Supplemental digital content is available in the text.
Key Words: human papillomavirus, HPV genotyping, HPV assay, cervical cancer screening, risk-based screening, risk discrimination, risk stratification, colposcopy, baseline, follow up
Objective
Thirteen human papillomavirus (HPV) genotypes are associated with the highest risk of cervical disease/cancer; however, the risk of disease progression and cancer is genotype dependent. The objective of this systematic review was to examine evidence for high-grade cervical intraepithelial neoplasia (≥CIN 3) risk discrimination using HPV genotyping.
Materials and Methods
A systematic review of English and non-English articles through MEDLINE, Cochrane, clinicaltrials.gov, and abstracts presented at relevant professional society conferences were searched from 2000 to 2019. Search terms included: cervical cancer screening, HPV genotyping, CIN, HPV persistence, humans, and colposcopy; prospective, controlled trials, observational studies, and retrospective studies of residual specimens; evidence included HPV genotyping (beyond genotypes 16/18/45) results. Data were obtained independently by authors using predefined fields. Risk of bias was evaluated with a modified Newcastle-Ottawa Scale. The Grading of Recommendations, Assessment, Development and Evaluation methodology facilitated overall quality of evidence evaluation for risk estimation. The study protocol was registered with the PROSPERO International Prospective Register of Systematic Reviews (CRD42018091093). The primary outcome was CIN 3 or worse risk both at baseline and at different follow-up periods.
Results
Of 236 identified sources, 60 full texts were retrieved and 16 articles/sources were included. Risk of bias was deemed low; the overall quality of evidence for CIN 3 or worse risk with negative for intraepithelial lesions or malignancies or low-grade squamous intraepithelial cytology was assessed as moderate; that with atypical squamous cells-undetermined significance and “all cytology” was assessed as high. Clinical and methodological heterogeneity precluded meta-analysis. Human papillomavirus genotyping discriminated risk of CIN 3 or worse to a clinically significant degree, regardless of cytology result.
Conclusions
The evidence supports a clinical utility for HPV genotyping in risk discrimination during cervical cancer screening.
Advanced molecular methods to characterize human papillomavirus (HPV) allows for the precise distinction between individual HPV genotypes.1,2 The individual oncogenic risk of HPV genotypes have been acknowledged ever since the definitive association was recognized between HPV as the etiological agent requisite for the development of cervical intraepithelial neoplasia (CIN) and cervical cancer3; international nomenclature defines HPV genotypes from high oncogenic risk to low risk.2,4 Today, HPV diagnostics has its main application in cervical cancer screening with a growing number of countries replacing cervical cytology with molecular HPV testing as the primary screening modality. In cervical cancer, 13 HPV genotypes are definitively associated with oncogenic risk and defined as high risk by the International Agency for Research on Cancer.4 Whereas most screening programs treat the finding of any of these 13 high-risk genotypes as one group with respect to screening diagnostics, some programs acknowledge that HPV 16 and 18 comes with elevated risk and therefore should be managed differently than the remaining 11 genotypes. However, using risk estimates for development of disease on all high-risk genotypes allows increased differentiation at the individual genotype level, enabling a much more detailed risk continuum from the highest-risk genotype toward the lowest-risk genotype with respect to oncogenicity.2,4 Originally, risk stratification in cervical screening of patients, based on the underlying HPV genotype was suggested in 2003 when the primary clinical HPV assays for screening indicated the presence or absence of high-risk HPV viruses—genotype detection was solely performed either in a research setting or as an in-house test. Clifford et al5 suggested that HPV genotypes 16, 18, and 45 would merit closer surveillance than women infected with other high-risk HPV genotypes. Subsequently, large-scale studies of cervical cancers established the contribution of different HPV genotypes to squamous cell carcinoma and adenocarcinoma, which established the hierarchy of high-risk HPV genotypes.2 Throughout the next decade, studies showed that genotypes 31, 33, 52, and 58 confer risks similar to HPV 18 and 45, thereby establishing impetus for contemplating more complex screening algorithms using genotype-specific risk stratification to allow for more precise colposcopy referral recommendations and reducing overtreatment.6–9 Until now, genotyping in various settings has been implemented across screening programs mainly to facilitate risk stratification of equivocal or low-grade cytology with the objective of reducing overtreatment. In this context, atypical squamous cells-undetermined significance (ASC-US) has been triaged by many HPV testing programs since the beginning of 2010, though only managing high-risk HPV genotypes as one pooled group. At the time, the rationale was not only to use genotype information per se but also to deselect women with ASC-US for colposcopy in the absence of HPV infection due to the very low risk of underlying disease resulting from an HPV-negative status in combination with ASC-US cytology.10 However, risk stratification by genotyping is gaining ground to improve triage of existing cytology-based screening, and proposals have been set forth that women with ASC-US or low-grade squamous intraepithelial (LSIL) cytology, who test negative for the 7 or 8 highest-risk genotypes, may not require immediate colposcopy.8,11–13 The first large-scale implementation of HPV genotype information to selectively focus attention on specific subgroups of cervical screening outcomes was the use of individual qualitative reporting of HPV genotypes 16 or 18 (with or without 45) as positive or negative in cervical cancer screening. This referral strategy was termed partial (or limited) genotyping,14 has been a component of cervical cancer screening guidelines since 2012,12,15 and will remain important as triage of light or moderate cytology abnormalities—as long as cytology remains the primary test for various national or regional cervical cancer screening programs. Thus, today's application of HPV diagnostics in screening distinguishes between a partial genotyping result for reporting of HPV 16 and 18, with the remaining high-risk HPV genotypes as a pooled result. A recent expert review by Xu et al,16 assessing the accuracy of HPV 16/18 genotyping to triage LSIL cytology, points out that although the partial genotyping strategy increases the positive predictive value, the specificity declines compared with cytology. A more complete differentiation between genotypes may improve this strategy. Compared with partial genotyping, extended genotyping requires assays reporting at least 6 individual genotypes, whereas full genotyping requires the assay to report all high-risk genotypes, individually.14
Before 2018, algorithms for both co-testing and primary HPV screening guidelines were based solely on partial genotyping and did not reflect the difference in risk for CIN 2 or worse (≥CIN 2) and CIN 3 or worse (≥CIN 3) among women with various non-16/18 HPV genotypes. In this systematic review, evidence is presented that describes assessment of oncogenic risk for individual HPV genotypes in the context of cervical cancer screening.
The PICO of this systematic review was as follows: (P) patients who underwent cervical screening to identify those at high risk for high-grade cervical disease or cervical cancer; (I) intervention was HPV genotyping; (C) comparator was pooled HPV result; and (O) outcome was risk for CIN 2/3 or worse.
METHODS
The study protocol was developed and the review performed in accordance with the Preferred Reporting Items for Systematic Reviews (PRISMA)17 and the Institutes of Medicine Standards for Systematic Reviews.18 No similar published systematic review and no similar study protocol was found; this study protocol was registered with the PROSPERO International Prospective Register of Systematic Reviews in 2018 (PROSPERO CRD42018091093).19 Eligible studies included prospective controlled trials and observational studies of women and retrospective studies of residual specimens that were screened or tested using HPV DNA or RNA assays reporting HPV genotyping results (beyond genotypes 16/18/45). The primary outcome was baseline or longitudinal CIN 3 or worse risk. The period for risk estimates was baseline, 1 year, 3 years, 5 years, or more than 5 years. The MEDLINE, Cochrane Database of Systematic Reviews, Health Technology Assessment, clinicaltrials.gov electronic databases, abstracts from several relevant professional society conferences (e.g., the American Society for Colposcopy and Cervical Pathology) were searched between January 2000 and April 2019. The search string was: (HPV[Title/Abstract] or “human papillomavirus”[Title/Abstract]) AND (genotyp*[Title/Abstract] AND screening[Title/Abstract] AND cervical[Title/Abstract] AND cancer[Title/Abstract] OR *cancer[Title/Abstract] OR carcinoma[Title/Abstract] OR lesion*[Title/Abstract] OR CIN[Title/Abstract] OR “Cervical intraepithelial neoplasia”[Title/Abstract] OR persistence[Title/Abstract])) AND ((“2000/01/01”[PDat]: “3000/12/31”[PDat]) AND Humans[Mesh]). All retrieved titles and abstracts were assessed for possible relevance by applying inclusion and exclusion criteria. Full-text review of the articles that passed abstract assessment was performed to identify studies that met inclusion and exclusion criteria. Supplemental Figure 1 reports the PRISMA flow diagram, http://links.lww.com/LGT/A125.
Data extraction tables were developed in Excel, piloted, and used for study characteristics and for risk estimates with 95% CI. Risk estimates for high-grade CIN, based on HPV and full individual genotyping, were analyzed irrespective of cytology (all cytology results, similar to a primary HPV screening paradigm) and subgrouped according to the concomitant patient cytology result: negative for intraepithelial lesions or malignancies (NILMs), ASC-US, or LSIL cytology with HPV positive (as for co-testing, primary HPV, and cytology paradigms). Because the risk for high-grade CIN associated with high-grade squamous intraepithelial lesion (HSIL) is considered to exceed the clinical action threshold for colposcopy regardless of HPV status in all published guidelines, this subgroup was not analyzed.
A modified Newcastle-Ottawa Scale was used to evaluate risk of bias (individual study quality)20; it included the following 6 domains: selection (eligibility criteria, forming the cohort, selection of participants), detection (measurement of test result), outcome (assessment, length of follow-up), attrition (loss to follow-up), reporting (failure to adequately control confounding, failure to measure all known prognostic factors), and other bias. Risk of bias summary assessment for individual studies, combining all evaluations from authors, was assessed as high, low, or unclear. Each author assessed the overall quality of evidence for the risk estimate outcomes (all included studies) using a modified Grading of Recommendations, Assessment, Development and Evaluation (GRADE)21 methodology for observational diagnostic studies and included the risk of bias summary assessment for the individual studies: indirectness, imprecision, inconsistency, publication bias, magnitude of effect, and whether all plausible confounders or other biases reduced confidence in the estimated effect. Summary levels of certainty, combining all authors' evaluations, were assessed as high, moderate, or low.
RESULTS
Characteristics of Study Populations
The search identified 236 unique abstracts; from those, 176 did not meet inclusion criteria or met exclusion criteria, and 60 were assessed for full-text review. Forty-four articles were excluded for reasons listed in the PRISMA flow diagram in Supplemental Figure 1, http://links.lww.com/LGT/A125. Data from 15 articles and one society abstract presentation were used for this synthesis and represented 10 cervical cancer screening research studies from the United States, United Kingdom, Sweden, Denmark, and the Netherlands (see Table 1)22–37 Of the 16 screening populations (henceforth referred to as “studies”), 10 were prospective studies,23,26–29,31,32,34,36,37 4 were retrospective analyses of a prospectively screened population,24,25,30,35 and 2 were post hoc analyses of a prospectively screened population.22,33 Combined, the 16 studies represent a screening population sample size of 1,406,328 women. A total of 240,674 samples were analyzed and contributed to the evidence synthesis. Seven studies contributed data with HPV genotyping of specimens from women across all cytology categories (n = 156,722),22–28 5 studies included genotyping information for women with either ASC-US or LSIL (n = 18,128),25,29,31–33 3 studies contributed information on genotyping for women with ASC-US cytology (n = 52,427),24,29,30 2 studies provided genotyping data for women with LSIL (n = 50,348),24,29 and 8 studies provided information on HPV genotyping from women with NILM cytology (n = 170,602).22,24,25,32,34–37 Because the absolute risks of CIN 2 or worse and CIN 3 or worse associated with a genotype are moderated by the co-existent cytology status, the narrative synthesis is grouped according to coincident cytology category.
TABLE 1.
Cervical Cancer Screening Studies Including HPV Genotyping and a Clinical End Point of ≥CIN 2 and ≥ CIN 3: Articles Providing Data for a Systematic Review of HPV Genotyping to Facilitate Risk-Based Screening
Genotype-Specific Risk in Women—Regardless of Cytology
Of the 7 studies that contributed data for HPV genotyping across all cytology categories, 3 were prospective, one was a post hoc analysis of a prospective study, one was a prospective cohort, and 2 were retrospective by design (see Table 1). Supplemental Table 1, http://links.lww.com/LGT/A126, and Figure 1 show results from genotyping of high-risk HPV and the associated CIN 3 or worse risk values during baseline screening and at longitudinal times following baseline.
FIGURE 1.
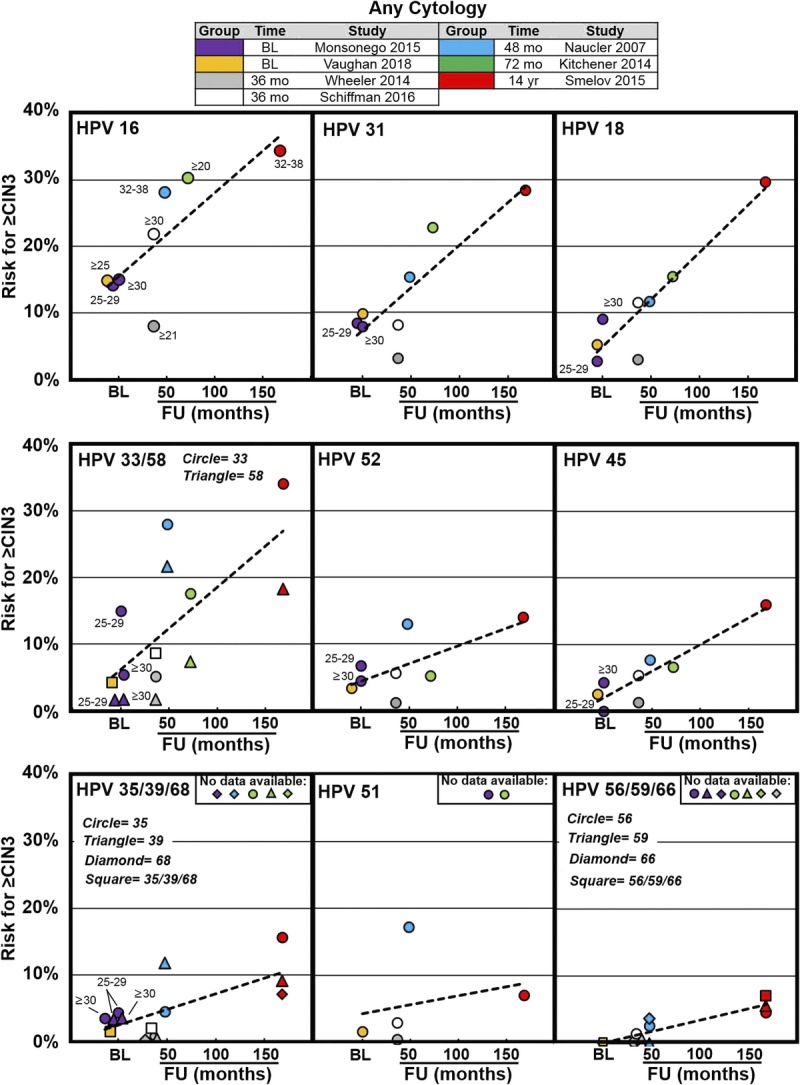
Cervical intraepithelial neoplasia 3 or worse risk values associated with individual HPV genotypes from previously described screening populations—regardless of cytology result. The x-axis represents time to follow-up in months or years (where indicated) and the y-axis represents increasing risk for CIN 3 or worse. Data were extracted from 7 articles that represent baseline results (Monsonego et al,22 2015; Vaughan et al,23 2018) and results at 36 months (Wheeler et al,24 2014; Schiffman et al,25 2016) 48 months (Naucler et al,26 2007), 72 months (Kitchener et al,27 2014), and 14 years (Smelov et al,28 2015) following baseline in each of the respective studies. Trend lines are superimposed across time points from baseline to 14 years to help visualize the increasing risk associated with long-term HPV infection. Abbreviation: BL, baseline.
A post hoc analysis of 40,901 women, 25 years or older, enrolled in the Addressing the Need for Advanced HPV Diagnostics (ATHENA) trial during screening and used Linear Array (Roche Molecular Systems Inc, Pleasanton, CA) for HPV genotyping of HPV-positive screening samples.22 Human papillomavirus genotype positivity was categorized by single-genotype infection and by hierarchical ranking.22
The Onclarity clinical trial enrolled women, 21 years or older, undergoing routine screening in the United States with the Onclarity HPV assay (Becton, Dickinson and Company, BD Life Sciences—Diagnostic Systems, Sparks, MD).23 Across 33,634 cytology results, Onclarity results for HPV genotypes (16, 31, 18, 33/58, 52, 45, 35/39/68, and 56/59/66) demonstrated clear stratification for CIN 3 or worse risk in women, 25 years or older.23
Five studies reported risk stratification in multiyear longitudinal studies (see Supplemental Table 1, http://links.lww.com/LGT/A126, and Figure 1). The 3-year genotype-specific risks for detection of CIN 3 or worse from 47,541 women undergoing opportunistic cervical cancer screening, as part of the New Mexico HPV and Pap Registry (NMHPVPR), was reported in 2014.24 A retrospective cohort study from the National Cancer Institute Kaiser Permanente Northern California (KPNC) Pap Cohort study of 8,664 samples from women in the United States, using the Onclarity HPV assay and cytology triage of HPV-positive samples, analyzed 18-month and 3-year CIN 3 or worse risks. Schiffman et al25 reported stratification of the HPV genotype results, into the following 5 tiers: HPV 16, else 18/45, else 31/33/58/52, else 51/35/39/56/59/66/68, else HPV negative (18-month data; not shown in Supplemental Table 1, http://links.lww.com/LGT/A126 or Figure 1). In the Swedish Screening randomized controlled trial (SWEDESCREEN) study, a population-based cohort of 5,696 women was followed for a mean of 4.1 years to assess the risk of CIN 2 or worse and CIN 3 or worse after type-specific HPV DNA positivity by GP5+/6+ at baseline.26 A Randomized Trial In Screening To Improve Cytology (ARTISTIC) study on cervical screening in primary care (Greater Manchester, United Kingdom) used a set-up with recalls for a third round of screening, 3 years after the second screening round, a total of 6 years after the cohort (n = 8,873) enrolled in the study.27 The SWEDESCREEN also estimated HPV type–specific risks for CIN 2 or worse and CIN 3 or worse with 14.6 years of follow-up using comprehensive nationwide registers after a prospective randomized primary HPV screening trial conducted in 5 Swedish regions and including a total 11,683 women, 32 to 38 years of age.28
Collectively, these longitudinal studies report cumulative CIN 3 or worse risk during follow-up periods spanning 3, 3.5, 4.1, 6, and 14 years. The risk hierarchy for HPV genotypes across these different longitudinal time points shows a pattern consistent with the 2 baseline studies described previously (see Supplemental Table 1, http://links.lww.com/LGT/A126 and Figure 1). In all the studies, HPV 16 was associated with the greatest risk for developing CIN 3 or worse. Human papillomavirus 31, 18, 33, and 58 were frequently the genotypes of next highest risk, and HPV 31 and 33 had similar or higher risks than HPV 18, including after 6 to 14 years of follow-up. Genotypes 35, 51, 56, 59, 66, and 68 were consistently associated with risks lower than the overall risk for pooled HPV positive and lower than the colposcopy threshold risk.
Risk by Genotype in the ASC-US/LSIL Cytology Population
Supplemental Table 2, http://links.lww.com/LGT/A127, and Figure 2 contain data from the included studies corresponding to CIN 3 or worse risk associated with individual genotyping results for the ASC-US/LSIL cytology population.
FIGURE 2.
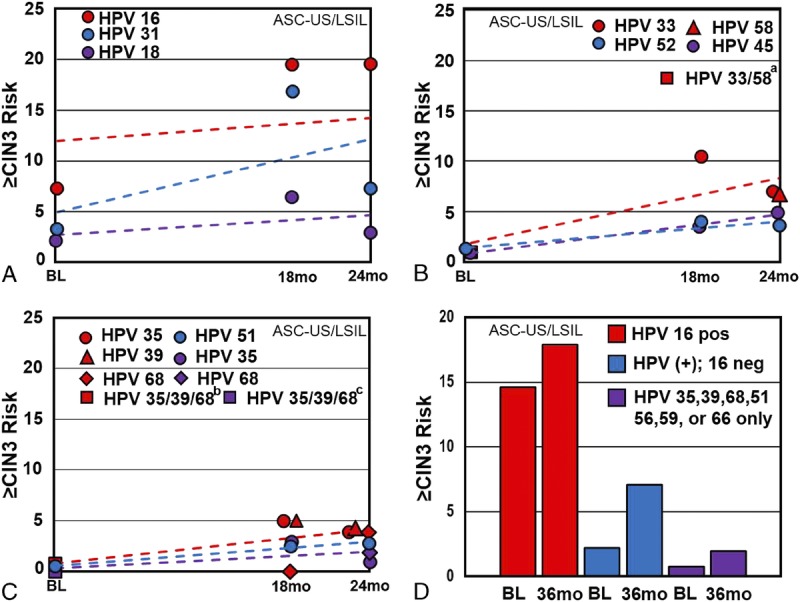
Individual genotype risk values in women with abnormal cytology. A–D, Cervical intraepithelial neoplasia 3 or worse risk values associated with individual HPV genotypes from previously described ASC-US/LSIL screening populations. In all panels, the x-axis represents time to follow-up in months and the y-axis represents CIN 3 or worse risk associated with individual HPV genotypes. A–C, The baseline values were obtained from Wright et al26 (2019); the 18-month values were obtained from Berkhof et al32 (2006); the 24-month values were obtained from Wheeler et al33 (2006). D, The 36-month values were obtained from Schiffman et al30 (2015). aPooled HPV 33/58 result. bPooled HPV 35/39/68 result. cPooled HPV 56/59/66 result.
Analyses from the baseline phase of the Onclarity trial included specimens from a total of 2,807 women with ASC-US or LSIL cytology and demonstrated clear risk stratification for CIN 3 or worse in this combined cytology population by genotype (see Figures 2A–D).29
A Dutch screening trial from 1999 to 2002 with 21,996 women enrolled in the intervention group determined the 18-month risk of CIN 3 or worse among 374 women, 29 years or older (mean age = 36.2 years), who tested positive for 14 HPV types and had borderline (ASC-US) or mild dyskaryosis (LSIL).32 The analysis at the time combined the cytology categories into what was termed borderline/mild dyskaryosis, and data do not allow for stratification of cytology categories. Human papillomavirus testing was done using GP5+/6+ polymerase chain reaction (PCR) enzyme immunoassay using a cocktail of the 14 high-risk types and subsequent typing via Southern blot analysis. By genotype, the hierarchical 18-month CIN 3 or worse risks with ASC-US/LSIL were as follows: 16 (37.0% [CI = 28.0–48.0]), 31 (27.0% [CI =14.0–46.0]), 33 (22.0% [CI = 9.0–44.0]), and 18 (11.0% [CI = 3.0–30.0]) (n = 374; data not shown in Supplemental Table 2, http://links.lww.com/LGT/A127). After excluding HPV 16 positive, the remaining average CIN 3 or worse risk was 12.0% (CI = 8.0–17.0) (data not shown in Supplemental Table 2, http://links.lww.com/LGT/A127); only HPV 31 and 33 (both HPV 16 negative) had hierarchical risks higher than the pooled average. Single-genotype infection of HPV 16, or 31, or 33 (n = 282) CIN 3 or worse risks are reported in tables and figures. (see Supplemental Table 2, http://links.lww.com/LGT/A127 and Figures 2A–C).32
In the ALTS trial (Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study Group), comparison of management strategies for 3,488 US women with ASC-US and 1,572 with LSIL with colposcopic biopsy and follow-up was done using a randomized trial design.33 The cumulative 2-year risks of CIN 2 or worse and CIN 3 or worse were reported for 14 carcinogenic genotypes, as detected by hybrid capture 2 (HC2; Qiagen Co, Gaithersburg, MD), followed by PCR. For single-type HPV infections with concurrent LSIL/ASC-US cytology, the rank ordered by 2-year cumulative CIN 3 or worse risks were as follows: HPV 16 (39.1% [32.9–45.7]), 31 (14.8% [8.1–23.9]), 33 (14.0% [5.3–27.9]), and non-16 HPV (7.9% [5.4–11.2]; data are not shown in Supplemental Table 2, http://links.lww.com/LGT/A127) (see Figures 2A–C).
In the retrospective National Cancer Institute KPNC Pap Cohort study, 8,664 were tested using the Onclarity assay in the ASC-US/LSIL cytology population at 18 months and 3 years to determine genotype risk associated with CIN 3 or worse. Here, the HPV genotype results were subsequently stratified into 5 tiers (HPV 16, else 18/45, else 31/33/58/52, else 51/35/39/56/59/66/68, else HPV negative).25 The 3-year hierarchical CIN 3 or worse risk by HPV result in the ASC-US/LSIL population was as follows: HPV 16, 17.9% (CI = 16.2–19.5); else HPV 18/45, 7.1% (CI = 5.6–8.6); else HPV 31/33/58/52, 5.7% (CI = 5.0–6.4); and else HPV 35/39/68/51/56/59/66, 2.0% (CI = 1.7–2.4) (see Figure 2D); HPV negative carried a CIN 3 or worse risk value of 0.3% (CI = 0.2–0.4) (see Supplemental Table 2, http://links.lww.com/LGT/A127; some listed data not included).
Finally, recent work from Bonde et al31 (2019) evaluated the baseline risk of CIN 3 or worse by genotype in a colposcopy referral population (referrals based on ≥ASC-US cytology or positive HPV result) of 655 women from Denmark and Italy. Bayesian probability modeling was used to determine the individual HPV genotype–based CIN 3 or worse risk values. Relative risk ratios for individual genotypes to overall (any) HPV in women with abnormal baseline cytology or women referred to colposcopy after baseline screening are shown in Figure 3.
FIGURE 3.
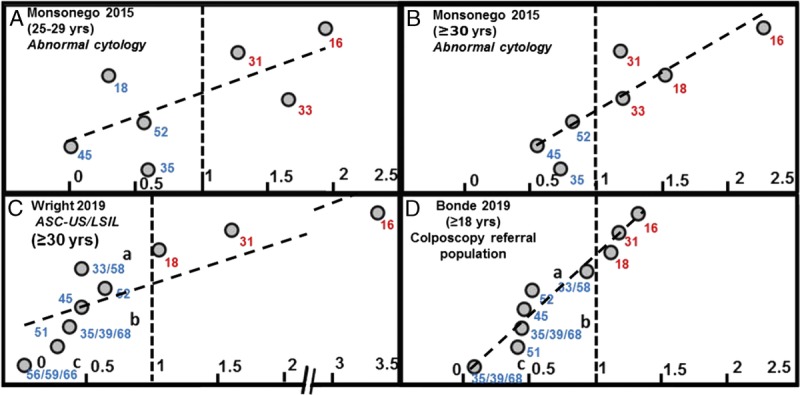
Individual genotype risk values in women with abnormal cytology or from a colposcopy referral population. A–D, Relative risk ratios of individual genotypes to overall (any) HPV in women with abnormal baseline cytology or women referred to colposcopy after baseline screening. Baseline CIN 3 or worse risk values are shown from 3 prospective cervical cancer screening trials.22,29,31 The populations shown here include any abnormal cytology in (E) and (F) (Monsonego et al, 2015), ASC-US or LSIL cytology in (G) (Wright et al,26 2019); in (H), data were obtained from a colposcopy referral population (based on abnormal cytology or positive HPV status; Bonde et al,31 2019). The risk ratios for each, individual genotypes, relative to the risk value for any HPV result, are plotted along the x-axis. The vertical, hashed line at x = 1 represents: individual genotypes risk value = risk value for any HPV result.
Data from 3 baseline, prospective screening populations with mixed, abnormal cytology,22,29,31 when normalized to overall HPV, reveal similar stratification of genotype-associated CIN 3 or worse risk (see Figures 3A–D).
Risk by Genotype in the ASC-US Cytology Population
Supplemental Table 2, http://links.lww.com/LGT/A127, contains data from the included studies corresponding to CIN 3 or worse risk associated with individual genotyping results for the ASC-US cytology population. Analyses from the baseline phase of the Onclarity trial included specimens from a total of 1,953 women with ASC-US cytology and demonstrated risk stratification for CIN 3 or worse, by genotypes.29
Wheeler et al24 (NMHPVPR) reported the genotype-specific 3-year risk of CIN 3 or worse for the combination of ASC-US and HPV positive.
A retrospective cohort study of 17,190 samples from women in the United States, using Onclarity HPV assay with 2,079 cases of ASC-US cytology, analyzed 3-year CIN 2 or worse and CIN 3 or worse risks.30 The overall 3-year CIN 3 or worse risk for ASC-US/HPV positive was 5.2%, with markedly varying risk assessment from 16.0% for HPV 16 to a 12-fold lower risk of 1.3% for the combined detection of HPV 56/59/66. Here, risk values for HPV 16, 18, 31, 33/58, 52, and 45 were above the overall US colposcopy referral risk threshold (5.2%); the confidence limits for HPV 52 overlapped this risk threshold.
Risk by Genotype in the LSIL Cytology Population
Supplemental Table 2, http://links.lww.com/LGT/A127, contains data from the included studies corresponding to CIN 3 or worse risk associated with individual genotyping results for the LSIL cytology population. The Onclarity trial consecutively enrolled women, 21 years or older, undergoing routine screening and included specimens from a total of 854 women with LSIL cytology. Baseline results demonstrated risk stratification for CIN 3 or worse in the LSIL cytology population by genotypes.29
During 3-year cervical cancer screening of 47,541 women as part of NMHPVPR, the CIN 3 or worse risk by genotype within the LSIL cytology population demonstrated definite risk stratification.24
Overall, with HPV positivity and low-grade cytology, HPV 16 had the highest CIN 3 or worse risk, with HPV 31, 18, 33, 58, and 52 ranked in the next tier. Human papillomavirus 31 and 33 had similar or higher risks than HPV 18, at 3 years of follow-up. Human papillomavirus 56, 59, 66, 68, and 51 ranked at the lowest-risk tier, with risks consistently below the colposcopy threshold.
Genotype-Specific Risk in Women With NILM Cytology
Supplemental Table 3, http://links.lww.com/LGT/A128, and Figure 4 contain data from the included studies corresponding to CIN 3 or worse risk associated with genotyping results for the NILM cytology population.
FIGURE 4.
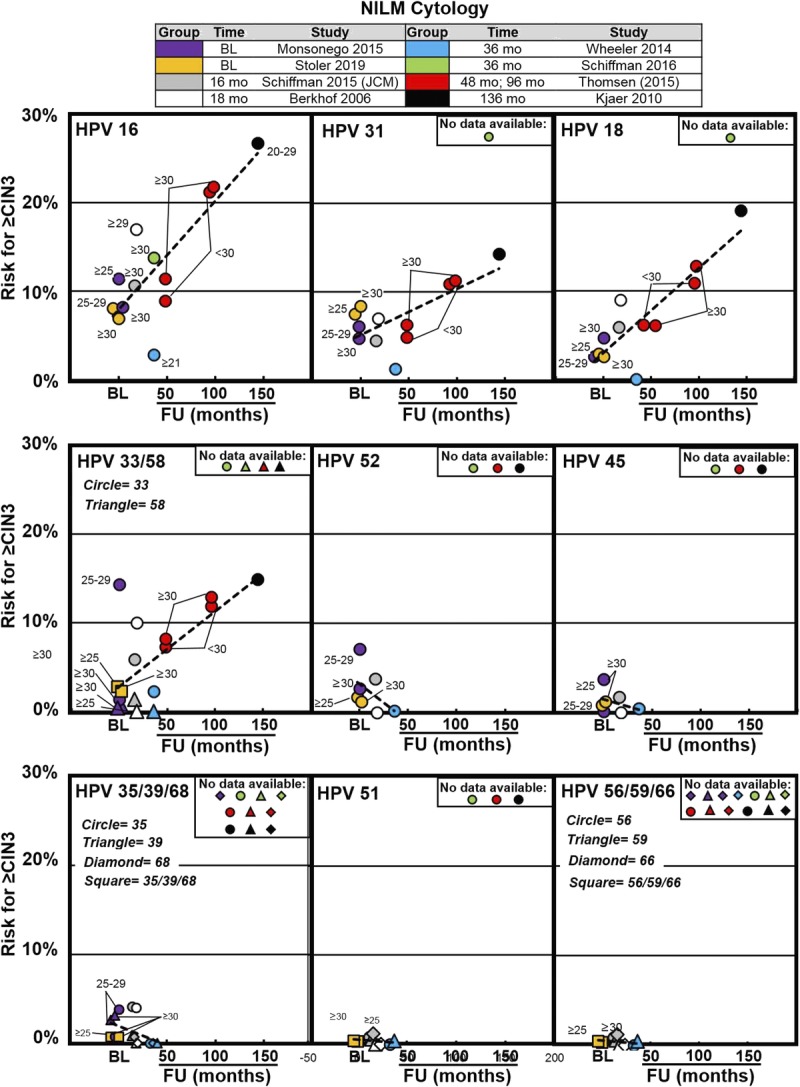
Cervical intraepithelial neoplasia 3 or worse risk values associated with individual HPV genotypes in women with NILM cytology from previously described screening populations. The x-axis represents time to follow-up in months or years (where indicated) and the y-axis represents increasing risk for CIN 3 or worse. Data were extracted from 8 articles that represent baseline results (Monsonego et al,22 2015; Stoler et al,34 2019) and results at 16 months (Schiffman et al,30 [JCM], 2015) 18 months (Berkhof et al,32 2006), 36 months (Schiffman et al,25 2016; Wheeler et al,24 2014), 48 and 96 months (Thomsen et al,36 2015), and 136 months (Kjaer et al,37 2010) following baseline in each of the respective studies. Trend lines are superimposed across time points from baseline to 136 months to help visualize the increasing risk associated with long-term HPV infection. Abbreviation: BL, baseline.
The post hoc analysis of 40,901 women (≥25 years of age) from the ATHENA Trial (described previously) used Linear Array for HPV genotyping to characterize CIN 3 or worse risk, associated with single-genotype infection by hierarchical ranking, in the NILM population at baseline.22 Women were stratified between ages 25 and 29 years and 30 years or older.22
Data from the NILM cytology population of the Onclarity trial was analyzed to determine the impact of risk stratification via HPV genotyping during baseline screening.34
The Dutch screening trial (described previously) determined the 18-month risk of CIN 3 or worse among women, 29 years or older (mean age = 36.2 years), who tested positive for 14 HPV types and had NILM.32
For HPV positive and NILM, the NMHPVPR study reported the CIN 3 or worse risk in descending order as follows: HPV 16 (2.8%), 33 (2.3%), 31 (1.3%), and all other genotypes were 0% to 0.4%.24 The reported subpopulation was biased toward younger women with 45% of NILM results in women younger than 30 years and 8% of NILM results in women older than 30 years.24
A retrospective study of 4,602 women, 30 years or older, with NILM cytology and HPV testing by HC2 and subsequent Linear Array HPV genotyping estimated the overall 3-year CIN 3 or worse risk for HPV positive and NILM to be 4.6%.35 The hierarchical genotype rank order 3-year risk of CIN 3 or worse with NILM provided clear risk discrimination.35 Similarly, the National Cancer Institute KPNC Pap Cohort study, using Onclarity HPV assay and cytology triage, reported CIN 3 or worse risks by HPV genotype grouping, with concurrent NILM cytology, to be as follows: 13.8% for HPV 16, 4.4% for non-16, HPV 18/45–positive women, 4.0% for non-16/18/45, HPV 31/33/58/52–positive women, and 1.2% for women positive only for the remaining 7 high-risk genotypes at 12-month follow-up; HPV negative with NILM was associated with a CIN 3 or worse risk of 0.06% to 0.33%.25
Similar findings were reported from a cohort of 7,482 Danish women from the general population who were examined twice (mean age at baseline was 28 years).37 For women with NILM who were persistently genotype positive at the second examination, the estimated 12-year probability of developing CIN 3 or worse by single-genotype positivity was as follows: HPV 16 (26.0%), 18 (15.4%), 33 (12.8%), 31 (9.8%), 35 (9.1%), 58 (8.3%), 45 (6.4%), 52 (4.7%), 51 (6.9%), 56 (2.3%), and 39/59/68/53/66 (0%). By contrast, the risk of CIN 3 or worse after a negative HPV test was 3.0%.37
This study was followed by a prospective cohort study in Denmark, estimating the long-term CIN 3 or worse risk by HPV genotype among 33,288 women aged 14 to 90 years with NILM baseline cytology.36 The cohort was followed in the nationwide Danish pathology register for up to 11.5 years. In women 30 years or older at baseline, the rank ordered, 11.5-year absolute risk for CIN 3 or worse after baseline detection of a single HPV genotype infection was HPV 16 (23.3%), 33 (17.9%), 31 (11.3%), 18 (10.8%), 52 (6.0%), 45 (3.9%), 58 (3.4%), 59 (2.4%), 56 (2.2%), 39 (1.9%), 51 (1.4%), 68 (1.2%), and 66 (0%). In this study, the risks for CIN 3 or worse associated with HPV 18, 31, and 33 were very similar throughout the follow-up period showing CIN 3 or worse risks for HPV 33 at 17.9% and HPV 31 at 11.3%, which were slightly higher than HPV 18 positive at 10.8%. The pooled HPV-positive risk was found to be 9.7%. The CIN 3 or worse risks for HPV 52 positive (6%) were higher than those of HPV 45 positive (3.9%).36
A population-based cohort of 5,696 women in Sweden was followed for a mean of 4.1 years to assess the risk of CIN 2 or worse after type-specific HPV DNA positivity by GP5+/6+ and NILM cytology at baseline.26 Here, HPV 16, 31, and 33 conveyed the highest risks and were responsible for a proportion of 33.1%, 18.3%, and 7.7% of CIN 2 or worse cases in the NILM group, respectively, whereas women with HPV 18, 35, 39, 45, 51, 52, 56, 58, 59, and 66 genotype results had significantly lower risks of CIN 2 or worse than women with HPV 16. After adjustment for mixed infection with other HPV types, HPV 35, 45, 59, and 66 had no detectable association with CIN 2 or worse in women with NILM cytology.26
A different Swedish study estimated HPV type–specific risks for CIN 2 or worse and CIN 3 or worse with 14.6 years of follow-up using comprehensive nationwide registers after a prospective randomized primary HPV screening trial conducted in 5 Swedish regions of women 32 years or older.28 Here, multivariate analysis was used to adjust for mixed infections, resulting in a rank-ordered genotype-specific cumulative CIN 3 or worse risk at 14 years with HPV 16 (34.5%), 33 (34.1%), 18 (29.7%), 31 (28.4%), 58 (18.3%), 45 (16.0%), 35 (15.6%), 52 (14.0%), 39 (9.1%), 68 (7.1%), 51 (7.0%), 66 (5.9%), 59 (5.6%), and 56 (4.6%). In comparison, the cumulative CIN 3 or worse risk at 14 years for overall HPV positive was 20.7%.28
The ARTISTIC study on cervical screening in primary care (Greater Manchester, United Kingdom) used a set-up with recalls for a third round of screening 3 years after the second screening round, a total of 6 years after study enrollment.27 Here, 8,873 women underwent liquid-based cytology and HPV genotyping, with colposcopic biopsy for abnormal results at the third round. Women participants had NILM/HPV positive in the first and second rounds. Women who were HPV 16 positive at entry had a cumulative 6-year CIN 3 or worse rate of 30.4%, compared with 25.9% for 16/18 positive, 10.7% for non-HPV 16/18 but 31/33/45/52/58 positive, 2.7% for non-HPV 16/18 but 35/39/51/56/59/66/68 positive, and 0.3% for HPV negative.27 Human papillomavirus 16/18 was associated with a significantly greater cumulative rate of CIN 3 or worse compared with 31/33/45/52/58 positivity, and both groups were significantly greater than the lower 7 genotype group (all p < .0001). The hierarchical cumulative 6-year CIN 3 or worse rate was as follows: HPV 16 (30.3%), 33 (17.6%), 31 (15.4%), 18 (11.7%), 58 (7.4%), 45 (6.6%), 52 (5.2%), and the group of lower 7 genotypes was 2.7%.27
Overall, with HPV-positive and NILM cytology, HPV 16 had the highest CIN 3 or worse risk, and only HPV 16 was associated with risk more than the 10% risk threshold for colposcopy in the European Union Human papillomavirus 31 and 33 posed risks similar to HPV 18, and more than the 5% risk threshold for colposcopy in the United States. Human papillomavirus 35, 39, 51, 56, 59, 66, and 68 ranked at the lowest tier and consistently less than the 5% risk threshold for colposcopy.
Risk of Bias
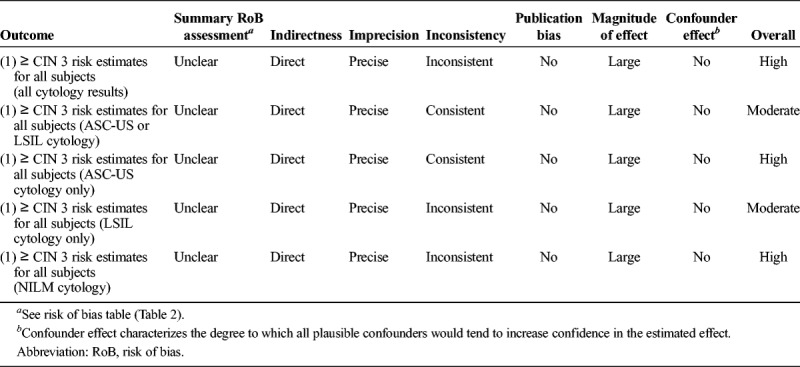
The risk of bias (individual study quality) was evaluated using a modified Newcastle-Ottawa Scale, which included the following 6 domains: selection (eligibility criteria, forming the cohort, selection of participants), detection (measurement of test result), outcome (assessment, length of follow-up), attrition (loss to follow-up), reporting (failure to adequately control confounding, failure to measure all known prognostic factors), and other bias. The risk of bias assessment was overall low or uncertain for the studies (see Table 2). The overall quality of evidence for the risk estimate outcomes (all included studies) was assessed using a modified GRADE methodology for observational diagnostic studies and judged to be moderate or high, depending on the categorization (see Table 3).
TABLE 2.
Modified Newcastle-Ottawa Scale (Risk of Bias Tool for Quality Assessment of Observational Studies)a
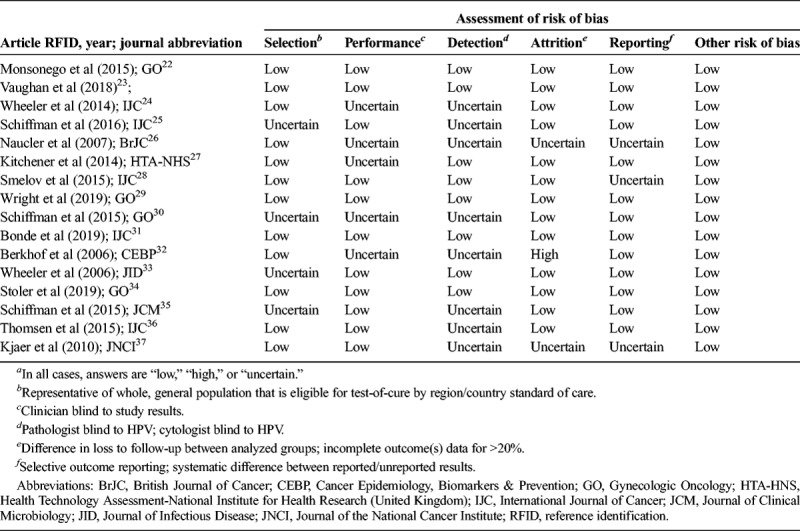
TABLE 3.
Overall Quality of Evidence for Outcomes (Modified GRADE)21
Summary of Findings
Individual HPV genotypes carry distinct risk values for high-grade cervical disease. Human papillomavirus 16 consistently carries the highest risk for CIN 3 or worse (approximately 15%–35% for any cytology and approximately 8%–25% for normal cytology), both during baseline screening, and through longitudinal follow-up—regardless of age, cytology result, or country of origin. Human papillomavirus 31, 18, and 33 carried intermediate-high CIN 3 or worse risk (ranging from approximately 8% to 20% in all cytology and approximately 5% to 10% in normal cytology). Beyond HPV 16, 31, 18, and 33, HPV 52, 58, and 45 carried moderate risks, with 35, 39, 51, 56, 59, 66, and 68 consistently having the lowest CIN 3 or worse risks, regardless of cytology. Risk for CIN 3 or worse was directly proportional to the time to follow-up across cytology for most genotypes, including HPV 16, 31, 18, 33, 58, 52, and 45. This pattern was less pronounced for HPV 35, 39, 51, 56, 59, 66, and 68, especially in the NILM population.
DISCUSSION
Collectively, the 16 studies included in this review emphasize that HPV genotyping can refine clinical management for women screened through the primary HPV paradigm and the co-testing paradigm by stratifying genotype-specific results and thereby assign women at highest risk for cervical disease to further testing (i.e., colposcopy) or treatment, while designating those with lowest risk to retesting at a shortened interval. Human papillomavirus 16 consistently carries the highest risk for CIN 3 or worse at baseline and during longitudinal follow-up and, depending on cytology, warrants clinical management ranging from colposcopy to immediate treatment. Human papillomavirus 31, 18, and 33 carried a lower risk for CIN 3 or worse compared with HPV 16 but consistently carried CIN 3 or worse risk values in the NILM population that would warrant very close monitoring. Human papillomavirus 52, 58, and 45 carried intermediate risks and would not warrant immediate colposcopy referral in the NILM population meet the risk threshold for colposcopy when combined with an abnormal cytology result. Human papillomavirus 35, 39, 51, 56, 59, 66, and 68 consistently had the lowest CIN 3 or worse risk, regardless of cytology; these genotypes consistently carried risk values that would warrant designation for retesting at a shortened time interval (e.g., 1 year) in the LSIL or less population. Stratification of genotype-specific risk could be an effective approach to reduce needless colposcopies in the ASC-US/LSIL population, while identifying women at high risk for cervical disease that should not be designated for repeat co-testing a shortened time interval (to preclude colposcopy) based on NILM cytology.
Early efforts to use HPV testing in screening was entirely focused on the test outcomes “HPV positive” or “HPV negative.” The concept of genotyping to stratify risk has been powered by clinical studies, such as those included in this synthesis. Another contributor is the development of clinical HPV assays with individual reporting of specific genotypes as part of the integrated test result. The latter's commercial use of the term “genotyping” to market HPV assays prompted an academic effort to streamline the nomenclature into partial or limited genotyping, extended genotyping, or full genotyping.14 The meaning of “partial” or “limited” genotyping is the ability to report HPV 16 and 18 individually and the remaining 12 high-risk genotypes in one group, whereas “extended” genotyping refers to assays that individually detect at least 6 high-risk HPV genotypes and the remaining in one or more groups. “Full” genotyping reflects assays that report all high-risk HPV genotypes individually.14 In addition, large-scale international consortia are conducting performance comparisons between assays with genotyping capability to present reliable data to decision-makers.14,38,39
Genotyping is an established method for HPV detection for research applications. Availability of Food and Drug Administration approved or Conformitè Europëenne-marked clinical assays, validated for extended or full genotyping, or to validated, clinical genotyping may be limited. Results for pooled detection and genotyping are usually reported simultaneously; therefore, the costs and resources for both detection methods are the same. Therefore, cost should be a negligible factor for integrating genotyping to improve clinical management during screening. A key stratagem is “similar management for similar risk,” indicating that it is the underlying risk of disease as determined by a combination of screening sample information that should drive the follow-up.13,25,40 Because cytology will act as the triage of HPV-positive screening samples for the foreseeable future, it is of interest to look at risk estimates by genotype in defined cytology outcomes. Assigning clinical actions to individual genotype findings in screening samples has not obtained international consensus, although several screening programs in the United States, Canada, Australia, and some European countries have implemented dedicated follow-up measures for screening samples positive for HPV 16 and 18.11,27,35
Main Findings
Applying the US threshold for colposcopy of approximately 5.2% (LSIL/unknown HPV result or ASC-US/HPV positive),41 ASC-US cytology combined with any of HPV 16, 18, 31, 33, 52, and 58 would merit direct referral. In contrast, assessment of HPV genotype–specific risk in women with ASC-US suggests that genotypes HPV 35, 39, 51, 56, 59, 66, and 68 represent lower risk that is clinically significant.24,29,30 Schiffman et al30 suggested in 2015 that women with ASC-US and these 7 genotype results could be at low enough risk to recommend a 12-month follow-up retesting regimen rather than direct colposcopy referral. In effect, this would defer 40% of women with ASC-US for retesting, half of whom would be cleared and half would be referred to colposcopy for retest 12 months later.30 Compared with the common practice of referring ASC-US/HPV-positive women to colposcopy today, this will represent a significant reduction in referrals. However, most non-US countries do not operate with a fixed colposcopy referral threshold. Here, a more “risk-tolerant” referral policy could be envisioned and eventually implemented where only the highest ranking genotypes would be directly referred (i.e., HPV 16, 31, 33, and 18), with the added security that the woman in question will be invited for a retest within a relatively short interval of typically no more than 12 months. Moreover, simply considering the HPV genotype and cytology result to create categorical management options is less effective than using genotype risk on a continuum and using other risk data (such as screening history) to generate a risk coefficient that could be applied against clinical action thresholds set by guideline panels.
Risk estimates by HPV genotype in women with LSIL were mainly derived from the NMHPVPR cohort and the Onclarity trial. Applying the United States threshold for colposcopy of 5.2%,13 LSIL combined with any of HPV 16, 18, 31, 33, 45, 52, and 58 (range = 4.6%–18.5%) would merit direct referral. Similar to women with ASC-US, the assessment of HPV genotype–specific risk in women with LSIL advocates that genotypes HPV 35, 39, 51, 56, 59, 66, and 68 (range = 0.0%–1.4%) represent lower risk that is clinically significant and these 7 genotype results could be followed up with retesting rather than warranting a direct colposcopy referral.24 Today, many cytology-based screening programs refer women with LSIL to one repeat test, and upon a repeat cytological abnormal sample, the women are referred for colposcopy. In this context, the benefit of HPV genotyping would be to conclude that these women are at highest risk after the first positive screening test, and they would be referred to colposcopy, rather than to wait 6 or 12 months. In conclusion, ASC-US or LSIL cytology with one or more of the lesser 7 genotypes carries a 3-year CIN 3 or worse risk of approximately 4%, which is below the United States standard of care threshold for colposcopy.
For women with NILM cytology, the genotypes that constitute or correspond with consensus risk thresholds remain to be firmly established. Schiffman et al35 noted that only NILM associated with HPV 16 had a 3-year CIN 3 or worse risk that clearly exceeded the US threshold for colposcopy, with genotypes HPV 33 and 18 approximating the threshold for colposcopy by 3-year CIN 3 or worse risk. However, the authors stated that this risk level would usually result in recall for retesting at 12 months.35 The results from ARTISTIC27 and the Danish prospective study36 showed that HPV 31 and 33 should be managed similar to HPV 18 and that HPV 58, 52, and 45 should be managed differently than the lowest 7 genotypes in women with NILM. The Swedish study by Smelov et al28 showed the strength of genotype information by noting that HPV 16/18/31/33 had risk estimations above the overall risk of other genotypes for a 14-year period. Together, these studies convincingly show that the use of nondescriptive HPV-positive/negative assays mask important information that can lead to unnecessary follow-up procedures, which lowers the efficacy of screening. Smelov et al28 proposed a subdivision of genotypes into different risk groups, clusters, or tiers: the highest risk oncogenic HPV types 16/18/31/33 with a CIN 3 or worse risk of 31.7%, the medium risk oncogenic HPV types, 35/45/52/58, with a CIN 3 or worse risk between 14% and 18%, and a large group (HPV 39/51/56/59/66/68) with limited risks, in which less than 10% of women developed CIN 3 or worse in the study follow-up period. The authors stated that these differences may be relevant for both clinical management and design of preventive strategies.28 The concept of grouping genotypes into bands, tiers, or clusters may ease the implementation of genotype information into screening algorithms as it simplifies the clinical management. Similarly, risk on a continuum can be a way to convert advanced study data into manageable units. In all studies, HPV 16 was always the highest risk tier. The next tier included HPV 31, 18, 33, 58, 52, and 45 in most of the studies that grouped by risk. The lesser-risk tier included HPV 39, 51, 56, 59, and 68 as well as 66—if included in the study. Human papillomavirus 35 was most commonly included in the lesser tier but also reported within the intermediate-risk tier. In effect, management can be proposed to 4 action bands corresponding to very high risk (colposcopy, with the ability to identify and treat cervical disease), moderate risk (colposcopy), low risk (12- or even 18- to 24-month follow-up), and very low risk (return to routine screening).
Referral to colposcopy is the highest level of intervention after positive screening; according to Schiffman et al25 (2016) the 3-year CIN 3 or worse risk of HPV 16 with HSIL is 60.6%, which is so high immediate treatment could be justified—without waiting for colposcopy and biopsy confirmation. Such an approach would reduce the overall number of colposcopies in women that would eventually receive conization; reducing time, health care costs, and the uncertainty women experience while waiting for the diagnostic outcome of their follow-up. Immediate treatment without colposcopy and biopsy confirmation, however, comes at the risk of over treating without due clinical reason. Similarly, non-HPV 16, HPV 18/45/31/33/55/52 positive with LSIL or ASC-US (range = 5.7%–7.1%), should be referred to colposcopy, whereas non-HPV 16/18/45/31/33/55/52 and ASC-US/LSIL (range = 1.2%–4.4%) can be referred to retest in a defined period. For the NILM cytology population, HPV 16–positive women should be referred to colposcopy, whereas those positive for the 7 lowest-risk genotypes should be designated for repeat co-testing at a shortened interval (e.g., 1 year); management of women positive for HPV 18, 31, 33, 45, 52, or 58 would depend on local or regional risk thresholds but could include retesting or colposcopy, based on a genotype risk continuum. Human papillomavirus–negative and NILM (0.06%–0.33%) should be returned to age-dependent screening interval.25
Strengths and Limitations
All studies included in this synthesis enrolled screening populations; however, given the inclusion of studies from various countries spanning more than a decade, the definition of screening population, and the HPV tests in use, varies. Most studies provided genotyping only for samples positive for HC2, as no genotyping assays at certain points in time had clinical cutoffs compatible with screening use. Hereby, the studies avoided the critique that the genotyping assay used did not have a clinical cutoff but at the same time introduced verification bias by HC2. The exceptions were studies using the Onclarity HPV assay, which has a validated clinical cutoff for both major liquid-based cytology collection media.29,34,42–45
A limitation of this analysis is that across the published literature, researchers have developed different methods for assigning genotype in the case of mixed infections.6,35,46,47 For useful genotype risk assessment, genotypes must be included in order from most discriminatory to least. To determine this order, variations of 3 different approaches may be used.6,34,35 In this analysis, the hierarchical method was preferred, where possible. The models for iterative attribution of risk rank were as follows: multivariate analysis,34,35 descending positive predictive values,6 and higher risk by single-genotype analysis. An alternative technique was to exclude all multiple infections and rank order risk for single-genotype results only. The simple proportional method of according equal risk to each genotype found in mixed infection results in totals exceeding 100%, and overestimation of risk for genotypes of lesser rank order. An underlying assumption for all the hierarchical models is that mixed infections do not involve synergism that leads to risk greater than that associated with either individual genotype.46
The period over which risk was estimated differed for many studies; 5 reported baseline risks, 6 reported risks between of 16 months and 4 years, and the remaining studies reported cumulative risks for a range of 4 to 14 years.
Rare cases of premalignant and invasive cervical lesions are related to non–high-risk HPV genotypes; these cases were not a focus of this systematic review but have been a confounding factor in some to the studies included in this synthesis.
Heterogeneity by PICO
In this systematic review, PICO heterogeneity could be considered both a strength and a limitation. Cervical cancer screening in the studies that constitute the data sources for this review were performed on different patient populations—which included differences in age, race, screening history, HPV genotype prevalence, disease prevalence, time to follow-up (from 16 months to 14 years), and clinical management at baseline screening and follow-up testing. Study populations also varied by size and cytology result, both of which impact the interpretation of results when considering the most appropriate risk thresholds for clinical management. In addition, HPV genotyping information was obtained from different methodology including PCR and sequencing—allowing for differences in sensitivity/specificity due to the intrinsic differences in the clinical cutoff values for respective assays. Nevertheless, all the tests showed excellent performances in terms of sensitivity and specificity. Therefore, the consistency of the genotyping results described here, despite the inherent clinical heterogeneity, supports the conclusion that HPV genotyping is a robust method for the triage and risk stratification of cervical cancer risk during baseline screening and follow-up. Moreover, the reproducibility of absolute and relative risk associated with HPV genotyping across clinically heterogeneous patient populations may be considered a strength of this review and better assures the applicability of these results to real-world scenarios.
CONCLUSIONS
Screening for cervical cancer prevents cancer and is recommended by clinical practice guidelines.12 Though limited, HPV 16 and 18 partial genotyping is implemented in a number of clinical screening guidelines, and accumulated evidence for more than a decade suggests that this definition should be expanded to include risk stratification on the full spectrum of high-risk HPV genotypes of women undergoing screening. Our analysis adds information about all 14 high-risk HPV genotypes and supports risk discrimination in screening paradigms, whether combined with NILM, ASC-US, or LSIL cytology or using cytology as triage of HPV-positive screening findings. Despite the different settings and methodologies in the studies evaluated, it is encouraging to observe nearly similar risk estimate profiles from many countries. For simplicity, stratification of risk by genotypes should likely be tiered, with HPV 16 at the highest, followed by 1 to 2 strata of intermediate risk, followed by a strata of lesser risk genotypes.
After high-risk HPV infection, the risk of progression to severe high-grade CIN and cancer is strongly associated with HPV genotype and genotype-specific persistence. Each HPV genotype has a specific associated risk for cervical cancer precursors and for cervical cancer. Using the continuum of risk, HPV 16 ranks highest and HPV 66 ranks the lowest. These genotype-specific risks may be further stratified when combined with cytology results. Under the principle of “similar management for similar risk,”12,13,41 genotype information can be used to support a more optimal risk-based management of patients. In conclusion, genotype information can be used as a triage method for HPV-positive women, reducing the cost of traditional colposcopy-based confirmation.48 However, before large-scale implementation, this use of genotype information would need formal evaluation and recommendations by groups issuing clinical testing guidelines. Recently, the UK National Institute for Health Research Health Technology Assessment concluded that HPV assays identifying not only HPV 16 and 18, but in addition HPV 31, 33, 45, 52, and 58, could be useful in triage as well as in primary HPV testing.49 Finally, the Danish National Health Authority Steering Committee on cervical screening has included use of genotyping and cytology as a combined triage of primary screening HPV-positive women for a defined implementation period starting 2020, becoming the first country to use not just HPV screening, but HPV genotype information in an advanced screening algorithm poised at reducing overtreatment while maintaining the sensitivity of HPV-based screening.
Footnotes
J.H.B. attended meetings with various HPV test manufacturers. J.H.B. has received honoraria from Hologic, Roche, Qiagen, Genomica, and BD Diagnostics for lectures and is the principal investigator on studies partly funded and/or received reagents at reduced or no cost by BD diagnostics, Biocartis, Qiagen, Self-screen, and Genomica SAU. The employer of J.H.B. has received free-of-charge reagents from test manufacturers involved in VALGENT (Arbyn et al., J Clin Virol 2016 76:S14-21, Bonde et al, J Clin Virol 2018 PM:30, 253, 376), EU VIPER Lt CE-IVD Study (Ejegod et al, Papillomavirus Res 2016;2:31-7, Ejegod et al, J Clin Microbiol 2016;54:2267-72). J.H.B. is furthermore subcontractor on EU HORIZON2020 No. 666-800. M.T.S. serves as a consultant for Becton Dickinson and Roche and has received honoraria as a speaker for Becton Dickinson. D.S.G. and J.C.A. are employees of Becton, Dickinson and Company, the sponsor of the study.
J.C.A. was responsible for the study concept, study design, search, and screening of titles and abstracts. J.C.A. and D.S.G. were responsible for data extraction. M.T.S., J.H.B., J.C.A., and D.S.G. were responsible for data interpretation, risk of bias assessment, overall quality of evidence assessment, and preparation of the manuscript. D.S.G. and J.C.A. did the data collection, data analysis, interpretation, manuscript preparation, and journal submission. J.H.B. and M.T.S. had final responsibility for the decision to submit this systematic review for publication.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.jlgtd.com).
REFERENCES
- 1.de Villiers EM, Fauquet C, Broker TR, et al. Classification of papillomaviruses. Virology 2004;324:17–27. [DOI] [PubMed] [Google Scholar]
- 2.de Sanjose S, Quint WG, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 2010;11:1048–56. [DOI] [PubMed] [Google Scholar]
- 3.zur Hausen H. Similarities of papillomavirus infections with tumor promoters. Princess Takamatsu Symp 1983;14:147–52. [PubMed] [Google Scholar]
- 4.International Agency for Research on Cancer Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum 2012;100B:255–314. [Google Scholar]
- 5.Clifford GM, Smith JS, Aguado T, et al. Comparison of HPV type distribution in high-grade cervical lesions and cervical cancer: a meta-analysis. Br J Cancer 2003;89:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuzick J, Ho L, Terry G, et al. Individual detection of 14 high risk human papilloma virus genotypes by the PapType test for the prediction of high grade cervical lesions. J Clin Virol 2014;60:44–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franceschi S, Clifford GM. Re: A study of the impact of adding HPV types to cervical cancer screening and triage tests. J Natl Cancer Inst 2005;97:938–9; author reply 9-41. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura Y, Matsumoto K, Satoh T, et al. HPV genotyping for triage of women with abnormal cervical cancer screening results: a multicenter prospective study. Int J Clin Oncol 2015;20:974–81. [DOI] [PubMed] [Google Scholar]
- 9.Xi LF, Schiffman M, Koutsky LA, et al. Lineages of oncogenic human papillomavirus types other than type 16 and 18 and risk for cervical intraepithelial neoplasia. J Natl Cancer Inst 2014;106:pii:dju270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arbyn M, Roelens J, Simoens C, et al. Human papillomavirus testing versus repeat cytology for triage of minor cytological cervical lesions. Cochrane Database Syst Rev 2013;Cd008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isidean SD, Mayrand MH, Ramanakumar AV, et al. Comparison of triage strategies for HPV-positive women: Canadian cervical cancer screening trial results. Cancer Epidemiol Biomarkers Prev 2017;26:923–9. [DOI] [PubMed] [Google Scholar]
- 12.Massad LS, Einstein MH, Huh WK, et al. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. J Low Genit Tract Dis 2013;17:S1–27. [DOI] [PubMed] [Google Scholar]
- 13.Katki HA, Schiffman M, Castle PE, et al. Five-year risks of CIN 2+ and CIN 3+ among women with HPV-positive and HPV-negative LSIL Pap results. J Low Genit Tract Dis 2013;17:S43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arbyn M, Depuydt C, Benoy I, et al. VALGENT: A protocol for clinical validation of human papillomavirus assays. J Clin Virol 2016;76:S14–21. [DOI] [PubMed] [Google Scholar]
- 15.Huh WK, Ault KA, Chelmow D, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Gynecol Oncol 2015;136:178–82. [DOI] [PubMed] [Google Scholar]
- 16.Xu L, Benoy I, Cuschieri K, et al. Accuracy of genotyping for HPV16 and 18 to triage women with low-grade squamous intraepithelial lesions: a pooled analysis of VALGENT studies. Expert Rev Mol Diagn 2019;19:543–51. [DOI] [PubMed] [Google Scholar]
- 17.Moher D, Liberati A, Tetzlaff J, et al. ; Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Institute of Medicine (IOM) Finding What Works in Health Care: Standards for Systematic Reviews. Washington, DC: The National Academies Press; 2011. [PubMed] [Google Scholar]
- 19.Booth A, Clarke M, Ghersi D, et al. An international registry of systematic-review protocols. Lancet 2011;377:108–9. [DOI] [PubMed] [Google Scholar]
- 20.Wells GA, Shea B, O'Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analysis. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. 2011. Accessed September 30, 2019.
- 21.Schunemann H, Brozek J, Guyatt G, Oxman A. GRADE Handbook for Grading Quality of Evidence and Strength of Recommendations. Available at: https://gdt.gradepro.org/app/handbook/handbook.html. 2013. Accessed September 30, 2019.
- 22.Monsonego J, Cox JT, Behrens C, et al. Prevalence of high-risk human papilloma virus genotypes and associated risk of cervical precancerous lesions in a large U.S. screening population: data from the ATHENA trial. Gynecol Oncol 2015;137:47–54. [DOI] [PubMed] [Google Scholar]
- 23.Vaughan L, Andrews J, Malinowski D, et al. If persistent HPV infection causes disease, why are we not measuring it [abstract]? In: 2018 American Society for Colposcopy and Cervical Pathology; 2018 Apr 18–21. Las Vegas, NV; Abstract 341. [Google Scholar]
- 24.Wheeler CM, Hunt WC, Cuzick J, et al. The influence of type-specific human papillomavirus infections on the detection of cervical precancer and cancer: a population-based study of opportunistic cervical screening in the United States. Int J Cancer 2014;135:624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiffman M, Hyun N, Raine-Bennett TR, et al. A cohort study of cervical screening using partial HPV typing and cytology triage. Int J Cancer 2016;139:2606–15. [DOI] [PubMed] [Google Scholar]
- 26.Naucler P, Ryd W, Tornberg S, et al. HPV type-specific risks of high-grade CIN during 4 years of follow-up: a population-based prospective study. Br J Cancer 2007;97:129–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C Kitchener H, Canfell K, Gilham C, et al. The clinical effectiveness and cost-effectiveness of primary human papillomavirus cervical screening in England: extended follow-up of the ARTISTIC randomised trial cohort through three screening rounds. Health Technol Assess 2014;18:1–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smelov V, Elfstrom KM, Johansson AL, et al. Long-term HPV type-specific risks of high-grade cervical intraepithelial lesions: a 14-year follow-up of a randomized primary HPV screening trial. Int J Cancer 2015;136:1171–80. [DOI] [PubMed] [Google Scholar]
- 29.Wright TC, Jr., Stoler MH, Parvu V, et al. Risk detection for high-grade cervical disease using Onclarity HPV extended genotyping in women, >21 years of age, with ASC-US or LSIL cytology. Gynecol Oncol 2019;54:360–7. [DOI] [PubMed] [Google Scholar]
- 30.Schiffman M, Vaughan LM, Raine-Bennett TR, et al. A study of HPV typing for the management of HPV-positive ASC-US cervical cytologic results. Gynecol Oncol 2015;138:573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonde J, Bottari F, Parvu V, et al. Bayesian analysis of baseline risk of CIN2 and ≥CIN3 by HPV genotype in a European referral cohort. Gynecol Oncol 2019;154:360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berkhof J, Bulkmans NW, Bleeker MC, et al. Human papillomavirus type-specific 18-month risk of high-grade cervical intraepithelial neoplasia in women with a normal or borderline/mildly dyskaryotic smear. Cancer Epidemiol Biomarkers Prev 2006;15:1268–73. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler CM, Hunt WC, Schiffman M, et al. Human papillomavirus genotypes and the cumulative 2-year risk of cervical precancer. J Infect Dis 2006;194:1291–9. [DOI] [PubMed] [Google Scholar]
- 34.Stoler MH, Wright TC, Jr., Parvu V, et al. Stratified risk of high-grade cervical disease using onclarity HPV extended genotyping in women, ≥25years of age, with NILM cytology. Gynecol Oncol 2019;153:26–33. [DOI] [PubMed] [Google Scholar]
- 35.Schiffman M, Burk RD, Boyle S, et al. A study of genotyping for management of human papillomavirus-positive, cytology-negative cervical screening results. J Clin Microbiol 2015;53:52–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomsen LT, Frederiksen K, Munk C, et al. Long-term risk of cervical intraepithelial neoplasia grade 3 or worse according to high-risk human papillomavirus genotype and semi-quantitative viral load among 33,288 women with normal cervical cytology. Int J Cancer 2015;137:193–203. [DOI] [PubMed] [Google Scholar]
- 37.Kjaer SK, Frederiksen K, Munk C, et al. Long-term absolute risk of cervical intraepithelial neoplasia grade 3 or worse following human papillomavirus infection: role of persistence. J Natl Cancer Inst 2010;102:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonde J, Ejegod DM, Cuschieri K, et al. The Valgent4 protocol: robust analytical and clinical validation of 11 HPV assays with genotyping on cervical samples collected in SurePath medium. J Clin Virol 2018;108:64–71. [DOI] [PubMed] [Google Scholar]
- 39.Polman NJ, Oštrbenk A, Xu L, et al. Evaluation of the clinical performance of the HPV-risk assay using the VALGENT-3 panel. J Clin Microbiol 2017;55:3544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castle PE, Aslam S, Behrens C. Cervical precancer and cancer risk by human papillomavirus status and cytologic interpretation: implications for risk-based management. Cancer Epidemiol Biomarkers Prev 2016;25:1595–9. [DOI] [PubMed] [Google Scholar]
- 41.Katki HA, Schiffman M, Castle PE, et al. Benchmarking CIN 3+ risk as the basis for incorporating HPV and Pap cotesting into cervical screening and management guidelines. J Low Genit Tract Dis 2013;17:S28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottari F, Sideri M, Gulmini C, et al. Comparison of onclarity human papillomavirus (HPV) assay with Hybrid Capture II HPV DNA assay for detection of cervical intraepithelial neoplasia grade 2 and 3 lesions. J Clin Microbiol 2015;53:2109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demarco M, Carter-Pokras O, Hyun N, et al. Validation of an HPV DNA cervical screening test that provides expanded HPV typing. J Clin Microbiol 2018;56;pii:e01910–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ejegod D, Bottari F, Pedersen H, et al. The BD Onclarity HPV assay on samples collected in SurePath medium meets the International Guidelines for Human Papillomavirus Test Requirements for Cervical Screening. J Clin Microbiol 2016;54:2267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ejegod DM, Junge J, Franzmann M, et al. Clinical and analytical performance of the BD Onclarity™ HPV assay for detection of CIN2+ lesions on SurePath samples. Papillomavirus Res 2016;2:31–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuschieri KS, Cubie HA, Whitley MW, et al. Multiple high risk HPV infections are common in cervical neoplasia and young women in a cervical screening population. J Clin Pathol 2004;57:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wentzensen N, Wilson LE, Wheeler CM, et al. Hierarchical clustering of human papilloma virus genotype patterns in the ASCUS-LSIL triage study. Cancer Res 2010;70:8578–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiffman M, Kinney WK, Cheung LC, et al. Relative performance of HPV and cytology components of cotesting in cervical screening. J Natl Cancer Inst 2018;110:501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilham C, Sargent A, Kitchener HC, et al. HPV testing compared with routine cytology in cervical screening: long-term follow-up of ARTISTIC RCT. Health Technol Assess 2019;23:1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


