EbMADS28, 35 and 39 were relevant to the flowering time. Six MADS-box genes of Mβ group which EbMADS1, 37 and 42 pertaining to PI subgroup and EbMADS6, 9 and 38 belong to AP3 subgroup and EbMADS5 of the Mγ and SEP group were related to floral organogenesis. The Mγ and FUL group including EbMADS11, 14, 17 and 32 affected the determination of floral meristem identity while the EbMADS22 of MIKC and TT16 group played an important role in seed pigmentation and endothelium development. EbMADS4, 12 and 36 in Mγ group were belong to unknown subgroups.
Fig 1. The phylogenetic tree construction of MADS-box genes in E. breviscapus and A. thaliana.
Maximum Likelihood (ML) phylogenetic tree of all detected MADS-box genes was constructed, using MEGA X 10.1 program with bootstrap analysis (1000 replicates). MADS-box genes in the phylogenetic tree were clustered into five distinct groups (Groups Mα, M β, M γ, M δ and MIKC).
Conserved motifs analysis of EbMADS gene families
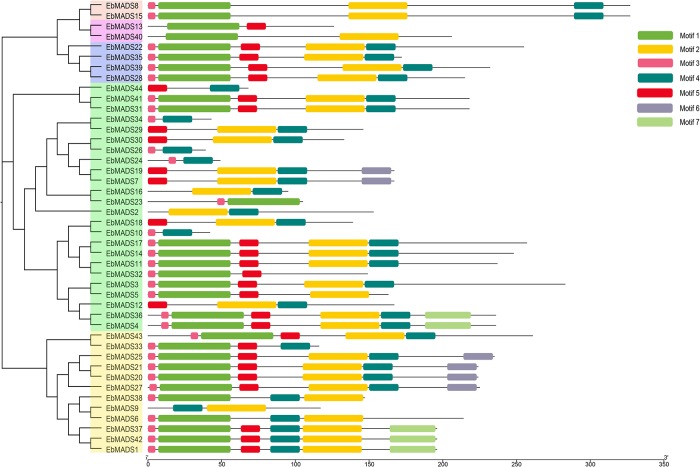
To assess the diversity and similarity of motif composition among the different E. breviscapus MADS-box genes, the MEME tool was employed to identify motifs within the 44 MADS-box protein sequences. A total of seven conserved motifs (denoted motifs 1–7; Fig 2) were identified in the MADS-box proteins and their consensus sequence information and logo are displayed in Table 2 and S1 Fig respectively. Given the phylogenetic tree and conserved motifs, we note that the EbMADS genes clustered in the same subgroup shared substantially consistent conserved motifs, which indicates that members of the same subgroup might possess functional similarities. Mδ-clade (EbMADS8 and EbMADS15) and MIKC-clade (EbMADS22, EbMADS35, EbMADS39 and EbMADS28) proteins of the type II family contained MADS domains with similar motif compositions. Both clades contained motif1, motif 2, motif 3 and motif 4, while the MIKC subgroup also included motif 5. Conversely, members of the type I MADS family displayed quite different motif composition. Mα, consisting of EbMADS13 and EbMADS40, only had two motifs, either motifs 1 and 5 or motifs 1 and 2. The MADS domains of the majority of the Mβ subfamily contained motif 1, motif 3 and motif 4 except for EbMADS9, which had neither motif 1 nor motif 6. The Mγ clade had the most members [29] and showed a complex motif profile. For example, eight gene members all coded for two varied motifs. The remaining gene domains contained at least three motifs, with most having motif 4 in the MADS domain. According to the homology comparison annotation in A. thaliana, motif 1 was related to DNA binding and motif 3 was found to concern with nuclear localization, which further illustrated the nuclear location of the EbMADS gene family. The functions of other motifs were unknown. Furthermore, the seven motifs and corresponding logos of A. thaliana were analyzed (S2 Fig). Each capital letter represented an amino acid in the motif logos. Same amino acid (or same capital letter) at the same position, suggesting the frequencies of amino acids used by motifs were conservative. As shown in the S1 and S2 Figs, motif distribution in A. thaliana was more conservative than E. breviscapus. However, the frequencies of amino acids used by motifs were not very conservative in A. thaliana, which was similar with E. breviscapus.
Fig 2. Motif analysis of MADS-box gene family in Erigeron breviscapus.
Motif compositions: protein sequences are indicated by thick gray lines, and the conserved motifs are represented by different colored boxes. The length (amino acids) of the protein and motif can be estimated using the scale bar at the bottom.
Table 2. The information of motif found in MEME.
| Motifs | Conserved amino acid sequences | E-value | Sites | Width |
|---|---|---|---|---|
| 1 | IKRIENNTNRQVTFSKRRNGLLKKAHELSVLCDAEVALIVFSSTGKLYEY | 1.0e-944 | 30 | 50 |
| 2 | RHLLGEDLGGLSLKELEQLEQQLEDGLSRVRSRKDQLMLEQ | 7.3e-317 | 35 | 41 |
| 3 | MGRGKI | 5.6e-067 | 33 | 6 |
| 4 | IENLQEKEKKLKEENEGLKKK | 5.6e-072 | 40 | 21 |
| 5 | MEDILERYQRHSKN | 4.4e-057 | 31 | 14 |
| 6 | SGPPPEDDGSDTSLKLGLPFS | 5.5e-031 | 6 | 21 |
| 7 | QQSEMNTMGEYQNHQPFSFRVQPLQPNLMERI | 1.8e-034 | 5 | 32 |
Analysis of EbMADS protein function link network
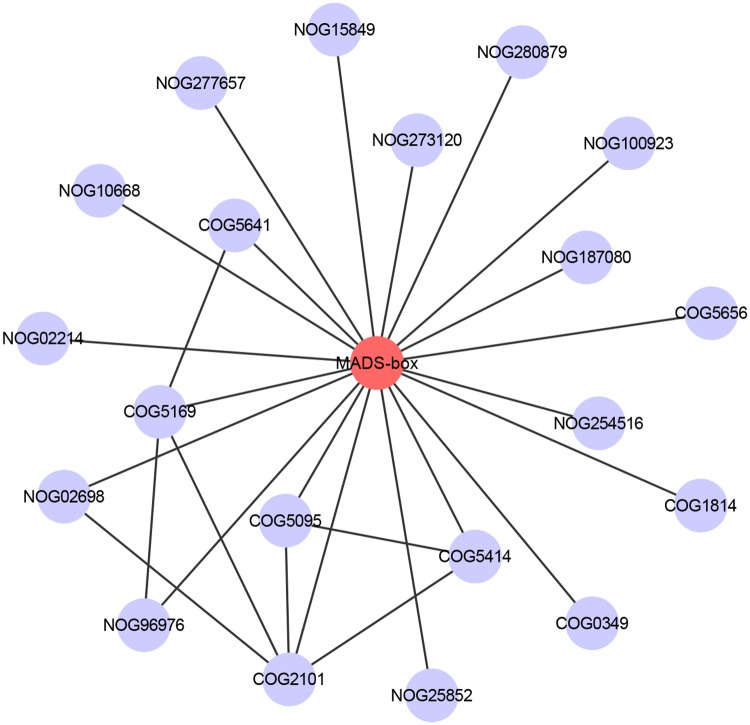
To investigate the potential molecular mechanisms of E. breviscapus MADS-box proteins, the protein patterns stored in the STRING database were used to construct the PPI network. From the results, we found that the EbMADS proteins exhibited a protein-protein interaction with 20 other proteins (Fig 3). Among the co-expression proteins, COG2101 (TATA-box binding protein, component of TFIID and TFIIIB), COG5169 (Heat shock transcription factor), COG5095 (Transcription initiation factor TFIID, subunit TAF6) and COG5414 (TATA-binding protein-associated factor) featured prominently in the protein-protein network, indicating that those proteins are vital to maintaining the protein interactions in the network. Moreover, NOG02698, NOG96976 and COG5641 (GATA Zn-finger-containing transcription factor) played a pivotal role in the network. While most of the key contacts in the PPI network are involved in transcriptional regulation, the function of NOG02698 and NOG96976 are unclear. Additional interactions were noted with non-transcription related proteins, such as COG5656 (Importin, protein involved in nuclear import) and COG0349 (Ribonuclease D). The results presented in this study have provided a way to identify the key proteins which could interact with EbMADS proteins, detailed information this PPI network are listed in S3 Table.
Fig 3. The analysis of EbMADS protein function link network.
The PPI data were preprocessed, including removing redundancy and self-loops. Targets with a high confidence score >0.7 were selected to construct the PPI networks. PPI networks are visualized in Cytoscape with the nodes representing the proteins/genes and the edges representing interactions between any two proteins/genes.
Tissue specific expression profiles for E. breviscapus MADS-box genes
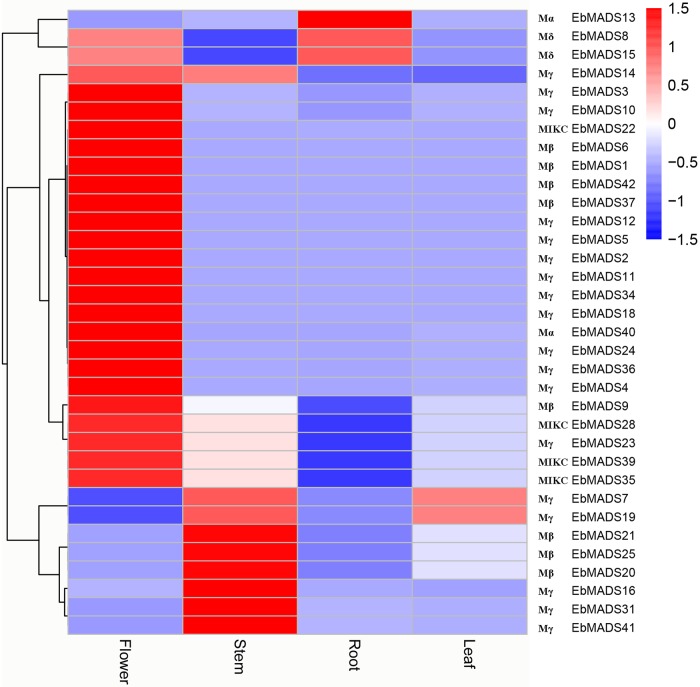
MADS-box genes are expressed in different plant organs, such as the vegetative organ roots, stems, leaves, reproductive organs, fruits and seeds, and play important regulatory roles in plant development, growth and reproduction [59, 60]. In order to gain insight into the tissue specific E. breviscapus MADS-box gene expression pattern and to elucidate their potential roles in tissue development, we utilized transcriptome data derived from Illumina RNA-Seq reads generated by Yang et al. [36]. The transcript abundance from each of the 34 EbMADS genes in four different tissues, including root, stem, leaf and flower, were analyzed and compared. As shown in Fig 4, EbMADS genes were expressed in all four E. breviscapus tissues studied. The majority of EbMADS genes showed high expression levels in flowers, consistent with MADS-box genes being originally identified as floral organ regulatory genes [4]. For instance, EbMADS1, 37 and 42 of PI subgroup and EbMADS6, 9 and 38 of AP3 subgroup were B-class genes in the ABC model, related to floral organogenesis. EbMADS8 and EbMADS15, belong to Mδ group, were expressed in both flowers and roots to similar levels, while the expression of EbMADS7 and EbMADS19 genes of Mγ group were noted in both leaves and roots. Three MIKC-type MADS-box genes, including EbMADS28, EbMADS35 and EbMADS39, were all expressed in both flowers and stems, however the transcript levels were less in stems than in flowers. Interestingly, ten EbMADS genes showed no expression in any of the tissue expression data studied. These genes played an important role in other plant tissues such as fruit development could be speculated [8]. To further confirm the expression profiles of the MADS-box genes in four tissues, six EbMADS genes were selected for qRT-PCR analysis (S3 Fig). EbMADS1, 4, 10 and 39 showed high expression levels in flower while EbMADS13 had maximal expression in root. EbMADS15 presented high levels of expression in flower and root. The results of qRT-PCR had strong consistency with those of transcriptome analysis.
Fig 4. Expression profiles for E. breviscapus MADS-box genes in different tissues.
The tissues included leaf, root, stem, and flower. Gene expression levels were calculated using the FPKM method. The bar at the right of each heat map represents relative expression values, thereby blue color representing low level expression, white shows medium level expression and red signifies high level expression.
Effect of pollination treatment on expression patterns of E. breviscapus MADS-box genes
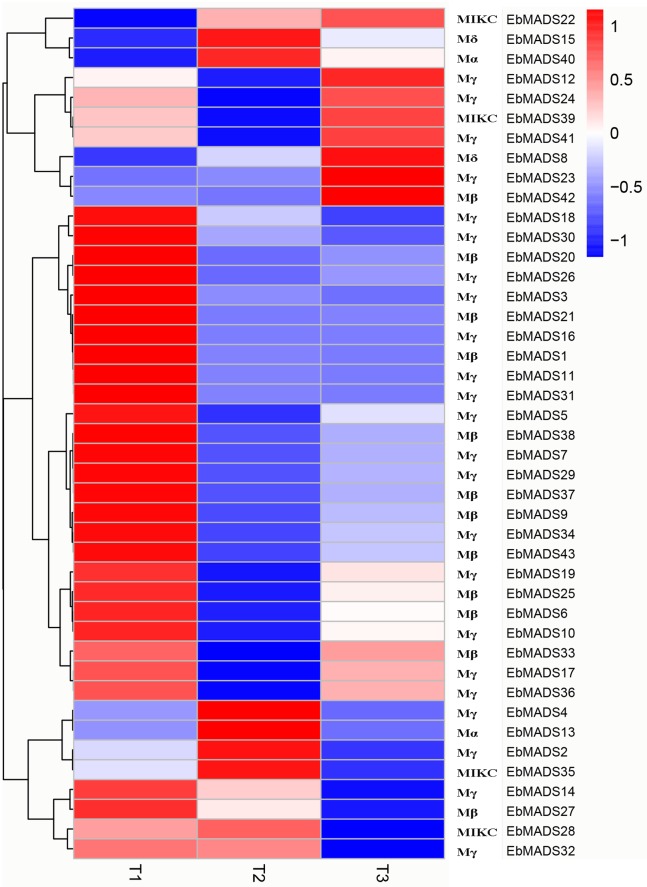
Self-incompatibility (SI) is an important mating system in many flowering plants, which ensures genetic diversity and is beneficial to plant evolution and adaptation to the environment [61, 62]. As a species of Asteraceae, E. breviscapus is self-incompatible. To further understand the potential functions of MADS-box genes in E. breviscapus SI responses, genome-wide transcriptome data, from three different pollination treatments, deposited by Zhang et al. [46], was analyzed. Heatmap representation of the expression profiles of the 43 EbMADS genes in non- (Sample T1), self- (Sample T2) and cross-pollination (Sample T3) treatments are shown in Fig 5, revealing that most of the MADS-box genes displayed a broad expression spectrum after non-pollination treatment. From the results, we found that a total of 26 MADS-box members exhibited maximal expression in this data set. Conversely, EbMADS24, EbMADS41, EbMADS17, EbMADS36 and EbMADS32 of Mγ subfamily and MIKC subfamily including EbMADS39 and EbMADS28 shared the characteristic of having low expression after non-pollination treatment. In the cross-pollination treatment data, eight MADS-box genes displayed high expression levels, with six additional genes, including EbMADS19, EbMADS10, EbMADS17 and EbMADS36 of Mγ group and Mβ group containing EbMADS25 and EbMADS33, showing low expression levels. However, compared with the expression patterns observed for non- and cross-pollination data, expression of the MADS-box genes was significantly down-regulated in self-pollination treatment. For example, while transcript abundances for EbMADS15, EbMADS40, EbMADS13, EbMADS4, EbMADS2 and EbMADS35 genes were all high, EbMADS22, EbMADS27, EbMADS14 and EbMADS32 showed relatively low transcript abundance. These results showed that MADS-box genes expression may be inhibited during self-pollination, causing the self-incompatibility of E. breviscapus reproduction. The results of qRT-PCR about the six EbMADS genes were significantly corroborated those of transcriptome analysis (S4 Fig). For example, EbMADS1 and EbMADS10 showed high expression levels under non-pollination treatment while EbMADS4 and EbMADS13 had maximal expression levels under self-pollination treatment. The EbMADS15 mainly expressed under self- and cross-pollination treatments. EbMADS39 showed higher expression levels in the treatments of non- and cross-pollination.
Fig 5. Expression patterns of E. breviscapus MADS-box genes in three pollination treatments.
T1, T2 and T3 indicated samples were respectively forced with non-pollinated, self-pollinated, and cross-pollinated treatment. The bar at the right of each heat map represents relative expression values, thereby blue color representing low level expression, white shows medium level expression and red signifies high level expression.
Discussion
E. breviscapus is an important traditional Chinese medicine. At present, it is widely used as raw material ingredients in remedies for cardiovascular and cerebrovascular diseases [63]. Use in the treatment of diabetes, nephropathy and senile diseases are also common [64]. In addition, it has been reported that E. breviscapus has an anti-cancer effect [65]. MADS-box genes, important transcription factors in plants, play significant roles in plant growth and development process. In flowering plants, MADS-box proteins have important biological significance in a wide range of processes, including the control of flowering time, the determination of floral meristem, the identification of floral organ, fruit development, endosperm development and nutritional development [59]. Recently, as successive advances are made in the genome sequencing of various species, the knowledge of the role of MADS-box genes in plant development is deepening. Increasingly, MADS-box gene families in numerous plants have been identified and analyzed from sequenced species using genome-wide bioinformatics analysis tools. MADS-box gene family identification and evolution analysis of A. thaliana [15], O. sativa [10], P. trichocarpa [26], V. vinifera [32], Cucumis sativus [66], Glycine max [9], P. mume [27], B. rapa [1], M. domestica [28] and G. hirsutum [29] have been successively completed. In the current study, these bioinformatics tools have been used to identify and predict the function of the MADS genes in the complete genome sequence of E. breviscapus.
MADS-box genes contain a highly conserved MADS-box domain composed of about 60 amino acids. A phylogenetic analysis of MADS-box genes from A. thaliana, fungi and animals performed by Alvarez-Buylla et al. [67] showed that the MADS-box genes underwent a gene duplication before the divergence of plants and animals, bringing about type I (SRF-like) and type II (MEF2-like) lineage. According to the MADS-box gene structure, duplication and motif analysis of A. thaliana, Parenicova et al. [15] suggested the type-I and type-II MADS-box genes can be further divided into five distinct subgroups, named Mα, Mβ, Mγ, Mδ and MIKC. Phylogenetic analysis of the MADS-box gene family in rice, determined that type I genes contained four subfamilies Mα, Mβ, Mγ and Mδ; while type II consisted of MIKC subgroups [10]. Of the 146 MADS-box genes identified in apple, 82 members could be unambiguously classified as MIKC type II, whereas the remaining 64 members were classified as type I (including Mα, Mβ, Mγ and Mδ) [28]. However, as shown in the phylogenetic dendrogram for P. mume MADS-box genes, while the genes of Mα, Mβ and Mγ subfamilies were type I MADS-box genes, the Mδ clade showed a similar phylogenetic tree to the type II genes [27]. This is consistent with the phylogenetic analysis of three cotton species (Gossypium raimondii, Gossypium arboreum and Gossypium hirsutum), determined that the type I lineage contained Mα, Mβ and Mγ groups while the type II lineage was comprised of both Mδ and MIKC [29]. Interestingly, studies of some species showed the absence of the Mδ subfamily. In this study, comparison of the phylogenetic trees of A. thaliana and E. breviscapus determined that E. breviscapus MADS-box genes were subdivided into five groups, including Mα, Mβ, Mγ, Mδ and MIKC. The MADS-box domains of Mα, Mβ, Mγ and MIKC were generally conserved, showing similar motif structures. The Mδ gene domains were simple and noted as components of the MIKC gene motifs. The Mδ and MIKC clades were closely related in the phylogenetic tree. Therefore, the type I MADS-box genes were confirmed to consist of three subgroups: Mα, Mβ and Mγ while the Mδ and MIKC clades formed type II MADS-box genes. The classification is similar to G. raimondii, G. arboreum and G. hirsutum [29].
MADS-box genes are widely expressed in plants, and known to be involved in multifarious and important aspects of vegetal development and differentiation. As key players in the regulation of developmental mechanisms at the molecular level, the function of MADS-box genes are extensively observed, not only for the flower organ [68], but also the regulation in fruit [69], root and leaf development [16]. In the present study, we use tissue specific transcriptomic data to compare the expression of E. breviscapus MADS-box genes in the flowers, stems, roots and leaves. The majority of MADS-box genes of E. breviscapus shared the same expression patterns in flowers, implying a functional redundancy in this organ and consistent with MADS-box genes originally being identified as flower related genes. The ABC model was widely known to explain the combined functions of A (AP1 and AP2), B (PI and AP3) and C (AG) classes genes to determine the Arabidopsis flower organs identity [15]. In E. breviscapus, there were only B-class genes (EbMADS1, 37 and 42 of PI, EbMADS6, 9 and 38 of AP3) further suggesting the six MADS-box genes were exactly related to floral organogenesis. In addition, the results of 25 EbMADS genes expressed in flowers most agreed with the categories by function, such as EbMADS28, 35 and 39 of MAF subgroup related to flowering time, EbMADS11 and 14 of FUL subgroup and flower meristem identity to be interrelated and EbMADS5 in SEP subgroup bound up with floral organogenesis. Furthermore, MADS-box genes are expressed in the flowers of many plants. For example, OsMADS3 controls terminal anther development in rice through regulating ROS homeostasis [70]. Nine out of 18 members of the MADS-box genes in cherry had expression profiles only in flower organs [71]. In Crocus sativus, the CsMADS genes, belonging to different MADS-box subfamilies direct the formation of floral organs by regulating the development of flower organs in different rounds [72]. In this study, more EbMADS genes were expressed in stems than in roots and leaves. Similarly, in sesame plants, SiMADS genes were highly expressed in both the flower buds and stem tips, where the first flower appears at the top of stem at the 10 or 12 leaf stage [31]. This may also be the reason why more gene members of E. breviscapus MADS-box family show specific and efficient expression in plant stems.
Studies have indicated that the MADS-box genes play an important role in the plant pollination process. Yang et al. [36] found the MADS-box gene in O. sativa, named OsMADS29, was highly expressed in development seeds after pollination [73]. The steady state expression of two maize MADS-box genes ZMM6 and ZMM27 increased in kernels after pollination [74]. And Ning et al. [75] suggested activated carbohydrate metabolism, cell division and expansion as well as the down-regulation of MADS-box could comprehensively regulate the plant pollination-dependent and parthenocarpic fruit set. E. breviscapus possesses the pollination system [76]. However, E. breviscapus is a member of the Asteraceae family, the archetypical plant that displays self-incompatible reproduction [77]. Such self-incompatibility systems widely exist in plants as a mechanism to maintain genetic diversity in their offspring [78]. Self-incompatibility or self-sterility, is the situation where a plant lacks the ability to self-pollinate. To understand the possible role of EbMADS genes in the self-incompatibility reaction, we performed an expression profile analysis on transcriptomic data from self-pollination and cross-pollination experiments. The results showed most of the EbMADS genes displayed high expression levels in the non-pollinated treatment data, which indicates that MADS-box gene family has a vital impact on E. breviscapus growth and development. Interestingly, the expression patterns of the genes from cross-pollinated plants were similar to that from non-pollinated plants, suggesting that cross-pollination plays an important role in the natural development progress of the E. breviscapus. Conversely, only seven genes (including EbMADS22, EbMADS15, EbMADS40, EbMADS4, EbMADS13, EbMADS2 and EbMADS35) displayed high expression levels in the self-pollinated treatment, indicating that MADS-box genes expression may be inhibited during self-pollination suggesting the molecular mechanism that may underlie the self-incompatibility of E. breviscapus.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(DOC)
(DOC)
(XLS)
Acknowledgments
The authors thank Yong Wang and Lijun Fu for critical reading of this manuscript.
Data Availability
All files of genome-wide transcriptome data of Erigeron breviscapus in different tissues and three pollination treatments are available from the NCBI SRA database (accession numbers PRJNA352312,SRA245957).
Funding Statement
This research was funded by National Natural Science Foundation of China (NSFC) (Grant no. 31460447 and 31700068), Doctor and master specific projects of Honghe University (XJ17B09), the project of young and middle-aged talent of Yunnan province (Grant No. 2017HB076).
References
- 1.Duan W, Song X, Liu T, Huang Z, Ren J, Hou X, et al. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Molecular Genetics & Genomics. 2015;290(1):239–55. [DOI] [PubMed] [Google Scholar]
- 2.Passmore S, Maine GT, Elble R, Christ C, Tye BK. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. Journal of Molecular Biology. 1988;204(3):593–606. 10.1016/0022-2836(88)90358-0 [DOI] [PubMed] [Google Scholar]
- 3.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55(6):989–1003. 10.1016/0092-8674(88)90244-9 [DOI] [PubMed] [Google Scholar]
- 4.Sommer H, Beltrán JP, Huijser P, Pape H, Nnig WE, Saedler H, et al. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. Embo Journal. 1990;9(3):605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346(6279):35–9. 10.1038/346035a0 [DOI] [PubMed] [Google Scholar]
- 6.Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(5):1560–5. 10.1073/pnas.1112871109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu Q, Huang Y, Wang Z, Chen F, Huang D, Lu Y, et al. Proteome Profile and Quantitative Proteomic Analysis of Buffalo (Bubalusbubalis) Follicular Fluid during Follicle Development. International Journal of Molecular Sciences. 2016;17(5):618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu W and Xi WP. MADS-box Transcription Factors are Involved in Regulation for Fruit Ripening and Quality Development. Acta Horticulturae Sinica. 2018; 45(09):1802–1812. [Google Scholar]
- 9.Fan CM, Wang X, Wang YW, Hu RB, Zhang XM, Chen JX, et al. Genome-wide expression analysis of soybean MADS genes showing potential function in the seed development. Plos One. 2013;8(4):e62288 10.1371/journal.pone.0062288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. Bmc Genomics. 2007;8(1):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development. 1998;125(8):1509–17. [DOI] [PubMed] [Google Scholar]
- 12.Heck GR, Perry SE, Nichols KW, Fernandez DE. AGL15, a MADS domain protein expressed in developing embryos. Plant Cell. 1995;7(8):1271–82. 10.1105/tpc.7.8.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrándiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289(5478):436–8. 10.1126/science.289.5478.436 [DOI] [PubMed] [Google Scholar]
- 14.Rounsley SD, Ditta GS, Yanofsky MF. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell. 1995;7(8):1259–69. 10.1105/tpc.7.8.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parenicova L, De FS, Kieffer M, Horner DS, Favalli C, Busscher J, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15(7):1538–51. 10.1105/tpc.011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Buylla ER, Liljegren SS, Gold SE, Burgeff C, Ditta. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant Journal. 2010;24(4):457–66. [DOI] [PubMed] [Google Scholar]
- 17.Becker A, Theiben G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics & Evolution. 2003;29(3):464–89. [DOI] [PubMed] [Google Scholar]
- 18.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229(1):1–13. [DOI] [PubMed] [Google Scholar]
- 19.Riechmann JL, Meyerowitz EM. MADS domain proteins in plant development. Biological Chemistry. 1997;378(10):1079–101. [PubMed] [Google Scholar]
- 20.Fan HY, Hu Y, Tudor M, Ma H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant Journal. 2010;12(5):999–1010. [DOI] [PubMed] [Google Scholar]
- 21.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409(6819):525–9. 10.1038/35054083 [DOI] [PubMed] [Google Scholar]
- 22.Lai X, Daher H, Galien A, Hugouvieux V, Zubieta C. Structural Basis for Plant MADS Transcription Factor Oligomerization. Comput Struct Biotechnol. 2019; 17: 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodt SD, Raes J, Florquin K, Rombauts S, Rouzé P, Theißen G, et al. Genomewide Structural Annotation and Evolutionary Analysis of the Type I MADS-Box Genes in Plants. Journal of molecular evolution. 2003;56(5):573–86. 10.1007/s00239-002-2426-x [DOI] [PubMed] [Google Scholar]
- 24.Stefanie DB, Jeroen R, Yves VdP, Günter T. And then there were many: MADS goes genomic. Trends in Plant Science. 2003;8(10):475–83. 10.1016/j.tplants.2003.09.006 [DOI] [PubMed] [Google Scholar]
- 25.Lucie P, Stefan dF, Martin K, David S H, Cristina F, Jacqueline B, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. The Plant Cell. 2003;15(7):1538–51. 10.1105/tpc.011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leseberg C, Li A, Kang H, Duvall M, Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene. 2006;378(1):84–94. [DOI] [PubMed] [Google Scholar]
- 27.Zongda X, Qixiang Z, Lidan S, Dongliang D, Tangren C, Huitang P, et al. Genome-wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume. Molecular Genetics & Genomics. 2014;289(5):903–20. [DOI] [PubMed] [Google Scholar]
- 28.Tian Y, Dong Q, Ji Z, Chi F, Cong P, Zhou Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene, 2014, 555(2):277–290. 10.1016/j.gene.2014.11.018 [DOI] [PubMed] [Google Scholar]
- 29.Nardeli SM, Artico S, Aoyagi GM, Moura SMD, Silva TDF, Grossi-De-Sa MF, et al. Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii). Plant Physiology & Biochemistry. 2018;127:169–184. [DOI] [PubMed] [Google Scholar]
- 30.Kong WL, Zhang KD, Wu JC, Zhang LP, Pan H, Tang JW, et al. Genome-wide identification and phylogenetic analysis of MADS-box family gene in Beta vulgaris. Acta Agriculturae Boreali-Sinica, 2018;33(1):86–95. [Google Scholar]
- 31.Wei X, Wang L, Yu J, Zhang Y, Li D, Zhang X. Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene. 2015;569(1):66–76. 10.1016/j.gene.2015.05.018 [DOI] [PubMed] [Google Scholar]
- 32.José DR, Diego L, José M MZ, María José C. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiology. 2009;149(1):354–69. 10.1104/pp.108.131052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan DF, Ding F, Zhuang D, Liang DD. Genome-wide sequence characterization analysis of MADS-box transcription factor gene family in cucumber (Cucumis sativus L.). Journal of Nuclear Agricultural Sciences. 2012;26(9):1249–1256. [Google Scholar]
- 34.Li HF, Jia HZ, Dong QL, Ran K, Wang HW. Cloning and Expression Analysis of Ten MADS-box Genes in Peach(Prunus persica var. nectarina ‘Luxing’). Scientia Agricultura Sinica. 2016; 49(23):4593–4605. [Google Scholar]
- 35.Sun JJ, Feng L, Wang DH, Liu XF, Xia L, Na L, et al. CsAP3: A Cucumber Homolog to Arabidopsis APETALA3with Novel Characteristics. Frontiers in Plant Science. 2016;7(e6144). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Zhang G, Zhang J, Liu H, Chen W, Wang X, et al. Hybrid de novo genome assembly of the Chinese herbal fleabane Erigeron breviscapus. Gigascience. 2017;6(6):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Zhang S, Yang Z, Song K, Yi T. Conservation genetics and population diversity of Erigeron breviscapus (Asteraceae), an important Chinese herb. Biochemical Systematics & Ecology. 2013;49(2):156–66. [Google Scholar]
- 38.Wang J, Chen Y, Liu B, Wang Q, Zhang L, Wang Z, et al. Systematic Investigation of the Erigeron breviscapus Mechanism for Treating Cerebrovascular Disease. Journal of ethnopharmacology. 2018;224:429–440. 10.1016/j.jep.2018.05.022 [DOI] [PubMed] [Google Scholar]
- 39.Zhang RW, Fan XE, Lai-Wei LI, Lin LZ, Harnly JM. Identification and Quantification of Phenolic Components of Erigeron breviscapus and Its Derived Drug Breviscapine. Natural Product Research & Development. 2015;27: 962–970. [Google Scholar]
- 40.Chu Q, Wu T, Liang F, Ye J. Simultaneous determination of active ingredients in Erigeron breviscapus (Vant.) Hand-Mazz. by capillary electrophoresis with electrochemical detection. Journal of Pharmaceutical & Biomedical Analysis. 2005;37(3):535–41. [DOI] [PubMed] [Google Scholar]
- 41.Li Li L, Ai Jun L, Jian Guo L, Xue-Hong Y, Lu Ping Q, Ding Feng S. Protective effects of scutellarin and breviscapine on brain and heart ischemia in rats. J Cardiovasc Pharmacol. 2007;50(3):327–32. 10.1097/FJC.0b013e3180cbd0e7 [DOI] [PubMed] [Google Scholar]
- 42.Li XL, Li YW, Li HY, Xu H, Zheng XX, Guo DW, et al. A study of the cardioprotective effect of breviscapine during hypoxia of cardiomyocytes. Planta Medica. 2004;70(11):1039–44. 10.1055/s-2004-832644 [DOI] [PubMed] [Google Scholar]
- 43.Tao YH, Jiang DY, Xu HB, Yang XL. Inhibitory effect of Erigeron breviscapus extract and its flavonoid components on GABA shunt enzymes. Phytomedicine. 2008;15(1):92–7. [DOI] [PubMed] [Google Scholar]
- 44.Yang S, Wang P, Yang J. Effects of Genotypic and Environmental on Yield and Scutellarin Content of Erigeron breviscapus. Chinese Agricultural Science Bulletin. 2011;27(8):140–3. [Google Scholar]
- 45.Zhao WJ, Yan SQ, Cui MK, Zhang YF, Guan HL. The Relationship between the Stage of Androgenesis and Flower Character in Erigeron breviscapus. Lishizhen Medicine & Materia Medica Research. 2010;21(5):1210–2. [Google Scholar]
- 46.Zhang W, Wei X, Meng H-L, Ma C-H, Jiang N-H, Zhang G-H, et al. Transcriptomic comparison of the self-pollinated and cross-pollinated flowers of Erigeron breviscapus to analyze candidate self-incompatibility-associated genes. Bmc Plant Biology. 2015;15(1):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Research. 2014;42(Database issue):222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution, 2018; 35(6):1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang W, Ouyang C, Liu L, Li H, Zeng C, Wang J, et al. Genome-wide identification of the fatty acid desaturases gene family in four Aspergillus species and their expression profile in Aspergillus oryzae. AMB Express. 2018;8:169 10.1186/s13568-018-0697-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akaike H. A new look at the statistical model identification. Automatic Control IEEE Transactions on. 1974;19(6): 716–723. [Google Scholar]
- 52.Posada D, Crandall K A. Selecting the best-fit model of nucleotide substitution. Systematic Biology. 2001; 50(4): 580–601. [PubMed] [Google Scholar]
- 53.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 54.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37(Web Server issue):202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He B, Gu Y, Xiang T, Cheng X, Wei C, Jian F, et al. De NovoTranscriptome Sequencing ofOryza officinalisWall ex Watt to Identify Disease-Resistance Genes. International Journal of Molecular Sciences. 2015;16(12):29482–95. 10.3390/ijms161226178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wankun D, Yongbo W, Zexian L, Han C, Yu X. HemI: a toolkit for illustrating heatmaps. Plos One. 2014;9(11):e111988 10.1371/journal.pone.0111988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He B, Ma L, Hu Z, Li H, Ai M, Long C, et al. Deep sequencing analysis of transcriptomes in Aspergillus oryzae in response to salinity stress. Applied Microbiology and Biotechnology. 2018; 102(2):897–906. 10.1007/s00253-017-8603-z [DOI] [PubMed] [Google Scholar]
- 58.Peter A. PASW Statistics by SPSS: A Practical Guide: Version 18.0. 2010.
- 59.Wang L. Advances in the Research of MADS-box Gene in Plant. Biotechnology Bulletin. 2010;8:12–19. [Google Scholar]
- 60.Cui Y, Zhang L, Huang M. Progress of MADS-Box Gene Research in Plant. Progress in Biotechnology. 2003; 23(9):50–54. [Google Scholar]
- 61.Nettancourt DD. Incompatibility and Incongruity in Wild and Cultivated Plants: Springer; Berlin Heidelberg: 2001. [Google Scholar]
- 62.Zhang Y, Zhao Z, Xue Y. Roles of Proteolysis in Plant Self-Incompatibility. Annual Review of Plant Biology. 2009; 60(1):21–42. [DOI] [PubMed] [Google Scholar]
- 63.Lu Q, Jia QL, Hong YU, Mei LC, Li HC, Min LA. Advances in studies on Erigeron breviscapus. Chinese Traditional & Herbal Drugs. 2005;36(1):141–144. [Google Scholar]
- 64.Wang Z, Wei QU, Liang JY. The research progress of Potentilla plants. Strait Pharmaceutical Journal. 2012;22(5):1–8. [Google Scholar]
- 65.Zhou Z, Zhang S, Hua L, Zhong W, Jin Z, Ding R. Study of Resisting Leukoplakia Canceration and Angiogenesis of Herba Erigerontis. Shanghai journal of stomatology. 2000;9(2):110 [PubMed] [Google Scholar]
- 66.Zhou Y, Hu L, Jiang L, Liu S. Genome-wide identification, characterization, and transcriptional analysis of the metacaspase gene family in cucumber (Cucumis sativus). Genome. 2018;61(3):187–94. 10.1139/gen-2017-0174 [DOI] [PubMed] [Google Scholar]
- 67.Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS et al. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(10):5328–33. 10.1073/pnas.97.10.5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405(6783):200–3. 10.1038/35012103 [DOI] [PubMed] [Google Scholar]
- 69.Moore S, Vrebalov J, Payton P, Giovannoni J. Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. Journal of Experimental Botany. 2002;53(377):2023 10.1093/jxb/erf057 [DOI] [PubMed] [Google Scholar]
- 70.Yamaguchi T, Lee D, Miyao A, Hirochika H, An G, Hirano H. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell. 2006;18(1):15–28. 10.1105/tpc.105.037200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu YY, Shao X, Wang Y, Li YQ, Guo WD. Bioinformatics analysis of MADS-box genes and their expression in cherry. Plant Physiology Journal. 2015;51(3):354–62. [Google Scholar]
- 72.Tsaftaris AS, Polidoros AN, Pasentsis K, Kalivas A. Cloning, structural characterization, and phylogenetic analysis of flower MADS-box genes from crocus (Crocus sativus L.). Scientific world journal. 2014;7(1):1047–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang X, Wu F, Lin X, Du X, Chong K, Gramzow L, et al. Live and Let Die. The Bsister MADS-Box Gene OsMADS29 Controls the Degeneration of Cells in Maternal Tissues during Seed Development of Rice (Oryza sativa). PLOS ONE. 2012;7 (12):e51435 10.1371/journal.pone.0051435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lid SE, Meeley RB, Min Z, Nichols S, Olsen OA. Knock-out mutants of two members of the AGL2 subfamily of MADS-box genes expressed during maize kernel development. Plant Science (Oxford). 2004;167(3):575–582. [Google Scholar]
- 75.Tang N, Deng W, Hu G, Hu N, Li Z. Transcriptome Profiling Reveals the Regulatory Mechanism Underlying Pollination Dependent and Parthenocarpic Fruit Set Mainly Mediated by Auxin and Gibberellin. PLOS ONE. 2015; 10(4): e0125355 10.1371/journal.pone.0125355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li L, Yang L, Wang X, Gu A, Yang B, Yan S, et al. Preliminary Studies on Breeding System and Visiting Insects of Erigeron breviscapus. Southwest China Journal of Agricultural Sciences. 2009;22(2):454–458. [Google Scholar]
- 77.Zhang W, Meng HL, Meng ZG, Liang Q, Wei X, Yang SC. Methylation Analysis on the Coding Region of Self-incompatibility SRK Gene of Erigeron breviscapus. Journal of West China Forestry Science. 2016;45(6):8–13. [Google Scholar]
- 78.Zhang YJ, Zhao ZH, Xue YB. Roles of proteolysis in plant self-incompatibility. Annual review of plant biology. 2009;60(1):21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(DOC)
(DOC)
(XLS)
Data Availability Statement
All files of genome-wide transcriptome data of Erigeron breviscapus in different tissues and three pollination treatments are available from the NCBI SRA database (accession numbers PRJNA352312,SRA245957).