Abstract
In both unicellular and multicellular organisms, long-tailed class I myosins function in clathrin-mediated endocytosis. Myosin 1e (Myo1e) in vertebrates and Myo1 in fission yeast have similar domain organization, yet whether these proteins or their individual protein domains are functionally interchangeable remains unknown. In an effort to assess functional conservation of class I myosins, we tested whether human Myo1e could replace Myo1 in fission yeast Schizosaccharomyces pombe and found that it was unable to substitute for yeast Myo1. To determine if any individual protein domain is responsible for the inability of Myo1e to function in yeast, we created human-yeast myosin-I chimeras. By functionally testing these chimeric myosins in vivo, we concluded that the Myo1e motor domain is unable to function in yeast, even when combined with the yeast Myo1 tail and a full complement of yeast regulatory light chains. Conversely, the Myo1e tail, when attached to the yeast Myo1 motor domain, supports localization to endocytic actin patches and partially rescues the endocytosis defect in myo1Δ cells. Further dissection showed that both the TH1 and TH2-SH3 domains in the human Myo1e tail are required for localization and function of chimeric myosin-I at endocytic sites. Overall, this study provides insights into the role of individual myosin-I domains, expands the utility of fission yeast as a simple model system to study the effects of disease-associated MYO1E mutations, and supports a model of co-evolution between a myosin motor and its actin track.
Keywords: endocytosis, myosin, actin, yeast, myosin-I, myosin 1e
Graphical Abstract
INTRODUCTION
Class I myosins are actin-binding motor proteins that typically function at sites of actin-based membrane deformation during cell shape changes. Humans have 8 class I myosins (Myo1a-h), whose expression and functions differ across tissues [1]. All class I myosins contain an N-terminal motor domain, a neck region that binds calmodulin or calmodulin-like light chains, and a short membrane-binding tail. While these domains are shared by all members of the myosin-I family, two myosins, Myo1e and Myo1f, known as “long-tailed”, contain an additional proline-rich TH2 domain and an SH3 domain at the C-terminus. “Long-tailed” class I myosins of similar structure also exist in lower eukaryotes, such as yeast, Dictyostelium, and Acanthamoeba. Class I myosins are evolutionarily ancient, with short-tailed myosins thought to represent the universal, ancestral class I myosin, and long-tailed class I myosins appearing in later eukaryotic lineages upon addition of C-terminal tail domains [2].
In both unicellular and multicellular organisms, long-tailed class I myosins seem to function in similar physiological processes, including clathrin-mediated endocytosis, pinocytosis, and phagocytosis [3-10]. Moreover, these myosins have similar binding partners and even appear at the same stage during endocytosis in diverse organisms. For example, at sites of clathrin-mediated endocytosis in both cultured animal cells and yeast, long-tailed class I myosins function within branched actin networks, assembled by the Arp2/3 complex, where they interact with both membrane phospholipids and members of the WASp family of Arp2/3 complex activators [7, 11-16]. Class I myosins are recruited to endocytic sites just ahead of actin assembly and are thought to generate force for membrane invagination and scission in both vertebrates and yeast [13, 15, 17-20].
The conservation of the overall body plan, binding partners, and functional activities between yeast and vertebrate long-tailed class I myosins suggests that the functions of individual protein domains may also be conserved. Given these similarities, we have previously used fission yeast Schizosaccharomyces pombe, which has only a single class I myosin, Myo1 [7, 8], to test the effects of point mutations in the Myo1 motor domain that are homologous to mutations found in the human Myo1e [21]. Patients homozygous for these MYO1E mutations develop kidney disease [22, 23]. Introducing these point mutations into yeast Myo1 replicated a loss of myosin-I function phenotype characterized by a severe endocytosis defect resulting in reduced cell growth. Thus, mutations in conserved motor domain residues appear to have similar effects in both human and yeast myosin-Is.
Despite structural and functional similarities, the fission yeast Myo1 and human Myo1e also differ in several respects. Myo1, like its budding yeast homologs Myo3/5, contains an additional region at the C-terminus of the tail, called the central-acidic domain, which can directly bind the Arp2/3 complex to initiate branched actin nucleation [7, 11-14]. While Myo1e contains a single light-chain binding motif that binds calmodulin [24], S. pombe Myo1 contains two IQ motifs, which bind two distinct light-chain proteins, calmodulin Cam1 and calmodulin-like Cam2 [8, 25]. Lastly, the motor domains of yeast and mammalian class I myosins differ at the TEDS site, a conserved amino acid residue located on a surface loop within the actin-binding site [26]. While mammalian myosin-Is have an acidic residue at this site, protozoan and fungal myosin-Is have a phosphorylatable threonine or serine residue at this location [27]. Phosphorylation of this threonine or serine by the PAK/Ste20 family kinases regulates myosin activity and function in amoeba and yeast [11, 14, 27-33]. Together, these differences suggest distinct modes of myosin-I regulation in yeast and mammalian cells.
Although yeast and human class I myosins perform similar functions and have a comparable protein structure, it is unknown whether these proteins or their individual protein domains are functionally interchangeable. Successful replacement of yeast Myo1 domains with respective domains from human Myo1e would provide insights into the roles of individual domains, support future studies exchanging protein domains with different biophysical properties to further dissect myosin-I function, and expand the utility of the yeast system for testing the effects of disease-associated mutations. In this study, we tested whether human Myo1e could replace Myo1 in S. pombe and found that the human myosin was unable to substitute for yeast Myo1 due to the inability of the Myo1e motor domain to function in yeast cells. However, we found that the Myo1e tail can successfully substitute for the Myo1 tail to support localization and function of myosin-I at endocytic sites.
MATERIALS AND METHODS
Yeast strain construction
Table S1 lists S. pombe strains used in this study. Table S2 lists primers used for strain and plasmid construction. Strains were constructed by homologous recombination-based genomic integrations following lithium acetate transformations [34] and by genetic crosses. To replace myo1+ CDS with MYO1E CDS, pBS-SpMyo1 construct, which contains a 5-kb EcoR1 fragment of S. pombe genomic DNA encompassing myo1+ gene in pBluescript [7] was linearized by PCR amplification with subM1utr5r and subM1utr3d primers. In parallel, MYO1E CDS flanked by 70 nt of myo1+ 5’ UTR and 70 nt of 3’ UTR was amplified from pEGFP-C1-myo1e-EcoRI-, in which EcoRI in MYO1E CDS was mutated without changing amino acid sequence. The amplified products were ligated using In-Fusion HD Cloning kit (Takara Bio USA, Mountain View, CA), and the resultant pBS-HsMyo1e construct contained MYO1E CDS flanked by myo1+ 5’ and 3’ UTRs. The HsMyo1e E337S TEDS site mutation was generated in pBS-HsMyo1e construct using the QuickChange kit (Agilent, Santa Clara, CA) according to the manufacturer’s instructions. To replace S. pombe myo1+ CDS with coding sequences for Myo1e or Myo1e(E337S) in the endogenous myo1 locus, the pBS constructs were digested with EcoR1, and 5-kb fragments containing HsMyo1e CDS or HsMyo1e(E337S) CDS flanked by myo1+ 5’ and 3’ UTRs were introduced via lithium acetate transformation into the myo1Δ::ura4+ strain (TP192), in which the myo1+ CDS was replaced with the ura4+ nutritional marker [13], followed by counter-selection against ura4+ on EMM agar plates containing 2 mg/ml 5-FOA (5-fluoroorotic acid) and 0.05 mg/ml uracil. Positive clones were identified by PCR and verified by sequencing. These strains express untagged Myo1e and Myo1e(E337S) from the native myo1 locus under control of the endogenous Pmyo1 promoter. To tag HsMyo1e and HsMyo1e(E337S) expressed from myo1 locus with mGFP at the C-terminus, the stop codon in the HsMyo1e coding sequence was replaced with mGFP-tagging cassette amplified from pFA6a-mGFP-kanMX6 (pJQW85-4) using primers m1e-GFP-int-F and ml-Kxr [35, 36]. The integrations were confirmed by PCR and sequencing.
Strains with different combinations of gene deletions and fluorescent protein-tagged alleles were generated by genetic crosses. Cells were mated on ME (Malt Extract) agar plates, followed by tetrad dissections on YES agar plates and screening for wanted gene combinations by replica plating onto appropriate selective plates, microscopy, and PCR diagnostics. To facilitate mating, in some cases myo1Δ cells were transformed with pUR-myo1 plasmid [7], which is subsequently lost upon sporulation.
Plasmid construction
Plasmids for expression of mGFP-tagged myosin constructs in S. pombe are listed in Table S3. For the purposes of making chimeras, S. pombe Myo1 domains were defined as follows: motor domain, aa 1-720; IQ1, aa 721-741; IQ2, aa 742-763; TH1, aa 764-966; TH2, aa 967-1112; SH3, aa 1113-1163; CA, 1164-1217. H. sapiens Myo1e domain boundaries were similarly defined: motor domain, aa 1-692; IQ, aa 693-717; TH1, aa 718-923; TH2, aa 924-1055; SH3, aa 1056-1108.
Chimeras HsM-SpT-1, HsM-SpT-2, HsM-SpT-3 and SpM-HsT-1, SpM-HsT-2 were first generated in pBluescript by In-Fusion cloning of inserts encoding different fragments of human Myo1e PCR-amplified from pEGFP-C1-myo1e-EcoR1-into the PCR-amplified segments of pBS-SpMyo1 vector using primers designed to replace selected SpMyo1 domain sequences with corresponding HsMyo1e sequences. For HsM-SpT-1, a fragment encoding the HsMyo1e motor and IQ domains was amplified with primers m1Myo1Ed and SpHs1hA and ligated into pBS-SpMyo1 amplified with primers subM1utr5r and SpHs1p1, thereby replacing S. pombe Myo1 motor, IQ1 and IQ2 domains. For HsM-SpT-2, a fragment encoding HsMyo1e motor domain was amplified with primers m1Myo1Ed and SpHs2hB and ligated into pBS-SpMyo1 amplified with primers subM1utr5r and SpHs2p2, thereby replacing S. pombe Myo1 motor domain. For HsM-SpT-3, a fragment encoding HsMyo1e motor and IQ domains was amplified with primers m1Myo1Ed and SpHs5hE and ligated into pBS-SpMyo1 amplified with primers subM1utr5r and SpHs5p5, thereby replacing S. pombe Myo1 motor and IQ1 domains. For SpM-HsT-1, a fragment encoding HsMyo1e tail consisting of the TH1, TH2, and SH3 domains was amplified with primers SpHs3hC and m1Myo1Er and ligated into pBS-SpMyo1 amplified with primers SpHs3p3 and subM1utr3d, thereby replacing S. pombe Myo1 tail consisting of TH1, TH2, SH3, and CA domains. For SpM-HsT-2, a fragment encoding HsMyo1e IQ and tail domains was amplified with primers SpHs4hD and m1Myo1Er and ligated into pBS-SpMyo1 amplified with primers SpHs4p4 and subM1utr3d, thereby replacing S. pombe Myo1 IQ1, IQ2 and tail domains.
To move into mGFP-tagging vector for yeast expression, SpMyo1, HsMyo1e, HsMyo1e(E337S), and Chimeras HsM-SpT-1, -2, -3 and SpM-HsT-1, -2 sequences were amplified using the appropriate primer pairs (selected from human primers Myo1e F, Myo1e R and yeast primers Myo1 F, SpTail R) containing NotI and NheI cloning sites, subcloned into pCR-BluntII-TOPO using the Zero Blunt TOPO PCR Cloning Kit (Thermo Fisher Scientific, Waltham, MA) and verified by sequencing. The inserts were then cut from the TOPO constructs using NotI and NheI and ligated into the pSGP-573 vector [37] that we had previously modified by replacing the GFP sequence with monomeric mGFP and adding GASGTGS linker at the mGFP C-terminus. All constructs in this modified pSGP-573 vector express N-terminally mGFP-tagged proteins under control of the strong thiamine-repressible 3xPnmt1 promoter [38]. To generate SpM-HsT-3, the IQ2 domain was removed from pSGP-573-SpM-HsT-1 by linearizing the plasmid using primers Chimera6 and m1D1IQ2L followed by ligation using In-Fusion cloning. The HsMyo1e tail construct was derived from pSGP-573-HsMyo1e using primers pSPG-m1etail-F and pSPG-m1etail-R followed by ligation using In-Fusion cloning. The SpMyo1ΔTH1 and SpMyo1ΔTH2-SH3-CA constructs were derived from pSGP-573-SpMyo1 by amplifying with primers SpMyo1deltaTH1 and IQ2rev to remove TH1 domain coding sequence and primers SpMyo1delta23CA and SpMyo1TH1rev to remove TH2-SH3-CA coding sequence followed by ligation using In-Fusion cloning. The SpM-HsT-1ΔTH1 and SpM-HsT-1ΔTH2-SH3 constructs were generated from pSGP-573-SpM-HsT-1 by amplifying with primers CHIM3deltaTH1 and IQ2rev to remove TH1 domain coding sequence and primers Sp1m-m1e-TH1-F and hmyo1edelTH2 to remove TH2-SH3 coding sequence followed by ligation using In-Fusion cloning. Empty modified pSGP-573 vector expressing mGFP alone was used as a negative control.
Yeast electroporation
For electroporation, cells were grown overnight in EMM5S medium to an OD595 of 0.4-1.5, pelleted at 3,000 rpm (2,000 rcf) in Beckman (Brea, CA) Allegra X-15R centrifuge at 4°C, washed twice with cold sterile water, once with cold sterile 1 M sorbitol, and re-suspended in sorbitol to a concentration of ~2 × 109 cells/ml. 1 μl of each plasmid at the concentration of 100-500 ng/μl was added to 40 μL of cells and incubated on ice for 7 minutes. Cells were then electroporated using the Bio-Rad (Hercules, CA) Micropulser Electroporator according to the instruction manual, plated onto EMM agar plates lacking uracil and containing 5 μg/ml thiamine, and incubated at 25°C.
Salt sensitivity assays
For Figure 1, stably integrated strains were streaked onto YES agar plates containing 1 M KCl and incubated for 3-5 days at 25°C. For Figure 2, cells were streaked on EMM agar plates lacking uracil with or without 5 μg/ml thiamine and with or without 1 M KCl and incubated at 36°C for 2 days. For Figures 7 and S4, cells transformed with constructs in pSGP-573 vector were grown over two days at 25°C in EMM medium lacking uracil (EMM-ura) and containing 5 μg/ml thiamine. Cells were washed once in EMM-ura, diluted to OD595=0.3 in EMM-ura without thiamine, and spotted, using 10 μl of liquid culture, onto EMM agar plates lacking uracil with or without 5 μg/ml thiamine and with or without 1 M KCl. Plates without 1 M KCl were incubated for 3 days and plates containing 1 M KCl were incubated for 5-6 days at 25°C.
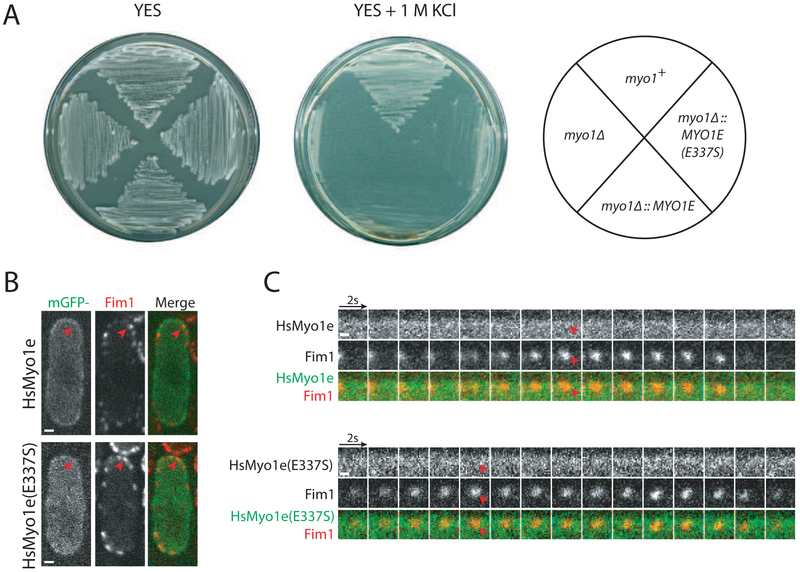
Figure 1: HsMyo1e cannot replace SpMyo1 and fails to localize to sites of endocytosis in S. pombe.
(A) Analysis of the salt sensitivity of the wild-type (myo1+), myo1Δ, myo1Δ::HsMYO1E, and myo1Δ::HsMYO1E(E337S) S. pombe cells. Cells were streaked onto YES agar plates with and without 1 M KCl and incubated for 3-5 days at 25°C.
(B, C) Colocalization analysis of mGFP-tagged HsMyo1e or HsMyo1e(E337S) (green) expressed off of a plasmid under control of 3xPnmt1 promoter in the absence of thiamine with Fim1-mCherry (red) in actin patches in myo1Δ cells. (B) Single confocal sections through the middle of the cells. Scale bars, 1 μm. (C) Montages of individual patches at 2-second intervals. Scale bars, 0.5 μm. Red arrowheads indicate lack of localization of HsMyo1e or TEDS site mutant HsMyo1e(E337S) with Fim1-mCherry in actin patches.
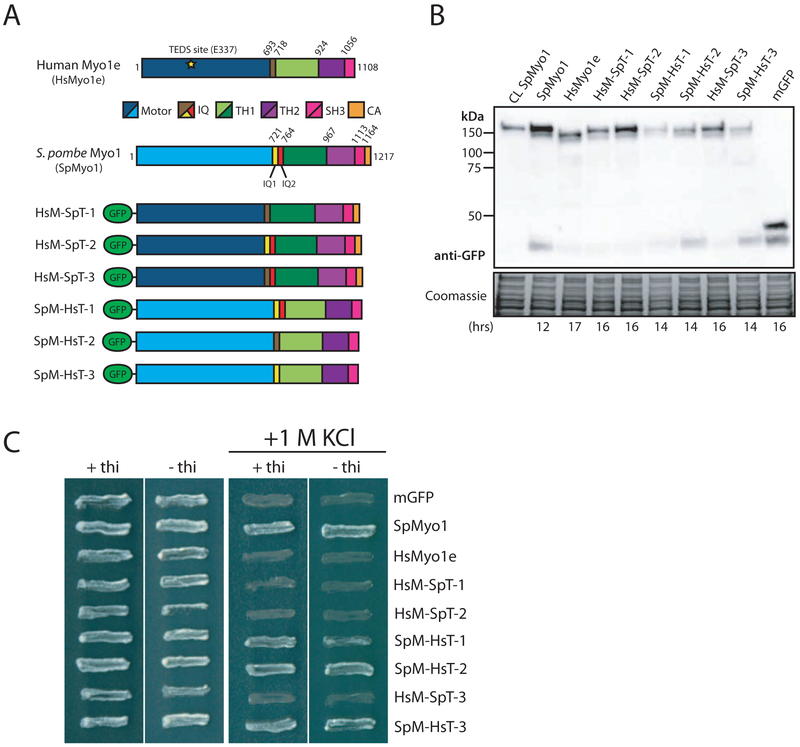
Figure 2: Human-yeast myosin-I chimeras require the SpMyo1 motor domain to rescue myo1Δ.
(A) Domain maps of H. sapiens Myo1e (HsMyo1e), S. pombe Myo1 (SpMyo1), and six human-yeast myosin-I chimeric constructs tagged with mGFP at the N-terminus. HsM-SpT chimeras 1, 2, and 3 contain the HsMyo1e motor domain and the SpMyo1 tail. SpM-HsT chimeras 1, 2, and 3 contain the SpMyo1 motor domain and the HsMyo1e tail. IQ, light chain binding IQ motif; TH, tail homology domain; SH3, Src Homology 3 domain; CA, central-acidic domain.
(B) Western blot analysis of total protein lysates of cells expressing mGFP-Myo1 (CL SpMyo1) from native myo1 locus under control of endogenous Pmyo1 promoter or myo1Δ cells expressing mGFP-tagged SpMyo1, HsMyo1e, human-yeast myosin-I chimeras, or mGFP alone from plasmids under control of 3xPnmt1 promoter. The times cells were grown in the absence of thiamine to induce protein expression are indicated at the bottom. Blots were probed with anti-GFP antibody. Coomassie stained gel serves as a loading control.
(C) Analysis of salt sensitivity of myo1Δ cells expressing mGFP alone, mGFP-tagged SpMyo1, HsMyo1e, or human-yeast myosin-I chimeras off of plasmid under control of 3xPnmt1 promoter in the presence and the absence of thiamine at 25°C on EMM agar plates with and without 1 M KCl. These cells also express actin patch marker Fim1-mCherry. Plates were incubated at 36°C for 2 days.
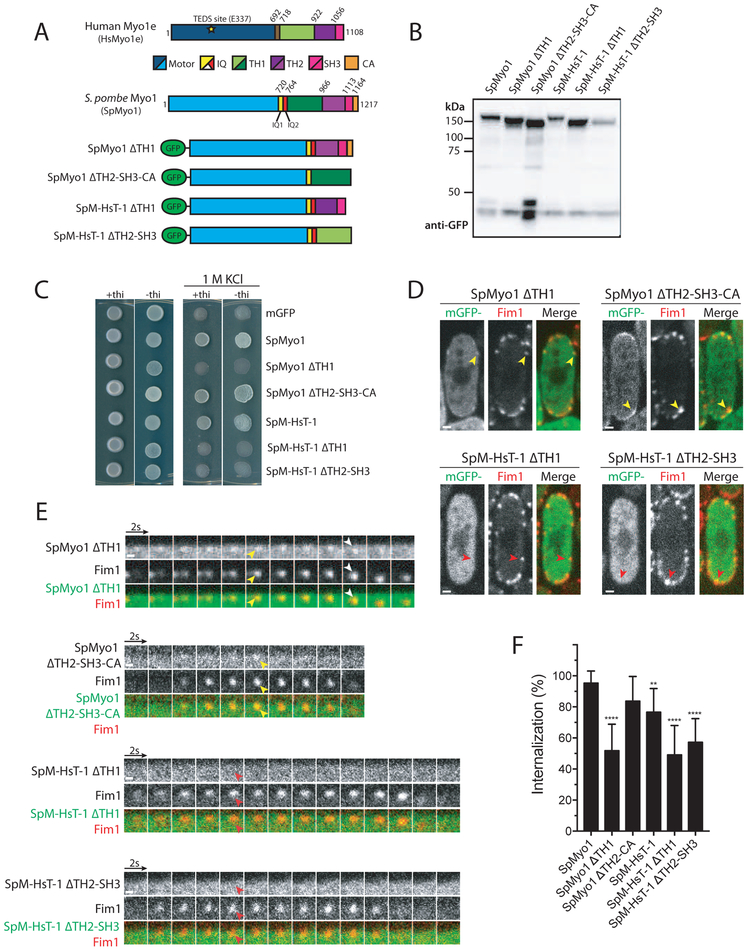
Figure 7: SpMyo1 motor - HsMyo1e tail chimera requires both Myo1e TH1 and TH2-SH3 domains to rescue myo1Δ and to localize to actin patches.
(A) Domain maps of H. sapiens Myo1e (HsMyo1e), S. pombe Myo1 (SpMyo1) and mGFP-tagged tail domain deletion constructs of SpMyo1 and SpM-HsT-3.
(B) Western blot of total protein lysates of myo1Δ cells expressing indicated mGFP-tagged myosin constructs. The constructs were expressed from plasmids under control of 3xPnmt1 promoter for 12-15 hours in the absence of thiamine. The blot was probed with an anti-GFP antibody.
(C) Analysis of salt sensitivity of myo1Δ cells expressing indicated mGFP-tagged myosin constructs from plasmids under control of 3xPnmt1 promoter in the presence or absence of thiamine at 25°C on EMM agar plates containing 1 M KCl. The cells also expressed Fim1-mCherry.
(D, E) Colocalization analysis of indicated mGFP-tagged myosin constructs (green) expressed from plasmids under control of 3xPnmt1 promoter in the absence of thiamine and Fim1-mCherry (red) in actin patches in myo1Δ cells. (D) Single confocal sections through the middle of the cells. Scale bars, 1 μm. (E) Montages of individual patches at 2-second intervals. Scale bars, 0.5 μm. Yellow arrowheads indicate colocalization and red arrowheads indicate lack of colocalization of myosin with Fim1-mCherry in actin patches. White arrows depict internalization of mGFP-SpMyo1ΔTH1 with Fim1-mCherry.
(F) Bar graph of percent internalization (± SD) of Fim1-mCherry patches in myo1Δ cells expressing indicated myosin constructs. N = 132-223 patches in 11-19 cells from 2 independent experiments. Values for SpMyo1 and SpM-HsT-1 are from Figure 4D. Asterisks indicate a significant difference from the mGFP-tagged SpMyo1 expressed from a plasmid under control of 3xPnmt1 promoter as determined by one-way ANOVA, ** p<0.01, ***p<0.001, ****p<0.0001.
Fluorescence microscopy
Stably integrated strains were grown to exponential phase in EMM5S medium at 25°C over two days. Fission yeast cells transformed with constructs in pSGP-573 vector were grown to exponential phase over two days at 25°C in EMM medium lacking uracil (EMM-ura) and containing 5 μg/ml thiamine. Cells were then washed 3 times in EMM-ura, seeded at OD595=0.08 into EMM-ura without thiamine, and incubated for 12-26 hours to induce protein expression. Generally, constructs containing the HsMyo1e motor domain needed longer induction times.
For imaging, cells were pelleted briefly using a tabletop microcentrifuge (1 minute at 2,000 rcf) and mounted on pads of 25% gelatin in EMM5S under coverslips sealed with Valap. Following 5 minutes of equilibration, cells were imaged at ambient room temperature using a PerkinElmer (Waltham, MA) UltraView VoX Spinning Disc Confocal system mounted on a Nikon Eclipse Ti-E microscope equipped with Hamamatsu C9100-50 EMCCD camera and a 100×/1.4 N.A. PlanApo objective, controlled by Volocity software. For Z-series through the entire cells, images were captured at 0.4 μm intervals. Time series of 2-color images were collected in the middle plane of the cells at 2-second intervals for 1 minute.
Image analysis
All image analysis was performed using Fiji (National Institutes of Health, Bethesda, MD) [39]. Whole-cell fluorescence intensities of mGFP-tagged proteins in individual cells, which represent protein expression levels, were averaged from frames 4-8 of a time series at the mid-cell section and subtracted for extracellular background. Only cells expressing exogenous myosin within 0.5-2-fold of the level of mGFP-SpMyo1 expressed from the native myo1 locus under control of the endogenous Pmyo1 promoter [13] were analyzed for tracking patch intensities and movement, percent internalization, and protein colocalization. For actin patch tracking, at least 10 patches from 3-5 cells were analyzed for each strain. Using a 10-pixel wide circular ROI, the patch fluorescence intensity and position were tracked for both myosin and Fim1-mCherry over the duration of each patch lifetime. Individual time courses of patch intensity and distance from the origin were aligned to the peak intensity of Fim1-mCherry at time zero and averaged at each time point. Percent internalization was measured by following patches over a 20-second time period in at least 8 cells. Internalization was considered successful if the patch moved at least 2 pixels (130 nm) from the cell membrane. Colocalization between mGFP-tagged myosin constructs and mCherry-tagged fimbrin Fim1, calmodulin Cam1, or calmodulin-related light chain Cam2 in patches was measured over a 20-second time period by using the Plot Z function to detect the presence of mGFP and mCherry signals within a 10-pixel wide ROI. At least 20 patches in at least 3 cells were analyzed for each strain. All strains were imaged at least twice to assess reproducibility.
Western blots
Cell cultures grown for live cell imaging or grown under the same conditions as for live cell imaging were harvested by centrifugation for 5 minutes at 3,000 rpm (2,000 rcf) in Beckman (Brea, CA) Allegra X-15R centrifuge at 4°C and processed for immunoblotting as previously described [40]. Pelleted cells were re-suspended in ice-cold lysis buffer U (50 mM HEPES pH7.5, 100 mM KCl, 3 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF and Complete (Roche, Branchburg, NJ) protease inhibitor cocktail), mixed with glass beads and lysed mechanically using the FastPrep-24 (MP-Bio, Santa Ana, CA), followed by addition of SDS-PAGE sample buffer. Lysates were then heated for 2 minutes at 100°C, followed by a 5-minute centrifugation at 7,000 rpm (4,600 rcf) in microcentrifuge. Total protein samples were adjusted for equal loading based on an OD595 reading prior to centrifugation, separated on 10-20% gradient SDS-PAGE gels, and transferred to Immobilon-P membranes (EMD Millipore, Billerica, MA). Membranes were incubated overnight at 4°C with primary anti-GFP antibody (67 MA5-15256, Pierce, Rockford, IL) diluted 1:3,000 in TBS-T containing 5% dry milk. The next day, the blot was washed three times with TBS-T for 5 minutes and incubated with secondary horseradish-peroxidase -conjugated antibody in TBS-T with 5% dry milk for 1 hour at room temperature. Blots were developed using Clarity™ Western ECL substrate (Bio-Rad, Hercules, CA) and imaged using Bio-Rad (Hercules, CA) ChemiDoc MP imager.
Statistical analysis
Graphpad Prism Software was used for graphing, linear regression and all other statistical analysis. For multiple comparisons, data were analyzed using a one-way ANOVA with Tukey’s Post-Hoc test, with statistical significance set at a p-value <0.05.
RESULTS
Human Myo1e cannot replace Myo1 in S. pombe
In order to test if human Myo1e, hereafter referred to as HsMyo1e, could functionally substitute for S. pombe Myo1, now abbreviated SpMyo1, we replaced the coding sequence of the wild-type yeast myo1+ with the sequence coding HsMyo1e at the endogenous myo1 locus in the yeast genome under control of the endogenous Pmyo1 promoter. These myo1Δ::HsMYO1E cells, like the myo1Δ cells [7, 8], were viable on regular YES medium at 25°C, but unable to grow in the presence of 1 M KCl (Fig. 1A and Table 1). Mutating the TEDS site in the motor domain of HsMyo1e, from glutamate to the phosphorylatable serine, did not improve its function in yeast and myo1Δ::HsMYO1E(E337S) cells exhibited the same sensitivity to high salt as myo1Δ::HsMYO1E or myo1Δ cells (Fig. 1A and Table 1).
Table 1.
Summary of salt sensitivity, localization and actin patch dynamics analyses of HsMyo1e, SpMyo1, and human-yeast myosin-I chimeras in myo1Δ cells.
| Name | Description | Promoter | Growth on 1 M KCl |
Localizes to Actin Patches |
Actin patch Internalizationa (%) |
Actin patch Lifetimea (sec) |
Colocalizes w/ Cam1 |
Colocalizes w/ Cam2 |
|---|---|---|---|---|---|---|---|---|
| CL SpMyo1b | S. pombe Myo1 | myo1+ | Y | Y | 94 ± 8 | 11.6 ± 1.3 | Y | Y |
| HsMyo1e | H. sapiens myosin 1e | myo1+ | N | N/Ac | N.D. | N.D | N/Ac | N/Ac |
| HsMyo1e (E337S) |
TEDS site mutant H. sapiens myosin 1e |
myo1+ | N | N/Ac | N.D. | N.D. | N/Ac | N/Ac |
| HsMyo1e | H. sapiens myosin 1e | nmt1 | N | N | N.D. | N.D. | N | N |
| HsMyo1e (E337S) |
TEDS site mutant H. sapiens myosin 1e |
nmt1 | N | N | N.D. | N.D. | N | N |
| HsMyo1e tail | HsMyo1e tail | nmt1 | N | N | N.D. | N.D. | N | N.D. |
| SpMyo1 | S. pombe Myo1 | nmt1 | Y | Y | 95 ± 8 | 13.4 ± 2.1 | Y | Y |
| HsM-SpT-1 | HsMyo1e motor-IQ SpMyo1 tail |
nmt1 | N | N | 50 ± 36 | 20.0 ± 4.4 | N | N |
| HsM-SpT-2 | HsMyo1e motor SpMyo1 IQ1-IQ2-tail |
nmt1 | N | N | 38 ± 26 | 20.2 ± 1.8 | N | N |
| HsM-SpT-3 | HsMyo1e motor-IQ SpMyo1 IQ2-tail |
nmt1 | N | N | 31 ± 12 | 20.8 ± 1.7 | N | N |
| SpM-HsT-1 | SpMyo1 motor-IQ1-IQ2 HsMyo1e tail |
nmt1 | Y | Y | 77 ± 15 | 14.8 ± 2.5 | Y | Y |
| SpM-HsT-2 | SpMyo1 motor HsMyo1e IQ-tail |
nmt1 | Y | Y | 78 ± 17 | 13.8 ± 2.4 | Y | N |
| SpM-HsT-3 | SpMyo1 motor-IQ1 HsMyo1e tail |
nmt1 | Y | Y | 80 ± 21 | 12.2 ± 0.6 | Y | N |
| mGFP | Empty vector | nmt1 | N | N | 33 ± 18 | 20.6 ± 3.4 | N | N |
Percent of internalization and lifetimes of actin patches in myo1Δ cells expressing indicated constructs were measured with Fim1-mCherry. Data are presented as ± SD.
CL SpMyo1 designates mGFP-Myo1 expressed from the native myo1 chromosomal locus under control of endogenous Pmyo1 promoter.
Data are not available because of the poor expression of Myo1e and Myo1e(E337S) from the myo1 locus.
Given the differences in codon preferences between yeast and humans [41], we hypothesized that the failure of HsMyo1e to replace SpMyo1 may be due to reduced protein expression in yeast cells. To estimate protein levels, we tagged HsMyo1e and HsMyo1e(E337S) expressed from the endogenous myo1 locus with monomeric GFP (mGFP) at the C-terminus. By fluorescence microscopy, we detected little to no signal for HsMyo1e-mGFP and HsMyo1e(E337S)-mGFP, compared to the whole cell intensity of wild-type SpMyo1, similarly tagged with mGFP and expressed from the native myo1 locus (Fig. S1A). Attempts to detect the mGFP-tagged HsMyo1e and HsMyo1e(E337S) in yeast lysates by western blot were also unsuccessful. The inability of yeast cells to express HsMyo1e from the endogenous myo1 locus at detectable levels impeded testing whether HsMyo1e can replace SpMyo1. To overcome this issue, we expressed mGFP-tagged HsMyo1e off of a multi-copy plasmid under control of the full strength, thiamine-repressible 3xPnmt1 promoter. This yeast plasmid contains an autonomously replicating sequence that propagates at a different rate than the yeast genome resulting in considerable variability of the plasmid copy number and consequently variability in protein expression levels amongst cells (Fig. S1B and S1C). To assess the ability of HsMyo1e to function in yeast, we transformed the mGFP-HsMyo1e construct into myo1Δ cells expressing fimbrin Fim1 tagged with mCherry as a marker for actin patches, which assemble at the sites of endocytosis in fission yeast. mGFP-HsMyo1e appeared mostly cytosolic and at elevated levels of expression formed protein aggregates on the membrane (Fig. 1B). We found that mGFP-tagged HsMyo1e and HsMyo1e(E337S) did not localize to sites of endocytosis marked by Fim1-mCherry (Fig. 1B and 1C, Table 1). Even after expression for several days in the absence of thiamine, HsMyo1e failed to rescue the inability of myo1Δ cells to grow on high salt (Fig. 2C and Table 1). Thus, while expression of HsMyo1e is not toxic since yeast cells expressing HsMyo1e are able to grow under normal growth conditions at 25 °C, HsMyo1e is not able to substitute for SpMyo1 in fission yeast cells to support growth under high salt conditions (Fig. 2C), even when expressed at levels greater than the endogenous SpMyo1 (Fig. S1B and S1C).
Human-yeast myosin-I chimeras require the SpMyo1 motor domain to rescue myo1Δ
Given the inability of HsMyo1e to replace SpMyo1, we next generated six chimeric constructs composed of varying combinations of HsMyo1e and SpMyo1 domains to determine which protein domain makes HsMyo1e unable to function in yeast (Fig. 2A). We defined the boundaries of HsMyo1e and SpMyo1 protein domains based on the alignment of their amino acid sequences (Fig. S2) and previous reports [7, 42]. Three of the chimeras, labeled as HsM-SpT-1, HsM-SpT-2, and HsM-SpT-3, contained the HsMyo1e motor domain paired with the SpMyo1 tail, and three others, abbreviated SpM-HsT-1, SpM-HsT-2, and SpM-HsT-3, contained the SpMyo1 motor domain and the HsMyo1e tail. To ascertain the functional importance of the IQ domains, we included the IQ domains from SpMyo1 and HsMyo1e in different combinations among the chimeras. All chimeras were tagged at the N-terminus with mGFP and expressed off of a multi-copy plasmid under control of the thiamine-repressible 3xPnmt1 promoter. The mGFP-tagged myosin chimeras were then transformed into a myo1Δ strain expressing Fim1-mCherry as a marker of actin patches and assessed for protein expression and the ability to support growth of myo1Δ cells on high salt (Fig. 2 and Table 1). We used mGFP-tagged SpMyo1 and mGFP alone expressed from the same multi-copy plasmids in myo1Δ cells as positive and negative controls, respectively. Western blot analysis revealed that all chimeric myosins were expressed as full-length proteins of expected molecular weights (Fig. 2B). Although western blotting could not be used to precisely measure the level of chimeric myosin expression due to the wide cell-to-cell variability of expression (see Fig. S1B and S1C), HsMyo1e and the chimeras containing the HsMyo1e motor domain required 2-3 hours longer expression than SpMyo1 and chimeras containing the SpMyo1 motor domain (Fig. 2B, see times indicated under the blot) to produce comparable signal on the western blot. All chimeras were able to grow on regular EMM medium; however, growth assays in the presence of 1 M KCl revealed that the chimeras containing the SpMyo1 motor domain with the HsMyo1e tail partially rescued the inviability of myo1Δ cells on high salt, similar to the SpMyo1 positive control but to a lesser degree (Fig. 2C). Curiously, these constructs were able to support growth on high salt even when expressed at low levels in the presence of thiamine. In contrast, the chimeras with the HsMyo1e motor domain failed to rescue the inviability of myo1Δ cells on high salt, similar to HsMyo1e and the mGFP control (Fig. 2C). These results suggest that the HsMyo1e motor domain is responsible for the inability of HsMyo1e to replace SpMyo1 and that the function of SpMyo1 requires the endogenous motor domain.
Human-yeast myosin-I chimeras require the SpMyo1 motor domain for localization to actin patches
To determine if the chimeras’ ability to rescue myo1Δ growth defects correlated with their ability to localize to actin patches, we examined their localization in myo1Δ cells expressing Fim1-mCherry (Fig. 3 and Table 1). Fluorescence microscopy revealed that indeed the rescuing chimeras SpM-HsT-1, -2, and -3, composed of the SpMyo1 motor domain and HsMyo1e tail, localized to dynamic actin patches marked by Fim1-mCherry (Fig. 3). However, unlike SpMyo1, SpM-HsT-1, -2, and -3 also strongly outlined the entire cell cortex. Accumulation of SpM-HsT-1, -2, and -3 over this enhanced cortical background was observed in ~80% of Fim1-marked actin patches (Fig. 3C). These chimeras are likely targeted to the cell cortex by the Myo1e tail since mGFP-tagged HsMyo1e tail alone similarly outlined the cell cortex but failed to localize to actin patches (Fig. S3A). Since the SpMyo1 motor domain alone also fails to localize to actin patches (Fig. S3B), both the SpMyo1 motor domain and HsMyo1e tail are needed for these chimeras to localize to actin patches.
Figure 3: Human-yeast myosin-I chimeras require the SpMyo1 motor domain for localization to actin patches.
(A, B) Analysis of colocalization of mGFP-tagged SpMyo1 or human-yeast myosin-I chimeras (green) with Fim1-mCherry (red) in actin patches in myo1Δ cells. In control, mGFP-Myo1 (CL SpMyo1) was expressed from the native myo1 locus under control of the endogenous Pmyo1 promoter in wild-type cells also expressing Fim1-mCherry. mGFP-tagged SpMyo1, mGFP alone, and mGFP-tagged human-yeast myosin-I chimeras were expressed from plasmids under control of 3xPnmt1 promoter for 12-18 hours in the absence of thiamine in myo1Δ cells expressing Fim1-mCherry. (A) Representative images in single confocal sections through the middle of the cells. Scale bars, 1 μm. (B) Montages of individual patches at 2-second intervals. Scale bars, 0.5 μm. Yellow and red arrowheads indicate the presence and the absence of myosin in Fim1-mCherry-labeled actin patches, respectively. Kymographs depict 32-seconds on the x-axis.
(C) Percent colocalization (mean ± SD) of Fim1 and mGFP, SpMyo1, and human-yeast myosin-I chimeras in dynamic actin patches. N=89-153 patches in 6-8 cells.
Chimeras HsM-SpT-1, -2, and -3, composed of the HsMyo1e motor domain and SpMyo1 tail, which failed to rescue growth of myo1Δ cells under high salt conditions, also failed to localize to sites of endocytosis, colocalizing with fimbrin in <10% of actin patches, similar to observations with mGFP alone (Fig. 3). All three chimeras exhibited mostly cytosolic localization and only mild accumulation at the cell cortex, compared to SpM-HsT-1, -2, and -3. While HsM-SpT-1, lacking the IQ2 motif of SpMyo1, lightly outlined the entire cortex, HsM-SpT-2 and -3, which both contain the IQ2 motif of SpMyo1 in addition to the SpMyo1 tail, localized to cortical thread-like structures reminiscent of eisosomes [43] in about 35-40% of the cells (Fig. S3C). To verify this, we transformed mGFP-tagged HsM-SpT-2 and -3 into myo1Δ cells expressing a known eisosome marker Fhn1 tagged with mCherry [43]. However, we could not detect convincing colocalization of the two proteins, suggesting that the cortical linear structures containing HsM-SpT-2 and -3 may not be eisosomes, or that tagging Fhn1 with mCherry interferes with the localization of these chimeras to eisosomes. Overall, this analysis revealed that the HsMyo1e motor domain combined with the SpMyo1 tail is not sufficient for localizing chimeric constructs to actin patches.
SpMyo1 motor - HsMyo1e tail chimeras support dynamics and internalization of actin patches
Since chimeras containing the SpMyo1 motor domain were capable of localizing to actin patches and rescuing the salt-sensitive growth defect of myo1Δ cells, we examined the effect of the human-yeast myosin-I chimeras on actin patch dynamics and endocytic internalization in myo1Δ cells expressing Fim1-mCherry (Fig. 4 and Table 1). We measured the time courses of intensities and distance traveled for mGFP-tagged myosin and Fim1-mCherry in individual actin patches in live cells and quantified the percent of internalizing actin patches (Fig. 4 and Fig. S4). Since not all patches in chimera-expressing cells internalized, it was not possible to align the individual time courses based on the time of internalization as done previously [44]. Therefore, we generated average time courses (Fig. 4A and Fig. S4A) by aligning individual time courses to the time Fim1-mCherry reached peak intensity (time zero), which in wild-type cells corresponds to the initiation of endocytic internalization [44]. Because the plasmid-based exogenous expression was highly variable from cell to cell (Fig. S1C), we compared the expression level of all constructs in individual cells to mGFP-tagged SpMyo1 expressed from the endogenous locus by measuring total cell intensities. Using the SpMyo1 control expressed off the plasmid, we determined whether varying levels of SpMyo1 expression have any impact on accumulation of SpMyo1 in actin patches and actin patch dynamics. This analysis revealed that SpMyo1 expression level correlated weakly with peak accumulation of SpMyo1 in patches, so that up to 2-fold more SpMyo1 accumulated in patches in cells expressing 1.5-3.5-fold more SpMyo1 than the endogenous level (Fig. S4B). This increased patch accumulation of SpMyo1 in higher expressing cells had, however, no effect on peak accumulation of Fim1-mCherry (Fig. S4C and S4D). On the other hand, in the complete absence of SpMyo1, peak accumulation of Fim1-mCherry was reduced by ~40% (Fig. 4B). To avoid potential artifacts due to variable expression levels, we chose to track actin patch dynamics only in cells that expressed myosin chimeras at the levels within the range of 0.5-2 fold the endogenous level of SpMyo1. Expression of the chimeras in myo1+ cells resulted in no growth defects in the presence or absence of high salt and 85-100% patch internalization rate, indicating that none of the myosin constructs exhibited dominant negative effects or were toxic to the cells (Fig. S4E and S4F).
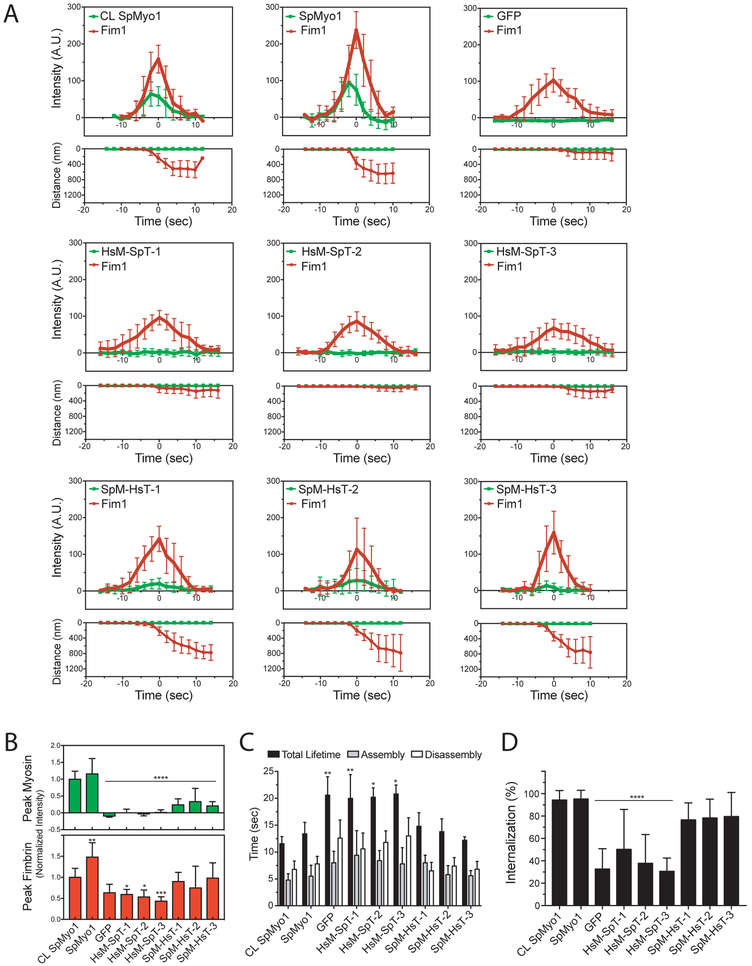
Figure 4: SpMyo1 motor - HsMyo1e tail chimeras partially rescue dynamics of endocytic internalization.
(A-D) Analysis of localization and dynamics of mGFP-tagged human-yeast myosin-I chimeras and Fim1-mCherry in actin patches in myo1Δ cells. mGFP-SpMyo1, mGFP alone, and mGFP-tagged myosin-I chimeras were expressed from plasmids under control of 3xPnmt1 promoter for 12-18 hours in the absence of thiamine in myo1Δ cells expressing Fim1-mCherry. In control (CL SpMyo1), the dynamics of mGFP-Myo1 expressed from endogenous myo1 locus and Fim1-mCherry were tracked in wild-type cells.
(A) Time courses of (upper panels) average fluorescence intensity (± SD) and (lower panels) average distance traveled (± SD) of mGFP-tagged SpMyo1, mGFP alone, or mGFP-tagged chimeras (green squares) and Fim1-mCherry (red circles) in actin patches in myo1Δ cells or, in control, wild-type cells. The time courses of cortical background-subtracted intensities and distances from the origin for individual patches were aligned to the peak of Fim1-mCherry patch intensity (time zero) and averaged at each time point.
(B) Bar graphs (mean ± SD) of peak intensities of mGFP-tagged myosin or mGFP alone (green) and Fim1-mCherry (red) in actin patches in myo1Δ cells from averaged plots (panel A) normalized to peak fluorescence intensities of mGFP-SpMyo1 (at −2 seconds) and Fim1-mCherry (at time zero) in CL SpMyo1 cells. Averaged intensities of SpMyo1 and SpM-HsT-3 in patches peaked at −2 seconds, while SpM-HsT-1 and -2 peaked at time zero. For mGFP alone and non-localizing chimeras HsM-SpT-1, -2, -3, the mGFP signal intensities at time zero were used as peak values. Asterisks indicate a significant difference from wild-type control as determined by a one-way ANOVA, *p<0.05, ** p<0.01, ***p<0.001, ****p<0.0001.
(C) Bar graph (mean ± SD) of assembly time (grey), disassembly time (white), and total lifetime (black) of Fim1-mCherry in actin patches in control wild-type cells or myo1Δ cells expressing mGFP-tagged SpMyo1, mGFP alone, or mGFP-tagged human-yeast myosin-I chimeras.
(D) Bar graph of percent internalization (± SD) of actin patches marked with Fim1-mCherry in control wild-type cells or in myo1Δ cells expressing mGFP-tagged SpMyo1, mGFP alone, or mGFP-tagged human-yeast myosin-I chimeras. N=9-12 patches in 3-6 cells in (A-C) and N=71-134 patches in 8-17 cells from 2-3 independent experiments in (D). Asterisks indicate a significant difference from wild-type control as determined by a one-way ANOVA, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
In agreement with the ability to localize to actin patches and support growth in the presence of high salt, SpM-HsT-1, -2, and -3 partially rescued the actin patch dynamics and endocytic internalization defects observed in myo1Δ cells. In myo1Δ cells expressing mGFP alone, the times of assembly, disassembly and total lifetime of Fim1-mCherry in patches were increased and only 33% of patches were able to internalize (Fig. 4). Expression of SpMyo1 off of the plasmid completely restored actin patch dynamics to wild-type levels: Fim1-mCherry lifetime in patches was 13.4 ± 2.1 seconds, similar to 11.5 ± 1.3 seconds observed in cells expressing SpMyo1 from the endogenous locus, and 90-100% of patches internalized (Fig. 4). Although SpM-HsT-1, -2, and -3 only accumulated in patches at 24%, 33%, and 21% levels of endogenously expressed SpMyo1, respectively (Fig. 4A and 4B), they partially rescued actin patch dynamics (Fig. 4C and 4D). In cells expressing these chimeras, peak accumulation and lifetimes of Fim1-mCherry in patches were similar to those observed in cells expressing SpMyo1 (Fig. 4C), and the percent of successfully internalized patches increased to 70-80%, greater than the 33% internalization rate in myo1Δ cells expressing mGFP (Fig. 4D). Similarly to SpMyo1, as actin patches internalized, SpM-HsT-1, -2, and -3 remained associated with the plasma membrane. Thus, the combination of the SpMyo1 motor domain with the HsMyo1e tail is able to support endocytosis in fission yeast.
In contrast, HsM-SpT-1, -2, and -3 that failed to localize to patches and rescue myo1Δ growth under high salt conditions did not rescue actin patch dynamics and internalization defects in myo1Δ cells (Fig. 4 and Table 1). Cells expressing these chimeras exhibited significantly longer Fim1-mCherry patch lifetimes (HsM-SpT-1: 20 ± 3.4 s; HsM-SpT-2: 20.2 ± 1.7 s; HsM-SpT-3:20.8 ± 1.6 s) and reduced accumulation of Fim1-mCherry in patches (Fig. 4A-C and Table 1). Furthermore, patch internalization in myo1Δ cells expressing these chimeras remained at 30-50%, similar to myo1Δ cells expressing mGFP (Fig 4D and Table 1). Thus, the SpMyo1 tail cannot support endocytic function when paired with the HsMyo1e motor.
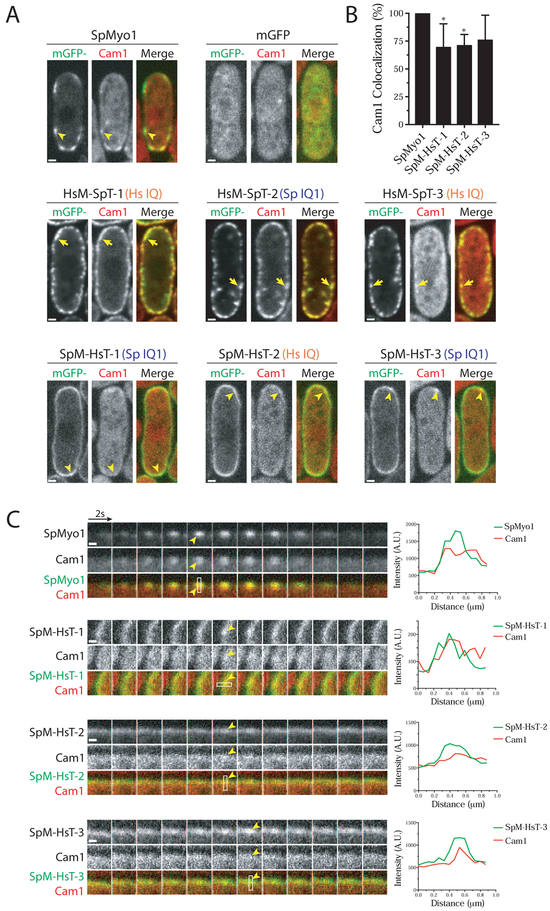
HsMyo1e IQ motif recruits Cam1 but not Cam2 in S. pombe
To further dissect the functionality of these human-yeast myosin-I chimeras, we tested whether they could bind and recruit myosin light chains, which regulate myosin activity. Vertebrate Myo1e has a single IQ motif bound by calmodulin that inhibits myosin ATPase activity in the presence of Ca2+ [24]. S. pombe Myo1 has two IQ motifs, IQ1 bound by calmodulin Cam1 and IQ2 bound by the calmodulin-like light chain Cam2 [8, 25]. The IQ motif of HsMyo1e shares greater amino acid sequence similarity with the first IQ motif of SpMyo1 (38%) than with the second IQ motif (23%) (Fig. S2). Moreover, human calmodulin is similar in amino acid sequence to S. pombe Cam1 (Fig. S5A). Thus, we hypothesized that yeast Cam1 may interact with the HsMyo1e IQ domain. To test if the HsMyo1e IQ motif can recruit Cam1, we transformed the mGFP-tagged chimeric constructs into a myo1Δ strain expressing Cam1 tagged with mCherry (Fig. 5, Fig. S6, and Table 1). As expected, the mGFP-SpMyo1 control and mCherry-Cam1 colocalized in 100% of endocytic patches (Fig. 5). Cam1 failed to localize to dynamic cortical patches in myo1Δ expressing mGFP (Fig. 5), indicating that Cam1 localization to actin patches relies solely on SpMyo1. The SpMyo1 motor – HsMyo1e tail chimeras, SpM-HsT-1, -2, and -3, previously observed to localize to endocytic actin patches, also showed colocalization with Cam1 in dynamic cortical patches (Fig. 5), regardless of whether they had the SpMyo1 IQ1 motif (SpM-HsT-1 and -3) or HsMyo1e IQ motif (SpM-HsT-2). On average, SpM-HsT-1, -2, and -3 colocalized with mCherry-Cam1 in ~73% of actin patches (Fig. 5B), which may be an underestimate due to reduced levels of these chimeras in patches and abundance of Cam1 in the cytoplasm. At high levels of expression, these three chimeras strongly outlined the cortex and recruited a significant amount of mCherry-Cam1 from the cytoplasm to the cortex (Fig. S5B). This recruitment was specific for the IQ motif-Cam1 interaction as cells overexpressing mGFP-HsMyo1e tail produced no such effect (Fig. S5B). The ability of the HsMyo1e IQ motif to recruit Cam1 was also apparent in HsM-SpT-1 and HsM-SpT-3, which contain the HsMyo1e IQ motif, and was similar to HsM-SpT-2, which contains the SpMyo1 IQ1 motif. At high level of expression, these chimeras recruited Cam1 from the cytoplasm to cortical aggregates, although HsM-SpT-3 did so less efficiently than HsM-SpT-1 and -2 (Fig. 5A). At lower levels of expression, HsM-SpT-2 and -3, which contain the SpMyo1 IQ2 motif but not HsM-SpT-1 lacking IQ2 motif recruited Cam1 to the cortical thread-like structures (Fig. S6A and S6B). Thus, the HsMyo1e IQ domain appears to be capable of binding Cam1 and recruiting it to actin patches and other cortical structures.
Figure 5: HsMyo1e IQ and SpMyo1 IQ1 motifs recruit calmodulin Cam1 in S. pombe.
(A-C) Colocalization analysis of mGFP-tagged human-yeast myosin-I chimeras (green) and mCherry-tagged calmodulin Cam1 (red) in myo1Δ cells. mGFP alone, mGFP-tagged SpMyo1 and indicated chimeras were expressed from plasmids under control of 3xPnmt1 promoter for 12-18 hours in the absence of thiamine.
(A) Single confocal sections through the middle of the cells. The orange Hs IQ and blue Sp IQ1 labels indicate the presence of the HsMyo1e IQ motif and the SpMyo1 IQ1 motif, respectively. Scale bars, 1 μm.
(B) Bar graph of the percent colocalization (mean ± SD) of Cam1 and SpMyo1 or SpMyo1 motor - HsMyo1e tail chimeras in cortical actin patches. N=19-31 patches in 5-7 cells. Asterisks indicate a significant difference from SpMyo1 as determined by a one-way ANOVA, * p<0.05.
(C) Montages of individual patches at 2-second intervals and line scans of fluorescence intensity in patches. White boxes mark areas used for line scans. Scale bar, 0.5 μm.
Yellow arrows indicate colocalization of Cam1 with overexpressed HsM-SpT-1, -2, and -3 in cortical aggregates. Yellow arrowheads indicate colocalization of Cam1 with SpMyo1 and SpM-HsT-1, -2, -3 in actin patches.
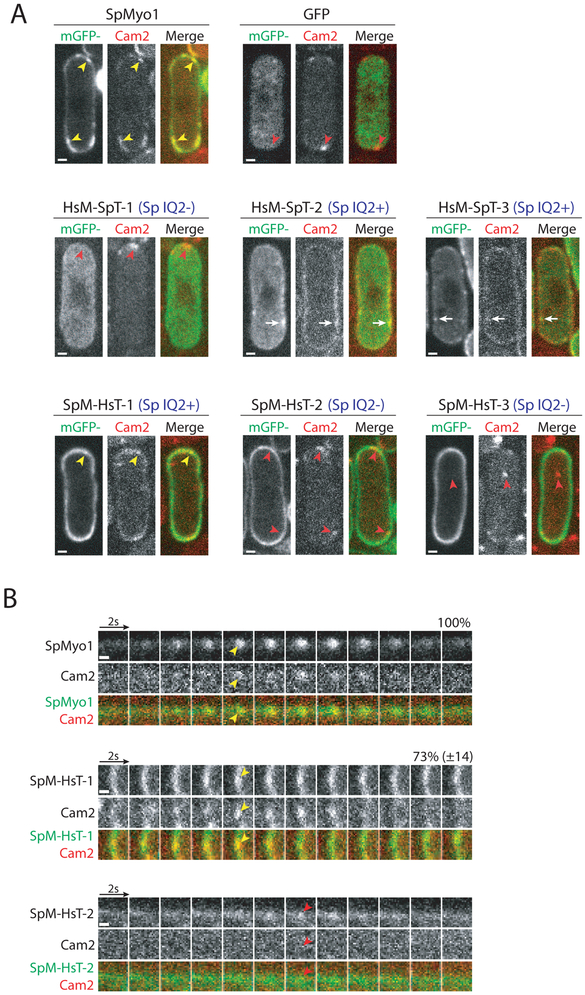
Cam2 binding to the lever arm of SpMyo1 has been found to maximize myosin motility in vitro [25]. Localization of Cam2 to actin patches depends solely on the SpMyo1 IQ2 motif [25]. In myo1Δ cells or when the binding of Cam2 to the IQ2 motif is disrupted, Cam2 does not localize to endocytic actin patches and instead moves to larger stationary puncta at the poles and a few mobile cytosolic puncta [25]. To determine whether the chimeric proteins were capable of recruiting Cam2, we transformed the mGFP-tagged myosin constructs into a myo1Δ strain expressing Cam2 tagged with mCherry (Fig. 6, Fig. S6C, and Table 1). Upon expression of mGFP-SpMyo1, Cam2 was recruited to dynamic actin patches and colocalized with SpMyo1 in 100% of endocytic patches (Fig. 6). As observed previously [25], Cam2 in myo1Δ cells expressing mGFP alone failed to localize to dynamic cortical patches and instead localized in bright puncta at the poles. Similar Cam2 puncta were observed at the poles of myo1Δ cells expressing HsM-SpT-1, SpM-HsT-2, and SpM-HsT-3 lacking the SpMyo1 IQ2 motif, and these chimeras did not recruit Cam2 and did not colocalize with Cam2 puncta (Fig. 6A). SpM-HsT-1, the only chimera to both localize to endocytic patches and contain the IQ2 domain, colocalized with Cam2 in 72% of dynamic cortical patches (Fig. 6A and 6B). HsM-SpT-2 and -3 that localized to cortical thread-like structures and contain the IQ2 motif recruited Cam2 to these eisosome-like structures, just as they did with Cam1 (Fig. S6C). Thus, Cam2 only bound chimeras that contained the SpMyo1 IQ2 motif.
Figure 6: SpMyo1 IQ2 motif recruits calmodulin-related light chain Cam2 in S. pombe.
(A, B) Colocalization analysis of mGFP-tagged human-yeast myosin-I chimeras (green) and mCherry-tagged calmodulin-like light chain Cam2 (red) in myo1Δ cells. mGFP alone, mGFP-tagged SpMyo1 and chimeras were expressed from plasmids under control of 3xPnmt1 promoter for 12-18 hours in the absence of thiamine.
(A) Single confocal sections through the middle of the cells. The blue Sp IQ+/− labels indicate the presence or the absence of the SpMyo1 IQ2 motif. Scale bars, 1 μm.
(B) Montages of individual patches at 2-second intervals. Scale bars, 0.5 μm. Numbers representing percent colocalization (mean ± SD) of Cam2 with SpMyo1 or SpM-HsT-1 in dynamic actin patches are shown above montages. N=41-63 patches in 3-6 cells.
Yellow arrowheads indicate colocalization of Cam2 with SpMyo1 and SpM-HsT-1 in actin patches. White arrows indicate colocalization of Cam2 with HsM-SpT-2 and HsM-SpT-3 in cortical aggregates. Red arrowheads indicate Cam2-only puncta that form in the absence of SpMyo1 IQ2 motif.
The SpMyo1 motor - HsMyo1e tail chimera requires the TH1 and TH2-SH3 domains for localization and function
Given the ability of the SpMyo1 motor - HsMyo1e tail chimeras to localize to endocytic actin patches and to rescue myo1Δ endocytosis defects, we sought to determine which HsMyo1e tail domains are minimally required for their localization and function. Using SpM-HsT-1, we created two tail domain deletion constructs: one lacking the membrane-binding TH1 domain and another lacking the TH2 and SH3 domains involved in protein-protein interactions (Fig. 7A). For comparison, we made similar deletions in SpMyo1, one removing the TH1 domain and another removing the C-terminal TH2, SH3, and CA domains. These mGFP-tagged constructs were transformed into the myo1Δ strain expressing Fim1-mCherry and tested for salt sensitivity, localization to actin patches, and the ability to rescue the myo1Δ patch internalization defect. Immunoblotting confirmed that all constructs were expressed at the expected molecular weights (Fig. 7B). Testing for growth in the presence of high salt revealed that both SpM-HsT-1 tail domain deletion mutants failed to rescue the growth of myo1Δ cells (Fig. 7C). In line with this finding, SpM-HsT-1ΔTH1 and SpM-HsT-1ΔTH2-SH3 failed to localize to actin patches marked with Fim1-mCherry and were mostly cytosolic (Fig. 7D and E). The fraction of internalizing patches in cells expressing these constructs remained at ~50%, similar to myo1Δ cells (Fig. 7F). Conversely, analogous deletions of tail domains from SpMyo1 resulted in drastically different behaviors. The SpMyo1ΔTH2-SH3-CA rescued the growth of myo1Δ cells in the presence of high salt, localized to dynamic actin patches, and partially rescued the patch internalization defect, increasing the fraction of internalizing patches to ~80% (Fig. 7C-F). In contrast, SpMyo1ΔTH1 failed to rescue salt-sensitive growth (Fig. 7C) and the patch internalization defect in myo1Δ cells (Fig. 7F), even though SpMyo1ΔTH1 clearly localized to actin patches marked with Fim1-mCherry (Fig. 7D, 7E, and S7A). Remarkably, unlike wild-type SpMyo1 that remains at the plasma membrane during actin patch internalization, SpMyo1ΔTH1 left the membrane and moved away together with Fim1-mCherry in a fraction of internalizing actin patches (Fig. 7E). This behavior was also observed in the presence of SpMyo1 in wild-type cells (Fig. S7B). Two color imaging confirmed that mGFP-tagged SpMyo1ΔTH1 internalized while mGFP- or mCherry-tagged full length SpMyo1 remained on the membrane (Fig. S7C and S7D). This finding emphasizes the importance of the TH1 domain for anchoring SpMyo1 at the membrane so that it can function in endocytic internalization. Intriguingly, while neither TH1 nor TH2-SH3-CA are required for SpMyo1 localization to endocytic patches, both the TH1 and TH2-SH3 domains in the HsMyo1e tail are needed for localization and function of the SpMyo1 motor - HsMyo1e tail chimera.
DISCUSSION
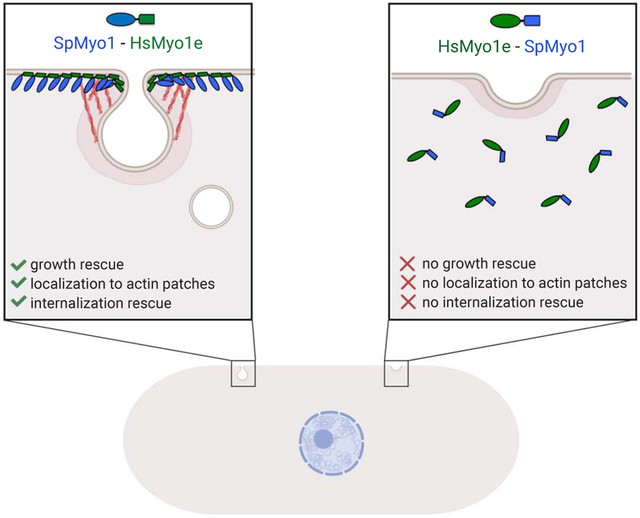
In this study, we set out to determine whether the conservation of overall functions and general protein organization between yeast and human myosin-Is reflects the conserved properties of individual protein domains or whether their protein domain properties have diverged while retaining a common function in endocytosis and overall protein organization. By creating human-yeast myosin-I chimeras, we tracked the inability of HsMyo1e to function in yeast to its motor domain. In contrast, HsMyo1e IQ motif and the tail can replace the tail of SpMyo1, although the specific tail domain requirements for localization and function differ between HsMyo1e and SpMyo1.
Evolutionary conservation of myosin-I motor domain with its actin track.
The inability of HsMyo1e motor domain to work properly with yeast actin rather than expression or folding issues appears to be the primary reason for the failure of HsMyo1e and HsMyo1e motor domain containing chimeras in S. pombe. Indeed, when expressed off of a plasmid, HsMyo1e and all chimeric constructs reached expression levels comparable or exceeding the endogenous level of SpMyo1. Moreover, all constructs were expressed as full-length proteins of expected molecular weights, unlike the unstable SpMyo1 motor domain mutants that are susceptible to degradation [21]. Furthermore, HsMyo1e and HsMyo1e motor chimeras did not colocalize with the myosin chaperone Rng3 (data not shown), which has been previously observed to bind destabilizing SpMyo1 motor domain mutants [21, 45]. Thus, while the possibility of folding defects cannot be completely ruled out, our results suggest that poor expression or misfolding were not the major factors contributing to the failure of the HsMyo1e motor in yeast.
The HsMyo1e motor domain may not function in S. pombe due to differences in regulation of myosin-I activity in yeast and mammals by phosphorylation at the TEDS site, association with calmodulin light chains, or by actin binding proteins, namely tropomyosins. While phosphorylation at the TEDS site is critical for SpMyo1 localization and function [33], HsMyo1e contains a glutamate residue at the TEDS site and therefore is expected to be constitutively active. The Glu-to-Ser mutation at the TEDS site failed to improve HsMyo1e functionality in yeast, suggesting that the lack of regulation at the TEDS site is not the contributing factor compromising HsMyo1e function in yeast, although yeast kinases may not be able to recognize the phosphorylation site on the HsMyo1e motor. Another mechanism regulating myosin-I activity is the binding of calmodulin or calmodulin-like myosin light chains to the IQ motifs in the myosin neck region, which stabilizes the myosin lever arm for efficient force production [46-48]. All of our constructs recruited Cam1 via either SpMyo1 first IQ motif or HsMyo1e IQ motif, suggesting that they have a fully functional lever arm. This is consistent with earlier work demonstrating that budding yeast and vertebrate calmodulins are functionally interchangeable [49]. In budding yeast, calmodulin has also been proposed to stabilize autoinhibitory intramolecular interactions within the myosin-I tail [50], thus calmodulin dissociation is required for myosin-I localization to patches. Since SpMyo1 and all functional chimeric constructs in our study recruited Cam1 to actin patches, Cam1 does not appear to play this role in S. pombe. Among actin binding proteins, tropomyosins are particularly important for regulating myosin activity and also serve as gatekeepers controlling the localization of myosins to actin filaments [51]. However, since both mammalian and fungal myosin-Is are excluded from tropomyosin-occupied actin filaments, and tropomyosin is not present at actin patches in yeast, it is unlikely that differences in tropomyosins would affect myosin-I activity [52-54].
Therefore, it is most likely that the inability of HsMyo1e motor domain to function in yeast is due to species-specific differences in the interaction of actin and myosin. S. pombe actin differs from mammalian actin kinetically, having a faster nucleotide exchange rate, phosphate dissociation rate, and F-actin nucleation rate by the Arp2/3 complex [55]. Most relevant to this study, fission yeast and budding yeast actin is significantly less efficient than mammalian skeletal muscle actin at activating conventional myosin-II [56-58]. This species specificity of myosins and actins has also been seen for budding yeast myosin-II and myosin-V in in vitro studies with purified actin and myosins that demonstrated that these motors exhibit greater actin-activated ATPase activity with yeast actin than with muscle actin [59]. The present study provides the first in vivo evidence of species specificity for a molecular motor in yeast. Furthermore, unlike myosin-II and myosin-V, molecular motors associated with the cytokinetic rings and actin cables, which are composed of formin-nucleated linear actin, myosin-I interacts with a distinct component of the yeast actin cytoskeleton, actin patches composed of branched actin filaments nucleated by the Arp2/3 complex. Thus, past in vitro and our current in vivo study both suggest that all myosins found in yeast appear to have evolved to work optimally with yeast-specific actin. The inability of HsMyo1e to function in yeast joins the list of other human actin binding proteins that fail to replace budding or fission yeast proteins either due to incompatibility with yeast actin as is the case with cofilin [60] or with yeast orthologs of normal binding partners as reported for profilin [61] and WASP [62]. Future in vitro experiments with purified HsMyo1e, SpMyo1, and yeast and mammalian actin will be needed to determine the precise mechanism of this species specificity.
Myosin-I tail domain contributions to myosin localization and function.
Our work indicates that both head and tail of myosin-I are required for localization at endocytic sites and that each domain makes a distinct contribution to myosin-I function. We have found that the HsMyo1e tail is partially functional in yeast when paired with the SpMyo1 motor domain. These chimeras were recruited to actin patches and partially rescued patch internalization defects in myo1Δ cells, but also exhibited enhanced cortical localization compared to SpMyo1. The exaggerated membrane localization of the HsMyo1e tail-containing chimeras could be the result of a stronger interaction of HsMyo1e tail with membrane lipids compared to the tail of SpMyo1 or different lipid specificity of HsMyo1e and SpMyo1 tails. Indeed, the tail of HsMyo1e has been shown to bind anionic phospholipids with high affinity and broad specificity [63], but the lipid affinity and specificity of SpMyo1 tail are currently unknown. Stronger cortical binding by HsMyo1e tail may also be due to the lack of myosin regulation, through intra- or intermolecular interactions, which were proposed to coordinate the timely arrival and departure of myosin-I at the endocytic sites in budding yeast [50]. In any case, the tail domain of HsMyo1e, when combined with the SpMyo1 motor domain, provides sufficient localization cues to target myosin chimeras to actin patches and support patch internalization.
To determine the roles of individual tail domains, we examined the effects of deletions of the membrane binding TH1 domain and the distal tail, which consists of TH2-SH3 domains in HsMyo1e and TH2-SH3-CA domains in SpMyo1. The TH1 domain is thought to act as a lipid anchor, positioning myosin so that it can efficiently promote membrane deformation [64, 65]. Supporting the role of the TH1 domain as a lipid anchor, deletion of the HsMyo1e TH1 domain from SpM-HsT-1 resulted in loss of localization to actin patches and failure to rescue endocytosis defects in myo1Δ cells. Removal of the TH1 domain from the full length SpMyo1 also resulted in an inability to rescue endocytosis defects of myo1Δ cells, even though SpMyo1ΔTH1, unlike SpM-HsT-1ΔTH1, was able to localize to endocytic patches. However, in contrast to SpMyo1 that remains at the base of endocytic invagination during patch internalization, in a portion of actin patches SpMyo1ΔTH1 internalized with F-actin, indicating a weaker attachment to the cortex. These results are similar to the observations made in S. cerevisiae [65, 66], where deletion of the TH1 domain decreased cortical retention of myosin-I, and HsMyo1e TH1 domain was able to replace the TH1 domain of yeast myosin-I [66]. Thus, in both budding and fission yeast, membrane anchoring by the TH1 domain is critical for myosin-I function in promoting endocytic internalization.
The distal tail domains of both SpMyo1 and HsMyo1e are known to be important for protein-protein interactions. In S. pombe, the SH3 domain of SpMyo1 binds Vrp1, Wsp1, Cdc15 and Bbc1 [13, 67, 68]. Yet despite this, deletion of the TH2-SH3-CA domains of SpMyo1 decreases but does not abolish SpMyo1 localization and function at the endocytic sites, in agreement with earlier studies [7]. Interestingly, deletion of the TH2-SH3 domains from the HsMyo1e tail of SpM-HsT-1 resulted in complete loss of localization to actin patches and the inability to rescue endocytosis in myo1Δ cells, suggesting that both the TH1 and TH2-SH3 domains of HsMyo1e are required for endocytic function. HsMyo1e has been shown to bind dynamin, synaptojanin, FAK, ZO-1, CARMIL, LSP1, and WASP via its SH3 domain, and SH3P2 via a combination of the TH2 and SH3 domains [9, 16, 69-73]. The human and frog Myo1e TH2 domain also weakly binds F-actin ([74] and our unpublished observations), unlike the SpMyo1 TH2 domain that lacks this activity [7]. In addition to these protein-protein interactions, the TH2 domain of HsMyo1e also contains a large number of basic residues, which may contribute to membrane localization [75]. The existence of multiple interactions in this portion of the tail is consistent with the results from our previous studies using mammalian cells where the deletion of the TH2 domain prevented HsMyo1e localization at cell-cell junctions, invadopodia and the phagocytic cup [69, 76]. Thus, in both yeast and mammalian cells, the distal tail of HsMyo1e appears to play a more important role in myosin-I localization and function than in its yeast counterpart.
Overall, our tail domain deletion analysis can be explained by a hypothetical model for the mechanism of myosin-I localization that is based on the modular organization of myosin-I molecule. Each myosin-I domain contributes an affinity for a distinct binding partner: the motor domain binds F-actin, the TH1 domain binds membrane, and the distal tail binds protein ligands. The motor domain is essential but by itself is not sufficient for localization to patches. However, as observed here and in budding yeast [65], combining motor domain with either of the two tail modules (TH1 or distal tail) is sufficient for localization of yeast myosin-I to patches. The deletion analysis of HsMyo1e tail in our chimeras suggests that the contributions of binding affinities from individual tail domains in human myosin are different. Specifically, we postulate that the HsMyo1e TH2 domain contributes to membrane binding. Based on this model for HsMyo1e tail properties, deletion of the distal tail would not only disrupt interactions with ligands but also weaken the interaction with the membrane. Furthermore, we speculate that, unlike the distal tail of yeast myosin-I, the distal tail of HsMyo1e does not provide additional binding interactions with SpMyo1 ligands that are strong enough to support patch localization when membrane binding of HsMyo1e tail is compromised by TH1 domain deletion.
The utility of yeast system to study myosin-I function.
The inability of HsMyo1e motor domain to function in yeast places certain limits on the utility of yeast system both for study of human myosin-I functions and the effects of disease-associated mutations. In particular, caution is advised when attempting to switch myosin motor domains with different mechanochemical properties as they may not function properly due to the incompatibility with yeast actin or the lack of normal binding partners or actin binding proteins, such as tropomyosins. On the other hand, the ability of human Myo1e tail to function in yeast provides a foundation for future work aiming at characterization of the effects of disease-associated mutations in a model system with a robust functional readout. Unlike yeast cells, where SpMyo1 is required for endocytosis, mammalian endocytosis does not always have an absolute requirement for actin or myosin-I, which makes testing of the effects of mutations in Myo1e in mammalian cells challenging, with no clear functional readout. By demonstrating that chimeras consisting of SpMyo1 motor and HsMyo1e tail functionally complement myo1Δ while retaining localization requirements similar to those observed for HsMyo1e in mammalian cells, this study provides information needed to use S. pombe as a simple model to effectively study disease-associated mutations residing in the HsMyo1e tail.
Supplementary Material
Human class 1 myosin Myo1e cannot replace fission yeast myosin-I Myo1
Chimeric analysis reveals domain requirements for myosin-I function in endocytosis
Human Myo1e motor domain cannot replace yeast Myo1 motor domain
Human Myo1e tail can replace yeast Myo1 tail to support endocytosis in yeast
The TH1 domain in the tail anchors Myo1 at the base of the endocytic invagination
ACKNOWLEDGMENTS
This work was supported by an AHA 18PRE34070066 fellowship to S.R.B., AHA SDG 11SDG5470024 to V.S. and the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH under Award R01DK083345 to M.K. We thank Robert Carroll and Cameron MacQuarrie for helpful discussions. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS AND NOMENCLATURE
- Myo1e
myosin 1e
- HsMyo1e
human myosin 1e
- SpMyo1
S. pombe myosin-I
- Fim1
fimbrin 1
- Cam1
calmodulin
- Cam2
calmodulin-related light chain 2
- TH1
tail homology 1
- TH2
tail homology 2
- SH3
Src homology 3
- CA
central-acidic
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].McIntosh BB, Ostap EM, Myosin-I molecular motors at a glance, Journal of cell science 129 (2016) 2689–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kollmar M, Muhlhausen S, Myosin repertoire expansion coincides with eukaryotic diversification in the Mesoproterozoic era, BMC Evol Biol 17 (2017) 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jung G, Hammer JA 3rd, Generation and characterization of Dictyostelium cells deficient in a myosin I heavy chain isoform, J Cell Biol 110 (1990) 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Novak KD, Peterson MD, Reedy MC, Titus MA, Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis, The Journal of cell biology 131 (1995) 1205–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Geli MI, Riezman H, Role of type I myosins in receptor-mediated endocytosis in yeast, Science 272 (1996) 533–535. [DOI] [PubMed] [Google Scholar]
- [6].Jung G, Wu X, Hammer JA 3rd, Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions, The Journal of cell biology 133 (1996) 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee WL, Bezanilla M, Pollard TD, Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp, The Journal of cell biology 151 (2000) 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Toya M, Motegi F, Nakano K, Mabuchi I, Yamamoto M, Identification and functional analysis of the gene for type I myosin in fission yeast, Genes to cells : devoted to molecular & cellular mechanisms 6 (2001) 187–199. [DOI] [PubMed] [Google Scholar]
- [9].Krendel M, Osterweil EK, Mooseker MS, Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis, FEBS Lett 581 (2007) 644–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Barger SR, Reilly NS, Shutova MS, Li Q, Maiuri P, Heddleston JM, Mooseker MS, Flavell RA, Svitkina T, Oakes PW, Krendel M, Gauthier NC, Membrane-cytoskeletal crosstalk mediated by myosin-I regulates adhesion turnover during phagocytosis, Nat Commun 10 (2019) 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lechler T, Shevchenko A, Li R, Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization, J Cell Biol 148 (2000) 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Evangelista M, Klebl BM, Tong AH, Webb BA, Leeuw T, Leberer E, Whiteway M, Thomas DY, Boone C, A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex, The Journal of cell biology 148 (2000) 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sirotkin V, Beltzner CC, Marchand JB, Pollard TD, Interactions of WASp, myosin-I, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast, The Journal of cell biology 170 (2005) 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun Y, Martin AC, Drubin DG, Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity, Dev Cell 11 (2006) 33–46. [DOI] [PubMed] [Google Scholar]
- [15].Taylor MJ, Perrais D, Merrifield CJ, A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis, PLoS biology 9 (2011) e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheng J, Grassart A, Drubin DG, Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis, Mol Biol Cell 23 (2012) 2891–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jonsdottir GA, Li R, Dynamics of yeast Myosin I: evidence for a possible role in scission of endocytic vesicles, Curr Biol 14 (2004) 1604–1609. [DOI] [PubMed] [Google Scholar]
- [18].Kaksonen M, Toret CP, Drubin DG, A modular design for the clathrin- and actin-mediated endocytosis machinery, Cell 123 (2005) 305–320. [DOI] [PubMed] [Google Scholar]
- [19].Idrissi FZ, Grotsch H, Fernandez-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI, Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane, J Cell Biol 180 (2008) 1219–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Galletta BJ, Chuang DY, Cooper JA, Distinct roles for Arp2/3 regulators in actin assembly and endocytosis, PLoS biology 6 (2008) e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bi J, Carroll RT, James ML, Ouderkirk JL, Krendel M, Sirotkin V, Effects of FSGS-associated mutations on the stability and function of myosin-1 in fission yeast, Disease models & mechanisms 8 (2015) 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mele C, Iatropoulos P, Donadelli R, Calabria A, Maranta R, Cassis P, Buelli S, Tomasoni S, Piras R, Krendel M, Bettoni S, Morigi M, Delledonne M, Pecoraro C, Abbate I, Capobianchi MR, Hildebrandt F, Otto E, Schaefer F, Macciardi F, Ozaltin F, Emre S, Ibsirlioglu T, Benigni A, Remuzzi G, Noris M, C. PodoNet, MYO1E mutations and childhood familial focal segmental glomerulosclerosis, The New England journal of medicine 365 (2011) 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Al-Hamed MH, Al-Sabban E, Al-Mojalli H, Al-Harbi N, Faqeih E, Al Shaya H, Alhasan K, Al-Hissi S, Rajab M, Edwards N, Al-Abbad A, Al-Hassoun I, Sayer JA, Meyer BF, A molecular genetic analysis of childhood nephrotic syndrome in a cohort of Saudi Arabian families, J Hum Genet 58 (2013) 480–489. [DOI] [PubMed] [Google Scholar]
- [24].Stoffler HE, Bahler M, The ATPase activity of Myr3, a rat myosin I, is allosterically inhibited by its own tail domain and by Ca2+ binding to its light chain calmodulin, J Biol Chem 273 (1998) 14605–14611. [DOI] [PubMed] [Google Scholar]
- [25].Sammons MR, James ML, Clayton JE, Sladewski TE, Sirotkin V, Lord M, A calmodulin-related light chain from fission yeast that functions with myosin-I and PI 4-kinase, Journal of cell science 124 (2011) 2466–2477. [DOI] [PubMed] [Google Scholar]
- [26].Bement WM, Mooseker MS, TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head, Cell Motil Cytoskeleton 31 (1995) 87–92. [DOI] [PubMed] [Google Scholar]
- [27].Barylko B, Binns DD, Albanesi JP, Regulation of the enzymatic and motor activities of myosin I, Biochim Biophys Acta 1496 (2000) 23–35. [DOI] [PubMed] [Google Scholar]
- [28].Wu C, Lee SF, Furmaniak-Kazmierczak E, Cote GP, Thomas DY, Leberer E, Activation of myosin-I by members of the Ste20p protein kinase family, J Biol Chem 271 (1996) 31787–31790. [DOI] [PubMed] [Google Scholar]
- [29].Wu C, Lytvyn V, Thomas DY, Leberer E, The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast, J Biol Chem 272 (1997) 30623–30626. [DOI] [PubMed] [Google Scholar]
- [30].Lechler T, Jonsdottir GA, Klee SK, Pellman D, Li R, A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast, J Cell Biol 155 (2001) 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ostap EM, Lin T, Rosenfeld SS, Tang N, Mechanism of regulation of Acanthamoeba myosin-IC by heavy-chain phosphorylation, Biochemistry 41 (2002) 12450–12456. [DOI] [PubMed] [Google Scholar]
- [32].Grosshans BL, Grotsch H, Mukhopadhyay D, Fernandez IM, Pfannstiel J, Idrissi FZ, Lechner J, Riezman H, Geli MI, TEDS site phosphorylation of the yeast myosins I is required for ligand-induced but not for constitutive endocytosis of the G protein-coupled receptor Ste2p, J Biol Chem 281 (2006) 11104–11114. [DOI] [PubMed] [Google Scholar]
- [33].Attanapola SL, Alexander CJ, Mulvihill DP, Ste20-kinase-dependent TEDS-site phosphorylation modulates the dynamic localisation and endocytic function of the fission yeast class I myosin, Myo1, Journal of cell science 122 (2009) 3856–3861. [DOI] [PubMed] [Google Scholar]
- [34].Keeney JB, Boeke JD, Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe, Genetics 136 (1994) 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR, Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe, Yeast 14 (1998) 943–951. [DOI] [PubMed] [Google Scholar]
- [36].Wu JQ, Kuhn JR, Kovar DR, Pollard TD, Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis, Developmental cell 5 (2003) 723–734. [DOI] [PubMed] [Google Scholar]
- [37].Siam R, Dolan WP, Forsburg SL, Choosing and using Schizosaccharomyces pombe plasmids, Methods 33 (2004) 189–198. [DOI] [PubMed] [Google Scholar]
- [38].Maundrell K, nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine, J Biol Chem 265 (1990) 10857–10864. [PubMed] [Google Scholar]
- [39].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nat Methods 9 (2012) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wu JQ, Pollard TD, Counting cytokinesis proteins globally and locally in fission yeast, Science 310 (2005) 310–314. [DOI] [PubMed] [Google Scholar]
- [41].Forsburg SL, Codon usage table for Schizosaccharomyces pombe, Yeast 10 (1994) 1045–1047. [DOI] [PubMed] [Google Scholar]
- [42].Bement WM, Wirth JA, Mooseker MS, Cloning and mRNA expression of human unconventional myosin-IC. A homologue of amoeboid myosins-I with a single IQ motif and an SH3 domain, J Mol Biol 243 (1994) 356–363. [DOI] [PubMed] [Google Scholar]
- [43].Kabeche R, Baldissard S, Hammond J, Howard L, Moseley JB, The filament-forming protein Pil1 assembles linear eisosomes in fission yeast, Mol Biol Cell 22 (2011) 4059–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD, Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast, Mol Biol Cell 21 (2010) 2894–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stark BC, James ML, Pollard LW, Sirotkin V, Lord M, UCS protein Rng3p is essential for myosin-II motor activity during cytokinesis in fission yeast, PloS one 8 (2013)e79593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Greenberg MJ, Ostap EM, Regulation and control of myosin-I by the motor and light chain-binding domains, Trends in cell biology 23 (2013) 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Houdusse A, Silver M, Cohen C, A model of Ca(2+)-free calmodulin binding to unconventional myosins reveals how calmodulin acts as a regulatory switch, Structure 4 (1996) 1475–1490. [DOI] [PubMed] [Google Scholar]
- [48].Jontes JD, Milligan RA, Brush border myosin-I structure and ADP-dependent conformational changes revealed by cryoelectron microscopy and image analysis, J Cell Biol 139 (1997) 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Davis TN, Thorner J, Vertebrate and yeast calmodulin, despite significant sequence divergence, are functionally interchangeable, Proc Natl Acad Sci U S A 86 (1989) 7909–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Grotsch H, Giblin JP, Idrissi FZ, Fernandez-Golbano IM, Collette JR, Newpher TM, Robles V, Lemmon SK, Geli MI, Calmodulin dissociation regulates Myo5 recruitment and function at endocytic sites, EMBO J 29 (2010) 2899–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP, Tropomyosin - master regulator of actin filament function in the cytoskeleton, Journal of cell science 128 (2015) 2965–2974. [DOI] [PubMed] [Google Scholar]
- [52].Collins K, Sellers JR, Matsudaira P, Calmodulin dissociation regulates brush border myosin I (110-kD-calmodulin) mechanochemical activity in vitro, J Cell Biol 110 (1990) 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tang N, Ostap EM, Motor domain-dependent localization of myo1b (myr-1), Current biology : CB 11 (2001) 1131–1135. [DOI] [PubMed] [Google Scholar]
- [54].Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M, Differential regulation of unconventional fission yeast myosins via the actin track, Current biology : CB 20 (2010) 1423–1431. [DOI] [PubMed] [Google Scholar]
- [55].Ti SC, Pollard TD, Purification of actin from fission yeast Schizosaccharomyces pombe and characterization of functional differences from muscle actin, J Biol Chem 286 (2011) 5784–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Takaine M, Mabuchi I, Properties of actin from the fission yeast Schizosaccharomyces pombe and interaction with fission yeast profilin, J Biol Chem 282 (2007) 21683–21694. [DOI] [PubMed] [Google Scholar]
- [57].Cook RK, Root D, Miller C, Reisler E, Rubenstein PA, Enhanced stimulation of myosin subfragment 1 ATPase activity by addition of negatively charged residues to the yeast actin NH2 terminus, J Biol Chem 268 (1993) 2410–2415. [PubMed] [Google Scholar]
- [58].McKane M, Wen KK, Meyer A, Rubenstein PA, Effect of the substitution of muscle actin-specific subdomain 1 and 2 residues in yeast actin on actin function, J Biol Chem 281 (2006) 29916–29928. [DOI] [PubMed] [Google Scholar]
- [59].Stark BC, Wen KK, Allingham JS, Rubenstein PA, Lord M, Functional adaptation between yeast actin and its cognate myosin motors, J Biol Chem 286 (2011) 30384–30392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kang H, Bradley MJ, Cao W, Zhou K, Grintsevich EE, Michelot A, Sindelar CV, Hochstrasser M, De La Cruz EM, Site-specific cation release drives actin filament severing by vertebrate cofilin, Proc Natl Acad Sci U S A 111 (2014) 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ezezika OC, Younger NS, Lu J, Kaiser DA, Corbin ZA, Nolen BJ, Kovar DR, Pollard TD, Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin: insights from crystal structures of fission yeast profilin, J Biol Chem 284 (2009) 2088–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rajmohan R, Meng L, Yu S, Thanabalu T, WASP suppresses the growth defect of Saccharomyces cerevisiae las17Delta strain in the presence of WIP, Biochemical and biophysical research communications 342 (2006) 529–536. [DOI] [PubMed] [Google Scholar]
- [63].Feeser EA, Ignacio CM, Krendel M, Ostap EM, Myo1e binds anionic phospholipids with high affinity, Biochemistry 49 (2010) 9353–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].McConnell RE, Tyska MJ, Leveraging the membrane - cytoskeleton interface with myosin-1, Trends in cell biology 20 (2010) 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pedersen RTA, Drubin DG, Type I myosins anchor actin assembly to the plasma membrane during clathrin-mediated endocytosis, J Cell Biol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lewellyn EB, Pedersen RT, Hong J, Lu R, Morrison HM, Drubin DG, An Engineered Minimal WASP-Myosin Fusion Protein Reveals Essential Functions for Endocytosis, Developmental cell 35 (2015) 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Carnahan RH, Gould KL, The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe, J Cell Biol 162 (2003) 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].MacQuarrie CD, Mangione MC, Carroll R, James M, Gould KL, Sirotkin V, Adaptor protein Bbc1 regulates localization of Wsp1 and Vrp1 during endocytic actin patch assembly, J Cell Sci 132 (2019) doi: 10.1242/jcs233502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bi J, Chase SE, Pellenz CD, Kurihara H, Fanning AS, Krendel M, Myosin 1e is a component of the glomerular slit diaphragm complex that regulates actin reorganization during cell-cell contact formation in podocytes, American journal of physiology. Renal physiology 305 (2013) F532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Heim JB, Squirewell EJ, Neu A, Zocher G, Sominidi-Damodaran S, Wyles SP, Nikolova E, Behrendt N, Saunte DM, Lock-Andersen J, Gaonkar KS, Yan H, Sarkaria JN, Krendel M, van Deursen J, Sprangers R, Stehle T, Bottcher RT, Lee JH, Ordog T, Meves A, Myosin-1E interacts with FAK proline-rich region 1 to induce fibronectin-type matrix, Proc Natl Acad Sci U S A 114 (2017) 3933–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Maxeiner S, Shi N, Schalla C, Aydin G, Hoss M, Vogel S, Zenke M, Sechi AS, Crucial role for the LSP1-myosin1e bimolecular complex in the regulation of Fcgamma receptor-driven phagocytosis, Mol Biol Cell 26 (2015) 1652–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tanimura S, Hashizume J, Arichika N, Watanabe K, Ohyama K, Takeda K, Kohno M, ERK signaling promotes cell motility by inducing the localization of myosin 1E to lamellipodial tips, J Cell Biol 214 (2016) 475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Liang Y, Niederstrasser H, Edwards M, Jackson CE, Cooper JA, Distinct roles for CARMIL isoforms in cell migration, Mol Biol Cell 20 (2009) 5290–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Yu HY, Bement WM, Multiple myosins are required to coordinate actin assembly with coat compression during compensatory endocytosis, Mol Biol Cell 18 (2007) 4096–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhang Y, Cao F, Zhou Y, Feng Z, Sit B, Krendel M, Yu CH, Tail domains of myosin-1e regulate phosphatidylinositol signaling and F-actin polymerization at the ventral layer of podosomes, Mol Biol Cell (2019) mbcE18060398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ouderkirk JL, Krendel M, Myosin 1e is a component of the invadosome core that contributes to regulation of invadosome dynamics, Experimental cell research 322 (2014) 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.