Pathways that converge for photosystem biogenesis occur in a specific region of the chloroplast called the T-zone in Chlamydomonas.
Abstract
Intracellular processes can be localized for efficiency or regulation. For example, localized mRNA translation by chloroplastic ribosomes occurs in the biogenesis of PSII, one of the two photosystems of the photosynthetic electron transport chain in the chloroplasts of plants and algae. The biogenesis of PSI and PSII requires the synthesis and assembly of their constituent polypeptide subunits, pigments, and cofactors. Although these biosynthetic pathways are well characterized, less is known about when and where they occur in developing chloroplasts. Here, we used fluorescence microscopy in the unicellular alga Chlamydomonas reinhardtii to reveal spatiotemporal organization in photosystem biogenesis. We focused on translation by chloroplastic ribosomes and chlorophyll biosynthesis in two developmental contexts of active photosystem biogenesis: (1) growth of the mature chloroplast and (2) greening of a nonphotosynthetic chloroplast. The results reveal that a translation zone is the primary location of the biogenesis of PSI and PSII. This discretely localized region within the chloroplast contrasts with the distributions of photosystems throughout this organelle and, therefore, is likely a hub where anabolic pathways converge for photosystem biogenesis.
INTRODUCTION
Intracellular processes can be localized for a variety of benefits (e.g., enhanced efficiency, regulation, and the prevention of deleterious side reactions). Within the cells of plants and algae, photosynthesis and other essential processes are performed in chloroplasts. These processes include photosynthetic electron transport (PET) and ATP synthesis, which occur in the membranes of vesicles called thylakoids within this organelle. The PET system includes three major complexes: PSII, the cytochrome b6f complex (Cytb6f), and PSI. The photosystems act as solar-powered molecular battery chargers by using light energy to drive electrons from low-energy to high-energy redox half-reactions, analogous to the charging of the half-cells of a battery. The high energy half-reactions are then used to drive downstream redox reactions in photosynthesis (Nelson and Ben-Shem, 2004).
The biogenesis of the photosystems requires the concerted synthesis and assembly of their component protein subunits, pigments, and cofactors. The spatial organization of these biogenic processes is complex due to the evolution of the plastids from a cyanobacterial endosymbiont (Elias and Archibald, 2009). Many subunits are encoded by the organellar genome and synthesized by bacterial-type chloroplastic ribosomes (Jarvis and López-Juez, 2013). Other subunits are synthesized in the cytoplasm and imported into the chloroplast. The PSII subunit PsbA (also known as D1) is synthesized into thylakoid membranes exposed to the chloroplast stroma for the repair of photodamaged PSII complexes (Mattoo and Edelman, 1987; van Wijk et al., 1996; Chotewutmontri and Barkan, 2018).
In plants, subunit synthesis for de novo photosystem biogenesis occurs in association with membranes that are presumed to be stroma-exposed thylakoid membranes (Zoschke and Barkan, 2015; Legen and Schmitz-Linneweber, 2017). However, in the unicellular green alga Chlamydomonas (Chlamydomonas reinhardtii), a translation zone (T-zone) is the primary location of the synthesis of the PSII subunits that are encoded by the plastid genome (Uniacke and Zerges, 2007, 2009; Schottkowski et al., 2012). The T-zone is more discretely localized than the broad distribution of PSII in the membranes of thylakoids throughout the chloroplast. It is unknown whether the T-zone is also the location of the biogenesis of other PET complexes.
A similar narrative describes our understanding of the biosynthesis of chlorophyll, the major photopigment in PSI, PSII, and their light-harvesting complexes (LHCs). The enzymes in the chlorophyll branch of the tetrapyrrole biosynthetic pathway are well characterized and many are membrane associated (Czarnecki and Grimm, 2012). However, precisely where these enzymes act within chloroplasts is controversial (Joyard et al., 2009). In the cyanobacterium Synechocystis PCC 6803, chlorophyll synthase is physically associated with the machinery that drives cotranslational insertion of nascent photosystem polypeptides into the thylakoid membrane (Chidgey et al., 2014). At least one enzyme in the chlorophyll biosynthetic pathway is associated with polysomes in the chloroplasts of barley (Hordeum vulgare; Kannangara et al., 1997). It remains to be determined whether chlorophyll biosynthesis and photosystem biogenesis are colocalized in chloroplasts.
Chloroplast biogenesis occurs in different developmental contexts. It is required for the growth of mature chloroplasts prior to their division by binary fission, for example, in expanding green tissues of leaves and stems (Sun and Zerges, 2015). Another mode of biogenesis occurs as proplastids differentiate to chloroplasts in young green tissues (Solymosi and Schoefs, 2010). Finally, cells of etiolated angiosperm seedlings contain etioplasts that, upon illumination, differentiate to chloroplasts in a process called greening (Hoober, 2007).
The proper timing of protein synthesis is important in many cell biological processes. For example, chloroplast development in both maize (Zea mays) and Chlamydomonas involves complex dynamics of the expression of the chloroplast genome and nuclear genes encoding trans-acting factors that regulate posttranscriptional steps of chloroplast gene expression (Idoine et al., 2014; Zones et al., 2015; Chotewutmontri and Barkan, 2016; Strenkert et al., 2019). Leaf development in maize involves complex patterned changes in the translation of chloroplast mRNAs (Chotewutmontri and Barkan, 2016). It is currently of interest to determine how the levels of the photosynthesis complexes and chloroplastic ribosomes change with respect to these dynamics in the transcriptome and translatome.
Chlamydomonas is an amenable model organism for studying chloroplast biogenesis. Spatial patterns are readily identifiable due to the stereotypic anatomy of the chloroplast and its several prominent cytological markers (Uniacke et al., 2011). Moreover, liquid cultures yield ample quantities of cells in defined stages of chloroplast biogenesis. For example, the growth of a photosynthetic chloroplast can be studied in wild-type cultures entrained to a 12/12-h light-dark (diel) cycle (Sun and Zerges, 2015; Zones et al., 2015). Under these conditions, cells synchronously grow in size in the light phase and undergo mitosis in the dark phase (Supplemental Figure 1). Alternatively, the greening process can be modeled with the yellow-in-the-dark-1 (y1) mutant (Ohad et al., 1967a, 1967b; Malnoë et al., 1988). y1 lacks the light-independent protochlorophyllide oxidoreductase (POR), an enzyme in the chlorophyll biosynthetic pathway (Cahoon and Timko, 2000). y1 cells cultured in the dark cannot synthesize chlorophyll and, therefore, lack PSI and PSII. Illumination restores chlorophyll synthesis by activating a light-dependent POR, thereby initiating the biogenesis of PSI and PSII in a process that resembles greening in angiosperms.
Here, we characterized the spatiotemporal organization of photosystem biogenesis in two developmental contexts that are associated with chloroplast biogenesis: (1) rapid chloroplast growth in the light phase of the diel cycle and (2) rapid photosystem biogenesis in greening y1 cells. We used indirect immunofluorescence (IF) staining and fluorescence in situ hybridization (FISH) to reveal distributions of endogenous proteins and mRNAs, respectively. To increase the objectivity of our fluorescence microscopy image analyses, we developed a macro and protocol to determine average intrachloroplastic distributions of IF and FISH signals in complete data sets. Therefore, our conclusions are based on both qualitative visual analyses and quantitative analyses by this macro. In addition, we performed live-cell imaging of chlorophyll fluorescence as a marker for the location of its biosynthesis. The results extend the role of the T-zone to the biogenesis of PSI and the biosynthesis of chlorophyll.
RESULTS
Photosynthesis Complexes and Chloroplastic Ribosomes Accumulate during Chloroplast Growth in the Light Phase of the Diel Cycle
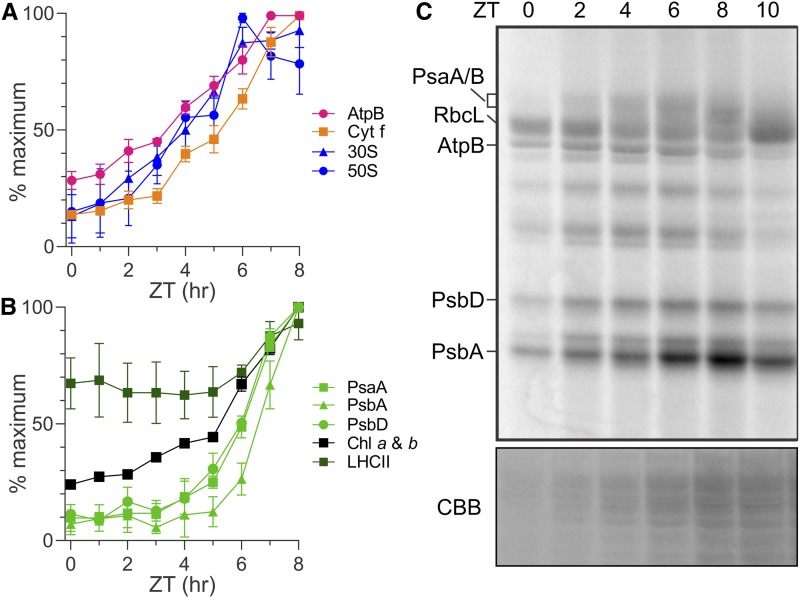
In order to characterize spatiotemporal patterns of PET complex biogenesis in growing chloroplasts, we sought culture conditions under which Chlamydomonas (1) undergoes high rates of chloroplast biogenesis for cellular growth, (2) yields samples at defined stages of cellular growth, and (3) is not undergoing mitosis, which likely affects biogenesis. Therefore, we entrained cultures of a wild-type strain to the diel cycle with a 12/12-h light/dark regime (Supplemental Figure 1). Time in the 24-h diel cycle is measured in Zeitgeber time (ZT) hours. The time points ZT0 and ZT12 are the transitions from dark to light and light to dark, respectively. To reveal temporal changes in the steady state levels of the PET complexes and the chloroplastic ATP synthase during the light phase, we monitored changes in the levels of representative subunit proteins with immunoblot analysis (Figures 1A and 1B; Supplemental Figures 2A–2C). In the PET system, each subunit represents the level of its complex because unassembled subunits are rapidly degraded (de Vitry et al., 1989). We also monitored the levels of marker proteins of the 30S and 50S subunits of the chloroplastic ribosome, S-21 and L7/L12, respectively, and LHCII (Randolph-Anderson et al., 1989).
Figure 1.
Temporal Changes of Protein Synthesis and Accumulation in the Growing Chloroplast during the Light Phase of the Diel Cycle.
(A) and (B) Results of immunoblot analyses of total protein samples from ZT time points in the ZT0-ZT8 interval of the light phase reveal the relative levels of marker proteins (parentheses) of the following complexes: ATP synthase (AtpB), Cytb6f (Cytf), and the 30S and 50S subunits of the chloroplastic ribosome (S-21 and L7/L12, respectively; [A]) and PSI (PsaA), PSII (PsbA and PsbD), the LHCII proteins, and chlorophyll (B). Results are from three biological replicates using independent cultures. Error bars indicate 1 se. Identical protein samples were used for all analyses in each panel. Immunoblots from one replicate are shown in Supplemental Figure 2A. Supplemental Figures 2B and 2C show the same plots as in (A) and (B), respectively, but with all three data points for each ZT time point.
(C) Protein synthesis rates in the chloroplast are revealed by 10-min in vivo pulse 35S labeling of PsaA/B, AtpB, PsbD, and PsbA. Also indicated is the 35S-labeled large subunit of Rubisco (RbcL). Preferentially elevated rates of PsbA synthesis for the PSII damage-repair cycle were detected in the ZT6-ZT8 interval. The doublet bands of PsbD represent phosphorylated and nonphosphorylated forms (Herrin et al., 1992). Cycloheximide inhibited cytoplasmic ribosomes to reveal products of chloroplastic ribosomes. A portion of the gel with proteins was stained with Coomassie Brilliant Blue (CBB).
As cells grew in size during the light phase, the levels of the photosynthesis complexes and ribosomal subunits increased (Figures 1A and 1B; Supplemental Figures 2A–2C). The levels of the chloroplastic ATP synthase and the 30S and 50S subunits of the chloroplastic ribosome increased in the ZT0-ZT6 interval (Figure 1A). The synthesis of AtpB (also known as CF1-β), a subunit of the F1 particle of ATP synthase, increased in the ZT0-ZT2 interval, based on results of a radioisotope pulse-labeling assay (Figure 1C; Supplemental Figure 2D). Cytb6f level increased slowly in the ZT0-ZT3 interval and more rapidly thereafter (Figure 1A). (Our pulse-labeling experiments did not detect the synthesis of ribosomal proteins or Cytb6f subunits.) Finally, the levels of PSI and PSII increased in the ZT5-ZT8 interval, although the synthesis of PSI and PSII subunits increased earlier (i.e., at the beginning of the light phase; Figures 1B and 1C; Supplemental Figures 2A, 2C, and 2D). LHCII and chlorophyll also increased in the ZT5-ZT8 interval, but to lesser degrees than did PSI and PSII, consistent with the large proportion of total chlorophyll that is in this complex (Figure 1B; Supplemental Figures 2A and 2C; Minagawa and Takahashi, 2004). These temporal patterns of accumulation are consistent with the programmed differentiation in chloroplast development revealed by previous results of transcriptome and ribosome profiling (Idoine et al., 2014; Zones et al., 2015; Chotewutmontri and Barkan, 2016).
Finally, the synthesis of the PSII subunit PsbA preferentially increased in the ZT6-ZT8 interval in the light phase of the diel cycle, relative to that of PsbD (also known as D2; Figure 1C; Supplemental Figure 2D). This preferentially elevated PsbA synthesis is well known to replace PsbA subunits damaged by aberrant photochemical reactions (Theis and Schroda, 2016).
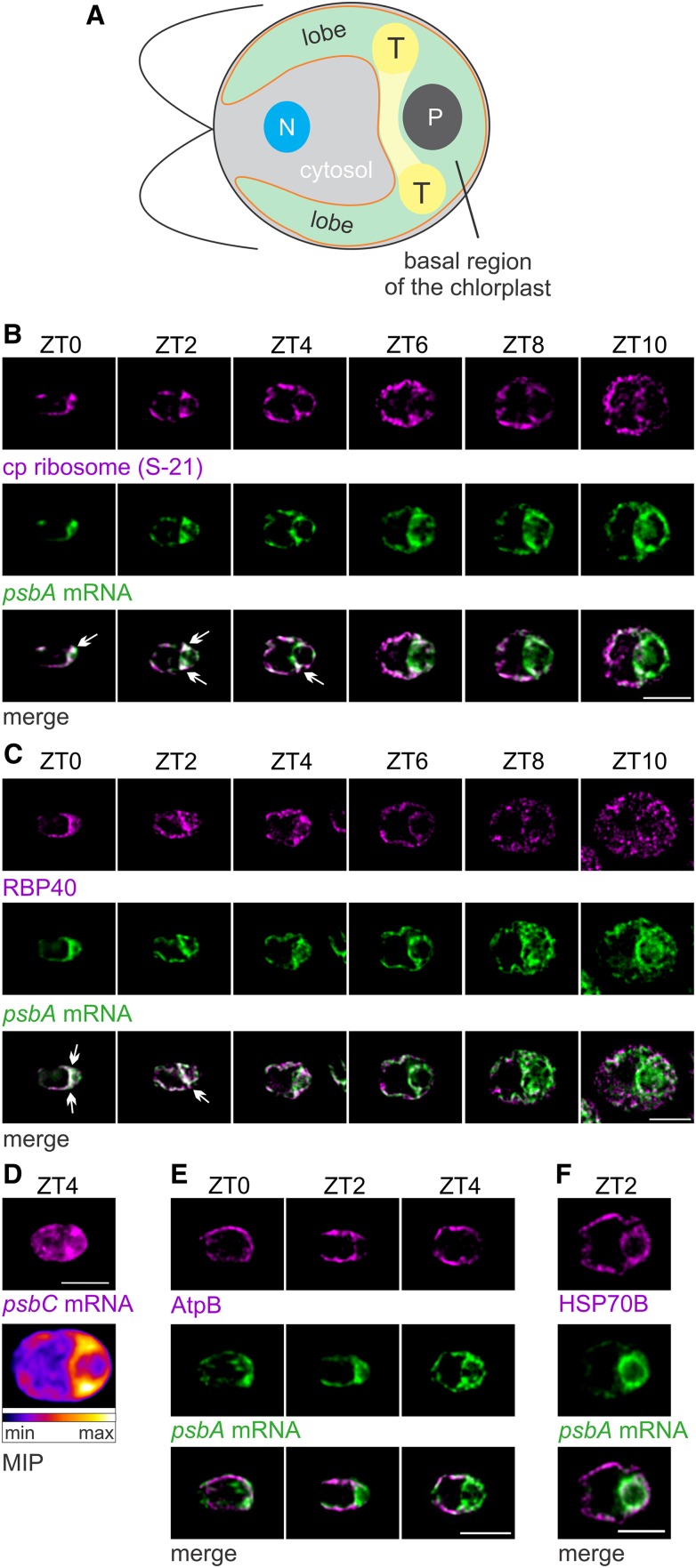
PSII Translation Markers Localize to the T-Zone Early in the Light Phase of the Diel Cycle
Chlamydomonas shows temporally complex patterns of gene expression for chloroplast biogenesis in the light phase of the diel cycle (Zones et al., 2015; Strenkert et al., 2019). Therefore, we asked whether PSII subunit synthesis is also spatially organized and, if so, how the localization patterns relate to the kinetics of PSII subunit synthesis and accumulation. From cultures entrained to the diel cycle, cells were sampled at 2-h intervals between ZT0 and ZT10 and then analyzed for their intrachloroplastic distributions of three translation markers: (1) the psbA mRNA, (2) the chloroplastic ribosomal protein S-21, and (3) RBP40 (also known as RB38), a translation factor required for early steps in the translation of PsbD (Barnes et al., 2004; Schwarz et al., 2007). PsbA and PsbD are encoded by the chloroplastic genome and translated by 70S chloroplastic ribosomes (Erickson et al., 1984; Rochaix et al., 1984). An illustration of a Chlamydomonas cell shows the relevant compartments (Figure 2A). These analyses revealed in most cells from the ZT0-ZT4 interval that the psbA mRNA and S-21 were enriched in patches in the basal region of the chloroplast near lobe junctions (Figure 2B). Many cells also showed a band of these colocalized signals extending between opposing lobe junctions. We designated this region as the T-zone in the growing chloroplast. This T-zone is located slightly anterior to the location of the T-zone in cells of asynchronous cultures under other conditions (Uniacke and Zerges, 2007). Later in the light phase, beginning at ZT6, the S-21 IF signal increased in the lobes such that it became distributed throughout the chloroplast (except in the pyrenoid). The psbA mRNA remained enriched in the T-zone throughout the light phase. The strongest RBP40 IF signal was from the T-zone in the ZT0-ZT4 interval and dispersed later in the ZT6-ZT10 interval (Figure 2C). Thus, the three translation markers were enriched specifically in this T-zone in the ZT0-ZT4 interval, when we also observed the major increases in PSII subunit synthesis (Figure 1C; Supplemental Figure 2D). T-zone localization of translation marker signals was not evident while the rates of PsbD synthesis declined in the ZT8-ZT10 interval. The increased level of S-21 in the lobes of the chloroplast in the ZT6-ZT8 interval may reflect PsbA synthesis for PSII repair because these events coincided and the latter is known to occur on thylakoid membranes throughout the chloroplast (Figures 1C and 2B; Mattoo and Edelman, 1987; van Wijk et al., 1996).
Figure 2.
Growing Chloroplasts during the Light Phase of the Diel Cycle Show Spatiotemporal Patterning of Markers of PSII Biogenesis.
(A) An illustration of a Chlamydomonas cell shows the chloroplast (green) with its T-zone (T, yellow), basal region, lobes, envelope (orange) and pyrenoid (P). Also shown are the cytosol and the nucleus (N).
(B) Epifluorescence microscopy images reveal the distributions of chloroplastic ribosomal protein S-21 and the psbA mRNA enriched in the T-zone of ZT0 cells (n = 27), ZT2 cells (n = 28), in approximately half of the ZT4 cells (n = 45), but not in most ZT6 cells (n = 78), ZT8 cells (n = 44), or ZT10 cells (n = 41). Arrows indicate the strongest overlaps. Each signal was manually adjusted to similar brightness across all images in each panel to allow comparisons of in situ distributions.
(C) RBP40 and the psbA mRNA localized to the T-zone in approximately half of the ZT0 cells (n = 123), in most ZT2 cells (n = 136) and ZT4 cells (n = 129), but not in most ZT6 cells (n = 117), ZT8 cells (n = 101), or ZT10 cells (n = 113).
(D) The psbC mRNA was enriched in the T-zone of ZT4 cells (top image). The heat map shows average signal intensities in a maximal intensity projection (MIP) of all cells in this data set (n = 56).
(E) The thylakoid membrane complex ATP synthase (AtpB) was not localized to the T-zone in most ZT0 cells (n = 18), ZT2 cells (n = 64), and ZT4 cells (n = 83).
(F) A protein of the chloroplast stroma (HSP70B) was not localized to the T-zone (marked by the psbA mRNA) in ZT2 cells (n = 79).
Bright-field images of all cells shown here are presented in Supplemental Figure 3 to show their proper anterior-posterior orientations (from left to right). Bars = 5.0 µm.
As the psbA mRNA is translated for both the biogenesis and repair of PSII, it does not serve as a definitive marker for the location of PSII biogenesis. Therefore, we asked whether the psbC mRNA, which encodes a PSII subunit that does not undergo a damage and repair cycle (CP43), also localizes to the T-zone in ZT4 cells (Chotewutmontri and Barkan, 2018). These ZT4 cells showed the highest psbC mRNA FISH signal in the T-zone, hence providing further support of the T-zone as the primary location of PSII biogenesis in the growing chloroplast (Figure 2D).
The localization of the translation markers to the T-zone does not reflect a feature of chloroplast anatomy, for example, densely populated thylakoid lamella or pockets of stroma in the T-zone, because it was not seen for AtpB, a subunit of the ATP synthase in thylakoid membranes, or a marker protein for the chloroplast stroma, HSP70B (Figures 2E and 2F). Moreover, our thin (0.2 µm) optical sections minimize contributions of local differences in chloroplast volume to signal intensities. In addition, transmission electron microscopy (TEM) images revealed that cells early in the light phase (ZT0 to ZT3) had the expected chloroplast ultrastructure and morphology, including thylakoid lamellae and stroma throughout the chloroplast (Supplemental Figure 4). Therefore, the enrichment of the translation markers in the T-zone likely reflects their localization and not some anatomical feature of the chloroplast unrelated to biogenesis.
Average Signal Distributions Confirm Localization of Translation Markers to the T-Zone
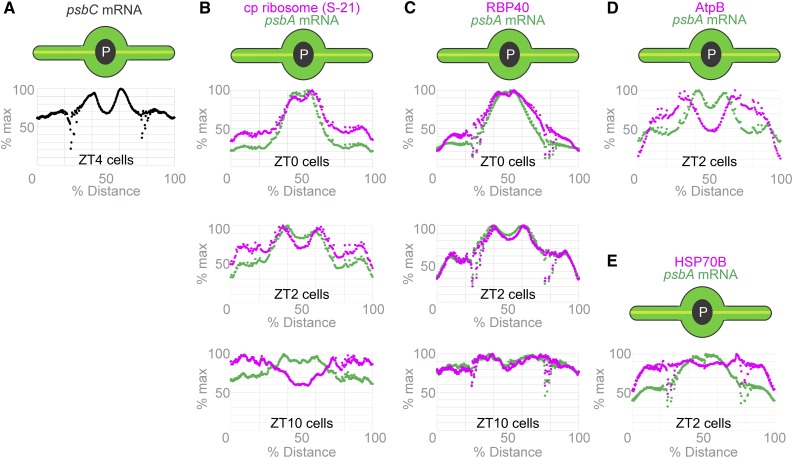
The precision of visual analyses for localization patterns at suborganellar levels in fluorescence microscopy images can be affected by the researcher’s unconscious bias and limited ability to simultaneously compare many images. To increase the objectivity of our analyses and discern localization patterns in Chlamydomonas, we developed a method to analyze fluorescence microscopy images by averaging signal intensity over many cells (Supplemental File). This macro takes a maximum intensity projection for each cell, collects the projections from all cells in a data set, and then rescales each cell such that its long and short axes are equal in length to those of the largest cell in the data set. It then superimposes the projections to generate an image of the average signal intensities (Figure 2D; Supplemental File). This analysis can further convert the average FISH signal intensity from the psbC mRNA in Figure 2D to the plot in Figure 3A of the percentages of the maximum value versus position on the chloroplast axis. The chloroplast axis was designated as a line from the tip of one lobe across the pyrenoid to the tip of the opposing lobe (see illustrations in Figure 3). The resulting plot shows two peaks of maximal average intensity where the axis traverses the T-zone on either side of the pyrenoid (seen as a dip of low average intensity).
Figure 3.
Plots of Average Fluorescence Signal Intensity along the Chloroplast Axis from All Cells of Each Data Set Support T-Zone Localization of the Translation Markers Early in the Light Phase of the Diel Cycle.
Chloroplasts are illustrated as their axis (yellow line) is presented on the horizontal axis of each graph. Our macro determines the average signal intensity along this axis.
(A) The average signal intensity of the psbC mRNA FISH signal in ZT4 cells (Figure 2D; n = 56) is plotted versus position on the chloroplast axis.
(B) to (E) The average signal intensities of the psbA mRNA FISH signal (green) and the IF signals of the S-21 chloroplastic ribosomal protein (ZT0, n = 27; ZT2, n = 28; and ZT10, n = 41; [B]), RBP40 (ZT0, n = 111; ZT2, n = 118, and ZT10, n = 123; [C]), the AtpB subunit of ATP synthase (n = 65; [D]), and the stromal marker HSP70B (n = 93; [E]) are plotted along the chloroplast axis (as percentage of the total length).
To better show the relationship between two signals, we determined the average signal intensity of each in the chloroplast lobes, the T-zone, and the pyrenoid (Supplemental File) and then plotted the percentages of their maximum values versus position on the chloroplast axis (Figure 3). Results of these analyses revealed that the signals of the three translation markers displayed maxima overlapping in the T-zones of cells from ZT0 and ZT2 (Figures 3B and 3C). At ZT0, a single peak of overlapping translation marker signals was seen where the axis crosses the T-zone. The nonstaining pyrenoid is small then (i.e., in the hours following mitosis) and, thus, it makes only a slight dip at the apex of this peak (compare Supplemental Figures 4B and 4C; Freeman Rosenzweig et al., 2017). At ZT2, the larger pyrenoid is now at the center of the chloroplast axis and separates two peaks of overlapping translation marker signals where the axis crosses the T-zone in opposing lobe junctions. By ZT10, the maxima had diminished for the psbA mRNA and were no longer seen for S-21 and RBP40, as the latter signals increased in the lobes (Figures 3B and 3C). The average signals of the control markers for thylakoid membranes and stroma, AtpB and HSP70B, respectively, did not show maxima in the T-zone (marked by the psbA mRNA maxima) at any of the time points examined (Figures 3D and 3E). These results confirm the results of visual analyses in the previous subsection and, thereby, provide further support of the T-zone being the primary location of PSII subunit synthesis early in the light phase.
The lobes of the chloroplast in some cells showed a gap in all fluorescent signals, which was located immediately anterior to the junction with the basal region (Figure 2B, ZT2 and ZT4 cells; Figures 3A, 3C, and 3E). Each of these gaps is caused by a constriction of the entire lobe, as can be seen in the TEM image in Supplemental Figure 4B. These constrictions appear on both lobes in some cells for unknown reasons. Additional work is required to understand their structure and functions.
Localization of the psbA mRNA and Chloroplastic Ribosome to the T-Zone Is Established following Mitosis
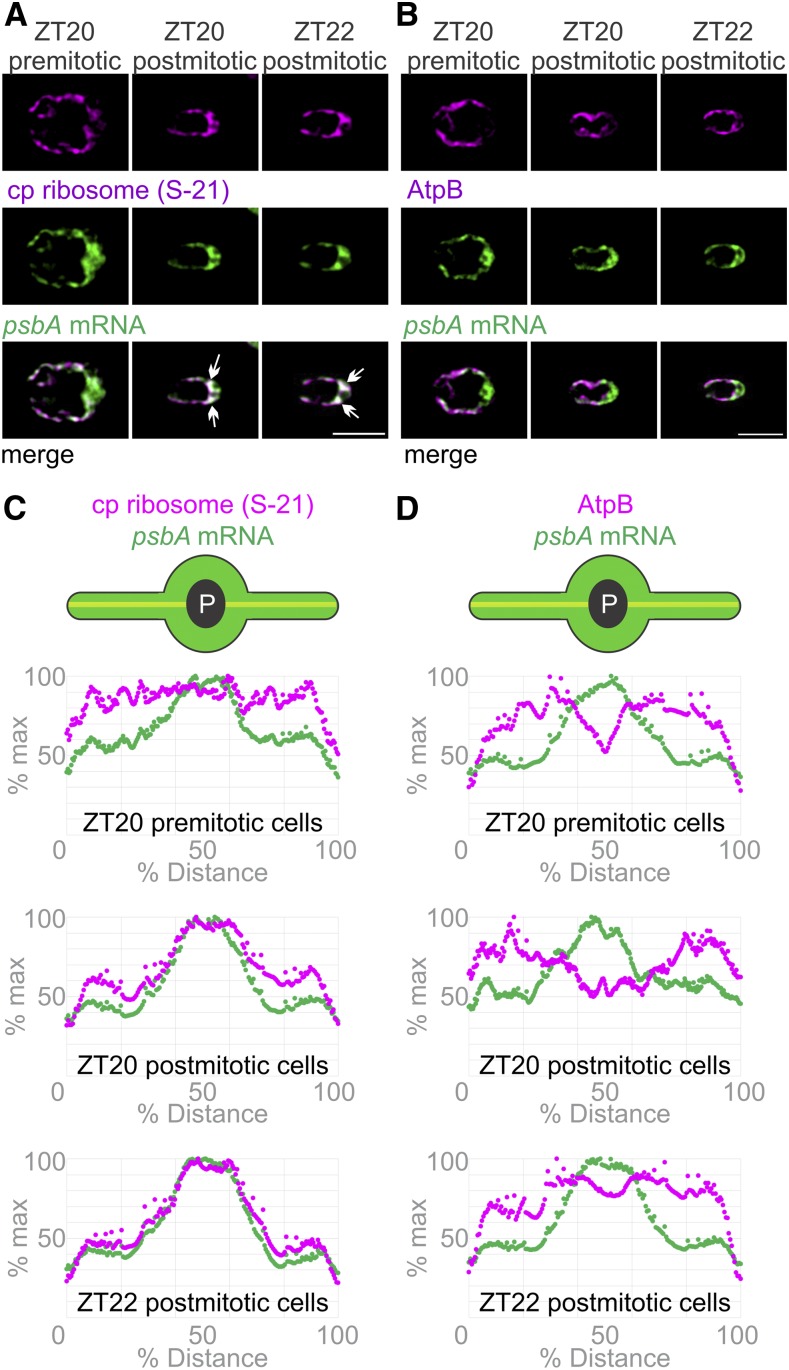
T-zone localization of the translation markers was established sometime during the dark phase; it was absent at ZT10 and present at ZT0 (Figures 2B, 2C, 3B, and C3C). To determine when this localization is established, cells at the end of the dark phase (ZT20 and ZT22) were analyzed for their in situ distributions of the psbA mRNA and the chloroplastic ribosomal protein S-21. At ZT20, we observed large and small cells. The large cells were premitotic while the small cells were postmitotic; they were similar in size to the daughter cells that predominated at ZT22 and constituted all cells at ZT0 (Supplemental Figure 1A). Premitotic ZT20 cells did not show T-zone localization of the psbA mRNA or S-21 (Figures 4A and 4C). The former was enriched in the basal region but not localized specifically in the T-zone, while the latter was dispersed throughout the chloroplast, except in the pyrenoid. Postmitotic ZT20 and ZT22 cells, however, showed T-zone localization of the psbA mRNA and S-21 (Figures 4A and 4C). This localization pattern was not seen for AtpB, revealing that it does not reflect the distribution of thylakoid membranes, the accepted location of psbA translation (Zoschke and Bock, 2018; Figures 4B and 4D). Therefore, localization of the psbA mRNA and the chloroplastic ribosome to the T-zone is established soon after mitosis and maintained throughout the remainder of the dark phase and during the initial 4 h of the light phase (Figures 2B and 2C). Previous reports describe increases in the synthesis rates of chloroplast genome-encoded subunits of PSI and PSII in the dark phase, when we observed the establishment of this localization pattern (∼ZT22; Howell et al., 1977; Lee and Herrin, 2002). Our pulse-labeling assay was not sufficiently sensitive to monitor these rates of synthesis in the dark phase. Thus, the chloroplast translation machinery and mRNAs localize to the T-zone when photosystem subunit synthesis is activated near the end of the dark phase.
Figure 4.
The Chloroplastic Ribosome Marker Protein and the psbA mRNA Localize to the T-Zone after Cell Division in the Dark Phase of the Diel Cycle.
(A) Epifluorescence microscopy images revealed in the premitotic cells at ZT20 (n = 15) that the psbA mRNA (green) was higher in the basal region than the lobes while the chloroplastic ribosomal protein S-21 (magenta) was distributed throughout the chloroplast. In most postmitotic ZT20 cells (n = 103) and postmitotic ZT22 cells (n = 72), the strongest signals from the psbA mRNA and the chloroplastic ribosomal protein S-21 colocalized to the T-zone (arrows). Bar = 5.0 µm.
(B) The AtpB subunit of the chloroplastic ATP synthase (magenta) was not localized with the psbA mRNA in premitotic ZT20 cells (n = 9), in postmitotic ZT20 cells (n = 39), or in ZT22 cells (n = 103). Bar = 5.0 µm.
(C) and (D) The patterns in (A) and (B) were confirmed by plots of the average signal intensities versus the chloroplast axis. Cell numbers were, for the psbA mRNA and S-21, ZT20 premitotic (n = 15), ZT20 postmitotic (n = 54), and ZT22 (n = 72; [C]), and for the psbA mRNA and AtpB, ZT20 premitotic (n = 9), ZT20 postmitotic (n = 54), and ZT22 (n = 112; [D]).
Chlorophyll Biosynthesis Is Localized to the T-Zone
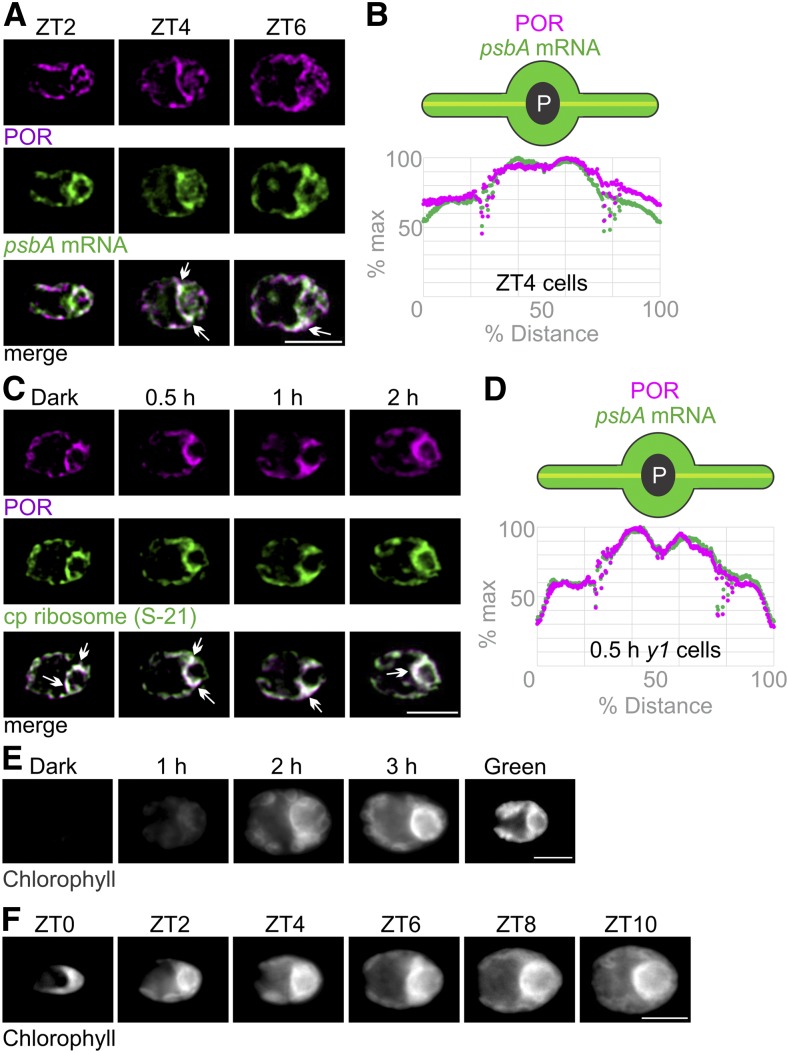
Newly synthesized chlorophyll is incorporated into PSI and PSII within seconds in Chlamydomonas (White and Hoober, 1994). This suggests that chlorophyll biosynthesis is colocalized with the synthesis of chlorophyll binding proteins, as has been shown in cyanobacteria (Chidgey et al., 2014). In addition, the oxidative cyclase CRD1, an enzyme in the chlorophyll biosynthesis pathway, is localized to the pyrenoid perimeter (i.e., in the T-zone; Allen et al., 2008). Therefore, we asked whether chlorophyll biosynthesis is localized to the T-zone by IF staining cells in the light phase of the diel cycle for the light-dependent POR. Epifluorescence microscopy images show the POR IF signal throughout the chloroplast in ZT2 cells. However, ZT4 and ZT6 cells showed strong POR IF signal in the T-zone (marked by the FISH signal of the psbA mRNA; Figures 5A and 5B). Therefore, the localization of POR to the T-zone coincided with the accelerated increase in chlorophyll level at ZT5 and with the increases in the levels of PSI and PSII (Figure 1B; Supplemental Figures 2A and 2C).
Figure 5.
The T-Zone Is the Primary Location of Chlorophyll Biosynthesis.
(A) Epifluorescence microscopy images show that the fluorescent signals of POR (magenta) and the psbA mRNA (green) were throughout the chloroplast in most ZT2 cells (n = 95) and colocalized in the T-zone (arrows) in most ZT4 cells (n = 73) and ZT6 cells (n = 47). Bar = 5.0 µm.
(B) For ZT4 cells, this pattern was confirmed by plots of average signal intensities versus position on the chloroplast axis (n = 112).
(C) Images show the IF signals of POR (magenta) and S-21 (green) colocalized in the T-zones of most dark-y1 cells (n = 111), 0.5 h-y1 cells (n = 62), 1 h-y1 cells (n = 71), and 2 h-y1 cells (n = 56). Bar = 5.0 µm.
(D) This pattern was confirmed for 0.5 h-y1 cells by plots of average signal intensities versus position on the chloroplast axis (n = 33).
(E) In live-imaged dark-y1 cells (n = 62), no autofluorescence was seen because they lack chlorophyll. The strongest chlorophyll fluorescence was seen in the T-zones of most live 1 h-y1 cells (n = 52), 2 h-y1 cells (n = 40), and 3 h-y1 cells (n = 40) and throughout the chloroplast of all green-y1 cells (n = 55). Bar = 5.0 µm.
(F) At ZT0 (n = 14), chlorophyll fluorescence was higher in the basal region than in the lobes, but not particularly localized to the T-zone. The strongest chlorophyll autofluorescence was seen in the T-zones of most cells at ZT2 (n = 16), ZT4 (n = 15), ZT6 (n = 19), ZT8 (n = 17), and ZT10 (n = 13). Bar = 5.0 µm.
We asked whether chlorophyll biosynthesis is localized to the T-zone in greening y1 cells. We confirmed that y1 cells cultured in the dark (dark-y1 cells) have low levels of PSI and PSII, low synthesis rates of PSI and PSII subunits (PsaA, PsbA, and PsbD), and that these levels and rates increase during greening due to the activation by light of chlorophyll biosynthesis and, consequently, photosystem biogenesis (Supplemental Figure 5; Ohad et al., 1967a; Malnoë et al., 1988). When we analyzed the in situ distribution of the POR, we found that it colocalized with S-21 in the T-zone in dark-y1 cells and in y1 cells after 0.5, 1, and 2 h of greening (hereafter 0.5 h-y1 cells, 1 h-y1 cells, and 2 h-y1 cells, respectively; Figures 5C and 5D). This result suggests that the T-zone is a primary location of chlorophyll biosynthesis in greening y1 cells.
To test this possibility further, we exploited the fact that dark-y1 cells begin greening without chlorophyll and then accumulate it during greening (Ohad et al., 1967a). Chlorophyll fluorescence should first appear in the T-zone if it is a principal location of chlorophyll biosynthesis. In live-imaged dark-y1 cells, only weak autofluorescence was seen because they lack chlorophyll. As was expected, green-y1 cells (y1 cells cultured in constant light) exhibited chlorophyll fluorescence throughout their chloroplast (i.e., in the known distribution of thylakoids; Figure 5E). By contrast, in y1 cells after 1, 2, or 3 h of greening, the strongest chlorophyll fluorescence was seen along the anterior perimeter of the pyrenoid (i.e., the T-zone). Similarly, in individual greening y1 cells followed in time-lapse movies of maximum intensity projections, chlorophyll fluorescence was seen first in the T-zone (Supplemental Video). Later, the strongest chlorophyll fluorescence expanded anteriorly and was seen throughout the chloroplast by the end of greening. These results provide further support that the T-zone is the primary location of chlorophyll biosynthesis in the greening chloroplast.
Similarly, we visualized chlorophyll fluorescence in live cells from ZT0 to ZT10 in the diel cycle (Figure 5F). At ZT0, chlorophyll fluorescence was higher in the basal region than in the lobes, but it was not particularly localized to the T-zone. Later, and throughout the light phase, the strongest chlorophyll fluorescence was seen along the anterior perimeter of the pyrenoid (i.e., the T-zone), as was observed in greening y1 cells (Figures 5E and 5F). This strong chlorophyll fluorescence appeared before the accelerated increase in chlorophyll level at ZT5 (Figure 1B), suggesting that it is from newly synthesized chlorophyll whose excitation is not yet quenched by processes in the photosystems and their LHCs (Müller et al., 2001). These results support the T-zone as being the primary location of chlorophyll biosynthesis in the growing chloroplast.
PSII Translation Markers Localize to the T-Zone of Greening y1 Cells
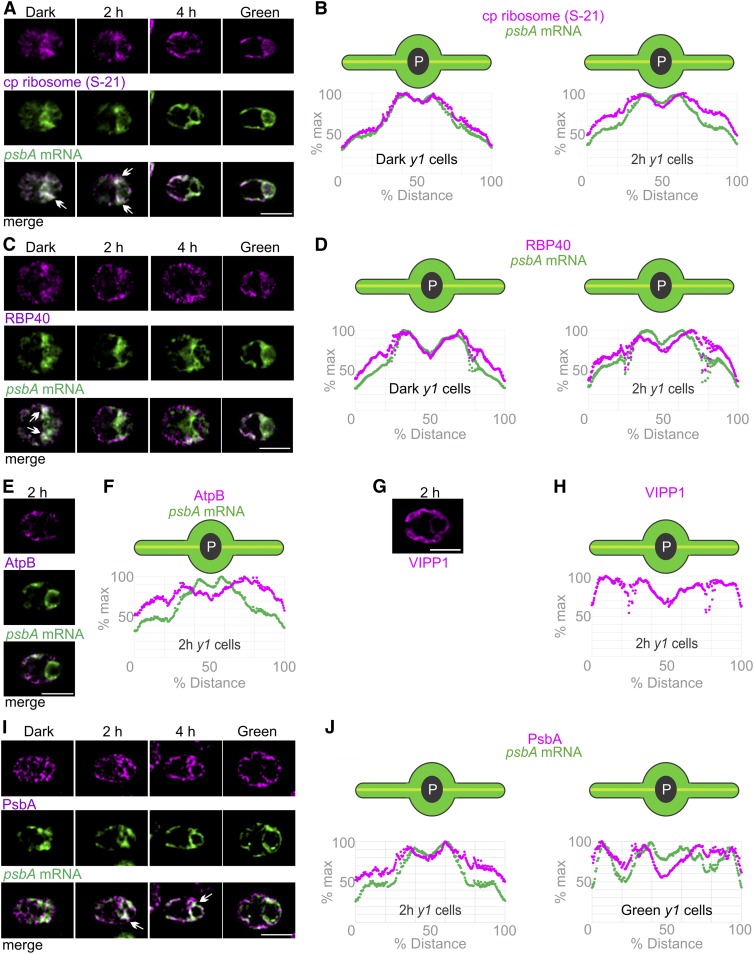
We then analyzed the in situ distributions of the PSII translation markers in the chloroplast of greening y1 cells (Figure 6). In dark-y1 cells and 2 h-y1 cells, the strongest signals from the psbA mRNA and S-21 overlapped in the T-zone (Figures 6A and 6B). In 4 h-y1 cells or green-y1 cells, the psbA mRNA FISH signal was still enriched in the T-zone while S-21 was more broadly distributed throughout the chloroplast, except in the pyrenoid. Similarly, RBP40 and the psbA mRNA localized to the T-zone in dark-y1 cells and 2 h-y1 cells (Figures 6C and 6D). This pattern was less evident in 4 h-y1 cells and not evident in green-y1 cells. These results provide evidence that the T-zone is the primary location of PSII subunit synthesis early in y1 greening.
Figure 6.
The T-Zone in Greening y1 Cells Revealed by the in Situ Localization of Translation Markers.
(A) Images show the strongest signals from chloroplastic ribosomal protein S-21 (magenta) and the psbA mRNA (green) colocalized in the T-zone in most dark-y1 cells (n = 91) and 2 h-y1 cells (n = 145) and approximately half of 4 h-y1 cells (n = 138) but not in most green-y1 cells (n = 89). Arrows indicate where the strongest signals from both channels overlap in the T-zone. Bar = 5.0 µm.
(B) Plots of the average signal intensities versus position on the chloroplast axis confirm these patterns in dark-y1 cells (n = 52) and 2 h-y1 cells (n = 95).
(C) Cell images show RBP40 (magenta) and the psbA mRNA (green) colocalized in the T-zone in approximately half of dark-y1 cells (n = 41) and in minorities of 2 h-y1 cells (n = 75) and 4 h-y1 cells (n = 103) but not in green-y1 cells (n = 82). Bar = 5.0 µm.
(D) Plots of average intensities of these signals showed peaks in the T-zone of dark-y1 cells (n = 65) and 2 h-y1 cells (n = 109).
(E) In 2 h-y1 cells, AtpB (magenta) was maximal in the lobes and not localized to the T-zone with the psbA mRNA (green) in most 2 h-y1 cells (n = 76). Bar = 5.0 µm.
(F) This was confirmed by a plot of the AtpB IF signal intensity versus position on the chloroplast axis (n = 54).
(G) VIPP1, a marker for thylakoids and stroma, also was not localized to the T-zone in most 2 h-y1 cells (n = 51). Bar = 5.0 µm.
(H) This was confirmed by a plot of the VIPP1 IF signal intensity versus position on the chloroplast axis (n = 44).
(I) PsbA is a marker for newly synthesized PSII proteins early in y1 greening (see text); it was T-zone localized (arrows) in most 2 h-y1 cells (n = 95) and 4 h-y1 cells (n = 40) but not in most dark-y1 cells (n = 79) or any green-y1 cells (n = 36). The PsbA signal was manually enhanced in the dark-y1 cell to allow comparisons of its in situ distributions but not relative levels across the conditions. Bar = 5.0 µm.
(J) The average intensity of the PsbA IF signals was maximal in the T-zone in 2 h-y1 cells (n = 89) but not in green-y1 cells (n = 34).
The localization of these translation markers to the T-zone does not reflect local enrichments of thylakoids and stroma in y1 cells because neither AtpB nor a marker for stoma and thylakoids, VIPP1 (Liu et al., 2005), was localized in this pattern in 2 h-y1 cells (Figures 6E–6H). As was mentioned above, the thin (0.2 µm) optical sections minimize the effects on signal intensity of any differences in volume between the T-zone and other parts of the chloroplast. Therefore, the enrichment of the PSII translation markers in the T-zone in dark-y1 and 2 h-y1 cells reflects their localization and not a local enrichment of thylakoids or stroma compared with elsewhere in the chloroplast.
Since the translation markers localized to the T-zone in dark-y1 cells (i.e., before PSII subunit synthesis was induced by illumination; Figures 6A–6D; Supplemental Figures 5C and 6D), we sought a direct marker for PSII subunit synthesis to determine whether the T-zone is a primary location of active translation. We were able to use the PsbA protein because dark-y1 cells have only trace amounts such that most of the PsbA pool early in greening is newly synthesized (Supplemental Figures 5A and 5B). Therefore, the first PsbA to appear in greening should mark its location of synthesis. Consistent with this rationale, the PsbA IF signal in dark-y1 cells was very weak and not in any particular localization pattern (Figure 6I). (The brightness of the PsbA IF signal in the dark-y1 cell image was enhanced to show its distribution.) In 2 h-y1 cells and 4 h-y1 cells, the PsbA IF signal initially increased in the T-zone, where it colocalized with the psbA mRNA FISH signal. In green-y1 cells, PsbA was seen throughout the chloroplast, consistent with this known distribution of thylakoids (Figure 6I). These patterns are supported by the average signal distributions along the chloroplast axis in 2 h-y1 cells and green-y1 cells (Figure 6J). T-zone localization of PsbA in 2 h-y1 cells does not represent the distribution of thylakoid lamellae or stroma because it was not seen for AtpB or VIPP1 (Figures 6E–6H). Therefore, PsbA marks the T-zone as the primary location of its synthesis during y1 greening.
This localized synthesis of PsbA in the T-zone suggests that, during greening, the newly synthesized protein migrates into the lobes where PSII is abundant in mature green-y1 cells (Figures 6I and 6J). In support of this hypothesis, the average PsbA IF signal in 2 h-y1 cells extended slightly farther into the lobes than did the psbA mRNA in the T-zone.
PSI Subunit Synthesis Occurs in the T-Zone of Greening y1 Cells
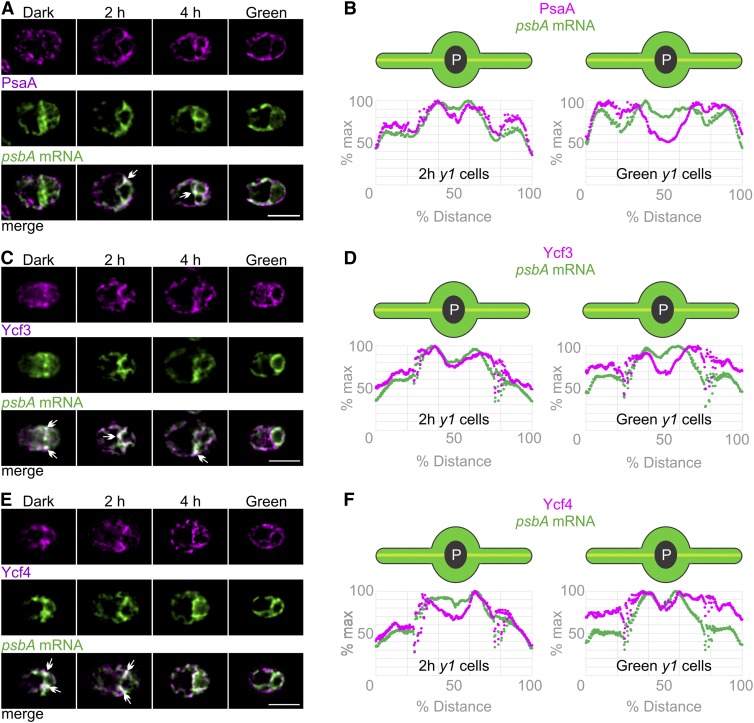
To determine whether subunits of PSI also are synthesized in the T-zone, we IF stained y1 cells for PsaA, a subunit of the PSI reaction center encoded by the plastid genome. Like PsbA, PsaA serves as a marker for the location of its own synthesis in greening y1 cells because most of its pool is newly synthesized (Supplemental Figures 5A and 5B). The PsaA IF signal was weak and nonlocalized in dark-y1 cells, consistent with their trace amounts of PsaA (Figure 7A). During greening, the PsaA localized to the T-zone of 2 h-y1 cells along with the psbA mRNA (Figure 7A). By 4 h of greening, the PsaA IF signal had increased in the lobes, although it still colocalized with the psbA mRNA in the T-zone in approximately half of these cells. Eventually, all green-y1 cells had the PsaA IF signal throughout the chloroplast, the known distribution of PSI in thylakoid membranes (Figure 7A). These patterns are supported by plots of the average signal intensity versus position on the chloroplast axis in 2 h-y1 cells and green-y1 cells (Figure 7B). These results provide evidence that the T-zone is the primary location of PSI subunit synthesis in greening y1 cells.
Figure 7.
Localization of PSI Subunit Synthesis and Assembly to the T-Zone of Greening y1 Cells.
(A) Overlap of the strongest signals of PSI subunit PsaA (magenta) and the psbA mRNA (green) in the T-zone was seen in most 2 h-y1 cells (n = 59) and in approximately half of 4 h-y1 cells (n = 30) but not in dark-y1 cells (n = 75) or green-y1 cells (n = 52). PsaA signal was enhanced in the dark-y1 cell to allow comparisons of its distributions. Arrows indicate where the strongest signals from both channels overlap in the T-zone. Bar = 5.0 µm.
(B) The colocalization in the T-zone of 2 h-y1 cells (n = 73) but not green-y1 cells (n = 43) was confirmed by plots of the average signal intensities versus the chloroplast axis.
(C) Epifluorescence microscopy images show the strongest signals from Ycf3 (magenta) and the psbA mRNA (green) colocalized in the T-zone in most dark-y1 cells (n = 38), in approximately half of 2 h-y1 cells (n = 84) and 4 h-y1 cells (n = 136), but not in green-y1 cells (n = 35). Bar = 5.0 µm.
(D) T-zone localization of both signals in 2 h-y1 cells (n = 90) but not green-y1 cells (n = 32) was confirmed by plots of the average signal intensities versus position on the chloroplast axis.
(E) The strongest signals of Ycf4 (magenta) and the psbA mRNA (green) colocalized in the T-zone in dark-y1 cells (n = 40), in 2 h-y1 cells (n = 55), and in approximately half of 4 h-y1 cells (n = 57) but not in green-y1 cells (n = 64). Bar = 5.0 µm.
(F) T-zone localization of both signals in 2 h-y1 cells (n = 57) but not green-y1 cells (n = 50) was confirmed by plots of the average signal intensities.
As was observed for PsbA, the average PsaA IF signal in 2 h-y1 cells extended into the lobes from the T-zone (Figure 7B). This supports the migration of the newly synthesized PsaA from the T-zone into the lobes during chloroplast greening.
PSI Assembly Factors Localize to the T-Zone in Greening y1 Cells
We addressed the location where newly synthesized PSI subunits are assembled to form the PSI reaction center by characterizing the in situ distributions of two PSI-specific assembly factors. Ycf3 and Ycf4 interact with the newly synthesized PSI subunits and promote their assembly into the PSI reaction center, but they are not present in fully assembled and functional PSI (Nellaepalli et al., 2018). Both Ycf3 and Ycf4 localized to the T-zone (marked by the psbA mRNA FISH signal) in 2 h-y1 cells but not in green-y1 cells (Figures 7C to 7F). Therefore, these results provide evidence that the T-zone is the primary location of PSI assembly.
DISCUSSION
Our results reveal the T-zone as a hub for the biogenesis of PSI and PSII under developmentally relevant conditions associated with rapid chloroplast biogenesis for cellular growth or greening. Such a region where multiple pathways converge for thylakoid membrane biogenesis has been described as a “biogenesis center” (Nickelsen and Zerges, 2013; Rast et al., 2015). The T-zone is among a growing number of examples of localized translation for biogenesis and pattern formation in diverse organisms. In the cytoplasm of yeast, neurons, and developing embryos, the translation of specific mRNAs is localized to RNA granules, membrane-bound organelles, or synapses for diverse functions (Lui et al., 2014; Hughes and Simmonds, 2019; Panasenko et al., 2019). An analogous region to the T-zone of the chloroplast was recently reported in mitochondria, the other semiautonomous organelle, for the biogenesis of the respiratory electron transport chain complexes (Stoldt et al., 2018).
The T-zone under the developmentally relevant conditions examined here is slightly more anterior than the T-zone described previously in cells that were briefly shifted from dark to light to induce PSII subunit synthesis (Uniacke and Zerges, 2007). The T-zone here is adjacent to domains of the chloroplast envelope that are enriched in protein import translocons (Schottkowski et al., 2012). The cytoplasm neighboring the T-zone is enriched in cytosolic ribosomes and an mRNA encoding a subunit of the LHC of PSII (Colón-Ramos et al., 2003; Uniacke and Zerges, 2009). Invaginations of the inner envelope membrane adjacent to the T-zone were observed in cryo-electron microscopy images (Engel et al., 2015). These results suggest an intercompartmental spatial coordination in the synthesis of nuclear genome-encoded photosystem subunits in the posterior cytoplasm, protein import into the chloroplast, and the synthesis and assembly of chloroplast genome-encoded subunits and chlorophyll in the T-zone within the chloroplast (e.g., a “thylakoid membrane biogenesis center”) and analogous to an intramitochondrial “zone” and “ER membrane contact sites” with other organelles (Rast et al., 2015; Wu et al., 2018; Shimizu, 2019).
Functions of localized photosystem biogenesis to the T-zone could include any of the known roles of intracellular compartmentalization. For example, elevated local concentrations of substrates, intermediates, and biochemical factors favor forward reactions. The sequestration of substrates, intermediates, enzymes, and factors prevents deleterious side reactions, such as the aggregation of nonnative proteins or singlet oxygen production by newly synthesized chlorophyll. Finally, compartmentalization can enhance efficiency by facilitating the channeling of intermediates and spatially coordinating pathways of a network.
It is unknown how newly synthesized chlorophyll is transported from its accepted site of synthesis at the chloroplast envelope, through the stroma, to thylakoid membranes for photosystem biogenesis (Joyard et al., 2009). The colocalization of the biosynthesis of chlorophyll and chlorophyll binding apoproteins in the T-zone suggests that chlorophyll transport is unnecessary.
The ordered increase in the photosynthesis complexes suggested by our results is consistent with the programmed differentiation seen at the levels of the transcriptome and, in maize, the translatome (Figures 1A and 1B; Zones et al., 2015; Chotewutmontri and Barkan, 2016; Strenkert et al., 2019). The increase in chlorophyll levels was intermediate to the increases in the complexes that bind most of the pool of this photopigment, the photosystems and LHCII (Figure 1B).
The localization of the translation markers to the T-zone arose soon after mitosis at ZT20 in the dark phase, whereupon it was maintained until approximately ZT4 (Figures 2B, 2C, 3A to 3C, 4A, 4C, and 8). Previous reports showed that the synthesis rates of PSII subunits by chloroplastic ribosomes began to increase late in the dark phase, approximately when we observed the localization of the translation markers to the T-zone (Howell et al., 1977; Lee and Herrin, 2002). Therefore, localization of the translation markers to the T-zone after mitosis coincides with the activation of PSII subunit synthesis.
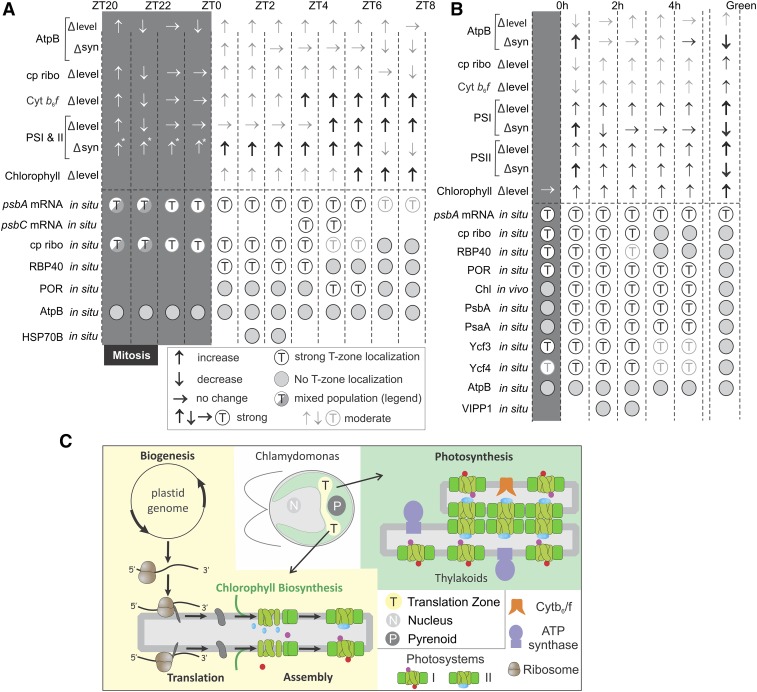
Figure 8.
Summaries of the Results.
(A) For the diel cycle interval ZT20-ZT8, arrows indicate changes in steady state level (∆ level) and synthesis rate (∆syn) of the marker proteins for the listed complexes as follows: ↑, increase; ↓, decrease; thick ↑, drastic increase; gray ↑, slight increase; →, no change. Below the broken line, for each mRNA or marker protein, T-zone localization is indicated by open circles and nonlocalization by shaded circles. Sectored circles indicate mixed populations of premitotic and postmitotic cells showing nonlocalization or T-zone localization, respectively. Faded circles indicate weak T-zone localization. The intervals of the dark and light phases examined have shaded and white backgrounds, respectively. Absence of a circle indicates not determined. Abbreviations and marker proteins in situ (in parentheses) are as follows: ATP synthase (AtpB), AtpB; chloroplastic ribosome (S-21), cp ribo; Cytb6f complex (Cyt f), Cytb6f; PSI and PSII (PsaA and PsbD, respectively), PSI and II. Asterisks indicate that data are from Howell et al. (1977) and Lee and Herrin (2002).
(B) The results obtained with greening y1 are summarized as described for (A). Changes in protein level and synthesis rates of the marker proteins (top arrows) are presented for reference to our in situ results (bottom circles). Levels and synthesis rates of the markers of ATP synthase, PSI, and PSII were reported previously (Malnoë et al., 1988).(C) Our model shows the role of the T-zone (yellow) as a compartment in the chloroplast which is the location of photosystem subunit translation and assembly as well as chlorophyll biosynthesis and distinct from the distribution of photosynthetic thylakoid membranes throughout the chloroplast (green).
The translation markers localized to the T-zone until ZT6 (Figures 2B, 2C, and 8A). Similarly, in y1, the translation markers localized to the T-zone in the dark and the first 2 h of greening, but not thereafter (Figures 6A to 6D, 6I, 6J, and B8B). Therefore, in both cellular growth and greening, translation markers localized to the T-zone early but not late in these different modes of chloroplast biogenesis, even though translation rates of these subunits were sustained throughout these processes (Figure 1C; Supplemental Figures 2D, 5C, and 5D). These results support a role of the T-zone as a hub for photosystem biogenesis early in chloroplast growth and greening. Later in these processes, the redistribution of the translation markers throughout the chloroplast coincided with the preferentially elevated PsbA synthesis rates for PSII damage-repair cycle on stroma-exposed thylakoid membranes throughout the chloroplast (Figure 1C; Jagendorf and Michaels, 1990). Thus, PsbA repair synthesis on stroma-exposed thylakoid membranes, which is believed to occur throughout the chloroplast, could have masked sustained localization of subunit synthesis in the T-zone for de novo PSII biogenesis. Alternatively, de novo photosystem biogenesis might redistribute from the T-zone to stroma thylakoids throughout the chloroplast beginning at approximately ZT4 in the diel cycle and after 2 h in the greening process of y1.
METHODS
Culture Conditions
The Chlamydomonas (Chlamydomonas reinhardtii) wild-type strain CC-125 (137c) was synchronized to the 12/12-h light/dark cycle by culturing in high-salt minimal (HSM) medium (Harris, 1989) with 0.5 to 1% CO2 at a flow rate of 300 to 400 mL/min and illuminated from the four sides and below by five banks of red and blue LEDs at 250 to 280 µE m−2 s−1 at 23°C in the day and 27°C at night, with a ramp down over the course of 1 h during the first hour of the light phase. Cultures were entrained under alternating cycles of 12 h of light/12 h of dark for 2 to 3 d to 1 × 106 to 2 × 106 cells/mL measured with a hemocytometer. Cultures were diluted with fresh HSM medium between ZT1 and ZT3 of each of the subsequent 3 d, thereby reducing the cell density to 1 × 105 to 2 × 105 cells/mL. On the final day, cultures were not diluted and samples were collected at the ZT points indicated in the text. Cells were immediately frozen at −80°C (e.g., for SDS-PAGE) or chemically fixed (i.e., for IF and FISH). ZT0 and ZT12 cells were collected ∼3 min following the respective transition.
The Chlamydomonas mutants for ycf3 and ycf4 (from Yuichiro Takahashi, Okayama University), y1, and psbA (FuD7; CC-1168 and CC-4147, respectively; www.chlamycollection.org) were cultured in Tris-acetate-phosphate (TAP) medium (Harris, 1989) in the dark at 24°C with orbital shaking to 1 × 106 to 2 × 106 cells/mL. Light-induced chloroplast differentiation was obtained by illuminating dark-grown y1 cultures (∼30 µE m−2 s−1) for the times indicated in the text.
Immunoblot Analysis
For the immunoblots with synchronized cells, equal volumes of the culture were centrifuged (4000g, 5 min) at 4°C. For the immunoblots with y1 cells, an equal number of cells were centrifuged at each time point. The same cell samples were used for the immunoblot analyses, although the 45-min denaturation in SDS-PAGE loading buffer was at 24°C for PsaA (to prevent it from forming large insoluble aggregates) and at 65°C for all other proteins. Proteins were resolved by SDS-PAGE (12% [w/v] acrylamide:bis-acrylamide at 29:1; Sambrook and Russell, 2001) and then were transferred to PVDF membranes (Bio-Rad) and reacted with primary and secondary antibodies (Sambrook and Russell, 2001). The primary antibodies were as follows: αPsbA (1:5000; Agrisera AS111786), αAtpB (1:5000; from André Jagendorf, Cornell University), αPsaA (1:60,000; from Kevin Redding, Arizona State University), αCytf (1:100,000), αLHCII, αcytf, and αPsbD (1:5000, 1:2000, and 1:5000, respectively; from Francis-Andre Wollman, Institut de Biologie Physico-Chimique, Paris), αcyL4, αS-21, and αL-7/L-12 (1:6,000, 1:4,000, and 1:10,000, respectively; we have the remaining stocks of these antisera of Nicholas Gilham; Fleming et al., 1987; Randolph-Anderson et al., 1989), and αPOR (1:50,000; Katrin Philippar and Jurgen Soll, Ludwig Maximilian University, Munich). The secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (KPL). Signals were detected using an ECL substrate (Thermo Fisher Scientific) with an Amersham Imager 600 (GE) according to the manufacturer’s protocols.
In Vivo 35S-Pulse-Labeling Experiments
At each time point, a 5-mL aliquot of a culture (∼1.2 × 107 cells) was centrifuged at 4000g for 2 min. Each cell pellet was resuspended in 1 mL of medium lacking sulfate (HSM-S for wild-type cells in the diel cycle and TAP-S for y1). When the cell pellet was fully resuspended, cycloheximide was added to 10 µg/mL. Cells were incubated for 5 min with shaking under the conditions described for each time point. Then, 90 µCi of [35S]H2SO4 (1050–1600 Ci/mM; Perkin-Elmer) was added, and labeling was performed for 10 min. Cells were pelleted by centrifugation at 4000g for 2 min, resuspended and lysed in 200 μL of SDS-PAGE loading buffer, and incubated at room temperature for 1 h. Then, 15 μL of each sample was loaded onto a SDS-PAGE gel (12% acrylamide:bis-acrylamide at 29:1 and8 M urea). Following electrophoresis, the gels were dried, and 35S-labeled proteins were revealed with a phosphoimager (Typhoon).
FISH, IF Staining, Live-Cell Imaging, and Microscopy
The FISH and IF staining procedures and the psbA FISH probes were described previously by Uniacke and Zerges, (2007) and Uniacke et al., (2011). The primary antibodies and the dilutions used for IF were as follows: αS-21 (1:1000), αRBP40 (1:1000; from Jörg Nickelsen, Ludwig Maximilian University), αHSP70B and αVIPP1 (both at 1:1000; from Michael Schroda, University of Kaiserslautern), αAtpB (1:1000), αPsbA (1:1000; Agrisera), αPsaA (1:2000), αPOR (1:1000), and αYcf3 and αYcf4 (both at 1:400). Sources of antibodies that were also used in immunoblot analyses are stated above.
Fluorescent secondary antibody used was AlexaFluor568 conjugated to goat anti-rabbit IgG (Thermo Fisher Scientific). Staining with the secondary antibody alone revealed only weak signal throughout the cells (Supplemental Figure 6). For the dual IF staining in Figure 5C, cells were first reacted with αPOR, which was subsequently indirectly IF labeled by excess AffiniPure Fab fragment donkey anti-rabbit IgG (H+L) conjugated to AlexaFluor488 (Jackson ImmunoResearch). These cells were then reacted with αS-21, which was subsequently indirectly IF labeled by goat anti-rabbit IgG conjugated to AlexaFluor568 (Thermo Fisher Scientific). High specificities of the psbA FISH signal and the PsaA IF signal were demonstrated previously by Uniacke and Zerges, (2007). The IF signals from the antisera against PsbA, Ycf3, and Ycf4 were specific because they were absent in deletion mutants for the respective chloroplastic gene (Supplemental Figure 6).
Microscopy was performed with a Leica DMI6000B inverted epifluorescence microscope with a 63× Plan Apo objective (numerical aperture 1.4) and further magnified by a 1.6× tube lens. Images were acquired on a Hamamatsu Orca R2 C10600-10B camera controlled by Volocity (Improvision) software. Filters used were as follows: Texas Red (562/40 nm excitation, 624/40 nm emission) for protein labeling and GFP (472/30 nm excitation, 520/35 nm emission) for probed mRNA message psbA. Acquired images were taken using Z plane stacks with a spacing of 0.2 µm per section; exposure settings, gain, and excitation intensity were constant between samples. Deconvolution of IF and FISH signals was performed with AutoQuant X3 software (Bitplane) using settings for the appropriate optics, glycerol-based sample medium, and Prolong Gold Antifade (Molecular Probes). An adaptive point-spread function was applied for deconvolution of 15 iterations, using low background removal for the IF signals and medium background removal for the FISH signal from the psbA mRNA. The same settings were used for the acquisition and deconvolution of each IF and FISH signal in all cell images in each figure panel. However, post acquisition, the IF and FISH signals were adjusted manually to similar brightness across all images in each figure panel to allow comparisons of in situ distributions, which would have been precluded by different signal levels between time points.
Chlorophyll fluorescence was imaged by embedding cells in 3% (w/v) low-melting-point agarose (Bethesda Research Laboratories), made with TAP medium upon a 35-mm cell culture imaging dish (Grenier Bio-one). This gel was submerged in TAP medium to prevent it from drying. Images were acquired using the same microscope, peripheral equipment, software, and settings described above and a CY5 filter cube (628/40 nm excitation, 692/40 nm emission). For time-lapse movies, dark-y1 cells were subjected to 100 µmol m−2 s−1 white light from a 100-W halogen bulb for the 8-h duration of the experiment. Images were captured every 10 min using a 100× (numerical aperture 1.3) lens on a Nikon Ti microscope fitted with a Photometrics Evolve EMCCD device. Chlorophyll fluorescence was stimulated using a 405-nm LED (Heliphor), passing through a Quad filter cube (405/485/555/640 excitation, 450/520/595/710 emission; Chroma). These images were not deconvolved.
Average Cell Signal Analysis
To determine the average distribution of fluorescent signals in cells at a specific time point, the macro was developed to operate within ImageJ (Supplemental File; Abramoff et al., 2004). The protocol is available at https://www.protocols.io/view/cell-harvester-macro-for-flourescence-microscopy-i-rjgd4jw. The macro is available at https://github.com/Zergeslab/cellHarvester. The macro finds cells in a maximum intensity projection of the Z-stack and fits an ellipse to the outline of each. Each cell is then rotated so that its long axis lay horizontally and the brighter end of the cell (the presumed location of the T-zone) was at the right; this is verified by the user. Each cell is then scaled in X and Y so that all cells were the same size, and their fluorescence signals are normalized to their own maximum intensity so that each cell contributed equally to the final image. A mean intensity projection is then performed on all of the normalized cells, resulting in the image in Supplemental File.
TEM
Samples were collected from cultures entrained to the diurnal cycle as described above and processed as described previously, with one additional step after infiltration: embedding with Epon and polymerization at 68°C for 48 h (Elimam et al., 2016). Images were acquired on an FEI Tecnai 12 120kv transmission electron microscope using the Tecnai User Interface software and an AMTv601 CCD camera. Settings used were an aperture of 3, a spot size of 2, and variable magnifications ranging from 2900× to 68,000×.
Measurement of Chlorophyll
Chlorophyll extractions were quantified spectrophotometrically as described previously by Porra, (2002).
Accession Numbers
The fully annotated Chlamydomonas chloroplast genome sequence is available from GenBank (accession number BK000554) for the following genes mentioned in this study: atpB, psaA, psbA, psbC, psbD, S-21/rps14 (Randolph-Anderson et al., 1989), ycf3, and ycf4. Accession numbers (Phytozome v12.1; https://phytozome.jgi.doe.gov/pz/portal.html#) for nuclear genes mentioned in this study are as follows: HSP70B (Cre06.g250100), RPLL7/L12 (Cre13.g581650), LHCB family (LHCBM1, Cre01.g066917; LHCBM2, Cre12.g548400; and LHCBM7, Cre12.g548950) PORA (Cre01.g015350), RBP40 (also known as RB38, Cre12.g483700), and VIPP1 (Cre13.g583550).
Supplemental Data
Supplemental Figure 1. Cultures entrained to the 12:12 h light-dark cycle had cells that were synchronized in growth during the light phase and mitosis during the dark phase.
Supplemental Figure 2. Steady state levels and synthesis rates of marker subunits.
Supplemental Figure 3. Bright field images of the cells in the figures reveal their orientations.
Supplemental Figure 4. TEM images reveal ultrastructures of cells from time points of the diel cycle when the T-zone colocalization of translation makers was seen.
Supplemental Figure 5. Temporal patterns of protein synthesis in the chloroplast of the y1 mutant before, during and after greening.
Supplemental Figure 6. Controls for background and signal specificities in the fluorescence microscopy images.
Supplemental Video. Live cell imaging of chlorophyll fluorescence during y1 greening.
Supplemental File. Protocol for determining average fluorescent signal intensity along the axis of the chloroplast with the macro.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
All light microscopy was carried out at the Concordia Center for Microscopy and Cell Imaging. For generous gifts of antibodies, we thank Kevin Redding (αPsaA; Arizona State University); Yves Choquet and Francis-Andre Wollman (αcytf and αLHCII; IBPC, Paris); Elizabeth Harris (αAtpB, αcyL4, αS-21, and αL7/L12; Duke University); Katrin Philippar (αPOR; Ludwig Maximilian University, Munich); Michael Schroda (αHSP70B and αVIPP1; University of Kaiserslautern), Jörg Nickelsen (αRBP40; Ludwig Maximilian University, Munich); and Yuichiro Takahashi (αYcf3 and αYcf4 as well as the ycf3 and ycf4 mutants; Okayama University). For critical reading of the article, we thank James Dhaliwal, Patrick Gulick, Ursula Oberholzer, and Alisa Piekny. For technical assistance and support, we thank Kelly Sears and Jeannie Mui and the Faculty for Electron Microscopy Research at McGill University. This work was supported by the Natural Sciences and Engineering Research Council of Canada (Discovery Grant 217566 to W.Z.).
AUTHOR CONTRIBUTIONS
Y.S., M.V.-P., and W.Z. designed the research; Y.S., M.V.-P., S.B., and Y.Z. performed the research; M.V.-P. and C.L. contributed new analytic and computational tools; Y.S., M.V.-P., S.B., and W.Z. analyzed data; Y.S., M.V.-P., and W.Z. wrote the article.
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. (2004). Image processing with ImageJ. Biophoton. Int. 11: 36–42. [Google Scholar]
- Allen M.D., Kropat J., Merchant S.S. (2008). Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem. Photobiol. 84: 1336–1342. [DOI] [PubMed] [Google Scholar]
- Barnes D., Cohen A., Bruick R.K., Kantardjieff K., Fowler S., Efuet E., Mayfield S.P. (2004). Identification and characterization of a novel RNA binding protein that associates with the 5′-untranslated region of the chloroplast psbA mRNA. Biochemistry 43: 8541–8550. [DOI] [PubMed] [Google Scholar]
- Cahoon A.B., Timko M.P. (2000). yellow-in-the-dark mutants of Chlamydomonas lack the CHLL subunit of light-independent protochlorophyllide reductase. Plant Cell 12: 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey J.W., Linhartová M., Komenda J., Jackson P.J., Dickman M.J., Canniffe D.P., Koník P., Pilný J., Hunter C.N., Sobotka R. (2014). A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Barkan A. (2016). Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 12: e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Barkan A. (2018). Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet. 14: e1007555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Ramos D.A., Salisbury J.L., Sanders M.A., Shenoy S.M., Singer R.H., García-Blanco M.A. (2003). Asymmetric distribution of nuclear pore complexes and the cytoplasmic localization of β2-tubulin mRNA in Chlamydomonas reinhardtii. Dev. Cell 4: 941–952. [DOI] [PubMed] [Google Scholar]
- Czarnecki O., Grimm B. (2012). Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 63: 1675–1687. [DOI] [PubMed] [Google Scholar]
- de Vitry C., Olive J., Drapier D., Recouvreur M., Wollman F.A. (1989). Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109: 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M., Archibald J.M. (2009). Sizing up the genomic footprint of endosymbiosis. BioEssays 31: 1273–1279. [DOI] [PubMed] [Google Scholar]
- Elimam H., Papillon J., Kaufman D.R., Guillemette J., Aoudjit L., Gross R.W., Takano T., Cybulsky A.V. (2016). Genetic ablation of calcium-independent phospholipase A2γ induces glomerular injury in mice. J. Biol. Chem. 291: 14468–14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B.D., Schaffer M., Kuhn Cuellar L., Villa E., Plitzko J.M., Baumeister W. (2015). Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4: e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.M., Rahire M., Rochaix J.D. (1984). Chlamydomonas reinhardii gene for the 32 000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 3: 2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming G.H., Boynton J.E., Gillham N.W. (1987). Cytoplasmic ribosomal proteins from Chlamydomonas reinhardtii: Characterization and immunological comparisons. Mol. Gen. Genet. 206: 226–237. [DOI] [PubMed] [Google Scholar]
- Freeman Rosenzweig E.S., et al. (2017). The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171: 148–162.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use.. (San Diego, CA: Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Herrin D.L., Battey J.F., Greer K., Schmidt G.W. (1992). Regulation of chlorophyll apoprotein expression and accumulation: Requirements for carotenoids and chlorophyll. J. Biol. Chem. 267: 8260–8269. [PubMed] [Google Scholar]
- Hoober J.K. (2007). Chloroplast development: Whence and whither In The Structure and Function of Plastids, Wise R.R.H.J.K., ed (Dordrecht, The Netherlands: Springer; ), pp. 7–55. [Google Scholar]
- Howell S.H., Posakony J.W., Hill K.R. (1977). The cell cycle program of polypeptide labeling in Chlamydomonas reinhardtii. J. Cell Biol. 72: 223–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S.C., Simmonds A.J. (2019). Drosophila mRNA localization during later development: Past, present, and future. Front. Genet. 10: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idoine A.D., Boulouis A., Rupprecht J., Bock R. (2014). The diurnal logic of the expression of the chloroplast genome in Chlamydomonas reinhardtii. PLoS One 9: e108760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagendorf A.T., Michaels A. (1990). Rough thylakoids: Translation on photosynthetic membranes. Plant Sci. 71: 137–145. [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2009). Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2: 1154–1180. [DOI] [PubMed] [Google Scholar]
- Kannangara C.G., Vothknecht U.C., Hansson M., von Wettstein D. (1997). Magnesium chelatase: Association with ribosomes and mutant complementation studies identify barley subunit Xantha-G as a functional counterpart of Rhodobacter subunit BchD. Mol. Gen. Genet. 254: 85–92. [DOI] [PubMed] [Google Scholar]
- Lee J., Herrin D.L. (2002). Assessing the relative importance of light and the circadian clock in controlling chloroplast translation in Chlamydomonas reinhardtii. Photosynth. Res. 72: 295–306. [DOI] [PubMed] [Google Scholar]
- Legen J., Schmitz-Linneweber C. (2017). Stable membrane-association of mRNAs in etiolated, greening and mature plastids. Int. J. Mol. Sci. 18: E1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Willmund F., Whitelegge J.P., Hawat S., Knapp B., Lodha M., Schroda M. (2005). J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell 16: 1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J., Castelli L.M., Pizzinga M., Simpson C.E., Hoyle N.P., Bailey K.L., Campbell S.G., Ashe M.P. (2014). Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 9: 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoë P., Mayfield S.P., Rochaix J.D. (1988). Comparative analysis of the biogenesis of photosystem II in the wild-type and Y-1 mutant of Chlamydomonas reinhardtii. J. Cell Biol. 106: 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo A.K., Edelman M. (1987). Intramembrane translocation and posttranslational palmitoylation of the chloroplast 32-kDa herbicide-binding protein. Proc. Natl. Acad. Sci. USA 84: 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa J., Takahashi Y. (2004). Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth. Res. 82: 241–263. [DOI] [PubMed] [Google Scholar]
- Müller P., Li X.-P., Niyogi K.K. (2001). Non-photochemical quenching: A response to excess light energy. Plant Physiol. 125: 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellaepalli S., Ozawa S.I., Kuroda H., Takahashi Y. (2018). The photosystem I assembly apparatus consisting of Ycf3-Y3IP1 and Ycf4 modules. Nat. Commun. 9: 2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N., Ben-Shem A. (2004). The complex architecture of oxygenic photosynthesis. Nature Reviews Molecular Cell Biology 5: 971–982. [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Zerges W. (2013). Thylakoid biogenesis has joined the new era of bacterial cell biology. Front. Plant Sci. 4: 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I., Siekevitz P., Palade G.E. (1967a). Biogenesis of chloroplast membranes. I. Plastid dedifferentiation in a dark-grown algal mutant (Chlamydomonas reinhardi). J. Cell Biol. 35: 521–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I., Siekevitz P., Palade G.E. (1967b). Biogenesis of chloroplast membranes. II. Plastid differentiation during greening of a dark-grown algal mutant (Chlamydomonas reinhardi). J. Cell Biol. 35: 553–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasenko O.O., et al. (2019). Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol. 26: 110–120. [DOI] [PubMed] [Google Scholar]
- Porra R.J. (2002). The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73: 149–156. [DOI] [PubMed] [Google Scholar]
- Randolph-Anderson B.L., Gillham N.W., Boynton J.E. (1989). Electrophoretic and immunological comparisons of chloroplast and prokaryotic ribosomal proteins reveal that certain families of large subunit proteins are evolutionarily conserved. J. Mol. Evol. 29: 68–88. [DOI] [PubMed] [Google Scholar]
- Rast A., Heinz S., Nickelsen J. (2015). Biogenesis of thylakoid membranes. Biochim. Biophys. Acta 1847: 821–830. [DOI] [PubMed] [Google Scholar]
- Rochaix J.D., Dron M., Rahire M., Malnoe P. (1984). Sequence homology between the 32K dalton and the D2 chloroplast membrane polypeptides of Chlamydomonas reinhardii. Plant Mol. Biol. 3: 363–370. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. (2001). Molecular Cloning: A Laboratory Manual.. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schottkowski M., Peters M., Zhan Y., Rifai O., Zhang Y., Zerges W. (2012). Biogenic membranes of the chloroplast in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 109: 19286–19291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C., Elles I., Kortmann J., Piotrowski M., Nickelsen J. (2007). Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19: 3627–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S. (2019). Organelle zones in mitochondria. J. Biochem. 165: 101–107. [DOI] [PubMed] [Google Scholar]
- Solymosi K., Schoefs B. (2010). Etioplast and etio-chloroplast formation under natural conditions: The dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 105: 143–166. [DOI] [PubMed] [Google Scholar]
- Stoldt S., Wenzel D., Kehrein K., Riedel D., Ott M., Jakobs S. (2018). Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat. Cell Biol. 20: 528–534. [DOI] [PubMed] [Google Scholar]
- Strenkert D., et al. (2019). Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. USA 116: 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zerges W. (2015). Translational regulation in chloroplasts for development and homeostasis. Biochim. Biophys. Acta 1847: 809–820. [DOI] [PubMed] [Google Scholar]
- Theis J., Schroda M. (2016). Revisiting the photosystem II repair cycle. Plant Signal. Behav. 11: e1218587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J., Colón-Ramos D., Zerges W. (2011). FISH and immunofluorescence staining in Chlamydomonas. Methods Mol. Biol. 714: 15–29. [DOI] [PubMed] [Google Scholar]
- Uniacke J., Zerges W. (2007). Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 19: 3640–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J., Zerges W. (2009). Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc. Natl. Acad. Sci. USA 106: 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk K.J., Andersson B., Aro E.M. (1996). Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem. 271: 9627–9636. [DOI] [PubMed] [Google Scholar]
- White R.A., Hoober J.K. (1994). Biogenesis of thylakoid membranes in Chlamydomonas reinhardtii y1 (A kinetic study of initial greening). Plant Physiol. 106: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Carvalho P., Voeltz G.K. (2018). Here, there, and everywhere: The importance of ER membrane contact sites. Science 361: eaan5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones J.M., Blaby I.K., Merchant S.S., Umen J.G. (2015). High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Barkan A. (2015). Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc. Natl. Acad. Sci. USA 112: E1678–E1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Bock R. (2018). Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell 30: 745–770. [DOI] [PMC free article] [PubMed] [Google Scholar]