Arabidopsis ORANGE, a DnaJ-like zinc finger domain protein, represses chloroplast biogenesis in dark-germinated cotyledons by interacting with the transcription factor TCP14 in the nucleus.
Abstract
The conversion of etioplasts into chloroplasts in germinating cotyledons is a crucial transition for higher plants, enabling photoautotrophic growth upon illumination. Tight coordination of chlorophyll biosynthesis and photosynthetic complex assembly is critical for this process. ORANGE (OR), a DnaJ-like zinc finger domain-containing protein, was reported to trigger the biogenesis of carotenoid-accumulating plastids by promoting carotenoid biosynthesis and sequestration. Both nuclear and plastidic localizations of OR have been observed. Here, we show that Arabidopsis (Arabidopsis thaliana) OR physically interacts with the transcription factor TCP14 in the nucleus and represses its transactivation activity. Through this interaction, the nucleus-localized OR negatively regulates expression of EARLY LIGHT-INDUCIBLE PROTEINS (ELIPs), reduces chlorophyll biosynthesis, and delays development of thylakoid membranes in the plastids of germinating cotyledons. Nuclear abundance of OR decreased upon illumination. Together with an accumulation of TCP14 in the nucleus, this derepresses chloroplast biogenesis during de-etiolation. TCP14 is epistatic to OR and expression of ELIPs is directly regulated by the binding of TCP14 to Up1 elements in the ELIP promoter regions. Our results demonstrate that the interaction between OR and TCP14 in the nucleus leads to repression of chloroplast biogenesis in etiolated seedlings and provide new insights into the regulation of early chloroplast development.
INTRODUCTION
Chloroplasts are the specialized organelles in plant cells in which photosynthesis occurs. In higher plants, chloroplasts develop following illumination via the active synthesis of pigments and proteins of photosystems, assembly of the thylakoid membrane network (Kobayashi et al., 2012), and proper incorporation of the pigments and photosystems into these networks. Because chlorophylls are essential for light harvesting, plants must precisely coordinate their synthesis with their incorporation into photosystems to avoid the accumulation of free chlorophylls, which are strong photosensitizers that produce highly active singlet oxygen and cause photodamage in the light. It has been reported that both red and blue photoreceptors and numerous transcription factors regulate chlorophyll biosynthesis in germinating cotyledons (Sullivan and Deng, 2003; Huq et al., 2004; Waters et al., 2009). For example, PHYTOCHROME INTERACTING FACTOR1 (PIF1) and PIF3 both negatively regulate the expression of glutamyl tRNA reductase (HEMA1), an enzyme required for tetrapyrrole biosynthesis (Stephenson et al., 2009). By contrast, Golden2-Like activates the expression of HEMA1 and genes for Mg-chelatase subunit ChlH and chlorophyllide a oxygenase in the tetrapyrrole pathway in the presence of light (Waters et al., 2009). Furthermore, ELONGATED HYPOCOTYL5, REVEILLE1, CIRCADIAN CLOCK ASSOCIATED1, ETHYLENE-INSENSITIVE3, and DELLAs, together with PIF1, were found to regulate the expression of the gene encoding NADPH:protochlorophyllide oxidoreductase (POR), which catalyzes the conversion of protochlorophyllide (Pchlide) to chlorophyllide (Chlide; Yuan et al., 2017).
Chloroplast biogenesis in dark-germinated cotyledons has been intensively studied as part of the de-etiolation process that enables the juvenile seedlings to grow photoautotrophically (Mochizuki et al., 1996; Pogson and Albrecht, 2011; Rudowska et al., 2012; Pogson et al., 2015). Different from true leaves in which chloroplasts directly develop from proplastids, dark-germinated cotyledons have a special intermediate type of plastids termed etioplasts. Etioplasts may be regarded as a checkpoint stage in preparation for immediate chlorophyll synthesis and photosynthetic competence once the seedlings emerge from the soil into the light (Sundqvist and Dahlin, 1997). In dark-germinated cotyledons, etioplasts accumulate both carotenoids and Pchlide in special internal membranous structures known as prolamellar bodies (PLBs; Jarvis and López-Juez, 2013).
Upon illumination, light triggers the conversion of Pchlide into chlorophylls on the picosecond timescale and the subsequent assembly of photosystems when chlorophylls are available (Oliver and Griffiths, 1982; Paulsen, 1997; Sytina et al., 2008). However, a rapid chlorophyll synthesis also increases the probability of photodamage by free chlorophylls before sufficient photosynthetic proteins are available for their incorporation. It was found that during the transition from etioplasts to chloroplasts, EARLY LIGHT-INDUCIBLE PROTEINS (ELIPs) that share striking structural similarities with chlorophyll a/b binding proteins (CABs) in light-harvesting complexes (LHCs) accumulate (Kolanus et al., 1987; Grimm et al., 1989). ELIPs temporarily bind free chlorophylls and then are replaced by CABs for LHCs when CABs are synthesized (Casazza et al., 2005). Different studies have revealed that the expression of ELIPs is affected by various factors, including the repression by the COP9 signalosome in dark and the induction by ELONGATED HYPOCOTYL5 in light, both of which are essential components in regulating chloroplast biogenesis during de-etiolation (Harari-Steinberg et al., 2001; Hayami et al., 2015).
Previously, we identified a DnaJ-like zinc finger domain-containing protein ORANGE (OR) from an orange curd cauliflower (Brassica oleracea var botrytis) mutant (Lu et al., 2006). OR is highly conserved in all land plants. Ectopic overexpression of Arabidopsis (Arabidopsis thaliana) OR, At-OR (also known as AtOR), triggers the development of nonpigmented plastids into chromoplasts (Lopez et al., 2008; Yuan et al., 2015) and enhances carotenoid accumulation in a number of plant species (Li et al., 2012; Park et al., 2015; Bai et al., 2016; Berman et al., 2017). This makes OR a useful gene for carotenoid enhancement in food crops (Giuliano and Diretto, 2007; Cazzonelli and Pogson, 2010; Sun et al., 2018). OR is localized in both chloroplasts and nuclei (Zhou et al., 2011, 2015; Kim et al., 2013; Sun et al., 2016). In chloroplasts, it interacts with phytoene synthase (PSY), a key enzyme for carotenoid biosynthesis, and posttranscriptionally regulates PSY protein level and catalytic activity (Zhou et al., 2015; Welsch et al., 2018). In the nucleus, OR was reported to interact with eukaryotic release factor eRF1-2 to regulate petiole development (Zhou et al., 2011). Recently, we demonstrated that OR predominantly localizes in the nucleus in etiolated cotyledons of germinating Arabidopsis seedlings and that protein abundance decreases upon illumination. This suggests a yet unknown function of OR during germination (Sun et al., 2016).
TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) transcription factors modulate a number of cellular and developmental processes, including branching, leaf shape, and seedling development (Tatematsu et al., 2008; Martín-Trillo and Cubas, 2010; Kieffer et al., 2011). All TCP proteins share a common TCP domain that is predicted to form a noncanonical basic helix-loop-helix (bHLH) structure for protein or DNA binding (Aggarwal et al., 2010). Their transcriptional activities are frequently found to be regulated by their protein partners in a context-depending fashion. For example, TCP14 interacts with and is modified by the O-linked N-acetylglucosamine transferase SPINDLY in response to cytokinins (Steiner et al., 2012). It interacts with the DELLA proteins GA-INSENSITIVE (GAI) and REPRESSOR OF GA1-3 (RGA) in response to gibberellins (Davière et al., 2014; Resentini et al., 2015). It also partners with the ubiquitin receptors DA1 and DA1-related proteins (DAR1 and DAR2) in the regulation of endoreduplication during leaf development (Peng et al., 2015). Moreover, TCP14 specifically regulates the embryo potential by inducing the expression of germination-associated genes in vascular tissues in germinating seeds (Tatematsu et al., 2008).
Here, we report that OR physically and genetically interacts with TCP14 in the nucleus of germinating Arabidopsis cotyledons. We demonstrate that such interactions regulate the expression of ELIPs and control the development from etioplasts to chloroplasts during de-etiolation.
RESULTS
OR Negatively Regulates Cotyledon Greening in Arabidopsis
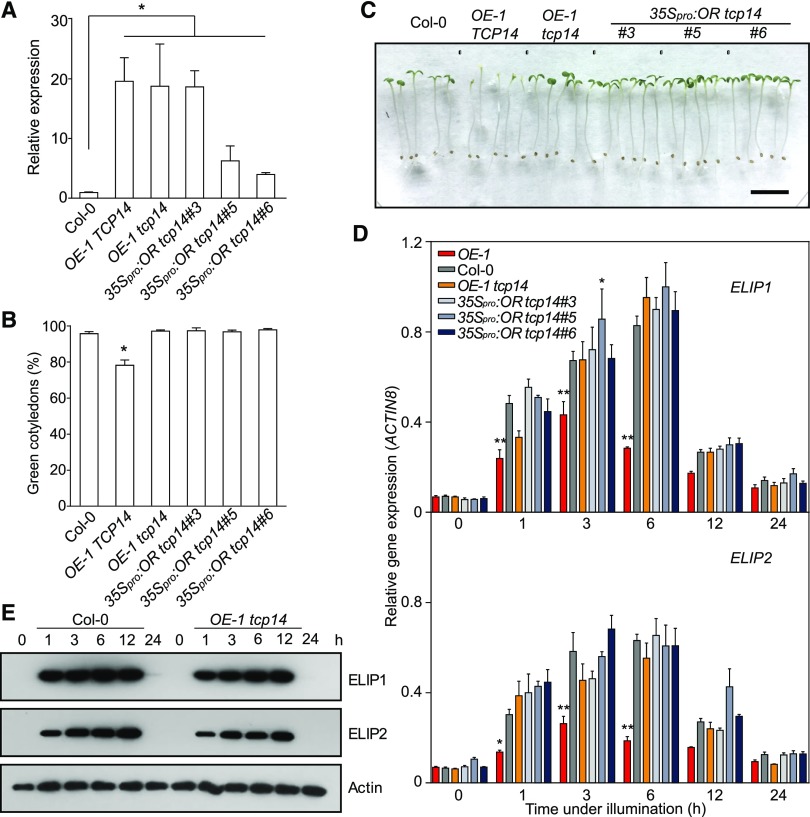
Cotyledon greening involves both pigment biosynthesis and the assembly of photosynthetic machinery into the developing thylakoid network. The greening rate is an index for assessing the potential of chloroplast biogenesis during germination (Park et al., 2002; Rosso et al., 2009). To examine the functions of OR in developing cotyledons, we generated transgenic Arabidopsis plants with OR either overexpressed (OE) or silenced by RNA interference (RNAi). We also obtained a T-DNA insertion line (or-1) from GABI-Kat (Supplemental Figure 1). When these lines were germinated in the dark for 4 d and then exposed to growth light for 12 h, OE seedlings developed significantly lower percentages of green cotyledons compared with the Columbia (Col-0) wild-type and OR-silencing plants (Figures 1A and 1B; Supplemental Figure 2). Among OE plants, the lines with relatively higher OR expression levels were more severely prevented from turning green as shown for OE-1 and OE-2 (Figure 1B; Supplemental Figures 1 and 2).
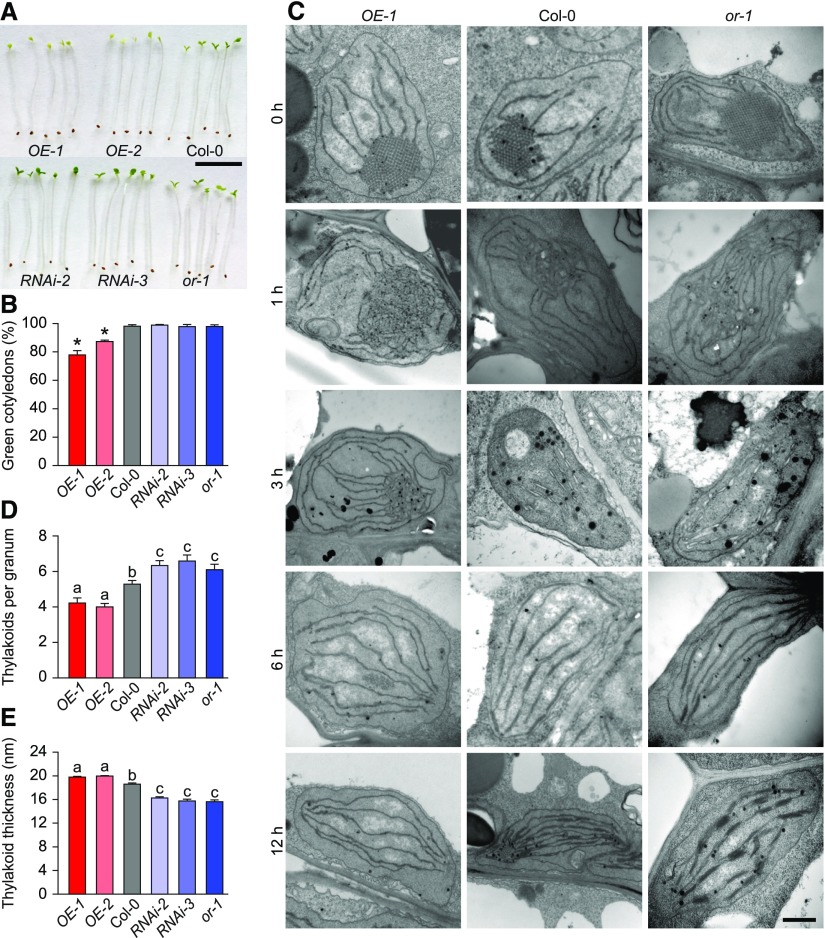
Figure 1.
OR Regulates the Greening of Cotyledons.
(A) Representative seedlings with different OR expression levels. Photographs show seedlings of OR-overexpressing (OE) and OR-silencing (RNAi) lines, Col-0, and the or-1 mutant germinated in darkness for 4 d and transferred to light for 12 h. Bar = 10 mm.
(B) Percentages of green cotyledons in seedlings with different OR expression levels. Seedlings of the same lines used in (A) were germinated in darkness for 4 d and then illuminated for 12 h. For each measurement, at least three plates with more than 100 seedlings per plate were counted. Data are means ± se (n > 3). Asterisks indicate significant differences between the transgenic lines and Col-0 (Student’s t test, P < 0.05).
(C) Electron micrographs of cotyledon plastids of 4-d-old etiolated seedlings of OE-1, Col-0, and the or-1 mutant. Images were taken during the transition from dark (0 h) to light conditions for 1, 3, 6, and 12 h. Bar = 200 nm.
(D) and (E) Number of thylakoids per granum (D) and average thickness of granal thylakoid (E) in cotyledon chloroplasts of OE, RNAi, Col-0, and the or-1 mutant. Values were calculated by dividing the height of each granum by thylakoid number. At least 20 grana in five chloroplasts from two independent cotyledon sections were measured. Data are means ± se. Letters above bars represent significance groups as determined by the Newman–Keuls multiple comparison test, P < 0.05 or better.
To help elucidate the nature of the greening process in these plants, we observed the ultrastructure of plastids using transmission electron microscopy (TEM). Before illumination, etioplasts from cotyledons of all lines contained PLBs and unstacked membranes (Figure 1C; Supplemental Figure 3). Illumination triggered chloroplast biogenesis in all lines, but it did so at different rates. In both wild-type and OR-silencing lines, PLB sizes were significantly reduced in 1 h. When prothylakoid membranes were observed in the wild type and OR-silencing lines after 3 h of illumination, residual PLBs were still visible in the OE lines. Noticeable stacking of grana thylakoids was observed in plastids of both the wild type and OR-silencing lines after 6 h of illumination, while granal stacking was not found in the OE lines. After a 12-h illumination, both the stroma and granal thylakoids were best developed in the chloroplasts of OR-silencing lines, followed by the wild type and then OE lines (Figure 1C; Supplemental Figure 3). Compared with those in the wild-type cotyledons, chloroplasts in the OE lines had fewer stacks of loosely packed granal thylakoids, whereas chloroplasts in the OR-silencing lines had more tightly organized granal thylakoids (Figures 1D and 1E).
Cotyledons of all different lines accumulated no chlorophylls before illumination. After a 12-h growth under light, both chlorophyll a/b were synthesized. In comparison with the wild type, chlorophyll a/b levels were significantly lower in cotyledons of OE and higher in the OR-silencing lines (Figure 2A), consistent with the greening phenotypes of these lines. Cotyledons of all these lines accumulated similar amounts of total carotenoids after the 12-h illumination (Figure 2A). However, our HPLC analysis revealed that OE lines accumulated more lutein, but less β-carotene, than the wild type after the illumination (Figure 2A; Supplemental Figure 4). In our preliminary analysis, we compared the expression of genes in the chlorophyll biosynthesis pathway in the wild type and OE lines. While most of the genes examined had similar expression levels, the transcript abundance of PORB was found to be lower in the OE lines than in the wild type after a 12-h illumination (Supplemental Figure 5A). However, our detailed quantification demonstrated that its expression was not significantly different among all the lines at different time points after illumination (Supplemental Figure 5B). It was reported that illumination represses the accumulation of PORB (Reinbothe et al., 2010), and consistent with this report, our immunoblot analysis showed that PORB protein levels were rapidly reduced in both the wild-type and OE seedlings after a 1-h illumination (Figure 2C). A slightly lower protein abundance of PORB and a higher level accumulation of its substrate Pchlide were observed in the OE lines than in the wild type (Figures 2C and 2D).
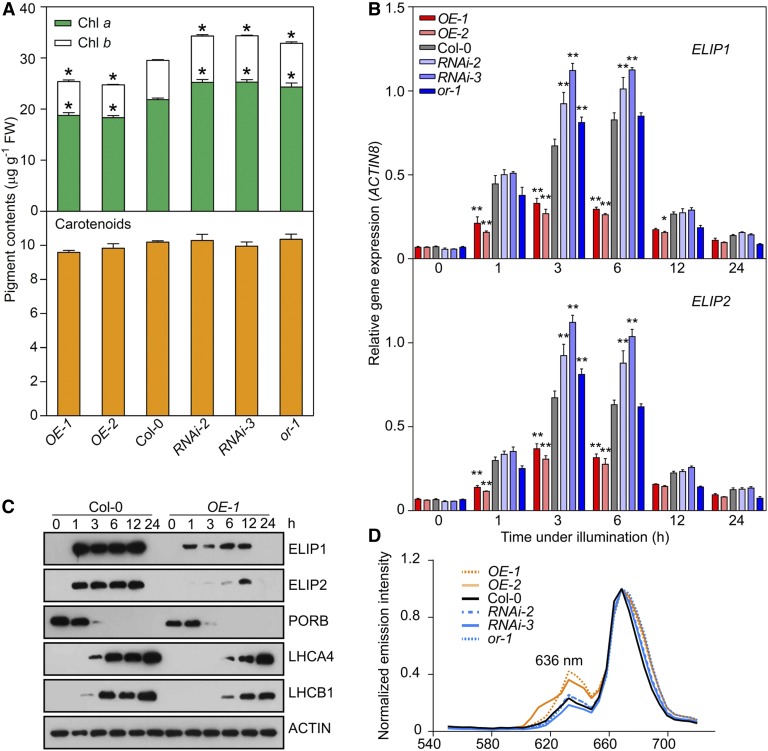
Figure 2.
OR Regulates the Biosynthesis of Chlorophylls and ELIPs.
(A) Chlorophyll and carotenoid contents in OR-overexpressing (OE) and OR-silencing (RNAi) lines, Col-0, and the or-1 mutant. Seedlings were germinated in dark for 4 d and then illuminated for 12 h. Data are means ± se (five pools of seedlings from separate plates were used). Asterisks indicate significant differences between the transgenic lines and Col-0 (Student’s t test, P < 0.05). FW, fresh weight.
(B) Expression of ELIP1 and ELIP2 during the transition from dark (0 h) to light conditions for 1, 3, 6, 12, and 24 h. Transcript abundance of each gene was quantified by qPCR. Relative expression was calculated as the ratio between the transcript abundance of the gene studied and that of ACTIN8 in the same sample. Data are means ± se (five pools of seedlings from separate plates were used). Asterisks indicate significant differences between the transgenic lines and Col-0 (two-way ANOVA followed by Dunnett’s multiple comparison test, *P < 0.05, **P < 0.01 or better).
(C) Immunoblot analysis showing the abundances of ELIPs, PORB, LHCA4, and LHCB1 in Col-0 and OR overexpression line OE-1 during the transition from dark (0 h) to light conditions for 1, 3, 6, 12, and 24 h. ACTIN was probed as a loading control.
(D) Fluorescence emission spectra showing relative fluorescence of Pchlide (fluorescence emission maximum at 636 nm) in transgenic lines and Col-0 in planta. For each sample, five cotyledon positions were measured to calculate an average spectrum, and the maximum emission value was set as 1.
Because the assembly of photosynthetic membrane structures in plastids involves the binding of chlorophylls, we also wanted to determine the transcript abundances of genes for ELIPs. Our quantitative real-time PCR (qPCR) analysis showed that both ELIP1 and ELIP2 transcripts were upregulated when dark-germinated seedlings were exposed to the light and then returned to their original levels in 24 h in wild-type cotyledons (Figure 2B). However, stronger induction of ELIP1 and ELIP2 was observed in the OR-silencing lines and weaker expression was found in the OE lines in comparison with the wild type in the first 6 h under illumination (Figure 2B).
We further compared protein abundances of ELIPs during de-etiolation. Immunoblot analysis revealed the accumulation of ELIPs in both wild-type and OE lines. However, their accumulation was at relatively lower levels in the OE lines than in the wild type, consistent with their differences at the transcriptional level (Figure 2C). The immunoblot analysis also demonstrated that the accumulation of both ELIPs occurred prior to that of LHCA4 and LHCB1 (Figure 2C), supporting the postulated function of ELIPs as a structure proxy for binding chlorophylls before the synthesis of CABs and the assembly of LHCs (Casazza et al., 2005). The accumulation of LHCA4 and LHCB1 also occurred at slightly lower abundances in the OE lines than in the wild type (Figure 2C). Taken together, these results demonstrated a negative regulatory role of OR on chloroplast biogenesis.
Nuclear Localized OR Is Dramatically Reduced upon Illumination
OR localized in both chloroplasts and nuclei (Zhou et al., 2011; Kim et al., 2013; Sun et al., 2016). In etiolated cotyledons, the full-length OR (34 kD) is predominantly targeted to the nucleus, with its abundance decreasing upon illumination (Sun et al., 2016). Sequence analysis revealed a twin-arginine (Arg34Arg35) motif at the N terminus of OR (Supplemental Figure 6). This suggests that OR is imported into chloroplast through the TOC/TIC pathway, similar to a vast majority of chloroplast proteins that are encoded by the nuclear genome and translated in cytosol (Robinson and Bolhuis, 2001; Andrès et al., 2010). As such, the N-terminal chloroplast transit peptide (ORcTP) prior to the twin-Arg motif is removed upon import into chloroplast, producing a mature protein (ORΔcTP) of 30 kD. To detect and distinguish the full-length (34-kD) and mature OR proteins, we raised antibodies against either ORcTP or ORΔcTP (Supplemental Figure 7).
We purified nuclei and plastids from cotyledons of 4-d-old dark-germinated seedlings at various time points after illumination and examined the OR protein levels. As shown in Figure 3A, both OR antibodies detected a single band at 34 kD from the nuclear proteins, indicating that only full-length OR was localized in the nucleus. Noticeably, the abundance of OR in the nuclear preparation dramatically decreased after a 24-h illumination, as we reported previously by Sun et al., (2016). By contrast, the antibody against ORΔcTP detected a band at 30 kD that corresponded to the mature OR protein in the plastid protein samples (Figure 3B). Its abundance increased during the progress of illumination (Figure 3B), suggesting an enhanced accumulation of the processed OR in chloroplasts.
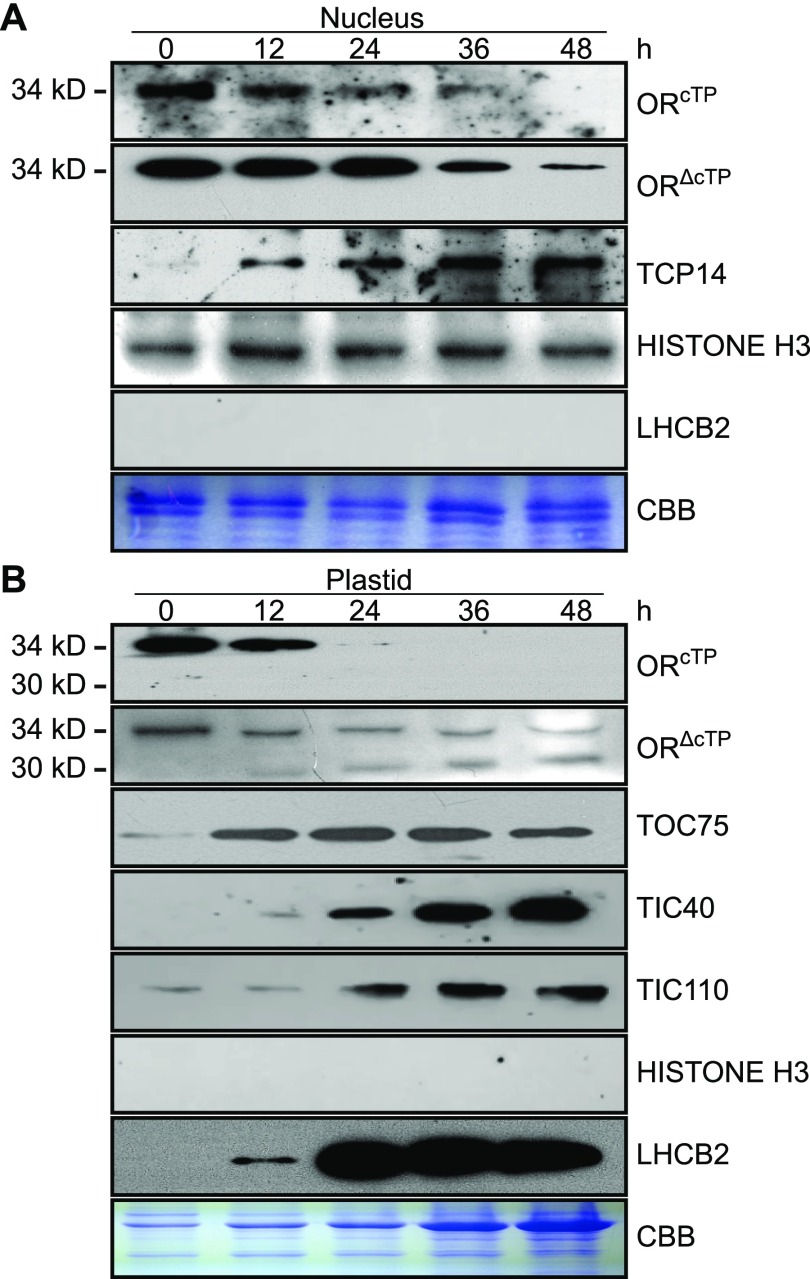
Figure 3.
Nucleus-Localized OR Is Reduced upon Illumination.
(A) and (B) Nuclear (A) and plastid (B) proteins were isolated from 4-d-old dark-germinated seedlings collected at 0, 12, 24, 36, and 48 h after illumination. Antibodies against either the putative transit peptide (ORcTP, Asp15-Ser27) or a peptide fragment beyond the transit peptide (ORΔcTP, Ile155-Thr168) of OR were used to probe both nuclear and plastid protein samples. Antibodies against TCP14, Histone H3, LHCB2, TIC40, TIC110, and TOC75 were also used to determine the levels of these proteins. OR protein masses are indicated on the left side of the panels. Duplicate gels stained by Coomassie Brilliant Blue were used as loading controls. CBB, Coomassie Brilliant Blue.
To our surprise, the full-length OR was also found in our plastid samples. Its abundance decreased under illumination (Figure 3B). When we probed the plastid protein samples with the antibody against HISTONE H3, no band was detected (Figure 3B), indicating this full-length OR was not due to the contamination of nuclear proteins. Indeed, when the antibody against ORcTP was used to probe the plastid samples, the full-length OR was only found in cotyledons up to 12-h illumination but was absent in the late stages (Figure 3B). The absence indicated a relatively rapid transit peptide removal from newly imported OR during chloroplast biogenesis. By examination of TOC75, TIC40, and TIC110 proteins in the plastid samples, we detected the accumulation of these three proteins from 12 to 24 h after illumination, indicating the assembly of the TIC/TOC machinery for chloroplast protein importing (Shi and Theg, 2013). Concomitantly, a rapid and significant accumulation of LHCB2 occurred in cotyledons 24 h after illumination (Figure 3B).
Overexpression of Nucleus-Targeted OR Is Sufficient to Repress De-Etiolation
OR was previously found to localize to the nucleus when its cTP is substituted with green fluorescent protein (GFP; Sun et al., 2016). We expressed this fusion protein (ORnuc) in the or-1 mutant and generated stable transgenic lines (ORnuc). In these lines, the GFP signal was detected only in the nucleus of cotyledon cells (Figure 4A). When we illuminated their dark-germinated seedlings, all lines showed distinctly delayed de-etiolation in comparison with the wild-type seedlings (Figure 4B). Pigment analysis revealed significantly lower chlorophyll contents in cotyledons of the ORnuc lines than in the wild type after a 12-h illumination (Figure 4C). Concomitantly, both ELIP1 and ELIP2 were expressed at lower levels in these transgenic lines than in the wild-type seedlings (Figure 4D), similarly as the OR overexpression lines OE-1 and OE-2 (Figure 2B). Moreover, ORnuc seedlings also accumulated more lutein, but less β-carotene, than the wild type in their cotyledons, although their total carotenoid amounts did not show significant variation from the wild-type level (Figure 4C; Supplemental Figure 8). Since overexpression of ORnuc was sufficient to phenocopy OE-1 and OE-2, we concluded that OR represses chloroplast biogenesis while it is in the nucleus.
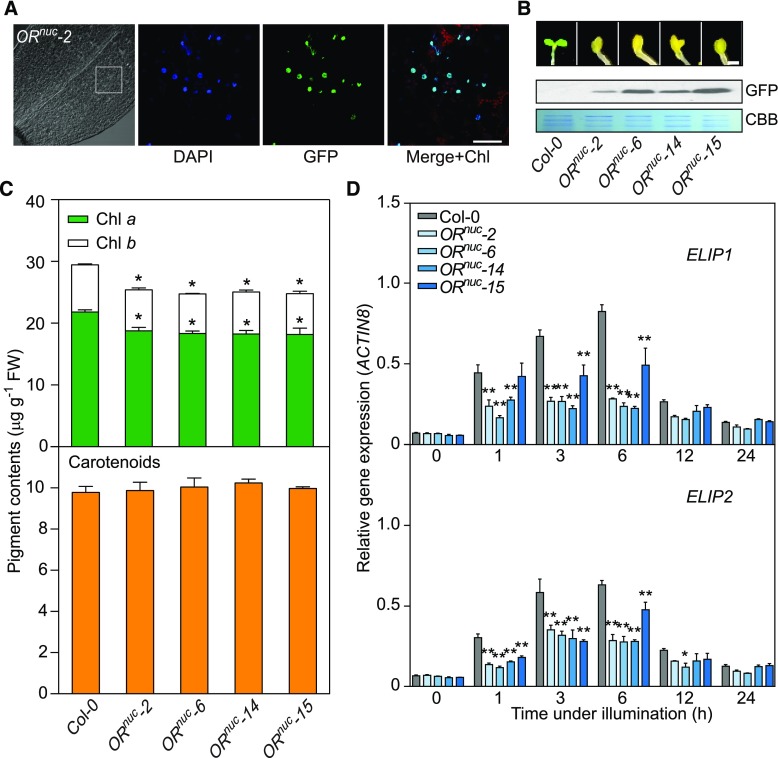
Figure 4.
Overexpression of the Nuclear Targeted OR Phenocopies OR Overexpression Lines.
(A) Nuclear targeting of OR. Localization of OR with its transit peptide substituted by GFP (ORnuc) was analyzed in cotyledons of stable transgenic lines. 4′,6-Diamidino-2-phenylindole stain indicates the nuclei. The 4′,6-diamidino-2-phenylindole and GFP signals were merged with the chlorophyll autofluorescence signal. Bar = 20 μm. Chl, chlorophyll; DAPI, 4′,6-diamidino-2-phenylindole.
(B) De-etiolation of seedlings overexpressing ORnuc. Seedlings were germinated in darkness for 4 d and transferred to light for 12 h. ORnuc abundances in transgenic lines were detected by immunoblot using a specific antibody against GFP. The loading amount was indicated by Coomassie Brilliant Blue stain. Bar = 1 mm. CBB, Coomassie Brilliant Blue.
(C) Chlorophyll and carotenoid contents in ORnuc lines. Seedlings were germinated in darkness for 4 d followed by a 12-h illumination. Data are means ± se (five pools of seedlings from separate plates were used). Asterisks indicate significant differences between the transgenic lines and Col-0 (Student’s t test, P < 0.05).
(D) Expression of ELIP1 and ELIP2 during the transition from dark (0 h) to light conditions for 1, 3, 6, 12, and 24 h. The transcript abundance of each gene was quantified by qPCR. Relative expression was calculated as the ratio between the transcript abundance of the gene studied and that of ACTIN8 in the same sample. Data are means ± se (five pools of seedlings from separate plates were used). Asterisks indicate significant differences between the transgenic lines and Col-0 (two-way ANOVA followed by Dunnett’s multiple comparison test, *P < 0.05, **P < 0.01 or better).
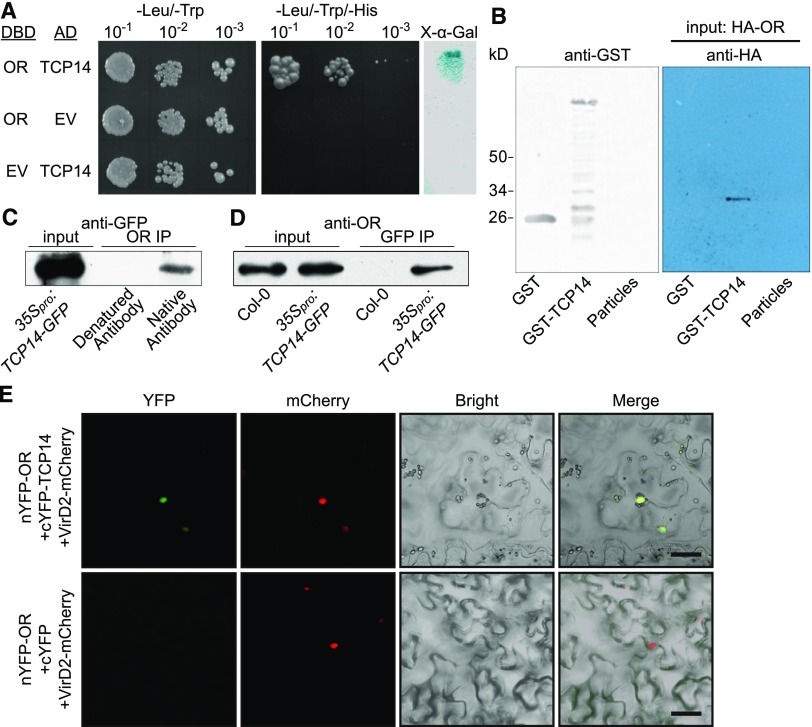
OR Interacts with TCP14 in the Nucleus
To elucidate the regulatory mechanism of the nuclear localized OR in repressing chloroplast biogenesis, we performed yeast two-hybrid library screening using the full-length OR as a bait. A library from a bulk of Arabidopsis seedlings at different stages was used. We identified a bHLH transcription factor TCP14 (At3g47620) that interacted with OR (Figure 5A). To verify this interaction, we performed a pull-down assay after expressing TCP14 as a glutathione S-transferase (GST)-TCP14 fusion protein in bacteria and producing OR with a hemagglutinin (HA)-tag at its N terminus by in vitro transcription and translation. Immunoblot analysis showed that OR was captured by GST-TCP14, but not by GST only or the beads alone (Figure 5B), demonstrating a direct in vitro interaction between OR and TCP14.
Figure 5.
OR Interacts with TCP14 in the Nucleus.
(A) Yeast two-hybrid assay. TCP14 was cloned into pDEST22, which encodes the activation domain, and OR was cloned into pDEST32, which encodes the DNA binding domain (DBD). Yeast AH109 cells were cotransformed with a combination of the indicated plasmids or empty vector and plated onto nonselective (−Leu/−Trp) and selective (−Leu/−Trp/−His) plates. A colony-lifting assay with 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside as the substrate was performed to confirm the interaction (right). AD, activation domain; EV, empty vector; X-α-Gal, 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside.
(B) Pull-down assay. GST-TCP14 fusion protein, GST, and glutathione particles were incubated with HA-OR that was translated in a wheat germ system. The bound protein was eluted, resolved by SDS-PAGE, blotted, and probed with the antibody against the HA tag.
(C) and (D) Co-IP assays using total protein extracted from the 35Spro:TCP14-GFP seedlings. (C) Co-IP using the antibody against ORcTP. The captured proteins were probed with the anti-GFP antibody to detect the presence of TCP14-GFP fusion protein that co-precipitated with OR. Denatured antibody against ORcTP was used as a negative control. (D) Co-IP using the anti-GFP antibody. The captured proteins were probed with the antibody against ORcTP. Col-0 seedlings were used as a negative control.
(E) BiFC observation showing that OR and TCP14 bind each other in the nucleus. C-terminal fragment of yellow fluorescent protein (cYFP) was used as a negative control. The YFP signal (green) and the signal from the nuclear marker VirD2NLS-mCherry (red) indicating the nucleus are shown. Bar = 10 μm. nYFP, N-termnial fragment of YFP.
We further examined the interaction between OR and TCP14 by co-immunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assays. In Co-IP assay, we used the 35Spro:TCP14-GFP transgenic plants. These plants were confirmed to have functional TCP14 in the nucleus that both activates the expression of ARR5 and also results in dark leaves and siliques together with an increased number of trichomes on sepals (Supplemental Figure 9; Steiner et al., 2012, 2016; Wang et al., 2013). When the antibody against ORcTP was used to precipitate full-length OR from the protein extract of the cotyledons, the TCP14-GFP fusion protein was detected by the anti-GFP antibody (Figure 5C). Similarly, when the anti-GFP antibody was used to precipitate TCP14-GFP from the same extract, OR was detected by anti-ORcTP (Figure 5D). In BiFC assay, the full-length OR and TCP14 were transiently expressed as fusion proteins of the N- and C-halves of enhanced yellow fluorescent protein (eYFP), respectively, in tobacco (Nicotiana benthamiana) leaves by infiltration (Citovsky et al., 2006). The reconstituted fluorescence signal was observed in the nucleus and merged with the signal of the nuclear marker VirD2NLS-mCherry (Figure 5E; Lee et al., 2008). These in vivo and in planta experiments confirmed the specific interaction between OR and TCP14 in the nucleus of germinating cotyledon cells.
In Arabidopsis, TCP14 shares the highest sequence identity with TCP15 (Supplemental Figure 10A). However, a previous study reported that TCP14 and TCP15 are not expressed in the same spatiotemporal pattern (Tatematsu et al., 2008). Consistent with this report, we found that TCP15 expressed at a significantly lower level than TCP14 in germinating cotyledons (Supplemental Figure 11). No direct interaction between TCP15 and OR was detected in yeast two-hybrid analysis either (Supplemental Figure 10B); therefore, TCP15 does not seem to be involved in the OR-TCP14 interaction in cotyledons. Moreover, although OR was also found to interact with eRF1-2 in the nucleus, eRF1-2 is predominantly expressed in young leaves and flowers (Zhou et al., 2011), and we could not detect its expression in cotyledons (Supplemental Figure 11). There appears to be no interplay between eRF1-2 and TCP14 or between eRF1-2 and OR during cotyledon greening.
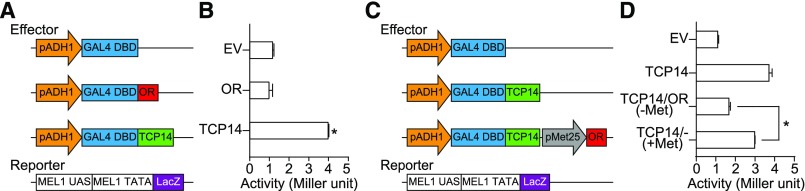
OR Represses the Transcriptional Activity of TCP14 through Protein–Protein Interaction
To examine whether OR and TCP14 had transcriptional activities, we performed an assay using a GAL4-responsive system in yeast strain AH109 (Friedman et al., 2004). The full-length open reading frame (ORF) of OR TCP14 was fused to the DNA binding domain (DBD) of GAL4 in pDEST32 as an effector, and the empty vector was used as a negative control. The LacZ gene driven by a fusion of the GAL4 upstream activating sequence and a minimal promoter (MEL1 TATA) in AH109 was used as a reporter (Figure 6A). Quantification of the β-galactosidase activity revealed the transcriptional activity only for TCP14 (Figure 6B). We then tested whether OR affected TCP14 activity. We used the pBridge vector to constitutively express TCP14 driven by the ADH1 promoter and to conditionally express OR under the control of the MET25 promoter. Therefore, OR was only expressed in a medium without Met (Figure 6C). We saw a significant reduction in β-galactosidase activity when OR was coexpressed with TCP14 in Met-free medium than in medium with Met to repress OR expression (Figure 6D). These results suggest that OR represses the transcriptional activity of TCP14.
Figure 6.
OR Represses the Transactivation Activity of TCP14.
(A) and (B) Transactivation activity assay for OR and TCP14. (A) As effectors, either the OR or the TCP14 full-length ORFs were fused to DNA encoding the GAL4-DBD in the pDEST32 vector. GAL4pro:LacZ in the yeast strain AH109 was used as a reporter. (B) β-Galactosidase activities were quantified (in Miller units) to determine the activation of the GAL4pro:LacZ reporter gene by OR and TCP14, separately. Yeast transformed with empty pDEST32 vector served as a negative control. Three independent transformants were measured for each construct. Data are means ± se. The asterisk indicates a significant difference between TCP14 and the empty vector control (Student’s t test, P < 0.05). EV, empty vector.
(C) and (D) Transactivation activity assay for the OR and TCP14 interaction. (C) TCP14 was fused to DBD in pBridge as the positive control (TCP14), and then OR was cloned to this vector driver by the MET25 promoter (TCP14+OR), which is suppressed by Met in the growth medium. TCP14, TCP14+OR, and the empty pBridge vector were used as effectors. The GAL4pro:LacZ in the yeast strain AH109 was used as a reporter. (D) β-Galactosidase activity was quantified in yeast cells transformed with the empty vector, TCP14, and TCP14+OR (with and without 1 mM Met), separately. Three independent transformants were measured for each construct. Data are means ± se. The asterisk indicates a significant difference between the TCP14+OR transformed yeast cultures with and without Met (Student’s t test, P < 0.05). EV, empty vector.
TCP14 Binds to the Up1 Elements in ELIP1 and ELIP2 Promoters
The Up1 element (GGCCCAWW) is over-represented in the promoter regions of germination-related genes that are regulated by TCP14 (Tatematsu et al., 2008). We identified this element in the upstream promoter regions of both ELIP1 and ELIP2. To determine whether TCP14 directly bound to the Up1 element, an electrophoretic mobility shift assay (EMSA) was performed. A biotin-labeled probe containing 4× Up1 elements was incubated with the affinity-purified GST-TCP14 fusion protein. Unlabeled probe at increasing concentrations was used as a competitor. As shown in Figure 7A, shifted bands were observed when the labeled DNA probe was mixed with the GST-TCP14 fusion protein, indicating a binding of the protein to the Up1 element. By contrast, no shifted band was detected when GST alone was used. The decreased binding capability with higher competitor concentrations demonstrated that TCP14 specifically binds to the Up1 element in vitro.
Figure 7.
TCP14 Directly Binds to the Promoter Regions of ELIP1 and ELIP2.
(A) EMSA confirming the binding of TCP14 to the Up1 element. Heterologously expressed and purified GST-TCP14 fusion protein was incubated with a biotin-labeled probe containing 4× Up1 elements. In each lane, 20 femtomoles of biotin-labeled probe was used, and unlabeled probe at different concentrations as indicated was added as a competitor. GST was used as a control. Shifted bands are indicated by a triangle.
(B) ChIP-qPCR quantification of the enrichment of indicated regions (short horizontal lines) in the 2000-bp promoter regions of ELIP1 and ELIP2 in 35Spro:TCP14-GFP seedling. Asterisks indicate the Up1 element positions. ChIP samples were prepared using the anti-GFP antibody.
(C) Expression levels of ELIPs in Col-0, OE-1, OE-2, and 35Spro:TCP14-GFP seedlings after illumination of 4-d-old dark germinated seedlings for 6 h. Data are means ± se (five pools of seedlings from separate plates were used). Asterisks indicate significant differences (Student’s t test, P < 0.05).
(D) ChIP-qPCR quantification of the enrichment of the same promoter regions of ELIPs as in (B) in Col-0 and tcp14 seedlings. ChIP samples were prepared using the anti-ORcTP antibody.
For (B) and (D), 4-d-old dark-germinated seedlings were used. The chromatin before precipitation was used as an input control, and ACTIN7 was used as a negative control. Three independent experiments were performed, and ChIP enrichment was normalized to the input. Data are means ± se. Asterisks indicate significant differences (Student’s t test, P < 0.05).
(E) Transactivation assay. Luc+ constructs driven by ELIP promoters were used as reporters and pTA7002-TCP14 as an effector. Two days after infiltration, Dex (10 μM) dissolved in 0.01% (w/v) Tween 20 was sprayed 3 h prior to luciferin treatment (+Dex), with Tween 20 as a negative control (−Dex). The images were captured 10 min after the luciferin treatment.
(F) Quantification of luminescence strengths on tobacco leaf areas in (E). Data are means ± se (three different areas on tobacco leaves were measured). Asterisks indicate significant differences (Student’s t test, P < 0.05).
To further confirm the involvement of the Up1 element in the regulation of ELIP1 and ELIP2 expression by TCP14, we prepared chromatin samples from the etiolated 35Spro:TCP14-GFP seedlings and performed chromatin immunoprecipitation (ChIP) with an anti-GFP antibody. Subsequent qPCR determination showed that the fragments containing the Up1 element in both ELIP1 and ELIP2 promoter regions were significantly enriched in comparison with other fragments without Up1 in these two genes (Figure 7B). These findings indicate the direct binding of TCP14 to the Up1 elements in ELIP1 and ELIP2 promoters.
Repression of Cotyledon Greening by OR Relies on TCP14
The expression levels of both ELIP1 and ELIP2 were lower in the OE lines but much higher in 35Spro:TCP14-GFP seedlings than in the wild type under illumination (Figure 7C), supporting our finding that OR represses TCP14 transcriptional activity. Because OR interacts with TCP14 in the nucleus, we also used the anti-ORcTP antibody to precipitate chromatin samples from the dark-germinated wild-type seedlings. Similar enrichments of these promoter fragments with Up1 were also detected (Figure 7D); however, when we used this antibody to precipitate chromatin from dark-germinated tcp14-1 knockout seedlings, a similar enrichment of the Up1-containing fragments was not detected for either ELIP1 or ELIP2 (Figure 7D). These results suggest that OR affects the expression of ELIPs through its interaction with TCP14.
To further test the in planta activation of ELIPs by TCP14, we utilized the luciferase gene (Luc+) driven by a 2000-bp upstream flanking fragment of each of the ELIPs as a reporter and TCP14 after a dexamethasone (Dex)-inducible promoter as an effector in infiltrated tobacco leaves (Aoyama and Chua, 1997). After Dex treatment, a significant increase in luminescence signal strength was observed when either ELIP1 or ELIP2 promoter region was used (Figures 7E and 7F), clearly confirming the activation of ELIPs by TCP14.
To elucidate the relationship between OR and TCP14 in the regulation of cotyledon greening, we crossed OE-1 with tcp14-1 and also transformed the 35Spro:OR construct into the tcp14 line. The relative transcript abundances of OR in OE-1 and the homozygous cross and transgenic progeny lines are shown in Figure 8A. As observed before, OR overexpression significantly repressed greening in seedlings with a wild-type TCP14 background (Figure 8B; Supplemental Figure 12). A certain percentage of cotyledons were unable to turn green even after 12- or 24-h illumination (Figures 8B and 8C); however, when OR was overexpressed in homozygous tcp14 background generated by either crossing or transformation, the seedlings showed similar greening rates as those of the wild-type plants, suggesting that the repression on cotyledon greening by OR overexpression was rescued when TCP14 was silenced (Figures 8B and 8C). We further analyzed the expression of ELIPs that were found to be downregulated by OR overexpression in the OE lines (Figure 4D). Consistent with the phenotype, expression of both genes was restored to their corresponding wild-type levels when OR was overexpressed in tcp14 plants (Figure 8D). Immunoblot analysis also further demonstrated a rescued accumulation of ELIPs in de-etiolating cotyledons of the 35Spro:OR tcp14 lines (Figures 2C and 8E). These results confirmed that the repression of greening by OR relies on the existence of TCP14.
Figure 8.
Functions of OR on Greening Rely on the Presence of TCP14.
(A) Quantification of OR expression levels in seedlings with different OR and TCP14 backgrounds. Transcript abundance of OR in each line was normalized against the Col-0 level. Data are means ± se (three pools of seedlings from separate plates were used). Asterisks indicate significant differences between the transgenic lines and Col-0 (Student’s t test, P < 0.05).
(B) Percentage of green cotyledons with different OR and TCP14 backgrounds. For each measurement, at least three plates with more than 100 seedlings per plate were counted. Data are means ± se (n > 3). The asterisk indicates a significant difference between the transgenic line and Col-0 (Student’s t test, P < 0.05).
For (A) and (B), seedlings were germinated in the dark for 4 d and then illuminated for 12 h.
(C) Representative seedlings with different OR and TCP14 backgrounds after 24 h illumination. Bar = 10 mm.
(D) Expression of ELIP1 and ELIP2 during de-etiolation. Expression level of each gene was quantified by qPCR and calculated as the ratio between the transcript abundance of the gene studied and that of ACTIN8 in the same sample. Data are means ± se (five pools of seedlings from separate plates were used). Asterisks indicate significant differences between the transgenic lines and Col-0 (two-way ANOVA followed by Dunnett’s multiple comparison test, *P < 0.05, **P < 0.01 or better).
(E) Immunoblot analysis showing the abundances of ELIPs in seedlings with OR-overexpressing and TCP14-silencing background. ACTIN was probed as a loading control.
For (D) and (E), cotyledons were sampled during the transition from dark (0 h) to light conditions for 1, 3, 6, 12, and 24 h.
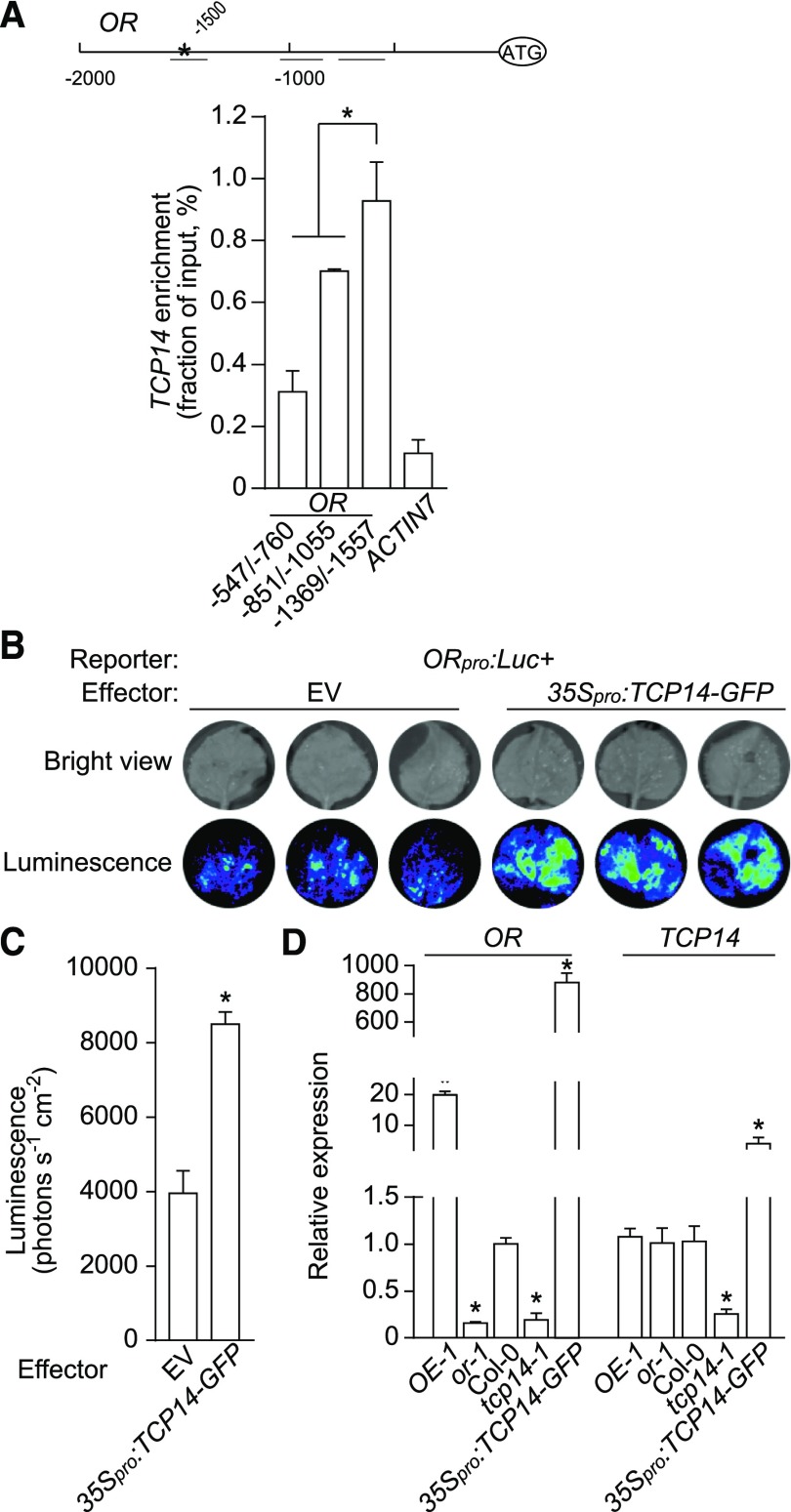
To our surprise, we also found an Up1 element at −1500 bp upstream of the ATG starting codon of OR (Figure 9A). We performed ChIP-qPCR using chromatin samples prepared from dark-germinated 35Spro:TCP14-GFP seedlings and the antibody against GFP. The fragment from −1557 to −1369 bp of OR containing this Up1 element was significantly enriched in comparison with the other two fragments quantified (Figure 9A). We also constructed a vector to express Luc under the control of a 2000-bp upstream promoter region of OR (ORpro:Luc+), and transformed it into tobacco leaves by infiltration, together with either 35Spro:TCP14 or pGWB5 empty vector (as a negative control). When substrate was supplied, the areas cotransformed with ORpro:Luc+ and 35Spro:TCP14 emitted stronger luminescence signals than those with ORpro:Luc+ and pGWB5 (Figures 9B and 9C). Moreover, we quantified the transcript levels of OR and TCP14 in lines with altered OR and TCP14 expression. Transcript abundance of OR was reduced to 19% of the wild-type level in tcp14-1 and induced to 880-fold of the wild-type level in 35Spro:TCP14-GFP seedlings, while the expression of TCP14 was not significantly affected by overexpression or silencing of OR (Figure 9D). The results showed that TCP14 positively regulates OR expression, but not vice versa.
Figure 9.
TCP14 Regulates OR Expression.
(A) ChIP-qPCR analysis of the enrichment of chromatin regions (short horizontal lines, the asterisk indicates the Up1 element position) in the 2000-bp promoter region of OR. ChIP samples were prepared from the 35Spro:TCP14-GFP seedlings germinated in the dark for 4 d using an anti-GFP antibody. The chromatin before precipitation was used as an input control, and ACTIN7 was used as a negative control. Three independent experiments were performed, and the ChIP enrichment was normalized to the input. Data are means ± se from three independent experiments. Asterisk indicates significant difference (Student’s t test, P < 0.05).
(B) and (C) Transactivation assay. Luc+ driven by the OR promoter was used as a reporter and pGWB2-35S:TCP14 as an effector. Empty vector (pGWB2) served as control. The images were captured 10 min after luciferin treatment at 2 d after infiltration (B) and quantified (C). Data are means ± se; three different leaves were measured. Asterisk indicates significant difference (Student’s t test, P < 0.05). EV, empty vector.
(D) Relative expression levels of OR and TCP14. Seedlings were collected at 12-h after illumination after a 4-d germination in the dark. Data are means ± se (three pools of seedlings from separate plates were used). Asterisks indicate significant differences between the transgenic lines and Col-0 (Student’s t test, P < 0.05).
DISCUSSION
The transition to light is a challenge for dark-germinated seedlings. To avoid photodamage, a number of key regulatory factors are required for the sophisticated manipulationof chlorophyll biosynthesis during the transition from etioplasts to chloroplasts (McCormac and Terry, 2002; Solymosi and Schoefs, 2010; Tang et al., 2012; Jarvis and López-Juez, 2013; Yuan et al., 2017). It has been reported that genes encoding several key enzymes for chlorophyll biosynthesis, such as HEMA1, POR, and chlorophyllide a oxygenase, are repressed by PIFs in the dark (Stephenson et al., 2009) but stimulated by Golden2-Like in the light (Waters et al., 2009). The gibberellin-regulated DELLA proteins accumulate in etiolated cotyledons and de-repress chlorophyll biosynthesis by repressing transcriptional activity of PIFs during de-etiolation (Cheminant et al., 2011). In this study, we demonstrate that the DnaJ-like zinc finger domain-containing protein OR and the bHLH transcription factor TCP14 interact to repress ELIPs expression, chlorophyll synthesis, and chloroplast biogenesis in etiolated Arabidopsis cotyledons.
Nuclear Localized OR Represses Chloroplast Biogenesis
During de-etiolation of the dark-germinated seedlings, chloroplast biogenesis via etioplasts is reflected by the distinct greening of cotyledons following chlorophyll synthesis and accumulation (von Arnim and Deng, 1996; Tanaka and Tanaka, 2007). Previously, OR was shown to localize in various plastids (e.g., chloroplasts and chromoplasts) and promote carotenoid accumulation in Arabidopsis (Zhou et al., 2015). Our recent study also identified its nuclear localization in germinating cotyledons (Sun et al., 2016). Here, we discovered that OR affects cotyledon greening (Figures 1A and 1B). The cotyledons of OE seedlings had significantly lower amounts of chlorophylls than those of the wild-type plants (Figure 2A), and their chloroplasts showed arrested thylakoid development upon illumination (Figures 1C to 1E). The OE seedlings also contained reduced expression of both ELIP genes to affect the de-etiolation process (Figure 2C; Casazza et al., 2005; Tzvetkova-Chevolleau et al., 2007).
Upon illumination and subsequent greening, the abundance of the nuclear localized OR decreased greatly (Figure 3A); however, overexpression of either OR or ORnuc, which is specifically targeted to the nucleus, was sufficient to repress the expression of ELIPs and the biosynthesis of chlorophylls (Figure 4). These results demonstrate that the nuclear localized OR imposes a negative effect on chloroplast biogenesis, and a decrease of its abundance in the nucleus is essential for chloroplast biogenesis during de-etiolation.
ELIPs Are the Targets of the OR-TCP14 Interaction
ELIPs are transiently expressed at early stages of greening and play an important role during chloroplast biogenesis upon illumination (Kolanus et al., 1987). They are known to be regulated by the COP9 signalosome and the Dof transcription factor DOF AFFECTING GERMINATION1 (DAG1) in Arabidopsis (Harari-Steinberg et al., 2001; Rizza et al., 2011). Both ELIP1 and ELIP2 were found here to be immediate targets of the OR-TCP14 interaction. By EMSA and ChIP-qPCR, we demonstrated that TCP14 directly binds to the Up1 elements in the promoter regions of both ELIP genes (Figure 7). TCP14 is also able to activate the expression of Luc driven by the promoters of both ELIPs in planta (Figures 7E and 7F). Because OR interacts with and represses the transcriptional activity of TCP14 (Figure 6), the OE lines exhibited downregulated expression of ELIPs and possessed arrested chloroplast biogenesis similar to that of elip mutants (Supplemental Figure 13; Casazza et al., 2005; Rossini et al., 2006). Although there are different hypotheses about how ELIPs affect chlorophyll biosynthesis, the accumulation of a high level of Pchlide in OE lines might be a consequence of the downregulation of both ELIPs (Figure 2E; Bruno and Wetzel, 2004; Castillon et al., 2007; Tzvetkova-Chevolleau et al., 2007).
We observed reciprocal regulation of OR and TCP14. While TCP14 activates the expression of OR, OR in turn represses the transcriptional activity of TCP14. Significantly, TCP14 is not the only TCP family transcription factor that shows reciprocal regulation. TCP8 interacts with PNM1, a mitochondrion and nucleus dual-targeted pentatricopeptide repeat protein, in nucleus to regulate gene expression. TCP8 may also bind to PNM1 promoter to fine-tune the PMN1 transcript level (Hammani et al., 2011). Thus, we speculate that the reciprocal regulation between OR and TCP14 helps to maintain a required homeostasis of both proteins and thus provides a plasticity in the regulation of ELIP expression to adjust chlorophyll biosynthesis and photoprotection during chloroplast biogenesis.
Members of the TCP transcription factor family are frequently found to function redundantly (Danisman et al., 2013). TCP14 and TCP15 not only share the highest sequence similarity but also share common functions in the regulation of internode length and leaf development (Kieffer et al., 2011). In our study, we found that TCP15 is expressed at a very low level in de-etiolating cotyledons and that there was no protein–protein interaction between TCP15 and OR (Supplemental Figures 10 and 11). To demonstrate that relationship between OR and TCP14, we overexpressed OR with a tcp14 background. In both OE-1 tcp14 and 35Spro:OR tcp14 lines, the greening of their cotyledons was rescued, and their ELIPs were not affected by OR overexpression at transcriptional and translational levels either (Figure 8; Supplemental Figure 12). Therefore, it is unlikely that another protein(s) functions redundantly with TCP14 in the regulation of de-etiolation. However, we cannot rule out the possibility that other TCP members are also modulated by OR through protein–protein interactions and/or have overlapping functions with TCP14 in germinating seedlings.
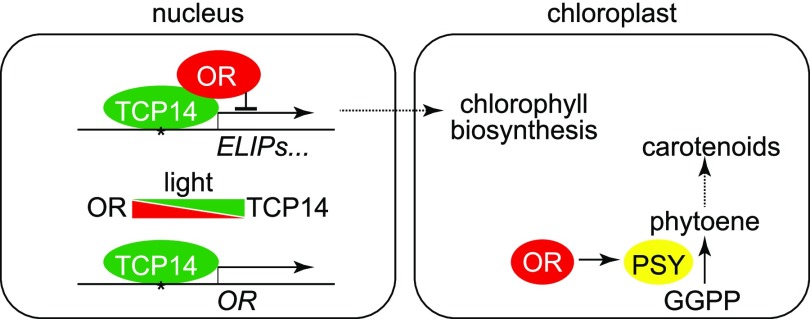
Based on these findings, we propose a working model of OR and TCP14 interaction in the regulation of early chloroplast development in Arabidopsis (Figure 10). Our data demonstrate that the nuclear localized OR represses ELIP gene expression and chloroplast biogenesis in germinating cotyledons through direct interaction with TCP14. Following illumination, abundance of the nuclear localized OR decreases, resulting in a de-repression of TCP14 transcriptional activity, together with the induction of TCP14 expression and a simultaneous accumulation of TCP14 in the nucleus (Figure 3A; Supplemental Figure 11). This interrupts the OR-TCP14 interaction and allows the etioplast-to-chloroplast transition. However, we do not yet know how the OR-TCP14 interaction integrates with other regulatory mechanisms, especially the phytochrome signaling pathway that is essential for photomorphogenesis regulation. Other protein components might also be involved in the OR-TCP14 interaction or their regulatory functions. Further studies with corresponding mutants should help to place this OR-TCP14 regulation into the regulatory network for chloroplast biogenesis and also help to better understand how ELIPs are managed to serve as a checkpoint during chloroplast biogenesis (Pfannschmidt, 2010).
Figure 10.
Postulated Working Model for the Regulation of Chloroplast Biogenesis by the Interaction between OR and TCP14.
In our model, TCP14 binds to the Up1 elements in promoter regions of ELIPs and OR and activates their expression. In dark-germinated seedlings, the interaction between OR and TCP14 in the nucleus represses ELIPs expression and chloroplast biogenesis. Upon illumination, the abundance of the nuclear localized OR decreases and that of TCP14 increases. The interruption of interaction between OR and TCP14 de-represses ELIPs expression and chloroplast biogenesis. The predominant localization of OR shifts from the nucleus to chloroplasts upon illumination. In chloroplasts, OR interacts with PSY to promote carotenoid biosynthesis, which protects chloroplasts from photodamage and favors the de-etiolation process. GGPP, geranylgeranyl diphosphate.
The plastid-localized OR has been demonstrated to promote carotenoid biosynthesis and accumulation in different plants, such as cauliflower (Brassica oleracea) and sweet potato (Ipomoea batatas; Lu et al., 2006, Kim et al., 2013). Carotenoids play an essential role in the protection against photo-oxidative damage (Davison et al., 2002; Toledo-Ortiz et al., 2010). The targeting of OR to chloroplasts during the etioplast-to-chloroplast transition likely enhances carotenoid biosynthesis and, as a result, favors the adaption of seedlings to the light. Recent studies reported that OR functions in chloroplasts in stabilizing and promoting PSY enzymatic activity for carotenoid biosynthesis (Zhou et al., 2015; Welsch et al., 2018). Such a function resembles that of the DnaJ cochaperone cell growth defect factor1 (CDF1) on POR as a holdase (Lee et al., 2013). Similar to CDF1, OR has a C-terminal DnaJ-like zinc finger domain that has been proven to possess the disulfide isomerase activity as the DnaJ proteins from Escherichia coli (Lu et al., 2011; Gray et al., 2015). Although we observed the presence of OR in full length in chloroplast preparations, it is unknown whether OR is also involved in the unfolding and translocation of protein precursors into chloroplasts as is CDF1 (Reinbothe et al., 2015; Gray et al., 2015). The elucidation of the mechanism governing the dual localization of OR in the nucleus and chloroplasts may reveal important new insights into the coordination of these two organelles during de-etiolation.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this study were in the Col-0 background. All seeds were stratified at 4°C for 3 d in the dark and then grown on plates with one-half strength Murashige and Skoog medium containing 1% (w/v) Suc. In general, the seeds were germinated at 22°C for 4 d in the dark and then de-etiolated under a growth light of 100 μmol m−2 s−1 for 12 h, unless otherwise indicated.
Seeds of the OR T-DNA insertion line, or-1 (GABI-Kat, no. 850E02), were ordered from GABI-Kat (Max Planck Institute for Plant Breeding Research, Germany). To create OR RNAi plants, a 584-bp fragment of OR cDNA (199 to 782 bp in the ORF) was amplified and inserted into pFGC5941 from the Arabidopsis Biological Resource Center (ABRC) in both forward and reverse orientations, separated by a CHSA intron, to generate pFGC5941-OR-i. For overexpressing OR, its ORF was first cloned after the 35S promoter in the pRTL2 vector, and then the entire cassette was digested from pRTL2 and cloned into the multiple cloning site of pCAMBIA1300 to form pCAMBIA1300-35S:OR construct and generate 35Spro:OR lines (OE). The seedlings that overexpress a truncated OR protein with its chloroplast transit peptide substituted by GFP (35Spro:GFP-ΔcTP), which is specifically targeted to the nucleus, were reported previously (Sun et al., 2016).
The TCP14 T-DNA insertion mutant, tcp14-1 (WiscDsLox445B10), was ordered from ABRC. To overexpress the TCP14-GFP fusion protein, the full-length TCP14 ORF without the stop codon was amplified from a cDNA pool of the Col-0 seedlings and fused upstream of the 5′ end of the GFP gene of pGWB5 to produce pGWB5-35S:TCP14-GFP (Nakagawa et al., 2007).
For overexpressing OR in different TCP14 backgrounds, the pCAMBIA1300-35S:OR vector was also used to transform tcp14-1 seedlings. We also crossed one of the homozygous OE lines (OE-1) with tcp14-1. Progeny lines with homozygous OR-overexpressing and TCP14-silencing genotype were screened.
Each of the constructs was transferred into the Agrobacterium tumefaciens strain GV3101 by electroporation (Lu et al., 2006) and transformed into Arabidopsis plants using the floral dip method (Clough and Bent, 1998). Transgenic plants were screened according to the SIGnAL iSect tool (http://signal.salk.edu/tdnaprimers.2.html) for homogeneity. Nicotiana benthamiana plants were grown from seeds as described previously by Sparkes et al. (2006).
Nucleic Acid Extraction, RT, and Gene Expression Quantification
Samples collected under different conditions were either used immediately or frozen in liquid nitrogen and stored at −80°C until use. Genomic DNA was extracted from leaves using the cetyltrimethylammonium bromide method (Murray and Thompson, 1980). Total RNA was isolated using the RNAiso reagent (TaKaRa), and cDNA was synthesized with a PrimeScript Double Strand cDNA Synthesis Kit (TaKaRa) following the manufacturer’s instructions. Gene expression levels were quantified by qPCR using SYBR Premix ExTaq II (TaKaRa) with a Thermal Cycler Dice Real-Time System TP800 (TaKaRa) following the manufacturer’s instructions. The expression values were calculated according to the comparative CT method (Schmittgen and Livak, 2008). For each sample, at least three biological replicates were analyzed, and each experiment was repeated three times. ACTIN8 and UBQ10 were used as reference genes for normalizing gene expression. Primers used in this study are listed in the Supplemental Table. Statistical analysis results are provided in the Supplemental File.
Microscopy
For TEM observation of plastid ultrastructure, cotyledons were fixed, embedded, and sectioned according to Faso et al. (2009). Briefly, cotyledons were fixed with 4.0% (w/v) formaldehyde, 2.5% (w/v) glutaraldehyde in 0.1 M phosphate buffer, pH 7.2, and then subsequently fixed with 1% (w/v) osmium tetroxide in 0.1 M phosphate buffer, pH 7.2, containing 1.5% (w/v) potassium ferricyanide. Samples were then embedded in Spurr’s low-viscosity resin. After resin polymerization, ultrathin sections were cut from each sample with a diamond knife on an ultramicrotome. The ultrathin sections were picked up on copper grids and stained with aqueous uranyl acetate and Reynolds lead citrate prior to observation. A Hitachi-7650 transmission electron microscope was used for observation and image capture.
Production and Purification of Antibodies
Two peptides corresponding to the transit peptide of OR (ORcTP, Asp15-Ser27) and to a fragment beyond the transit peptide (ORΔcTP, Ile155-Thr168) were synthesized and used as antigens. The dilution of ORcTP and ORΔcTP antibodies for immunoblotting were 1:500 and 1:1000, respectively. Full-length TCP14 protein was expressed in bacteria and purified as an antigen. The dilution of TCP14 antibody for immunoblotting was 1:1000. Peptide sequences from ELIP1 (CPLPKSPSPPPPMKP), ELIP2 (AQGDPIKEDPSVPSC), and PORB (CLLDDLKKSDYPSKR) were synthesized and used as antigens. The dilution of these three antibodies for immunoblotting was 1:1000. All antibodies were raised in rabbits (Oryctolagus cuniculus) by GenScript and further purified from immunoblots (Harlow and Lane, 1988).
Greening Analysis and Pigment Quantification
To calculate the greening rate, seedlings were germinated in the dark for 4 d and then moved to growth light for 12 h, after which the percentage of cotyledons that turned green was determined. For each measurement, at least three plates with more than 100 seedlings per plate were counted. Chlorophyll and carotenoid contents were determined according to Li et al. (2001).
The in planta visualization of chlorophyll intermediates was performed as described previously by Ankele et al., (2007), with minor modifications. Seeds were germinated on plates containing 5 mM 5-aminolevulinate (Sigma-Aldrich) feeding to enhance the accumulation of precursors for chlorophyll biosynthesis. After a 4-d etiolation treatment and 1-h light exposure, cotyledons were scanned from lambda 550 to 720 nm with 405 nm laser excitation using an SP5 laser microscope (Leica). Each emission spectrum represents an averaging measurement of five positions from cotyledons. Pchlide has a specific emission from 627 to 657 nm (Ankele et al., 2007).
Protein Extraction and Immunoblot Analysis
Total proteins were extracted by homogenization of 100 mg of seedlings in 300 μL of 2× SDS loading buffer (Green and Sambrook, 2012) containing 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 1× Protease Inhibitor Cocktail (NEB) and then centrifuged at 10,000g for 10 min at 4°C. Purification of the nuclear and chloroplast fractions from Arabidopsis cotyledons was conducted according to previous reports by Aronsson and Jarvis (2011) and Zheng et al. (2013).
For immunoblot analysis, protein samples were denatured and separated by SDS-PAGE and transferred to Protran BA 83 nitrocellulose membrane (GE Healthcare). In addition to the polyclonal antibodies we raised against OR, TCP14, ELIPs, and PORB, the antibody against TIC110 was a gift from Danny Schnell (Michigan State University), and antibodies against HISTONE H3 (no. AS10710, dilution 1:1000), TIC40 (no. AS10709, dilution 1:4000), LHCB1 (no. AS01004, dilution 1:5000), and LHCA4 (no. AS01008, dilution 1:5000) were purchased from Agrisera. Antibodies against GST (rabbit, no. CAB4169, dilution 1:5000), GFP (monoclonal from mouse, no. MA5-15256, dilution 1:1000), ACTIN (monoclonal from mouse, no. MA1-744, dilution 1:5000), and HA-tag (rabbit, no. 715500, dilution 1:1000) were purchased from Thermo Fisher Scientific. Horseradish peroxidase–conjugated secondary antibodies against rabbit/mouse IgG were purchased from Promega. Immobilon Western Horseradish Peroxidase substrate for chemiluminescent detection was from Merck Millipore. Common protocols (Green and Sambrook, 2012) and the manufacturer’s manuals for SDS-PAGE, semidry blotting, and immunodetection with chemiluminescent substrate were followed. For quantification of protein abundance, ImageQuant software (GE Healthcare) was used. Signals from three independent experiments were quantified.
Yeast Two-Hybrid Assay
For yeast two-hybrid screening, a ProQuest Two-Hybrid System (Life Technologies) was used. The full-length ORF of OR was amplified and fused after the DBD of pDEST32 to generate pDEST32-OR as the bait. The three-frame Arabidopsis cDNA library in pDEST22 was a gift from Ji-Rong Huang (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences). After cotransformation of the library and pDEST32-OR into the yeast strain Saccharomyces cerevisiae AH109, positive clones were screened on synthetic drop-out (SD)/−Leu/−Trp/−His plates according to the manufacturer’s instructions.
Full-length ORFs of TCP14 and its homolog gene TCP15 were fused after the DNA activation domain of pDEST22 to produce pDEST22-TCP14 and pDEST22-TCP15, respectively. AH109 cells cotransformed with different combinations of the pDEST32- and pDEST22-constructs were cultivated and spotted onto SD/−Leu/−Trp (nonselective) and SD/−Leu/−Trp/−His (selective) plates following the manufacturer’s instructions to identify possible interactions. Empty vectors were used as negative controls. A colony-lifting assay using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside as the substrate was subsequently performed according to the manufacturer’s instructions to confirm the interaction.
Pull-Down Assay
The pull-down assay was performed as described previously (Zhou et al., 2011). The TCP14 full-length ORF was subcloned in pGEX-4T1 (GE Healthcare) to yield pGEX-TCP14 for prokaryotic expression. After transformation into E. coli Rosetta 2(DE3)pLysS and induction with 1 mM isopropyl β-d-thiogalactoside at 37°C for 4 h, the recombinant protein GST-TCP14 was purified and immobilized on MagneGST glutathione particles (Promega) according to the manufacturer’s instructions. Because we were not able to express a full-length OR in E. coli, a TNT High-Yield Wheat Germ Protein Expression System (Promega) was used to express OR with an HA-tag at the N terminus (HA-OR) following the manufacturer’s instructions. After coupled transcription and translation, the total protein was incubated with GST-TCP14–bound MagneGST glutathione particles in binding/wash buffer containing 4.2 mM Na2HPO4, 2 mM KH2PO4, 140 mM NaCl, and 10 mM KCl, pH 7.2, at 4°C for 1 h. The particles were washed with binding/wash buffer three times and resuspended in 2× SDS loading buffer. Proteins captured by the particles were separated by SDS-PAGE and examined by immunoblotting using an antibody recognizing the HA-tag.
Co-IP Assay
For Co-IP assays, total protein was extracted from etiolated seedlings in an extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% (v/v) Triton X-100, 0.2% (v/v) Nonidet P-40, 0.1 mM phenylmethylsulfonyl fluoride, and 1× Protease Inhibitor Cocktail. Homogenates were centrifuged at 10,000g for 10 min at 4°C. The antibody was added to the supernatant, and the mixture was incubated overnight with Protein A beads (Abmart) at 4°C with gentle agitation. The beads were washed three times with the extraction buffer containing 0.1% (w/v) SDS and suspended in 2× SDS loading buffer. The proteins bound to the beads were separated by SDS-PAGE and examined by immunoblotting.
BiFC Assay
Fluorescent protein fusion vectors for BiFC (Citovsky et al., 2006) were ordered from ABRC. The full-length OR and TCP14 ORFs were cloned into pSAT4-nEYFP-C1 and pSAT1-cEYFP-C1 (ABRC), respectively. The two expression cassettes in these two constructs, together with the cassette from the pSAT6-mCherry-VirD2NLS vector, which carries a nuclear marker, were cloned into the I-SceI, AscI, and PI-PspI sites, respectively, of the binary vector pPZP-RCS2-Bar (ABRC). The construct was transformed into the A. tumefaciens strain GV3101 by electroporation, and N. benthamiana leaves were then infiltrated as described previously (Sparkes et al., 2006). Fluorescent fusion protein signals were observed at 24 h after infiltration using the FLUOVIEW FV1000 Laser Confocal Microscopy System (Olympus).
Transactivation Assay
To study the transactivation capability of OR and TCP14, we used the ProQuest Two-Hybrid System. In brief, the full-length TCP14 ORF was fused to the DBD of pDEST32 to create pDEST32-TCP14. The yeast strain AH109 was separately transformed with pDEST32-TCP14, pDEST32-OR, and the empty pDEST32 vector. Transactivation activity was quantified by measuring the β-galactosidase activity in liquid cultures of the transformants using O-nitrophenyl-β-D-galactopyranoside as a substrate according to the instruction of the ProQuest system. The transactivation activity (in Miller units) was calculated after normalization with the OD at 600 nm of the yeast culture.
To measure the effect of OR on the transactivation activity of TCP14, a pBridge vector (Clontech) that can express two proteins simultaneously in yeast was used. TCP14 was fused with the DBD to generate pBridge-TCP14, and OR was then cloned 3′ to the MET25 promoter to generate pBridge-TCP14+OR. MET25pro:OR in this construct is expressed in the absence of Met and is repressed in the presence of 1 mM Met. The empty pBridge vector was used as a control. Each of the constructs was transformed into AH109. The yeast cells were cultivated with or without Met in the medium, and O-nitrophenyl-β-D-galactopyranoside assays were conducted. Three independent transformants were quantified for the β-galactosidase activity assay for each of the constructs.
ChIP and EMSA Assays
ChIP was performed according to Saleh et al. (2008). Four grams of 4-d-old etiolated seedlings was used in the assay. An equal amount of chromatin sample without antibody precipitation was used as an input control, and ACTIN7 was used as a negative control. ChIP DNA was analyzed by qPCR, and the ChIP values were normalized against the values of respective input. The primers used for ChIP-qPCR are listed in the Supplemental Table.
For EMSA, a LightShift Chemiluminescent EMSA kit (Pierce) was used following the manufacturer’s instructions. The GST-TCP14 fusion protein was purified as described in MagneGST protein purification system (Promega) techincal manual. GST protein expressed from the empty pGEX-4T1 vector was purified and used as a control. The biotin-labeled Up1 probe (GGCCCAAAGGGCCCATTTGGCCCATTAGGCCCAAGT) was synthesized by GenScript. The unlabeled probe was used as a competitor.
Luc Reporter Assay
The Luc+ gene from pSP-Luc+NF (Promega) was cloned into pCAMBIA1390 (CAMBIA) to generate pCAMBIA1390-Luc+, and then a 2000-bp upstream flanking region of OR was inserted to drive the expression of Luc+ as a reporter. pGWB2-35S:TCP14 was used as effector and the pGWB2 empty vector served as a negative control.
For transactivation of ELIP1 and ELIP2, a 2000-bp upstream promoter region of each gene was cloned to pCAMBIA1390-Luc+ as reporter. Meanwhile, TCP14 was cloned to pTA7002 under the control of a Dex-inducible promoter (Aoyama and Chua, 1997).
Each of the constructs was transferred into the A. tumefaciens strain GV3101 by electroporation. Agrobacterium cells were collected by centrifugation at 8000g for 15 min and then resuspended in infiltration media (50 mM MES, pH 5.6, 0.5% [w/v] Glc, 2 mM NaPO4, and 100 μM acetosyringone). Leaves of N. benthamiana from uniformly grown plants at the same developmental stage were transformed using a mixture of equal amounts of resuspended Agrobacterium cultures harboring effector and reporter constructs. The transfected plants were kept in a growth chamber under a 16-h light/8-h dark light cycle for 2 d. Leaves were then detached, sprayed with 0.1 mM luciferin solution, and kept in darkness for 10 min. To determine transactivation assay of ELIP1 and ELIP2 by TCP14, leaves were sprayed with 10 μM Dex dissolved in 0.01% (w/v) Tween 20 3 h prior to luciferin treatment. Tween 20 solution was sprayed as mock control. The images were captured by Xenogen IVIS Imaging System (PerkinElmer) and analyzed using ImageJ software.
Statistical Analysis
To determine statistical significance, we used Student’s t test and an analysis of variance (ANOVA) followed by the Newman–Keuls multiple comparison test. For the de-etiolation time series gene expression analysis, two-way ANOVA and Dunnett’s multiple comparison test against Col-0 were used. Differences were considered significant at P < 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: OR (At5g61670); TCP14 (At3g47620); TCP15 (At1g69690); ELIP1 (At3g22840); ELIP2 (At4g14690); ACTIN7 (At5g09810); ACTIN8 (At1g49240); UBQ10 (At4g05320); HEMA1 (At1g58290); CHLD (At1g08520); CHLH (At5g13630); CHLI (At4g18480); CHLM (At4g25080); GUN4 (At3g59400); GGR (At1g74470); PORA (At5g54190); PORB (At4g27440); PORC (At1g03630). Accession numbers of other genes studied in this work are listed in the Supplemental Table.
Germplasm used includes or-1 (GABI-Kat, no. 850E02), tcp14-1 (WiscDsLox445B10, ABRC stock no. CS856207), porb mutants (SALK_008867 and SALK_043887), elip1 (ABRC stock no. CS435332), and elip2 (ABRC stock no. CS00148).
Supplemental Data
Supplemental Figure 1. Quantification of OR transcript abundances in different lines.
Supplemental Figure 2. OR overexpression lines showed delayed greening upon illumination.
Supplemental Figure 3. Electron micrographs of cotyledon plastids of the 4-d-old etiolated seedlings before and after a 12-h illumination.
Supplemental Figure 4. Contents of major carotenoid compounds in Col-0 and OE-1 cotyledons before and after a 12-h illumination.
Supplemental Figure 5. Expression of genes encoding enzymes for chlorophyll biosynthesis.
Supplemental Figure 6. Amino acid sequence of OR.
Supplemental Figure 7. Immunoblot analysis of antibody specificity.
Supplemental Figure 8. Contents of major carotenoid compounds in Col-0 and transgenic lines that overexpress the nuclear-targeted OR (ORnuc).
Supplemental Figure 9. TCP14-GFP is functionally identical to TCP14 in our study.
Supplemental Figure 10. OR does not interact with TCP15.
Supplemental Figure 11. TCP15 and eRF1-2 do not coexpress with OR and TCP14.
Supplemental Figure 12. The phenotypes of seedlings overexpressing OR with different TCP14 genotypes.
Supplemental Figure 13. Seedlings of both elip1 and elip2 mutants showed delayed greening.
Supplemental Table. Primers used in this study.
Supplemental File. Statistical analysis tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
POR Gramene: NADPH:protochlorophyllide oxidoreductase
POR Araport: NADPH:protochlorophyllide oxidoreductase
Acknowledgments
We thank the editor and reviewers for their constructive comments. We thank Xiao-Ya Chen and Ji-Rong Huang (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for their help with the TEM and yeast-two hybrid experiments, Danny Schnell (Michigan State University) for sharing the antibody against TIC110, and Yan Lu (Western Michigan University) for sharing the seeds of elip mutants. We also thank ABRC for providing seeds of different Arabidopsis lines and vectors. This study was supported by the State Key Basic Research Project of China from the Chinese Ministry of Science and Technology (grant 2013CB127004) and the National Natural Science Foundation of China (grants 90817002 and 30771167).
Acknowledgments
T.S. and S.L. designed the experiments. T.S., F.Z., X.-Q.H., M.-J.K., W.-C.C., C.-F.Z. and Z.Z. performed the research. T.S., F.Z., X.-Q.H., L.L., and S.L. analyzed the data. T.S., L.L., and S.L. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Aggarwal P., Das Gupta M., Joseph A.P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22: 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrès C., Agne B., Kessler F. (2010). The TOC complex: Preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803: 715–723. [DOI] [PubMed] [Google Scholar]
- Ankele E., Kindgren P., Pesquet E., Strand A. (2007). In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19: 1964–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Aronsson H., Jarvis R.P. (2011). Rapid isolation of Arabidopsis chloroplasts and their use for in vitro protein import assays In Chloroplast Research in Arabidopsis: Methods and Protocols, Jarvis R.P., ed (New York: Humana Press; ), pp. 281–305. [DOI] [PubMed] [Google Scholar]
- Bai C., Capell T., Berman J., Medina V., Sandmann G., Christou P., Zhu C. (2016). Bottlenecks in carotenoid biosynthesis and accumulation in rice endosperm are influenced by the precursor-product balance. Plant Biotechnol. J. 14: 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J., Zorrilla-López U., Medina V., Farré G., Sandmann G., Capell T., Christou P., Zhu C. (2017). The Arabidopsis ORANGE (AtOR) gene promotes carotenoid accumulation in transgenic corn hybrids derived from parental lines with limited carotenoid pools. Plant Cell Rep. 36: 933–945. [DOI] [PubMed] [Google Scholar]
- Bruno A.K., Wetzel C.M. (2004). The early light-inducible protein (ELIP) gene is expressed during the chloroplast-to-chromoplast transition in ripening tomato fruit. J. Exp. Bot. 55: 2541–2548. [DOI] [PubMed] [Google Scholar]
- Casazza A.P., Rossini S., Rosso M.G., Soave C. (2005). Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Mol. Biol. 58: 41–51. [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521. [DOI] [PubMed] [Google Scholar]
- Cazzonelli C.I., Pogson B.J. (2010). Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15: 266–274. [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.P., Erhardt M., Hayes S., Terry M.J., Genschik P., Achard P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Lee L.-Y., Vyas S., Glick E., Chen M.-H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Danisman S., van Dijk A.D.J., Bimbo A., van der Wal F., Hennig L., de Folter S., Angenent G.C., Immink R.G.H. (2013). Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64: 5673–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.-M., Wild M., Regnault T., Baumberger N., Eisler H., Genschik P., Achard P. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24: 1923–1928. [DOI] [PubMed] [Google Scholar]
- Davison P.A., Hunter C.N., Horton P. (2002). Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418: 203–206. [DOI] [PubMed] [Google Scholar]
- Faso C., Chen Y.-N., Tamura K., Held M., Zemelis S., Marti L., Saravanan R., Hummel E., Kung L., Miller E., Hawes C., Brandizzi F. (2009). A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21: 3655–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.S., Khanna H., Swain P.K., Denicola R., Cheng H., Mitton K.P., Weber C.H., Hicks D., Swaroop A. (2004). The minimal transactivation domain of the basic motif-leucine zipper transcription factor NRL interacts with TATA-binding protein. J. Biol. Chem. 279: 47233–47241. [DOI] [PubMed] [Google Scholar]
- Giuliano G., Diretto G. (2007). Of chromoplasts and chaperones. Trends Plant Sci. 12: 529–531. [DOI] [PubMed] [Google Scholar]
- Gray J., Rustgi S., von Wettstein D., Reinbothe C., Reinbothe S. (2015). Common functions of the chloroplast and mitochondrial co-chaperones cpDnaJL (CDF1) and mtDnaJ (PAM16) in protein import and ROS scavenging in Arabidopsis thaliana. Commun. Integr. Biol. 9: e1119343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R., Sambrook J. (2012). Molecular Cloning: A Laboratory Manual. (Cold Springer Harbor: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Grimm B., Kruse E., Kloppstech K. (1989). Transiently expressed early light-inducible thylakoid proteins share transmembrane domains with light-harvesting chlorophyll binding proteins. Plant Mol. Biol. 13: 583–593. [DOI] [PubMed] [Google Scholar]
- Hammani K., Gobert A., Hleibieh K., Choulier L., Small I., Giegé P. (2011). An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23: 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari-Steinberg O., Ohad I., Chamovitz D.A. (2001). Dissection of the light signal transduction pathways regulating the two early light-induced protein genes in Arabidopsis. Plant Physiol. 127: 986–997. [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Hayami N., Sakai Y., Kimura M., Saito T., Tokizawa M., Iuchi S., Kurihara Y., Matsui M., Nomoto M., Tada Y., Yamamoto Y.Y. (2015). The responses of Arabidopsis Early Light-Induced Protein2 to ultraviolet B, high light, and cold stress are regulated by a transcriptional regulatory unit composed of two elements. Plant Physiol. 169: 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Ahn Y.O., Ahn M.-J., Jeong J.C., Lee H.-S., Kwak S.-S. (2013). Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol. Biochem. 70: 445–454. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Obayashi T., Masuda T. (2012). Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal. Behav. 7: 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolanus W., Scharnhorst C., Kühne U., Herzfeld F. (1987). The structure and light-dependent transient expression of a nuclear-encoded chloroplast protein gene from pea (Pisum sativum L.). Mol. Gen. Genet. 209: 234–239. [DOI] [PubMed] [Google Scholar]
- Lee J.-Y., Lee H.-S., Song J.-Y., Jung Y.J., Reinbothe S., Park Y.-I., Lee S.Y., Pai H.-S. (2013). Cell growth defect factor1/chaperone-like protein of POR1 plays a role in stabilization of light-dependent protochlorophyllide oxidoreductase in Nicotiana benthamiana and Arabidopsis. Plant Cell 25: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.-Y., Fang M.-J., Kuang L.-Y., Gelvin S.B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Paolillo D.J., Parthasarathy M.V., Dimuzio E.M., Garvin D.F. (2001). A novel gene mutation that confers abnormal patterns of β-carotene accumulation in cauliflower (Brassica oleracea var. botrytis). Plant J. 26: 59–67. [DOI] [PubMed] [Google Scholar]
- Li L., Yang Y., Xu Q., Owsiany K., Welsch R., Chitchumroonchokchai C., Lu S., Van Eck J., Deng X.-X., Failla M., Thannhauser T.W. (2012). The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Mol. Plant 5: 339–352. [DOI] [PubMed] [Google Scholar]
- Lopez A.B., Van Eck J., Conlin B.J., Paolillo D.J., O’Neill J., Li L. (2008). Effect of the cauliflower Or transgene on carotenoid accumulation and chromoplast formation in transgenic potato tubers. J. Exp. Bot. 59: 213–223. [DOI] [PubMed] [Google Scholar]
- Lu S., et al. (2006). The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Hall D.A., Last R.L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23: 1861–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39. [DOI] [PubMed] [Google Scholar]
- McCormac A.C., Terry M.J. (2002). Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32: 549–559. [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Susek R., Chory J. (1996). An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol. 112: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Oliver R.P., Griffiths W.T. (1982). Pigment-protein complexes of illuminated etiolated leaves. Plant Physiol. 70: 1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Kreunen S.S., Cuttriss A.J., DellaPenna D., Pogson B.J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Kim S.H., Park S., Lee H.-U., Lee J.S., Park W.S., Ahn M.-J., Kim Y.-H., Jeong J.C., Lee H.-S., Kwak S.-S. (2015). Enhanced accumulation of carotenoids in sweetpotato plants overexpressing IbOr-Ins gene in purple-fleshed sweetpotato cultivar. Plant Physiol. Biochem. 86: 82–90. [DOI] [PubMed] [Google Scholar]
- Paulsen H. (1997). Pigment ligation to proteins of the photosynthetic apparatus in higher plants. Physiol. Plant 100: 760–768. [Google Scholar]
- Peng Y., Chen L., Lu Y., Wu Y., Dumenil J., Zhu Z., Bevan M.W., Li Y. (2015). The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T. (2010). Plastidial retrograde signalling–A true “plastid factor” or just metabolite signatures? Trends Plant Sci. 15: 427–435. [DOI] [PubMed] [Google Scholar]
- Pogson B.J., Albrecht V. (2011). Genetic dissection of chloroplast biogenesis and development: An overview. Plant Physiol. 155: 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Ganguly D., Albrecht-Borth V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847: 1017–1024. [DOI] [PubMed] [Google Scholar]
- Reinbothe C., El Bakkouri M., Buhr F., Muraki N., Nomata J., Kurisu G., Fujita Y., Reinbothe S. (2010). Chlorophyll biosynthesis: Spotlight on protochlorophyllide reduction. Trends Plant Sci. 15: 614–624. [DOI] [PubMed] [Google Scholar]
- Reinbothe S., Gray J., Rustgi S., von Wettstein D., Reinbothe C. (2015). Cell growth defect factor 1 is crucial for the plastid import of NADPH:protochlorophyllide oxidoreductase A in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 5838–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resentini F., Felipo-Benavent A., Colombo L., Blázquez M.A., Alabadí D., Masiero S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8: 482–485. [DOI] [PubMed] [Google Scholar]
- Rizza A., Boccaccini A., Lopez-Vidriero I., Costantino P., Vittorioso P. (2011). Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytol. 190: 896–905. [DOI] [PubMed] [Google Scholar]
- Robinson C., Bolhuis A. (2001). Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2: 350–356. [DOI] [PubMed] [Google Scholar]
- Rossini S., Casazza A.P., Engelmann E.C.M., Havaux M., Jennings R.C., Soave C. (2006). Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol. 141: 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D., Bode R., Li W., Krol M., Saccon D., Wang S., Schillaci L.A., Rodermel S.R., Maxwell D.P., Hüner N.P.A. (2009). Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21: 3473–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudowska L., Gieczewska K., Mazur R., Garstka M., Mostowska A. (2012). Chloroplast biogenesis - Correlation between structure and function. Biochim. Biophys. Acta 1817: 1380–1387. [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Shi L.-X., Theg S.M. (2013). The chloroplast protein import system: From algae to trees. Biochim. Biophys. Acta 1833: 314–331. [DOI] [PubMed] [Google Scholar]
- Solymosi K., Schoefs B. (2010). Etioplast and etio-chloroplast formation under natural conditions: The dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 105: 143–166. [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T.-S., Kieffer M., Eshed Y., Olszewski N., Weiss D. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Livne S., Kobinson-Katz T., Tal L., Pri-Tal O., Mosquna A., Tarkowská D., Mueller B., Tarkowski P., Weiss D. (2016). The putative O-linked N-acetylglucosamine transferase spindly inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 171: 1485–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J.A., Deng X.W. (2003). From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 260: 289–297. [DOI] [PubMed] [Google Scholar]
- Sun T., Yuan H., Cao H., Yazdani M., Tadmor Y., Li L. (2018). Carotenoid metabolism in plants: The role of plastids. Mol. Plant 11: 58–74. [DOI] [PubMed] [Google Scholar]
- Sun T.H., Zhou F., Liu C.-J., Zhuang Z., Lu S. (2016). The DnaJ-like zinc finger domain protein ORANGE localizes to the nucleus in etiolated cotyledons of Arabidopsis thaliana. Protoplasma 253: 1599–1604. [DOI] [PubMed] [Google Scholar]
- Sundqvist C., Dahlin C. (1997). With chlorophyll pigments from prolamellar bodies to light-harvesting complexes. Physiol. Plant 100: 748–759. [Google Scholar]
- Sytina O.A., Heyes D.J., Hunter C.N., Alexandre M.T., van Stokkum I.H.M., van Grondelle R., Groot M.L. (2008). Conformational changes in an ultrafast light-driven enzyme determine catalytic activity. Nature 456: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58: 321–346. [DOI] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., Lin R. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53: 42–52. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Rodríguez-Concepción M. (2010). Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 107: 11626–11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T., Franck F., Alawady A.E., Dall’Osto L., Carrière F., Bassi R., Grimm B., Nussaume L., Havaux M. (2007). The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J. 50: 795–809. [DOI] [PubMed] [Google Scholar]
- von Arnim A., Deng X.-W. (1996). A role for transcriptional repression during light control of plant development. BioEssays 18: 905–910. [DOI] [PubMed] [Google Scholar]
- Wang M.Y., Zhao P.M., Cheng H.Q., Han L.B., Wu X.M., Gao P., Wang H.Y., Yang C.L., Zhong N.Q., Zuo J.R., Xia G.X. (2013). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162: 1669–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Wang P., Korkaric M., Capper R.G., Saunders N.J., Langdale J.A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R., Zhou X., Yuan H., Álvarez D., Sun T., Schlossarek D., Yang Y., Shen G., Zhang H., Rodríguez-Concepción M., Thannhauser T.W., Li L. (2018). Clp protease and OR directly control the proteostasis of phytoene synthase, the crucial enzyme for carotenoid biosynthesis in Arabidopsis. Mol. Plant 11: 149–162. [DOI] [PubMed] [Google Scholar]
- Yuan H., et al. (2015). A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis. Plant Physiol. 169: 421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Zhao Y.-Q., Zhang Z.-W., Chen Y.-E., Ding C.-B., Yuan S. (2017). Light regulates transcription of chlorophyll biosynthetic genes during chloroplast biogenesis. Crit. Rev. Plant Sci. 36: 35–54. [Google Scholar]
- Zheng X., et al. (2013). Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Sun T.-H., Wang N., Ling H.-Q., Lu S., Li L. (2011). The cauliflower Orange gene enhances petiole elongation by suppressing expression of eukaryotic release factor 1. New Phytol. 190: 89–100. [DOI] [PubMed] [Google Scholar]
- Zhou X., Welsch R., Yang Y., Álvarez D., Riediger M., Yuan H., Fish T., Liu J., Thannhauser T.W., Li L. (2015). Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 112: 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]