Abstract
Background
Escherichia coli is an important aetiological agent of bovine mastitis worldwide.
Methods
In this study, 82 E. coli from bovine mastitis milk samples from 49 farms were analysed for their genetic diversity using phylogenetic grouping and multilocus sequence typing. The isolates were examined by PCR for a selection of virulence factors (VFs). Antimicrobial susceptibility profiles were assessed using the disk diffusion method.
Results
The most prevalent phylogroups were group B1 (41.5 per cent of the isolates) and group A (30.5 per cent). A variety of 35 different sequence types (STs) were identified, including ST1125 (11 per cent), ST58 (9.8 per cent), ST10 (8.5 per cent) and ST88 (7.3 per cent). Aggregate VF scores (the number of unique VFs detected for each isolate) ranged from 1 to 3 for 63.4 per cent of the isolates and were at least 4 for 12.2 per cent. For 24.4 per cent of the isolates, the score was 0. The three most frequent VFs were traT, fyuA and iutA. The majority (72 per cent) of the isolates harboured traT. The majority (68.3 per cent) of the isolates were fully susceptible to all antimicrobials tested, with 22 per cent resistant to ampicillin and 14.6 per cent to tetracycline. Resistance rates were low for gentamicin (3.7 per cent), amoxicillin/clavulanic acid (2.4 per cent) and ceftiofur (1.2 per cent), respectively.
Conclusion
Among the study’s sample population, E. coli strains were genotypically diverse, even in cows from the same farm, although some STs occurred more frequently than others. Susceptibility to clinically relevant compounds remained high.
Keywords: bovine mastitis, Escherichia coli, sequence types, virulence genes, antimicrobial resistance
Introduction
Escherichia coli occurs in the digestive tract of human beings and animals as a non-pathogenic, commensal part of the normal gut flora; however, some E. coli types may cause gastrointestinal disease and a range of extraintestinal infections.1
Many pathogenic E.coli are distinguished from commensal strains due to specific virulence features that increase their ability to cause disease in otherwise healthy individuals. Among the pathogenic features, certain virulence factors (VFs) are characteristic of intestinal pathogenic E. coli (IPEC), which comprise well-described and important categories such as enteropathogenic E. coli (EPEC), Shiga toxin-producing E.coli (STEC), and its subgroup enterohaemorrhagic E. coli (EHEC), enteroaggregative E. coli (EAEC) or enterotoxigenic E. coli (ETEC).1–3
Other virulence attributes are associated with extraintestinal pathogenic E. coli (ExPEC), which encompass uropathogenic E. coli, newborn meningitis E. coli and avian pathogenic E. coli (APEC), which causes respiratory tract infections and septicaemia in poultry.1
In cattle, E. coli is an important aetiological agent of mastitis, which is one of the most common and economically significant diseases in the dairy industry worldwide.4 Bovine mastitis E. coli—unlike many other pathogenic E. coli—appears to lack consistent genotypes and specific defining virulence profiles, making a differentiation from commensal E. coli challenging.5–7 Previous studies show that virulence traits typically identified in IPEC or ExPEC are not accountable for invasion and survival of E. coli inside mammary epithelial cells in vitro, nor do they appear to contribute to clinical severity of the disease.8 9 Bovine mastitis E. coli is generally thought to bear the same pathogenic potential as commensal or environmental E. coli.10 11 Nevertheless, some studies suggest that certain genotypes may be more prevalent among mastitis-associated E. coli compared with environmental strains.5 Therefore, further assessment of the genotypes of E. coli causing bovine mastitis is needed to fill current knowledge gaps.
Bovine mastitis is the most frequent reason for use of antimicrobials in dairy cattle.6 12 To promote the efficient and appropriate use of antimicrobials in veterinary medicine, the Swiss Veterinary Society (SVS), which is the official representing body of veterinarians, together with the Federal Food Safety and Veterinary Office have issued guidelines that provide practical recommendations for the prescription and application of antimicrobials for treating livestock. The recommendations are in accordance with the Swiss law on pharmaceutical and medicinal products and with international Good Clinical Practice standards.13
For mastitis cases requiring treatment, the SVS guidelines recommend gentamicin as first-line antimicrobial agent for intramammary treatment in cases of bovine mastitis caused by E. coli, and fourth-generation cephalosporins as second-line therapy.13 However, third-generation and fourth-generation cephalosporins belong to the highest priority critically important antimicrobials (HPCIA) for the use in human beings, and the emergence and global dissemination of antimicrobial resistance in general, and extended-spectrum ß-lactamase (ESBL) producing Enterobacteriaceae in particular, represent a threat to public health.14 ESBL producers have been described infrequently in E. coli isolated from bovine mastitis in Germany, France, Switzerland and the UK.15–18 Further, a study from Germany found a prevalence of 9.5 per cent ESBL-producing E. coli in bulk tank milk.19 Therefore, current data on the antimicrobial susceptibility profiles of bovine mastitis isolates are warranted.
The aims of the present study were to determine the genotypes and the virulence profiles of E. coli strains isolated from bovine mastitis, and to obtain data on their antimicrobial susceptibility profiles.
Materials and methods
Bacterial isolation and species identification
In this study, a total of 82 non-duplicate E. coli strains were collected from 82 dairy cows with clinical mastitis on 49 different farms during 2017. The farms were customers of the ambulatory veterinary hospital of the University of Zürich that services the canton of Zürich and the surrounding region. The client population consists of approximately 124 cattle farms, whereof 60 per cent are dairy farms. The average dairy herd size in the study region is 20–30 cows, and the most common dairy cow is the Brown Swiss. Clinical mastitis diagnosis was performed during farm calls by the veterinarian in charge according to a standardised procedure which included physical examination of the udder and the estimation of the somatic cells in the milk using the California mastitis test. Milk samples were taken from the affected quarter of each cow and were submitted to the routine mastitis diagnostic laboratory of the Institute for Food Safety and Hygiene in Zürich. Overall, during the study period, a total of 1281 milk samples from the farms in the client population were submitted for culture to the diagnostic laboratory. From samples yielding microbial growth, the range of pathogens identified as the cause of mastitis included staphylococci (16.6 per cent), Staphylococcus aureus (7 per cent), Streptococcus uberis (12.6 per cent), Streptococcus dysgalactiae (3 per cent), E. coli (11.9 per cent) and other pathogens (10 per cent).
Samples were cultured according to standard procedures.20 Briefly, using a sterile loop, the samples were streaked onto sheep blood agar base (Becton Dickinson, Allschwil, Switzerland), supplemented with 5 per cent sheep blood (Oxoid, Pratteln, Switzerland) and incubated at 37°C overnight. The strains were confirmed by colony morphology, Gram stain, and biochemical tests such as the mannitol fermentation test, O-nitrophenyl-beta-D-galactopyranoside (ONPG) test, tests for urease, indole and hydrogen sulfide (H2S) production, and the lysine decarboxylase test. The strains were stored at –80°C.
Phylogenetic and multilocus sequence typing
DNA from E. coli isolates was subjected to quadruplex PCR targeting arpA, chuA, yjaA and an unspecified DNA fragment termed TspE4.C2, as described previously.21 The isolates were classified as belonging to one of the eight phylogenetic groups A, B1, B2, C, D, E, F (E. coli sensu stricto), or Escherichia clade I.
For multilocus sequence typing (MLST), internal fragments of the seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA) were amplified by PCR as described by Wirth and colleagues.22 Sequencing of the amplification products was performed by Microsynth (Balgach). Sequences were imported into the E. coli MLST database website (https://pubmlst.org/escherichia/) to determine MLST types. Alleles and sequence types (STs) that had not been previously described were designated ‘new ST’, but not assigned numerical designations, since whole-genome sequencing was not performed. Data were visualised using the platform independent JAVA software Phyloviz 2.0 and the goeBURST algorithm.23
Virulence factor determination
All 82 isolates were screened for genetic markers of virulence associated with ExPEC by conventional PCR using primers and conditions described previously for targeting afa, papAH, papC, papEF, sfaS, fyuA, hlyA, iutA, KpsMII, PAI and traT,24vat and yfcV.25 The aggregate VF score was defined as the number of unique VF detected for each isolate, counting the PAI marker as one.
Strains were further screened by PCR for genetic markers characterising IPEC. Screening for stx1 and stx2 genes in STEC was performed by real-time PCR (LightCycler 2.0 Instrument, Roche Diagnostics Corporation, Indianapolis, Indiana, USA) using the QuantiFast Multiplex PCR Kit (Qiagen, Hombrechtikon, Switzerland) according to the guidelines of the European Union Reference Laboratory (EURL).26 The determination of stx1 subtypes was performed by conventional PCR amplification.27
The presence of subAB encoding subtilase cytotoxin SubAB was tested by conventional PCR using primers described previously.28
Screening for the intimin gene eae in EPEC, and for the heat-labile and heat-stabile enterotoxins LT, STp and STh in ETEC, was performed by real-time PCR according to the guidelines of the EURL.29 30 Screening for aggR, which encodes a transcriptional regulator of EAEC, was performed by conventional PCR using primers and conditions described previously.31
For all PCR assays, DNA from previously characterised isolates from the authors’ strain collection was used as positive or negative controls for strains harbouring VFs associated with ExPEC32 33 and VFs characteristic of IPEC.32 34 35
Antimicrobial susceptibility testing
The strains were subjected to antimicrobial susceptibility testing (AST) using the standard disk diffusion method according to the protocols recommended by the Clinical and Laboratory Standards Institute (CLSI),36 which is currently the only committee offering interpretive criteria for veterinary AST.37 The isolates were classified as susceptible, intermediate or resistant using the breakpoints listed by the VET01S of the CLSI.38 Multidrug resistance (MDR) was defined as resistance to three or more classes of antimicrobials, counting ß-lactams as one.
Susceptibility disks containing ampicillin (AM), amoxicillin/clavulanic acid (AMC), tetracycline (TE) and sulfamethoxazole/trimethoprim (SXT) were obtained from Becton Dickinson, and disks containing ceftiofur (EFT) and gentamicin (CN) were from Thermo Fisher Diagnostics (Pratteln, Switzerland). E. coli ATCC 25922 was used as a control during AST.
The antibiotics were chosen on the basis of their recommendations for intramammary application in Switzerland and of the panel included in the German national antibiotic resistance monitoring of veterinary pathogens from cattle (GermVet).13 39
To test for the presence of ESBL-producing E. coli, the isolates were cultured on Brilliance ESBL agar plates (Oxoid, Hampshire, UK) and incubated for 24 hours at 37°C.
Results
Bacterial isolates and herd data
For this study, a total of 82 E. coli strains originating from 82 different animals from 49 different herds were obtained. Twelve (24.5 per cent) of the herds were sampled more than once, whereby each sample derived from a case of mastitis diagnosed by the veterinarian in charge. The highest numbers of E. coli mastitis cases per farm were eight cases on farm F8 and F16, respectively, and seven cases on farm F14 (online supplementary table S1). Acute mastitis was diagnosed in 66 cases, subclinical in two cases and chronic mastitis in one case, respectively. For 13 cases, the type of mastitis was unspecified (online supplementary table S1).
vetreco-2019-000369supp001.pdf (1.1MB, pdf)
Phylogenetic groups, STs and VF distribution
Of the 82 E. coli isolates, 34 (41.5 per cent) belonged to phylogenetic group B1, followed by group A (n=25; 30.5 per cent), C (n=9; 11 per cent) and D (n=8; 9.8 per cent). The remaining isolates (n=2; 2.4 per cent each) belonged to phylogenetic groups B2, E and F (table 1 and online supplementary table S1).
Table 1.
Distribution of virulence factors among the phylogenetic groups of 82 Escherichia coli causing bovine mastitis
| Gene or marker* | Prevalence by phylogenetic group (n, %) | ||||||
| A | B1 | B2 | C | D | E | F | |
| (n=25) | (n=34) | (n=2) | (n=9) | (n=8) | (n=2) | (n=2) | |
| ExPEC-associated genes | |||||||
| papAH | 2 (8.0) | 2 (5.9) | – | 3 (33.3) | – | – | – |
| papC | 2 (8.0) | 2 (5.9) | – | 3 (33.3) | – | – | – |
| papEF | – | – | – | 1 (11.1) | – | – | – |
| yfcv | – | – | 2 (100) | – | – | – | |
| hlyA | 1 (4.0) | 12 (35.3) | 1 (50) | – | 1 (12.5) | – | – |
| vat | 1 (50) | – | – | – | 2 (100) | ||
| fyuA | 6 (24.0) | 8 (23.5) | 1 (50) | 6 (66.7) | – | – | 2 (100) |
| iutA | 3 (12.0) | 5 (14.7) | 1 (50) | 5 (55.6) | 1 (12.5) | – | 2 (100) |
| KpsMII | – | – | 1 (50) | – | 1 (12.5) | – | – |
| PAI | – | – | 1 (50) | – | – | – | – |
| traT | 14 (56.0) | 27 (79.4) | 2 (100) | 8 (88.9) | 4 (50) | 2 (100) | 2 (100) |
| Virulence factor score (median, range) | 1, 0–4 | 2, 0–5 | 5, 2–8 | 3, 0–6 | 0.5, 0–3 | 1, 1–1 | 4, 4–4 |
| IPEC-associated genes | |||||||
| stx1a | 2 (8.0) | – | – | – | – | – | – |
| subAB | – | 1 (2.9) | – | – | – | – | – |
*aggR, afa, eae, LT, sfa, STh, STp and stx2 genes were not identified in any of the isolates.
–, not detected; ExPEC, extraintestinal pathogenic E. coli;fyuA, ferric yersiniabactin uptake protein; hlyA, haemolysin; IPEC, intestinal pathogenic E. coli; iutA, aerobactin siderophore receptor; KpsMII, group 2 polysaccharide capsule; PAI, right-hand terminus of pathogenicity island; papAH, pyelonephritis-associated major pilin protein; papC, outer membrane usher protein; papEF, fimbrial protein subunit; stx1a, Shiga toxin subunit; subAB, subtilase cytotoxin; traT, lipoprotein involved in serum resistance; vat, vacuolating autotransporter toxin; yfcv, major subunit of a chaperone-usher fimbria.
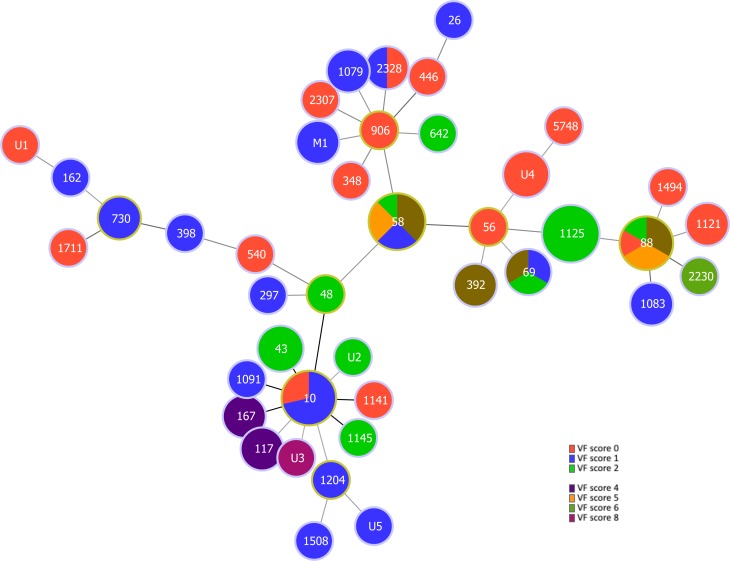
MLST identified 35 different STs, the four most common represented by ST1125 (n=9; 11 per cent of the isolates), ST58 (n=8; 9.8 per cent), ST10 (n=7; 8.5 per cent) and ST88 (n=6; 7.3 per cent). Twenty-one (26 per cent) of the isolates belonged to STs that occurred only once. Five (6 per cent) of the isolates belonged to human ExPEC lineages ST69 or ST117 (online supplementary table S1). Seven (8.5 per cent) of the isolates belonged to STs with a total of five new allelic profiles (online supplementary table S1). The new STs were not assigned numerical designations by the E. coli MLST database. Further, two isolates could not be typed due to failure to sequence the purA gene. The goeBURST analysis of the strains is shown in figure 1.
Figure 1.
MLST-based minimal spanning tree of 82 Escherichia coli isolated from bovine mastitis milk in Switzerland during 2017. The tree was calculated and generated using the goeBURST full MLST algorithm in Phyloviz 2.0. Node sizes reflect the number of isolates with specific MLST profile. Numbers within the nodes indicate the ST. Founder STs are encircled in yellow. Node colours refer to virulence scores within an ST. Nodes differing by one or two loci are linked by dark lines. M1, strains with incomplete profiles (the missing purA allele was treated as an own category); U1–U5, strains with new STs. New STs were not assigned numerical designations by the E. coli MLST database (https://pubmlst.org/escherichia/). MLST, multilocus sequence typing; ST, sequence type; VF, virulence factor.
On farms with multiple cases of mastitis, various STs were observed. On farms F8 and F16 (eight individual cases of E. coli mastitis each), a total of seven STs and on farm F14 (seven cases of mastitis) four different STs were noted (online supplementary table S1).
Among the 82 isolates, the prevalence of individual ExPEC associated VFs ranged from 0 per cent (afa and sfa, respectively) to 72 per cent (traT).
The frequency of VFs among the phylogenetic groups is shown in table 1. Median aggregate VF scores were highest for isolates belonging to phylogenetic group B2 (median VF score 5, range 2–8) and group F (median VF score 4, range 4–4); however, these two groups only included two isolates each. Median aggregate VF scores were lower for isolates belonging to phylogroup D (median VF score 0.5, range 0–3).
The distribution of VFs among the most frequently occurring STs is summarised in table 2. Median aggregate VF scores were highest for isolates belonging to ST58 (median VF score 3, range 1–5) and ST88 (median VF score 3, range 0–5), and low for isolates belonging to ST1125 (median VF score 2, range 2–2) and ST10 (median VF score 1, range 0–1). For all other STs, the median VF score was 1, with a range of 0–8 (table 2). Among these, one isolate with a new ST scored a VF factor of 8 (online supplementary table S1). The diversity of VF scores among all the isolates analysed in this study is illustrated in figure 1.
Table 2.
Distribution of virulence factors among four main sequence types of 82 Escherichia coli causing bovine mastitis
| Gene or marker* | Prevalence by sequence type (n, %) | ||||
| ST10 | ST58 | ST88 | ST1125 | Other | |
| (n=7) | (n=8) | (n=6) | (n=9) | (n=52) | |
| ExPEC-associated genes | |||||
| papAH | – | 2 (25) | 2 (33.3) | – | 3 (5.8) |
| papC | – | 2 (25) | 2 (33.3) | – | 3 (5.8) |
| papEF | – | – | – | – | 1 (1.9) |
| yfcv | – | – | – | – | 2 (3.8) |
| hlyA | – | – | – | 9 (100) | 6 (11.5) |
| vat | – | – | – | – | 3 (5.8) |
| fyuA | 3 (42.9) | 6 (75) | 5 (83.3) | – | 9 (17.3) |
| iutA | – | 5 (62.5) | 4 (66.7) | – | 8 (15.4) |
| KpsMII | – | – | – | – | 2 (3.8) |
| PAI | – | – | – | – | 1 (1.9) |
| traT | 2 (29.6) | 8 (100) | 5 (83.3) | 9 (100) | 35 (67.3) |
| Virulence factor score (median, range) | 1, 0–1 | 3, 1–5 | 3, 0–5 | 2, 2–2 | 1, 0–8 |
| IPEC-associated genes | |||||
| stx1a | – | – | – | – | 2 (3.8) |
| subAB | – | – | – | – | 1 (1.9) |
*aggR, afa, eae, LT, sfa, STh, STp and stx2 genes were not identified in any of the isolates.
–, not detected; ExPEC, extraintestinal pathogenic E. coli;fyuA, ferric yersiniabactin uptake protein; hlyA, haemolysin; IPEC, intestinal pathogenic E. coli; iutA, aerobactin siderophore receptor; KpsMII, group two polysaccharide capsule; PAI, right-hand terminus of pathogenicity island; papAH, pyelonephritis-associated major pilin protein; papC, outer membrane usher protein; papEF, fimbrial protein subunit; stx1a, Shiga toxin subunit; subAB, subtilase cytotoxin; traT, lipoprotein involved in serum resistance; vat, vacuolating autotransporter toxin; yfcv, major subunit of a chaperone-usher fimbria.
IPEC-associated VFs stx1a and subAB were detected in two (2.4 per cent) and one (1.2 per cent) of the isolates, respectively (table 1). None of strains tested positive for aggR, afa, eae, LT, sfa, STh, STp or stx2.
Antimicrobial susceptibility testing
The prevalence of resistant, intermediate and susceptible strains among the bovine mastitis E. coli isolates is summarised in figure 2. Overall, the majority (n=56; 68.3 per cent) of the isolates were fully susceptible to all antimicrobials tested in this study (online supplementary table S1). The highest rate of resistance was observed for AM (n=18; 22 per cent), followed by TE (n=12; 14.6 per cent), SXT (n=8; 9.8 per cent) and CN (n=3; 3.7 per cent). Resistance rates were low for AMC (n=2; 2.4 per cent) and EFT (n=1; 1.2 per cent), respectively. Notably, for EFT, seven strains were categorised intermediate resistant (figure 2). Eight strains (9.8 per cent) were resistant to three or more classes of antibiotics (MDR). The MDR phenotype AM/TE/SXT represented the most common pattern (six isolates), followed by AM/AMC/CN/TE and AM/CN/TE/SXT (one isolate each, respectively).
Figure 2.
Antimicrobial susceptibility percentages among 82 Escherichia coli isolated from bovine mastitis milk in Switzerland during 2017. AM, ampicillin; AMC, amoxicillin/clavulanic acid; CN, gentamicin; EFT, ceftiofur; I, intermediate; R, resistant; S, susceptible; SXT, sulfamethoxazole/trimethoprim; TE, tetracycline.
The prevalence of antimicrobial resistance was high among isolates belonging to ST58 and ST88 and low among ST10 and ST1125 (table 3). Among the remaining STs, antimicrobial resistance was found predominantly among ST117 and ST167, respectively (online supplementary table S1).
Table 3.
Distribution of antimicrobial resistance among four main sequence types of 82 Escherichia coli causing bovine mastitis
| Resistance* | Prevalence by sequence type (n, %) | ||||
| ST10 | ST58 | ST88 | ST1125 | Other | |
| (n=7) | (n=8) | (n=6) | (n=9) | (n=52) | |
| AM | – | 4 (50.0) | 3 (50.0) | 1 (11.1) | 10 (19.2) |
| AMC | – | 1 (12.5) | 1 (16.7) | – | – |
| EFT | 1 (14.3) | – | – | – | – |
| CN | – | 2 (25.0) | 1 (16.7) | – | – |
| TE | – | 4 (50.0) | 1 (16.7) | – | 7 (13.5) |
| SXT | – | 3 (37.5) | 1 (16.7) | – | 4 (7.7) |
| MDR | – | 3 (37.5) | 1 (16.7) | – | 4 (7.7) |
*Antimicrobial susceptibility was determined using the disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines and breakpoints.36
–, no resistance detected; AM, ampicillin; AMC, amoxicillin/clavulanic acid; CN, gentamicin; EFT, ceftiofur; MDR, multidrug resistant (resistant to three or more classes of antimicrobials); SXT, sulfamethoxazole/trimethoprim; TE, tetracycline.
None of the E. coli isolates yielded growth on ESBL plates.
Discussion
Bovine mastitis caused by E. coli is generally considered to include typically commensal strains of intestinal or environmental origin.40 41 Commensal strains have been shown by phylogenetic analyses to position within phylogenetic groups A, B1 or C, while virulent extraintestinal E. coli strains belong mainly to group B2, and to a lesser extent to groups D, E or F.21
In line with previous reports,5 6 10 42 the isolates from the present study were mainly of phylogenetic groups A and B1, with the exception of isolates belonging to phylogroup C, which were not identified in earlier studies that restricted classifications to four main phylogenetic groups A, B1, B2 and D.43 Phylogroup C is closely related to group B1, but before its recognition as a distinct phylogroup its members were classified within group A.44 Consequently, the prevalence of phylogroup A mastitis strains may have been overestimated in the past.45 Furthermore, mastitis isolates belonging to phylogroups E and F were detected in the current study. While phylogroup F comprises strains that were formerly classified group D, phylogroup E includes a small number of E.coli, to which EHEC O157:H7 and its ancestor O55:H7 belong.21 Similar to previously described mastitis strains belonging to phylogroup F, both isolates in the current study lacked virulence traits that characterise EHEC strains, and their significance among mastitis isolates, although remarkable, remains unclear.46
In agreement with previous studies that document the absence of specific genotypes to characterise mastitis E. coli, a wide variety of STs were observed, indicating high heterogeneity.6 7 10 Even most isolates from multiple cows on the same farms displayed diverse STs, with a maximum of two isolates from any one farm belonging to the same ST. Nevertheless, E. coli ST10, ST58 and ST1125, which were among the most common STs from this collection of isolates, were also frequent among mastitis strains isolated previously in Israel.5E. coli ST1125 has also been described among mastitis strains from cattle in Germany and Ireland.11 47 Taken together, these findings indicate that notwithstanding the high genetic variability, certain STs may predominate among bovine mastitis strains. E. coli ST10, which occurs frequently among livestock, food and healthy human beings, is significantly more prevalent among mastitis isolates compared with environmental isolates.5 32 ST58 and ST88, although also isolated globally from a wide variety of sources, may cause disease in human beings, including urinary tract infection and sepsis.48 49 Several STs less frequently identified in this study have also been described among human clinical isolates, diseased animals and livestock, and represent typical ExPEC. For instance, E. coli ST69, isolated from three mastitis cases in this study, is accountable for community-acquired and healthcare-associated urinary tract infections worldwide,50 and E. coli ST117, detected in two cases, has been identified among APEC strains causing colibacillosis in broilers.51 52 Nevertheless, an association of these STs with severity of mastitis was not investigated, which may be considered a limitation of this study. Interestingly, E. coli ST69 was recently identified in faecal samples of dairy cows in Washington state in USA, and in raw milk cheese in Egypt, suggesting that this pandemic human disease-associated ST may be circulating in dairy cattle worldwide.53 54
Although the pathogenesis mechanisms of many E. coli genotypes are well described for other epithelial systems, VFs enabling adherence and survival within the bovine mammary gland remain, to a large extent, undefined.55 56 In the present study, traT was the only VF to characterise the majority (72 per cent) of the strains. This plasmid-located determinant encodes an outer membrane protein thought to block the membrane attack complex present in the serum of the host.57 Although serum resistance has repeatedly been reported in E. coli mastitis isolates, it is currently considered to be an unspecific feature of mastitis isolates.58 Therefore, although the traT gene is prevalent among human, avian and porcine ExPEC,24 59 60 and has also been described in mastitis E. coli in previous studies, the significance of traT among mastitis E. coli remains unclear.58 61 In addition to traT, only two other VFs, fyuA and iutA, were found to be prevalent among the isolates from this study. Both genes encode ferric acquisition proteins that allow bacteria to grow in environments with limited concentrations of free iron, such as in tissues and fluids of the host.62 The fyuA gene encodes a ferric yersiniabactin uptake protein and is strongly associated with uropathogenicity in human beings.25 It has been suggested that the presence of fyuA is not essential for survival of E. coli in mammary glands, but that its presence may contribute to the ability to utilise iron from lactoferrin, one of the main iron sources available to bacteria in milk.63 Similarly, iutA, encoding for an aerobactin receptor, is frequently associated with ExPEC but not, so far, with mastitis strains.40 62 In this study, a limited number of VF genes were analysed. Despite this constraint, the wide variation of aggregate VF scores among the phylogenetically diverse strains in this study lends support to previous data that there exists no distinct bovine mastitis E. coli pathotype.40
The prevalence of IPEC-associated virulence genes was low among the isolates, which is remarkable because cattle are considered a major reservoir for EPEC, STEC and EHEC.64 65 Moreover, a recent study evaluating the global prevalence of STEC in bovine mastitis cases estimated that in Europe, STEC occurs in 0.5–13.7 per cent of mastitic milk, and suggested that the prevalence of STEC in mastitis may be underestimated.66 Only two (2.4 per cent) of the isolates in the present study, both belonging to phylogenetic group A and ST730, were stx1a-positive, and they did not encode the intimin gene eae. Interestingly, the occurrence of subAB in an isolate that did not contain stx1 or stx2 was observed. This is one of very few reports of non-STEC harbouring subAB.67
Overall, these data show that susceptibility to antimicrobials remains high among the study’s sample population of mastitis E.coli in Switzerland. Utmost caution should be applied when comparing AST results obtained across different strain collections, countries and settings.68 A comparison with earlier data from Switzerland obtained using identical methodologies and interpretive criteria showed no major shift in antibiotic resistance.16 Moreover, using identical CLSI interpretation criteria, resistance data from this study are similar to those from the German antimicrobial resistance monitoring programme GermVet, with the exception of EFT.39 Regarding this antimicrobial, a lower rate of resistance compared with that reported by GermVet for the year 2016 was identified (1.2 per cent v 7.6 per cent). Notably, in the study presented here, seven (8.5 per cent) of the isolates were categorised intermediate resistant. This may be indicative of a shift towards a resistant phenotype and warrants future observation. An increase of resistance to EFT from 0.4 per cent in 2006 to 2.4 per cent in 2016 has been reported for mastitis isolates in France.69 By contrast, resistance rates among mastitis isolates are reportedly low in other countries, for example, Denmark,70 with resistance to AM reported in only 11.3 per cent and resistance to EFT and CN in 0 per cent of E. coli mastitis, respectively. Given that EFT is among the category of HPCIA, it is crucial that this antimicrobial should only be used as a last resort, and first-line antimicrobial drugs should remain the treatment of choice.
Of the antimicrobials tested, EFT is the only one with a defined breakpoint for E. coli isolated from bovine mastitis.38 It is therefore important to note that the AST results for the other antimicrobials were interpreted using breakpoints relevant for the administration in human beings and thus may not be reflective of susceptibility, or be clinically predictive, in the context of bovine mastitis. Nevertheless, the results presented here provide a means by which to monitor trends in antimicrobial susceptibility and to identify emerging resistance. To this end, quantitative data, such as the zone diameter measures provided in online supplementary table S1, could be helpful. Finally, this study emphasises the need for veterinary clinical breakpoints to improve surveillance data, optimise treatment of animal disease and promote prudent antimicrobial use.
Conclusions
This work represents a study exploring the phenotypic and genotypic traits of E. coli involved in mastitis in Swiss dairy cows. The results highlight the clonal diversity of the isolates and suggest that certain STs such as ST58, ST88 and ST1125 may be more successful than others at colonising and infecting the mammary gland. Only a minority of the isolates represented typical ExPEC. Although no distinct virulence gene profile was detected among the isolates, traT was found in the majority of bovine E. coli mastitis cases. Therefore, traT may represent a virulence trait that favours pathogenesis in the bovine udder. Overall, antimicrobial susceptibility was high for ß-lactams, CN, TE and SXT. Moreover, no ESBL-producing E. coli were detected.
Acknowledgments
The authors thank Marc Stevens for assistance with bioinformatics and Valerie Wist for technical assistance.
Footnotes
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol 2004;2:123–40. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 2.Croxen MA, Law RJ, Scholz R, et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 2013;26:822–80. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karmali MA. Emerging Public Health Challenges of Shiga toxin-producing Escherichia coli related to changes in the pathogen, the population, and the environment. Clin Infect Dis 2017;64:371–6. [DOI] [PubMed] [Google Scholar]
- 4.Hogeveen H, Huijps K, Lam T. Economic aspects of mastitis: new developments. N Z Vet J 2011;59:16–23. 10.1080/00480169.2011.547165 [DOI] [PubMed] [Google Scholar]
- 5.Blum SE, Leitner G. Genotyping and virulence factors assessment of bovine mastitis Escherichia coli. Vet Microbiol 2013;163:305–12. 10.1016/j.vetmic.2012.12.037 [DOI] [PubMed] [Google Scholar]
- 6.Suojala L, Kaartinen L, Pyörälä S. Treatment for bovine Escherichia coli mastitis - an evidence-based approach. J Vet Pharmacol Ther 2013;36:521–31. 10.1111/jvp.12057 [DOI] [PubMed] [Google Scholar]
- 7.Singer RS. Urinary tract infections attributed to diverse ExPEC strains in food animals: evidence and data gaps. Front Microbiol 2015;6:28 10.3389/fmicb.2015.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dogan B, Klaessig S, Rishniw M, et al. Adherent and invasive Escherichia coli are associated with persistent bovine mastitis. Vet Microbiol 2006;116:270–82. 10.1016/j.vetmic.2006.04.023 [DOI] [PubMed] [Google Scholar]
- 9.Guerra ST, Dalanezi FM, de Paula CL, et al. Putative virulence factors of extra‐intestinal Escherichia coli isolated from bovine mastitis with different clinical scores. Lett Appl Microbiol 2019;68:403–8. 10.1111/lam.13113 [DOI] [PubMed] [Google Scholar]
- 10.Fernandes JBC, Zanardo LG, Galvão NN, et al. Escherichia coli from clinical mastitis: serotypes and virulence factors. J Vet Diagn Invest 2011;23:1146–52. [DOI] [PubMed] [Google Scholar]
- 11.Leimbach A, Poehlein A, Witten A, et al. Whole-genome draft sequences of six commensal fecal and six mastitis-associated Escherichia coli strains of bovine origin: TABLE 1. Genome Announc 2016;4:e00753–16. 10.1128/genomeA.00753-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krömker V, Leimbach S. Mastitis treatment-Reduction in antibiotic usage in dairy cows. Reprod Domest Anim 2017;52:21–9. 10.1111/rda.13032 [DOI] [PubMed] [Google Scholar]
- 13.Swiss Veterinary Society (SVS) and Federal Food Safety and Veterinary Office (FSVO) Umsichtiger Einsatz von Antibiotika bei Rindern und Schweinen: Therapieleitfaden für Tierärztinnen und Tierärzte. Therapieleitfaden für Tierärztinnen und Tierärzte, 2018. Available: https://www.blv.admin.ch/dam/blv/de/therapieleitfaden-de.pdf [Accessed July 2019].
- 14.World Health Organization (W H O) Critically important antimicrobials for human medicine - 5th rev, 2017. Available: http://wwwwhoint/foodsafety/publications/antimicrobials -fifth/en/ [Accessed July 2017].
- 15.Dahmen S, Métayer V, Gay E, et al. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet Microbiol 2013;162:793–9. 10.1016/j.vetmic.2012.10.015 [DOI] [PubMed] [Google Scholar]
- 16.Moser A, Stephan R, Corti S, et al. Resistance profiles and genetic diversity of Escherichia coli strains isolated from acute bovine mastitis. Schweizer Archiv für Tierheilkunde 2013;155:351–7. 10.1024/0036-7281/a000470 [DOI] [PubMed] [Google Scholar]
- 17.Timofte D, Maciuca IE, Evans NJ, et al. Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob Agents Chemother 2014;58:789–94. 10.1128/AAC.00752-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenberger D, Carl A, Balsliemke J, et al. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli isolates from milk samples of dairy cows with mastitis in Bavaria, Germany. Microbial Drug Resistance 2018;24:505–10. 10.1089/mdr.2017.0182 [DOI] [PubMed] [Google Scholar]
- 19.Odenthal S, Akineden Ömer, Usleber E. Extended-spectrum β-lactamase producing Enterobacteriaceae in bulk tank milk from German dairy farms. Int J Food Microbiol 2016;238:72–8. 10.1016/j.ijfoodmicro.2016.08.036 [DOI] [PubMed] [Google Scholar]
- 20.National Mastitis Council (NMC) Laboratory handbook on bovine mastitis Rev. ED. National mastitis Council Inc, 1999. [Google Scholar]
- 21.Clermont O, Christenson JK, Denamur E, et al. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 2013;5:58–65. 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- 22.Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006;60:1136–51. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francisco AP, Bugalho M, Ramirez M, et al. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinformatics 2009;10:152 10.1186/1471-2105-10-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000;181:261–72. 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- 25.Spurbeck RR, Dinh PC, Walk ST, et al. Escherichia coli Isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun 2012;80:4115–22. 10.1128/IAI.00752-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.European Union Reference Laboratory (EURL) Identification and characterization of verocytotoxin-producing Escherichia coli (VTEC) by PCR amplification of the main virulence genes, 2013a. Available: http://old.iss.it/binary/vtec/cont/EU_RL_VTEC_Method_01_Rev_0.pdf [Accessed July 2019].
- 27.Scheutz F, Teel LD, Beutin L, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing stx nomenclature. J Clin Microbiol 2012;50:2951–63. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Funk J, Stoeber H, Hauser E, et al. Molecular analysis of subtilase cytotoxin genes of food-borne Shiga toxin-producing Escherichia coli reveals a new allelic subAB variant. BMC Microbiol 2013;13:230 10.1186/1471-2180-13-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Union Reference Laboratory (EURL) Identification and characterization of verocytotoxin-producing Escherichia coli (VTEC) by real time PCR amplification of the main virulence genes and the genes associated with the serogroups mainly associated with severe human infections, 2013b. Available: http://old.iss.it/binary/vtec/cont/EU_RL_VTEC_Method_02_Rev_0.pdf [Accessed July 2019].
- 30.European Union Reference Laboratory (EURL) Detection of enterotoxigenic Escherichia coli in food by real time PCR amplification of the LT, STh, and STP genes, encoding the heat-labile and heat-stable enterotoxins, 2013c. Available: http://old.iss.it/binary/vtec/cont/EU_RL_VTEC_Method_08_Rev_0.pdf [Accessed July 2019].
- 31.Boisen N, Scheutz F, Rasko DA, et al. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis 2012;205:431–44. 10.1093/infdis/jir757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller A, Stephan R, Nüesch-Inderbinen M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci Total Environ 2016;541:667–72. 10.1016/j.scitotenv.2015.09.135 [DOI] [PubMed] [Google Scholar]
- 33.Nüesch-Inderbinen MT, Baschera M, Zurfluh K, et al. Clonal diversity, virulence potential and antimicrobial resistance of Escherichia coli causing community acquired urinary tract infection in Switzerland. Front Microbiol 2017;8:2334 10.3389/fmicb.2017.02334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fierz L, Cernela N, Hauser E, et al. Characteristics of Shigatoxin-producing Escherichia coli strains isolated during 2010–2014 from human infections in Switzerland. Front Microbiol 2017;8:1471 10.3389/fmicb.2017.01471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nüesch-Inderbinen MT, Funk J, Cernela N, et al. Prevalence of subtilase cytotoxin-encoding subAB variants among Shiga toxin-producing Escherichia coli strains isolated from wild ruminants and sheep differs from that of cattle and pigs and is predominated by the new allelic variant subAB2-2. Int J Med Microbiol 2015;305:124–8. 10.1016/j.ijmm.2014.11.009 [DOI] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 27th informal supplement. CLSI document M100S.. USA: Clinical and Laboratory Standards Institute, Wayne, PA, 2017. [Google Scholar]
- 37.Toutain P-L, Bousquet-Mélou A, Damborg P, et al. En route towards European clinical breakpoints for veterinary antimicrobial susceptibility testing: a position paper explaining the VetCAST approach. Front Microbiol 2017;8:2344 10.3389/fmicb.2017.02344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 3rd ED. CLSI supplement VET015. Clinical and Laboratory Standards Institute, Wayne, PA,2015. [Google Scholar]
- 39.Bundesamt für Verbraucherschutz und Lebensmittelsicherheit (BVL) BVL-Report 12.5. Bericht Zur Resistenzmonitoringstudie 2016: Resistenzsituation bei klinisch wichtigen tierpathogenen Bakterien, 2018. Available: https://www.bvl.bund.de/SharedDocs/Downloads/09_Untersuchungen/Resistenz-Monitoring-2016.pdf?__blob=publicationFile&v=2 [Accessed July 2019].
- 40.Leimbach A, Poehlein A, Vollmers J, et al. No evidence for a bovine mastitis Escherichia coli pathotype. BMC Genomics 2017;18:359 10.1186/s12864-017-3739-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klaas IC, Zadoks RN. An update on environmental mastitis: challenging perceptions. Transbound Emerg Dis 2018;65:166–85. 10.1111/tbed.12704 [DOI] [PubMed] [Google Scholar]
- 42.Fazel F, Jamshidi A, Khoramian B. Phenotypic and genotypic study on antimicrobial resistance patterns of E. coli isolates from bovine mastitis. Microb Pathog 2019;132:355–61. 10.1016/j.micpath.2019.05.018 [DOI] [PubMed] [Google Scholar]
- 43.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 2000;66:4555–8. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clermont O, Olier M, Hoede C, et al. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect Genet Evol 2011;11:654–62. 10.1016/j.meegid.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 45.Goldstone RJ, Harris S, Smith DGE. Genomic content typifying a prevalent clade of bovine mastitis-associated Escherichia coli. Sci Rep 2016;6:30115 10.1038/srep30115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kempf F, Slugocki C, Blum SE, et al. Genomic comparative study of bovine mastitis Escherichia coli. PLoS One 2016;11:e0147954 10.1371/journal.pone.0147954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keane OM. Genetic diversity, the virulence gene profile and antimicrobial resistance of clinical mastitis-associated Escherichia coli. Res Microbiol 2016;167:678–84. 10.1016/j.resmic.2016.06.011 [DOI] [PubMed] [Google Scholar]
- 48.Toval F, Köhler CD, Vogel U, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol 2014;52:407–18. 10.1128/JCM.02069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinnon J, Roy Chowdhury P, Djordjevic SP. Genomic analysis of multidrug-resistant Escherichia coli ST58 causing urosepsis. Int J Antimicrob Agents 2018;52:430–5. 10.1016/j.ijantimicag.2018.06.017 [DOI] [PubMed] [Google Scholar]
- 50.Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 2014;20:380–90. 10.1111/1469-0691.12646 [DOI] [PubMed] [Google Scholar]
- 51.Mora A, López C, Herrera A, et al. Emerging avian pathogenic Escherichia coli strains belonging to clonal groups O111:H4-D-ST2085 and O111:H4-D-ST117 with high virulence-gene content and zoonotic potential. Vet Microbiol 2012;156:347–52. 10.1016/j.vetmic.2011.10.033 [DOI] [PubMed] [Google Scholar]
- 52.Ronco T, Stegger M, Olsen RH, et al. Spread of avian pathogenic Escherichia coli ST117 O78:H4 in Nordic broiler production. BMC Genomics 2017;18:13 10.1186/s12864-016-3415-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hammad AM, Hoffmann M, Gonzalez-Escalona N, et al. Genomic features of colistin resistant Escherichia coli ST69 strain harboring mcr-1 on IncHI2 plasmid from raw milk cheese in Egypt. Infect Genet Evol 2019;73:126–31. 10.1016/j.meegid.2019.04.021 [DOI] [PubMed] [Google Scholar]
- 54.Afema JA, Ahmed S, Besser TE, et al. Molecular epidemiology of dairy cattle-associated Escherichia coli carrying bla CTX-M genes in Washington State. Appl Environ Microbiol 2018;84:e02430–17. 10.1128/AEM.02430-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Döpfer D, Almeida RA, Lam TJGM, et al. Adhesion and invasion of Escherichia coli from single and recurrent clinical cases of bovine mastitis in vitro. Vet Microbiol 2000;74:331–43. 10.1016/S0378-1135(00)00191-7 [DOI] [PubMed] [Google Scholar]
- 56.Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog 2019;11:10 10.1186/s13099-019-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miajlovic H, Smith SG. Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol Lett 2014;354:1–9. 10.1111/1574-6968.12419 [DOI] [PubMed] [Google Scholar]
- 58.Nemeth J, Muckle CA, Lo RYC. Serum resistance and the traT gene in bovine mastitis-causing Escherichia coli. Vet Microbiol 1991;28:343–51. 10.1016/0378-1135(91)90069-R [DOI] [PubMed] [Google Scholar]
- 59.Zhu Y, Dong W, Ma J, et al. Characterization and virulence clustering analysis of extraintestinal pathogenic Escherichia coli isolated from swine in China. BMC Vet Res 2017;13:94 10.1186/s12917-017-0975-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez-Siek KE, et al. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 2005;151:2097–110. 10.1099/mic.0.27499-0 [DOI] [PubMed] [Google Scholar]
- 61.Kaipainen T, Pohjanvirta T, Shpigel NY, et al. Virulence factors of Escherichia coli isolated from bovine clinical mastitis. Vet Microbiol 2002;85:37–46. 10.1016/S0378-1135(01)00483-7 [DOI] [PubMed] [Google Scholar]
- 62.Johnson JR, Porter S, Johnston B, et al. Host characteristics and bacterial traits predict experimental virulence for Escherichia coli bloodstream isolates from patients with urosepsis. Open Forum Infect Dis 2015;2:ofv083 10.1093/ofid/ofv083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olson MA, Siebach TW, Griffitts JS, et al. Genome-wide identification of fitness factors in mastitis-associated Escherichia coli. Appl Environ Microbiol 2018;84:e02190–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beutin L, Geier D, Steinrück H, et al. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Cin Microbiol 1993;31:2483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holland RE, Wilson RA, Holland MS, et al. Characterization of eae+ Escherichia coli isolated from healthy and diarrheic calves. Vet Microbiol 1999;66:251–63. 10.1016/S0378-1135(99)00013-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murinda SE, Ibekwe AM, Rodriguez NG, et al. Shiga Toxin–producing Escherichia coli in mastitis: An international perspective. Foodborne Pathog Dis 2019;16:229–43. 10.1089/fpd.2018.2491 [DOI] [PubMed] [Google Scholar]
- 67.Tozzoli R, Caprioli A, Cappannella S, et al. Production of the subtilase AB5 cytotoxin by Shiga toxin-negative Escherichia coli. J Clin Microbiol 2010;48:178–83. 10.1128/JCM.01648-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwarz S, Silley P, Simjee S, et al. Editorial: assessing the antimicrobial susceptibility of bacteria obtained from animals. J Antimicrob Chemother 2010;65:601–4. 10.1093/jac/dkq037 [DOI] [PubMed] [Google Scholar]
- 69.Boireau C, Cazeau G, Jarrige N, et al. Antimicrobial resistance in bacteria isolated from mastitis in dairy cattle in France, 2006–2016. J Dairy Sci 2018;101:9451–62. 10.3168/jds.2018-14835 [DOI] [PubMed] [Google Scholar]
- 70.Chehabi CN, Nonnemann B, Astrup LB, et al. In vitro antimicrobial resistance of causative agents to clinical mastitis in Danish dairy cows. Foodborne Pathog Dis 2019;16:562–72. 10.1089/fpd.2018.2560 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
vetreco-2019-000369supp001.pdf (1.1MB, pdf)