This study shows how elongation factor G helps to maintain the mRNA reading frame during protein synthesis on the ribosome.
Abstract
During translation, the ribosome moves along the mRNA one codon at a time with the help of elongation factor G (EF-G). Spontaneous changes in the translational reading frame are extremely rare, yet how the precise triplet-wise step is maintained is not clear. Here, we show that the ribosome is prone to spontaneous frameshifting on mRNA slippery sequences, whereas EF-G restricts frameshifting. EF-G helps to maintain the mRNA reading frame by guiding the A-site transfer RNA during translocation due to specific interactions with the tip of EF-G domain 4. Furthermore, EF-G accelerates ribosome rearrangements that restore the ribosome’s control over the codon-anticodon interaction at the end of the movement. Our data explain how the mRNA reading frame is maintained during translation.
INTRODUCTION
The ribosome makes proteins by decoding a sequence of codons in the mRNA. Once the open reading frame is selected during translational initiation, the ribosome decodes base triplets with the help of aminoacyl–transfer RNAs (tRNAs). The fidelity of protein synthesis is a fundamental parameter for the fitness of living cells. Sophisticated selection mechanisms safeguard the high fidelity of aminoacyl-tRNA selection (1) and ensure a low error frequency of amino acid incorporation (2). Each time an amino acid is incorporated into the growing peptide chain, the ribosome moves along the mRNA by exactly one codon. Maintaining the correct reading frame is even more crucial than preventing codon misreading, because frameshifting would result in synthesis of completely aberrant proteins. The error frequency of spontaneous frameshifting is <10−5 per codon (3). It is conceivable that for most sequences, the reading frame is sustained because of base pairing between the mRNA codon by its respective tRNA anticodon. However, when the ribosome translates a so-called slippery mRNA sequence, i.e., a sequence where codons in the 0 and −1 or +1 reading frames can be read by the same tRNA, a mechanism must exist to maintain the ribosome in the correct 0 reading frame. Spontaneous frameshifting increases when decoding of slippery sequence codons is slowed down, e.g., because of the shortage of the respective aminoacyl-tRNA (4–6). In special cases, frameshifting is stimulated by the mRNA sequences upstream and downstream of the slippery site, leading to the so-called programmed ribosome frameshifting (7–11). While the mechanism of programmed ribosome frameshifting is increasingly well understood, it remains unclear how the reading frame is maintained during canonical tRNA-mRNA translocation. No testable model has been proposed so far as to which component of the translation machinery controls the maintenance of the reading frame, what the roles of the ribosome and elongation factor G (EF-G) are, and how the dynamics of the ribosome and tRNAs during translocation ensures the codon-wise ribosome progression. Here, we unravel the essential features of the mechanism of reading frame maintenance by exploring spontaneous frameshifting in real time using a rapid kinetics approach.
RESULTS
As an example of a slippery sequence, we have chosen a heptanucleotide A AAA AAG (Fig. 1A), as it causes natural frameshifting in Escherichia coli dnaX gene (12). In E. coli, AAA and AAG codons are read by a single tRNALys. Frameshifting on the dnaX mRNA is stimulated by a downstream stem-loop structure and an upstream Shine-Dalgarno (SD)–like sequence (13). The stimulatory elements stall the ribosome at the slippery sequence, thereby providing the time for ribosome slippage into the −1 frame during translocation on the AAA AAG codons (4, 14–16). In the absence of the downstream mRNA stem loop, frameshifting on the A AAA AAG sequence is greatly reduced (4, 15, 17), suggesting that the translational machinery is intrinsically capable of maintaining the reading frame at the slippery sequences. As an alternative slippery sequence, we have chosen G GGA AAG, with tRNAGly reading GGA/GGG and tRNALys reading AAA/AAG, which can direct programmed ribosome frameshifting in vivo almost as well as A AAA AAG (17, 18). In contrast, sequences where only one codon is slippery, such as A AAG, are far less prone to frameshifting (17). To study how translational machinery ensures the reading frame maintenance, we carried out in vitro translation experiments in a fully reconstituted translation system and quantified the fraction of 0- and −1-frame products using the three different mRNAs (Fig. 1A). The mRNA AUG GCA AAA AAG is designed to mimic the dnaX gene, including the SD sequence upstream of the slippery site (fig. S1A) (4, 14, 16). For the AUG GGA AAG mRNA, the spacing to the SD sequence is different but still within an optimal range for frameshifting (13). The sequence AUG GAA AAG is unfavorable for slippage, because the tRNA reading the 0-frame GAA codon (tRNAGlu) would have to pair with the noncognate GGA codon in the −1 frame, resulting in a second-position mismatch; Lys-tRNA can pair with both 0-frame AAG and −1-frame AAA. None of the three mRNAs contains an mRNA secondary structure that might stimulate programmed frameshifting. We formed 70S initiation complexes (ICs) with fMet-tRNAfMet using purified translation components from E. coli and started translation by the addition of EF-G–GTP (guanosine triphosphate) and saturating amounts EF-Tu–GTP complexes with the aminoacyl-tRNAs required to synthesize the short peptides, as indicated. Translation was stopped after 5 min, and the appearance of the 0-frame and −1-frame translation products was analyzed by high-performance liquid chromatography (HPLC) (fig. S1B).
Fig. 1. Role of EF-G in mRNA reading frame maintenance.
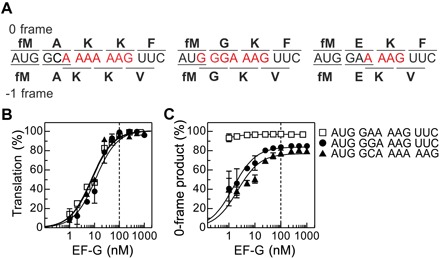
(A) Coding sequences of the mRNAs used in this work. The potential slippery sequence is colored red. Amino acids incorporated in 0 and −1 frame are indicated above and below the respective coding sequences. (B) Dependence of translation on EF-G concentration. Products were analyzed after 5 min of translation. Translation efficiency was normalized by setting the end level to 100% for better comparison between mRNAs. Lines are visual guides. The dashed line indicates the concentration at which ribosomes and EF-G are equimolar. Error bars represent SD of three experiments (n = 3). (C) Fraction of the 0-frame peptide depending on the EF-G concentration. Error bars represent SD of three experiments (n = 3).
To identify which component of the translation machinery—the ribosome with tRNAs or EF-G—ensures reading frame maintenance, we first quantified the translation efficiency at increasing concentrations of EF-G (Fig. 1B). With the sequence where only one tRNA can slip, mainly the 0-frame product is synthesized, regardless of the EF-G concentration (Fig. 1C). In contrast, for both slippery mRNAs, the ratio between 0- and −1-frame products changes with the EF-G concentration. At low catalytic EF-G concentrations, the −1-frame product is prevalent (>70%), whereas at saturating EF-G concentrations, mostly the 0-frame product is synthesized. The ratio between the 0- and −1-frame products does not change with time (fig. S2), indicating that the commitment to the alternative reading frame is rapid. These results suggest that the ribosome is inherently prone to slippage, whereas binding of EF-G restricts frameshifting during translocation.
EF-G is a five-domain guanosine triphosphatase (GTPase) that increases the translocation rate by about five orders of magnitude (19). Domain 4 of EF-G is critical for the catalysis of translocation; deletions or mutations at the tip of domain 4 slow down translocation (20–22) but have little effect on the GTPase activity of EF-G (fig. S2, B to D). Two conserved residues in domain 4 of EF-G, Q507 and H583 in E. coli, contact the tRNA during translocation (Fig. 2A and fig. S2A) (23–25). Spontaneous frameshifting is greatly increased when Q507 or H583 is mutated and the −1-frame product becomes prevalent even at saturating concentrations of mutant EF-G, particularly with EF-G(Q507D) (Fig. 2B). As with wild-type (wt) EF-G, the ratio of the 0- and −1-frame products does not change with time with mutant EF-G(Q507D) (fig. S2E). Amino acid substitutions at EF-G position Q507 that introduce a different charge or size of the amino acid side chain disrupt frame maintenance to a different extent, with D and E substitutions causing the maximum frameshifting and F, N, S, A, and H showing intermediate effects. Replacements of H583 with A or K have also intermediate effects (Fig. 2C). The tendency of a particular substitution to increase frameshifting is similar on the slippery sequence G GGA AAG and the less slippery sequence G GAA AAG, although the extent of frameshifting is different. These results suggest that the residues at the tip of domain 4 of EF-G have an active role in maintaining the reading frame during translation, also on the sequences that are normally not prone to slippage. A detailed structural comparison of ribosomes captured in an intermediate state of translocation in the presence and absence of EF-G suggests that domain 4 restricts the forward movement of the A-site tRNA into the −1 frame (26); our mutational data support this notion.
Fig. 2. Frameshifting effect of mutations at the tip of domain 4 of EF-G.
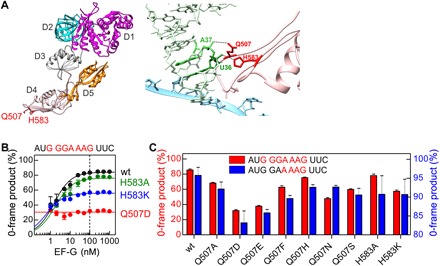
(A) Left: Crystal structure of EF-G [Protein Data Bank (PDB): 3J5X]. Domains (D) 1 to 5 and the positions of residues Q507 and H583 (red) in domain 4 are indicated. Right: Interaction of EF-G residues Q507 and H583 with the tRNA in the ap/P state of the ribosome (PDB: 4V5F). (B) Fraction of the 0-frame peptide depending on the concentration of wt EF-G and EF-G mutants. Lines are visual guides, and the dashed line indicates the concentration at which ribosomes and EF-G are equimolar. Error bars represent SD of three experiments (n = 3). (C) Effect of different substitutions of Q507 and H583 in EF-G on the fraction of 0-frame product. Error bars represent SD of three experiments (n = 3).
We next compared the rates of translocation and the frameshifting efficiency in the presence of EF-G domain 4 mutants. We estimated the rate of single-round translocation by a rapid puromycin assay (fig. S3, A and B) (27). The rates vary from 20 to 0.7 s−1, depending on the amino acid substitution. The amount of the 0-frame product increases with the translocation rate (Fig. 3 and fig. S3). This correlation not only suggests that the ability of EF-G to maintain the mRNA reading frame and to facilitate translocation are coupled through the action of domain 4 of EF-G but also allows us to estimate the intrinsic rates of frameshifting. In the simplest model, the ribosome can undergo reversible transitions between the 0 and −1 state with rate constants kf and kb in forward and backward directions, respectively (Fig. 3, scheme). These transitions are terminated by the completion of translocation with the rate constant kt, thereby committing the ribosome to translation in one of the frames. Because the frameshifting efficiency provides the ratio kf/kb and kt is known, we can estimate the rates of the frameshifting transitions on the three slippery sequences from fitting the analytic solution of the respective kinetic equations (Supplementary Materials) to the kinetic data (Fig. 3). The transition to the −1 frame, in the range of about 3 to 10 s−1, is faster than the return to the 0 frame, about 1 to 2 s−1. Because the ribosome starts translation in the 0 frame, the rate of translocation is crucial: At high translocation rates, the ribosome is committed to the 0 frame before it can slip to the −1 frame, whereas long pausing leads to equilibration that favors the −1 frame, depending on the mRNA sequence. This simple model does not take into account the potential changes in translocation rates at each given mRNA sequence but provides a lower estimate for the intrinsic rate of tRNA movements during frameshifting. To test whether the puromycin reaction rates used here are representative for translocation rates on slippery sequences, we analyzed the translation kinetics in real time by quench flow using the AUG GGA AAG mRNA (fig. S3D). The MGK peptide is converted to the MGKF peptide with the rate of 7 or 1 s−1 at saturating concentrations of wt EF-G and EF-G(Q507D), respectively. These values provide lower limit for translocation rates of the MGK-tRNA and are in the same range (20 and 0.8 s−1, respectively) as the rates of the rapid puromycin reaction, further supporting the analysis. This analysis of the frameshifting-prone EF-G mutants shows that reading frame maintenance requires rapid translocation and provides the first estimation for the rates of spontaneous frameshifting on different mRNAs.
Fig. 3. Correlation between the fraction of the correct reading frame and the rate of translation for three different slippery sequences.
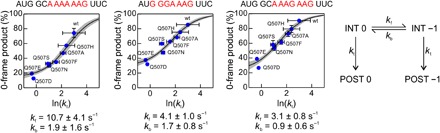
The mRNA coding sequences are indicated above the graphs. The rate of translation, kt, was measured by a rapid puromycin assay (fig. S3). Black lines represent fits to a frameshifting kinetics model shown to the right. INT is a translocation intermediate, and POST is posttranslocation state of the ribosome. Rate constants kf and kb are the rate constants of ribosome transitions to the −1 and 0 frames, respectively (for details, see text and the Supplementary Materials). Error bars represent SD of three experiments (n = 3).
We asked next why translocation becomes slow when the crucial residues at the tip of EF-G domain 4 are mutated. We used our toolbox of fluorescence and fluorescence resonance energy transfer (FRET) reporters established for wt EF-G (27, 28) to dissect the kinetics of translocation with frameshifting-prone EF-G mutants Q507D, H583K, and H583A. For wt EF-G, we used the model of translocation that combines the kinetic data with structural assignment of intermediates (Fig. 4A) (24–29). Rate constants of each step were estimated by global fitting of combined datasets for eight independent observables at five different EF-G concentrations (figs. S4 to S9 and tables S1 and S2).
Fig. 4. Choreography of translocation and reading frame maintenance by the residues at the EF-G domain 4 tip using FRET pairs and fluorescence observables extensively validated in previous work [see (28) and references therein; see also figs. S4 to S9 and tables S1 and S2].
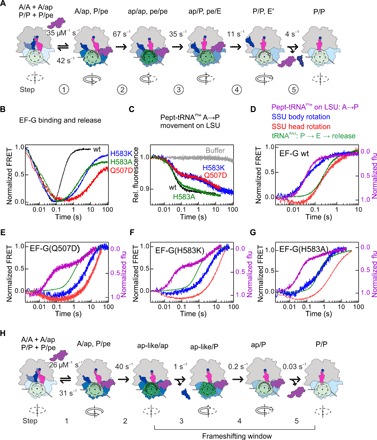
(A) Unifying model of translocation based on kinetics and structural data (25–28, 30). The SSU head domain is colored in different shades of green depending on the degree of rotation, with the maximum rotation (20°) indicated in dark green. Rotation of the SSU body domain is indicated by blue shades, with dark blue indicating the maximum rotation. Backward rotation of SSU head and body is gradual, as indicated by different color shades. Arrows below the scheme indicate the direction of SSU body rotation. The rate constants were obtained by global fitting of experimental data for wt EF-G carried out in this paper. (B) Time courses of EF-G binding and dissociation measured with the FRET pair attached to the ribosomal protein L12 and EF-G. (C) Formation of the ap/ap and ap/P states measured by fluorescence change of the A-site BOF-MetPhe-tRNAPhe. (D) Comparison of time courses of P site–tRNA dissociation (green), tRNAfMet movement (light blue), SSU body rotation (blue), and SSU head rotation (red) upon addition of wt EF-G. (E) As in (D), but with EF-G(Q507D). (F) As in (D), but with EF-G(H583K). (G) As in (D), but with EF-G(H853A). (H) Model of translocation for EF-G(Q507A) and EF-G(H583K) mutants. Color code as in (A).
EF-G binding and dissociation were visualized using FRET reporters attached to ribosomal protein L12 and EF-G. The fluorescence of the Alexa Fluor 488 reporter attached to L12 is quenched upon binding of EF-G labeled with QSY9 (Fig. 4B) (28). Mutations in EF-G domain 4 do not affect appreciably the rate of EF-G binding to the ribosome, whereas the dissociation is substantially delayed when Q507 or H583 is mutated. Binding of wt EF-G to the ribosome facilitates the displacement of the A-site peptidyl-tRNA into a so-called chimeric state on the small ribosomal subunit (SSU), with the tRNA anticodon positioned roughly between the A site of the 30S head and the P site of the body (25). On the large ribosomal subunit (LSU), the CCA end of the tRNA moves toward the P site (ap/ap state), which is then rapidly converted to the chimeric/hybrid ap/P state, which is puromycin reactive (Fig. 4A, fig. S3, and tables S1 and S2) (24, 25, 27, 29). This movement was monitored using a Bodipy FL (BOF) label attached to the N terminus of the nascent peptide (27). With wt EF-G, the formation of ap/ap and ap/P states is kinetically indistinguishable and appears as a single step accounting for >90% of the fluorescence change (Fig. 4C). With the EF-G(Q507D) and EF-G(H583K) mutant, the time courses are biphasic. The faster step, which reflects the transition to the ap/ap state, is not affected by the domain 4 mutations, whereas the second step is much slower, compared to wt EF-G (Fig. 4C). With EF-G(H583A), the effect on the second step is small. The results of the rapid puromycin reaction, which reflects the transition from the ap/ap to ap/P state, show the same trend (fig. S3). Thus, the initial steps of EF-G binding up to the formation of the ap/ap intermediate are not affected by the mutations, but the transition toward the ap/P state and the following reactions depend on the identity of the amino acid residues in domain 4 of EF-G.
A key element of translocation is the movement of the SSU body and head domains, which we monitored in real time by FRET using pairs of FRET reporters attached to ribosomal proteins S6 and L9 (SSU body rotation) or S13 and L33 (SSU head rotation, also called SSU swiveling) (fig. S4) (28). Movements of the P-site tRNAfMet were visualized monitoring FRET between tRNAfMet and L33. Within the first 0.01 s after wt EF-G addition, the SSU body domain starts to rotate backward relative to the LSU toward the nonrotated state, whereas the SSU head domain remains in a highly rotated state until almost 0.1 s (Fig. 4D). In the resulting intermediate, the SSU head is rotated by 20°, but the SSU body is rotated by only 2° relative to the LSU, and the tRNAs adopt ap/P and pe/E states (24, 25, 30). The release of Pi from EF-G initiates the backward movement of the SSU head domain and facilitates the completion of tRNA translocation to the P/P and E/E states, followed by the dissociation of the E-site tRNA and EF-G–GDP (guanosine diphosphate) from the ribosome (28); the latter is monitored by FRET between tRNAfMet(Flu) and the Atto 540Q quencher on ribosomal protein L33 (Fig. 4, A and D). In addition, we used the fluorescence changes of an mRNA labeled with fluorescein (Flu) at its +14 position to track mRNA translocation, as well as fluorescence changes of tRNAfMet and S13 as a source of additional kinetic information (Supplementary Materials). Global fitting yields rates of the elemental reactions for wt EF-G (Fig. 4A) that are similar within statistical error to previously reported values (28). Thus, with wt EF-G, the back rotation of the SSU body precedes the back rotation of the SSU head, whereas the dissociation of the E-site tRNA is concomitant with the backward movement of the SSU head and release of EF-G.
Mutations of EF-G residues Q507 and H583 affect rates of essentially every step following the formation of the ap/ap intermediate (Fig. 4, E to G). Binding of mutant EF-G to the ribosome induces the ap-like state of the SSU, as inferred from the rapid decrease in the S13-L33 FRET efficiency; however, the backward rotation of the SSU body is substantially delayed (Fig. 4E). The P-site tRNA likely moves into the pe/E-like state, as this intermediate can form spontaneously in the absence of EF-G (26, 31). Unexpectedly, the tRNA movement is not coupled to the SSU back rotation. With EF-G(Q507D) and EF-G(H583K), the P-site tRNA moves away from L33 even before the SSU body rotates backward; the rate of P-site tRNA movement is ~1 s−1 (Fig. 4, E and F, and table S1). We note that the tRNA-L33 quenching is expected to be >90% in the pre- and posttranslocation state (32, 33). Hence, the observed large fluorescence change indicates that the P-site tRNA leaves its posttranslocation state in the E site, i.e., moves further into the E′ site or is released from the ribosome. At about the same time, the A-site peptidyl-tRNA becomes puromycin reactive, suggesting that the 3′ end of the tRNA has now moved to the P site (table S1). EF-G starts to dissociate at this point (fig. S9). The subsequent back rotation of the SSU body and head domains proceed in the usual order but are extremely slow, about 0.2 and 0.05 s−1, respectively, compared to 67 and 35 s−1 with wt EF-G. With EF-G(H583A), the late steps of translocation are also slower than with wt EF-G, but the movements of the SSU body domain and the A-site tRNA appear concomitantly, followed by the SSU head back rotation (Fig. 4G). Thus, residues at the tip of EF-G domain 4 not only accelerate translocation by initiating rapid movements of the SSU domains and the tRNAs but also synchronize the back rotation of the SSU head and body domains with the forward movement of the P-site tRNA out of its posttranslocation state in the E site (Fig. 4H and table S2).
To slip, the ribosome must loosen the grip on the mRNA and the tRNAs. The structure of the ap/P state without EF-G shows that the interactions of the ribosome with the codon-anticodon complexes are disrupted, and mRNA and tRNA move apart and are misaligned by one-half nucleotide position, which creates the possibility for alternative base pairing (26). The structure shows a high degree of SSU head swiveling but a low degree of SSU body rotation, which, in our kinetic data, corresponds to the area between the blue and red curves in Fig. 4 (D to G). The comparison between the timing of the P site–tRNA movement and of the backward body rotation implies that the disruption of the P-site codon-anticodon complex, which would be required to release the P/E-site tRNA, occurs even earlier than the formation of the ap/P state. Moreover, the P-site tRNA release is independent of the EF-G domain 4 tip residues and of the backward movements of the SSU domains. With the EF-G(Q507D) and EF-G(H583K) mutants, the P-site tRNA release coincides with the transition from the ap/ap to the ap/P state; therefore, the tRNA must be destabilized in an earlier state, i.e., in the ap/ap state. This is consistent with the crystal structure of the ribosome in complex with a +1 frameshifting tRNA mutant, which suggests that the P-site tRNA can induce the formation of a chimeric state even in the absence of EF-G (31). When the subsequent steps are delayed and the specific interactions between the tip of domain 4 and the tRNA cannot form, the ribosome allows the tRNAs to sample the codons in either 0 or –1 frame and to establish interactions, depending on which one is more favorable thermodynamically. In the absence of EF-G, the P- and A-site tRNAs fluctuate toward the neighboring states but prevent each other from moving (33). Mutations of 23S ribosomal RNA residues at the E site affect frameshifting (34), which underscores the importance of the tRNA transiting from the P to E site in maintaining the reading frame.
DISCUSSION
In summary, our results provide a detailed model of how EF-G helps to avoid frameshifting. Key elements of reading frame maintenance are the specific interactions of EF-G domain 4 tip residues with the tRNA and the rapid reverse rotation of the SSU head facilitated by EF-G, which ensures rapid completion of translocation before the tRNAs can move into the −1 frame. In the intermediate state of translocation, the interactions between the tRNA and mRNA are destabilized. If key EF-G residues are mutated, the P-site tRNA leaves the P site before translocation of the A-site tRNA can be completed. In this state, the A-site tRNA is constrained neither by specific interactions with EF-G nor by the P-site tRNA that could limit its mobility and can rapidly sample the available reading frames to establish codon-anticodon interactions in the thermodynamically favorable frame. This reequilibration is normally slow compared to the completion of translocation; however, the delay in the SSU head closing creates a time window for spontaneous frameshifting. Thus, EF-G maintains the mRNA reading frame by guiding the A-site tRNA and accelerating rearrangements of the ribosome that restore the ribosome control over the codon-anticodon interaction. These results underscore the importance of rapid ribosome progression along the mRNA for reading frame maintenance and predict that pausing ribosomes are prone to frameshifting. Similarly, pausing at the downstream mRNA structure stimulates programmed ribosome frameshifting (4, 14, 16). Thus, the speed of the ribosome movement along the mRNA is a crucial parameter that determines the collinearity of the mRNA and the protein during translation.
MATERIALS AND METHODS
Component of the translation machinery
Ribosomes from E. coli, f[3H]Met-tRNAfMet, [14C]Phe-tRNAPhe, [14C]Lys-tRNALys, Glu-tRNAGlu, Gly-tRNAGly, Phe-tRNAPhe, Val-tRNAVal, BODIPY-[3H]Met-tRNAfMet, initiation factors, and EF-Tu were prepared as described (27, 35, 36). The gene coding for EF-G was cloned into pET24 with a C-terminal histidine tag, and mutations were introduced by site-directed mutagenesis. All mutations were verified by DNA sequencing. Wt EF-G and EF-G mutants were overexpressed in BL21 (DE3) cells and purified using Protino Ni-IDA (Macherey-Nagel) in gravity flow columns as described (37). All mRNAs were synthesized by IBA Lifesciences (Göttingen). Start codons are in bold, and slippery site are underlined. The following mRNA sequences were used to study reading frame maintenance: 5′-GUUAACAGGUAUACAUACUAUGGAAAAGUUCAUUACCUAA-3′, 5′-GUUAACAGGUAUACAUACUAUGGGAAAGUUCAUUACCUAA-3′, 5′-GGGAAUUGUGAGCGGAUAACAAUUCCCCUCUAGAGCAGUUGCAGCGCGUGCAGGGAGCAACCAUGGCAAAAAAGUUCUAG-3′, and 5′-GGGAAUUGUGAGCGGAUAACAAUUCCCCUCUAGAGCAGUUGCAGCGCGUGCAGGGAGCAACCAUGGCAAAGAAGUUCUAG-3′.
For the translocation experiments, the following mRNA was used:
5′-GUUAACAGGUAUACAUACUAUGUUUGUUAUUAC-3′ (mMF).
Ribosome complexes
To prepare ICs, 70S ribosomes were incubated with a 2-fold excess of mRNA, 1.7-fold excess of initiation factors, 3-fold excess of f[3H]Met-tRNAfMet, and 1 mM GTP in TAKM7 buffer [50 mM tris-HCl (pH 7.5) at 37°C, 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2] for 30 min at 37°C (38). Ternary complex (TC) was prepared by incubating EF-Tu (threefold excess over tRNA) with 1 mM GTP, 3 mM phosphoenolpyruvate, and 0.5% pyruvate kinase in TAKM7 buffer for 15 min at 37°C and subsequent addition of aminoacyl-tRNAs cognate to the mRNA coding sequence. Pretranslocation complex (PRE) was formed by mixing IC and TC (two- to fivefold excess over IC). Purification of IC and PRE was performed by centrifugation through a 1.1 M sucrose cushion in TAKM21 buffer [50 mM tris-HCl (pH 7.5) at 37°C, 70 mM NH4Cl, 30 mM KCl, and 21 mM MgCl2]. Pellets were dissolved in TAKM21 buffer, and the concentration of purified complex was determined by nitrocellulose filtration (21). Posttranslocation complex (POST) was prepared by mixing purified PRE and EF-G (4 μM) and incubated for a minute at 37°C before it was used. The magnesium concentration of purified complexes was adjusted to working concentration before use. All experiments were carried out in TAKM7, unless stated otherwise.
Biochemical assays
Reading frame maintenance assay
Purified IC and TC with corresponding tRNAs (Glu-tRNAGlu, Gly-tRNAGly, Ala-tRNAAla, Lys-tRNALys, [14C]Lys-tRNALys, Phe-tRNAPhe, [14C]Val-tRNAVal, or Val-tRNAVal) were prepared as described above. Translation was carried out by incubating purified IC (0.1 μM), TC (two to fivefold excess over IC), and EF-G (1 nM to 2 μM) at 37°C for 5 min. Reaction was then quenched with KOH (0.5 M), and peptides were released by hydrolysis at 37°C for 30 min. Samples were neutralized by adding 1/10 volume of glacial acetic acid and analyzed by reversed-phase HPLC (LiChrospher 100 RP-8 HPLC column, Merck) using a 0 to 65% acetonitrile gradient in 0.1% trifluoroacetic acid or 0.1% heptafluorobutyric acid. The 0- and −1-frame products were quantified by scintillation counting according to f[3H]Met, [14C]Lys, or [14C]Val radioactivity labels.
GTP hydrolysis by EF-G
Multiple turnover GTPase activity of EF-G was tested by incubating vacant ribosome (0.5 μM) and EF-G (1 μM) together with 1 mM GTP with a trace amount of [γ32P]GTP at room temperature. Reactions were quenched by adding the same volume of 40% formic acid. Samples were analyzed via thin-layer chromatography using 0.5 M potassium phosphate (pH 3.5) as mobile phase. The radioactivity was detected by the phosphor screen and analyzed by phosphorimager (21).
Time-resolved puromycin assay
The rate of tRNA translocation of EF-G was examined by time-resolved puromycin reaction. Purified PRE and POST containing the nonslippery mMF mRNA were prepared as described above. The experiment was performed by rapidly mixing PRE or POST (0.2 μM), as indicated, with EF-G (4 μM) and puromycin (10 mM) in a quench-flow apparatus. The reaction was quenched after a specific period of time by the addition of KOH (0.5 μM), and the products were released from the ribosomes at 37°C at 30 min. Samples were then neutralized with glacial acetic acid and analyzed by reversed-phase HPLC (Chromolith RP-8e column, Merck) using a 0 to 65% acetonitrile gradient in 0.1% trifluoroacetic acid. The amounts of reacted and unreacted peptides were quantified by [3H]Met and [14C]Phe radioactivity counting. The apparent rates of the puromycin reaction of PRE (kPRE) and POST (kPOST) were determined by one-exponential fitting using GraphPad Prism. The rate of tRNA translocation (kt) was calculated from the rate of PRE (kPRE) and the rate of POST (kPOST). The time required for PRE includes the puromycin reaction and tRNA translocation, while for POST, only the puromycin reaction is required. Deconvolution of the apparent rates from PRE and POST gives the rate of tRNA translocation (1/kt = 1/kPRE − 1/kPOST) (27).
Global fitting
Eight fluorescence reporters and FRET pairs were combined for the global fitting to a linear five-step kinetic model (28). The binding and dissociation of EF-G, P-site tRNA translocation to the E site and dissociation, SSU head swiveling, and SSU body rotation relative to LSU were monitored by FRET between L12–EF-G, L33-tRNAfMet, S13-L33, and S6-L9, respectively. Movement of the peptidyl-tRNA on the LSU was monitored using a fluorescence change of BODIPY-MetPhe-tRNAPhe. Additional data used for global fitting were fluorescence changes of S13, P-site tRNAfMet, and the mRNA labeled with fluorescein as the 3′ end at position +14 (fig. S4). Labeled proteins, tRNAs, and mRNA were prepared as described (27, 28, 39–41).
Stopped-flow experiments
To reconstitute 70S with labeled 30S and 50S, 30S subunit was heat-activated in TAKM20 at 37°C for 30 min and then incubated with MF mRNA, IF1, IF2, IF3, and f[3H]Met-tRNAfMet for an additional minute at room temperature. The 70S initiation complex was formed by incubating activated 30S initiation complex with a 1.5-fold molar excess of large 50S subunit at 37°C for 30 min in TAKM7. The preparation and purification of the PRE were carried out as described above. The purified PREs (0.05 μM) were mixed with different EF-G (0.5 to 4 μM) mutants in the stopped-flow apparatus at 37°C. Fluorescence of the respective reporter (or the donor in a FRET pair) was excited at 465/470 nm and detected after passing a KV500/590 cutoff filter. The changes in fluorescence/FRET were recorded with time, and six to eight technical replicates were averaged (n = 6 to 8).
Kinetic model
We used the five-step kinetic model that has been extensively validated in our previous work, including consideration of alternative models (28). To identify the number of kinetic steps for the reaction with EF-G mutants, we first performed exponential fitting of each individual time course. We identified five kinetic domains, starting from two that were indicative of very rapid steps that were similar for the wt and mutant EF-G (Fig. 4, B and C) and three very slow kinetically distinguishable steps of around 1, 0.2, and 0.01 s−1. We then analyzed the data by numerical integration with KinTeK Explorer (42). For each EF-G type, 32 independently measured time courses were combined into a dataset, which was used for global fitting. For six observables (L12–EF-G, L33–tRNAfMet, S13-L33, S6-L9, S13, and tRNAfMet), time courses were obtained at different EF-G concentrations from 0.5 to 4 μM; other observables were recorded at 4 μM EF-G. The fitted variables included rate constants as shown in Fig. 4, as well as intrinsic fluorescence intensities (IFIs) that reflect the fluorescence of each translocation intermediate for a given observable. For the wt EF-G dataset, all parameters were kept free; fitting resulted into a set of fully defined rate constants and IFIs with the SEM of the fit between 3 and 32% of the respective value. Because EF-G binding was generally similar for wt EF-G and the mutants, we reduced the complexity by coupling the k1 and k−1 values for the mutants to the ratio determined for the wt EF-G; all other parameters including the actual values of k1 and k−1 were free to change. This yielded very good fits for the mutant EF-G dataset with the SEM of the fit <6%. The fit space could not be calculated because of the large size of the dataset.
Estimation of frameshifting rates from the fraction of 0-frame peptides
To estimate the rates of slippage between 0 and –1 frame, we used a four-state kinetic model (Fig. 3). The vector contains the probability of being in each of the four states (INT 0, INT –1, POST 0, and POST −1) as a function of time t. The master equation of the kinetic model is then given by , where M is the matrix formed from the individual transition rates
The master equation is solved by with initial conditions . In the case of frameshifting, all ribosomes started in the 0 frame; therefore, . This initial condition results in the final probabilities . The probability of ending up in the 0-frame as a function of the translocation rate was then least-squares fitted against the measured ratios of 0-frame product formation including (fig. S3C) and excluding (Fig. 3) values corresponding to H583 mutations. The errors of the fit parameters kb and kf were determined via bootstrapping. To that aim, for each mutation, the fraction of 0-frame product and translocation rates were drawn from normal distributions using mean and SD of the measured values. Next, the function was fitted against these drawn values, yielding a kb and kf value. These two steps were repeated 105 times, and the mean values and SDs of kb and kf were calculated. Figure 3 and fig. S3C show the functions with the respective mean kb and kf values as black lines. The gray area indicates the resulting SD of the 0-frame product fractions.
Supplementary Material
Acknowledgments
We thank W. Wintermeyer for critical reading of the manuscript and A. Pfeifer, O. Geintzer, S. Kappler, C. Kothe, T. Niese, T. Wiles, F. Hummel, T. Hübner, V. Herold, and M. Zimmermann for expert technical assistance. Funding: The work was funded by the Max Planck Society and the grants of the Deutsche Forschungsgemeinschaft (SFB860 and Leibniz Prize to M.V.R.). Author contributions: B.Z.P. contributed to project design, conception, data collection, data analysis, manuscript writing, and figure creation. L.V.B. contributed to conception, data analysis, manuscript writing, and figure creation. R.B. provided materials and contributed to project design, conception, data analysis, figure creation, and manuscript writing. F.P., H.G., and M.V.R. contributed to project design, data analysis, conception, and manuscript writing. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaax8030/DC1
Table S1. Rate constants (s−1) of the key translocation steps with frameshifting EF-G mutants.
Table S2. Rate constants of elemental translocation rates for a five-step model A + EF-G = B→C→D→E→F.
Fig. S1. Analysis of 0- and −1-frame products of translation.
Fig. S2. Characterization of EF-G mutants.
Fig. S3. Activity of EF-G mutants in translocation and 0-frame maintenance.
Fig. S4. Schematics of the observables used to measure translocation kinetics.
Fig. S5. Examples of global fitting results with wt EF-G (4 μM).
Fig. S6. Examples of global fitting results with EF-G(Q507D) (4 μM).
Fig. S7. Examples of global fitting results with EF-G(H583K) (4 μM).
Fig. S8. Examples of global fitting results with EF-G(H583A) (4 μM).
Fig. S9. Comparison of the IFIs for wt EF-G (black) and EF-G mutants Q507D (red), H583K (blue), and H583A (green).
REFERENCES AND NOTES
- 1.Rodnina M. V., Fischer N., Maracci C., Stark H., Ribosome dynamics during decoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garofalo R., Wohlgemuth I., Pearson M., Lenz C., Urlaub H., Rodnina M. V., Broad range of missense error frequencies in cellular proteins. Nucleic Acids Res. 47, 2932–2945 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurland C. G., Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 26, 29–50 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Caliskan N., Wohlgemuth I., Korniy N., Pearson M., Peske F., Rodnina M. V., Conditional switch between frameshifting regimes upon translation of dnaX mRNA. Mol. Cell 66, 558–567.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Yelverton E., Lindsley D., Yamauchi P., Gallant J. A., The function of a ribosomal frameshifting signal from human immunodeficiency virus-1 in Escherichia coli. Mol. Microbiol. 11, 303–313 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korniy N., Goyal A., Hoffmann M., Samatova E., Peske F., Pöhlmann S., Rodnina M. V., Modulation of HIV-1 Gag/Gag-pol frameshifting by tRNA abundance. Nucleic Acids Res. 47, 5210–5222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atkins J. F., Loughran G., Bhatt P. R., Firth A. E., Baranov P. V., Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.E. P. Plant, in Viral Genomes—Molecular Structure, Diversity, Gene Expression Mechanisms and Host-Virus Interactions, M. Garcia, Ed. (InTech, 2012). [Google Scholar]
- 9.I. Brierley, R. Gilbert, S. Pennell, Pseudoknot-dependent programmed–1 ribosomal frameshifting: Structures, mechanisms, and models, in Recoding: Expansion of Decoding Rules Enriches Gene Expression, J. F. Atkins, R. F. Gesteland, Eds. (Springer, 2010), pp. 149–174. [Google Scholar]
- 10.Advani V. M., Dinman J. D., Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression. Bioessays 38, 21–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caliskan N., Peske F., Rodnina M. V., Changed in translation: mRNA recoding by −1 programmed ribosomal frameshifting. Trends Biochem. Sci. 40, 265–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blinkowa A. L., Walker J. R., Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase-III γ subunit from within the τ subunit reading frame. Nucleic Acids Res. 18, 1725–1729 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen B., Wills N., Gesteland R., Atkins J. F., rRNA-mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 176, 6842–6851 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J., Petrov A., Johansson M., Tsai A., O’Leary S. E., Puglisi J. D., Dynamic pathways of −1 translational frameshifting. Nature 512, 328–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsuchihashi Z., Brown P. O., Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 6, 511–519 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Kim H.-K., Liu F., Fei J., Bustamante C., Gonzalez R. L. Jr., Tinoco I. Jr., A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A. 111, 5538–5543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma V., Prère M.-F., Canal I., Firth A. E., Atkins J. F., Baranov P. V., Fayet O., Analysis of tetra- and hepta-nucleotides motifs promoting −1 ribosomal frameshifting in Escherichia coli. Nucleic Acids Res. 42, 7210–7225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin M. E., Hendrix R. W., Casjens S. R., A programmed translational frameshift is required for the synthesis of a bacteriophage λ tail assembly protein. J. Mol. Biol. 234, 124–139 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Katunin V. I., Savelsbergh A., Rodnina M. V., Wintermeyer W., Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 41, 12806–12812 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Rodnina M. V., Savelsbergh A., Katunin V. I., Wintermeyer W., Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385, 37–41 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Savelsbergh A., Matassova N. B., Rodnina M. V., Wintermeyer W., Role of domains 4 and 5 in elongation factor G functions on the ribosome. J. Mol. Biol. 300, 951–961 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Liu G., Song G., Zhang D., Zhang D., Li Z., Lyu Z., Dong J., Achenbach J., Gong W., Zhao X. S., Nierhaus K. H., Qin Y., EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon–anticodon duplex. Nat. Struct. Mol. Biol. 21, 817–824 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Gao Y.-G., Selmer M., Dunham C. M., Weixlbaumer A., Kelley A. C., Ramakrishnan V., The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J., Lancaster L., Donohue J. P., Noller H. F., Crystal structures of EF-G–ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramrath D. J., Lancaster L., Sprink T., Mielke T., Loerke J., Noller H. F., Spahn C. M., Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc. Natl. Acad. Sci. U.S.A. 110, 20964–20969 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J., Lancaster L., Donohue J. P., Noller H. F., Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state. Proc. Natl. Acad. Sci. U.S.A. 116, 7813–7818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holtkamp W., Cunha C. E., Peske F., Konevega A. L., Wintermeyer W., Rodnina M. V., GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 33, 1073–1085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belardinelli R., Sharma H., Caliskan N., Cunha C. E., Peske F., Wintermeyer W., Rodnina M. V., Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol. 23, 342–348 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Zhou J., Lancaster L., Donohue J. P., Noller H. F., How the ribosome hands the A-site tRNA to the P site during EF-G–catalyzed translocation. Science 345, 1188–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratje A. H., Loerke J., Mikolajka A., Brünner M., Hildebrand P. W., Starosta A. L., Dönhöfer A., Connell S. R., Fucini P., Mielke T., Whitford P. C., Onuchic J. N., Yu Y., Sanbonmatsu K. Y., Hartmann R. K., Penczek P. A., Wilson D. N., Spahn C. M. T., Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong S., Sunita S., Maehigashi T., Hoffer E. D., Dunkle J. A., Dunham C. M., Mechanism of tRNA-mediated +1 ribosomal frameshifting. Proc. Natl. Acad. Sci. U.S.A. 115, 11226–11231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brilot A. F., Korostelev A. A., Ermolenko D. N., Grigorieff N., Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. U.S.A. 110, 20994–20999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bock L. V., Blau C., Schröder G. F., Davydov I. I., Fischer N., Stark H., Rodnina M. V., Vaiana A. C., Grubmüller H., Energy barriers and driving forces in tRNA translocation through the ribosome. Nat. Struct. Mol. Biol. 20, 1390–1396 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Sergiev P. V., Lesnyak D. V., Kiparisov S. V., Burakovsky D. E., Leonov A. A., Bogdanov A. A., Brimacombe R., Dontsova O. A., Function of the ribosomal E-site: A mutagenesis study. Nucleic Acids Res. 33, 6048–6056 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodnina M. V., Wintermeyer W., GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl. Acad. Sci. U.S.A. 92, 1945–1949 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milon P., Konevega A. L., Peske F., Fabbretti A., Gualerzi C. O., Rodnina M. V., Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 430, 1–30 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Cunha C. E., Belardinelli R., Peske F., Holtkamp W., Wintermeyer W., Rodnina M. V., Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation 1, e24315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.B. Z. Peng, “The role of EF-G in translational reading frame maintenance on the ribosome,” thesis, Georg-August-Universität Göttingen, Göttingen, Germany (2018). [Google Scholar]
- 39.Sharma H., Adio S., Senyushkina T., Belardinelli R., Peske F., Rodnina M. V., Kinetics of spontaneous and EF-G-accelerated rotation of ribosomal subunits. Cell Rep. 16, 2187–2196 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Caliskan N., Katunin V. I., Belardinelli R., Peske F., Rodnina M. V., Programmed –1 frameshifting by kinetic partitioning during impeded translocation. Cell 157, 1619–1631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peske F., Savelsbergh A., Katunin V. I., Rodnina M. V., Wintermeyer W., Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA–mRNA translocation. J. Mol. Biol. 343, 1183–1194 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Johnson K. A., Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods Enzymol. 467, 601–626 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaax8030/DC1
Table S1. Rate constants (s−1) of the key translocation steps with frameshifting EF-G mutants.
Table S2. Rate constants of elemental translocation rates for a five-step model A + EF-G = B→C→D→E→F.
Fig. S1. Analysis of 0- and −1-frame products of translation.
Fig. S2. Characterization of EF-G mutants.
Fig. S3. Activity of EF-G mutants in translocation and 0-frame maintenance.
Fig. S4. Schematics of the observables used to measure translocation kinetics.
Fig. S5. Examples of global fitting results with wt EF-G (4 μM).
Fig. S6. Examples of global fitting results with EF-G(Q507D) (4 μM).
Fig. S7. Examples of global fitting results with EF-G(H583K) (4 μM).
Fig. S8. Examples of global fitting results with EF-G(H583A) (4 μM).
Fig. S9. Comparison of the IFIs for wt EF-G (black) and EF-G mutants Q507D (red), H583K (blue), and H583A (green).


