The FtsH1/3 protease complex is critical for acclimation of the model cyanobacterium Synechocystis to nutrient stress, specifically iron, phosphate, carbon, and nitrogen starvation.
Abstract
The membrane-embedded FtsH proteases found in bacteria, chloroplasts, and mitochondria are involved in diverse cellular processes including protein quality control and regulation. The genome of the model cyanobacterium Synechocystis sp PCC 6803 encodes four FtsH homologs designated FtsH1 to FtsH4. The FtsH3 homolog is present in two hetero-oligomeric complexes: FtsH2/3, which is responsible for photosystem II quality control, and the essential FtsH1/3 complex, which helps maintain Fe homeostasis by regulating the level of the transcription factor Fur. To gain a more comprehensive insight into the physiological roles of FtsH hetero-complexes, we performed genome-wide expression profiling and global proteomic analyses of Synechocystis mutants conditionally depleted of FtsH3 or FtsH1 grown under various nutrient conditions. We show that the lack of FtsH1/3 leads to a drastic reduction in the transcriptional response to nutrient stress of not only Fur but also the Pho, NdhR, and NtcA regulons. In addition, this effect is accompanied by the accumulation of the respective transcription factors. Thus, the FtsH1/3 complex is of critical importance for acclimation to iron, phosphate, carbon, and nitrogen starvation in Synechocystis.
INTRODUCTION
The membrane-embedded, ATP-dependent FtsH metalloproteases are conserved in bacteria, chloroplasts, and mitochondria. These enzymes form oligomeric complexes that function in a number of cellular processes including protein quality control, organelle biogenesis, and cytosolic protein homeostasis (Urech et al., 2000; Yu et al., 2007). The model cyanobacterium Synechocystis sp PCC 6803 (hereafter Synechocystis) possesses four FtsH homologs, designated FtsH1 to FtsH4 (Mann et al., 2000). FtsH4 (Sll1463) forms a homo-oligomeric complex of unknown function (Boehm et al., 2012). FtsH3 appears in two hetero-oligomeric complexes consisting of homologs FtsH2 (Slr0228) and FtsH3 (Slr1604) or FtsH1 (Slr1390) and FtsH3 (hereafter FtsH2/3 and FtsH1/3, respectively) in Synechocystis (Boehm et al., 2012; Krynická et al., 2014). The more abundant FtsH2/3 complexes are involved in the quality control of photosystem II (PSII) in thylakoid membranes (Komenda et al., 2006; Nixon et al., 2010). The FtsH1/3 heterocomplexes are localized to cytoplasmic membranes and are essential for cell viability (Yu et al., 2007; Krynická et al., 2014); however, their functions are still elusive. We recently showed that FtsH1/3 complexes are involved in acclimation to iron (Fe) deficiency by controlling the level of the ferric uptake regulator Fur (Sll0567; Krynická et al., 2014). Fur is a widespread bacterial transcriptional repressor responsible for maintaining Fe homeostasis (Crosa, 1997; Escolar et al., 1999; Dian et al., 2011; González et al., 2012) by controlling the expression of Fe-uptake and Fe-storage systems and switching on certain physiological responses to help bacteria cope with low intracellular Fe concentrations.
Similarly, the homeostasis of intracellular nutrients, such as inorganic carbon (Ci), nitrogen (N), or phosphate (P), is mainly achieved by regulating the expression of transportation proteins through dedicated transcription factors. Ci acclimation is controlled by two LysR-type transcriptional regulators, NdhR and CmpR, that operate as repressors and/or activators of the genes associated with the Ci assimilation pathway (Burnap et al., 2015; Klähn et al., 2015). The FtsH2/3 complexes are required for the proper function of NdhR in Synechocystis (Zhang et al., 2007). Similar to that of Ci assimilation, the homeostasis of N is regulated by NtcA, a transcription factor belonging to the CRP family (Schwarz and Forchhammer, 2005), that functions as a global regulator by activating genes involved in N assimilation. The activity of NtcA is influenced by two other factors: the signal transduction protein PII and the PII-interacting protein PipX (Espinosa et al., 2014). The balance between N and Ci metabolism is maintained by the availability of 2-oxoglutarate, a common cofactor for both NdhR and NtcA (Huergo et al., 2013; Burnap et al., 2015). Finally, P uptake is controlled by a two-component system consisting of the sensor kinase PhoB, which responds to P deficiency by phosphorylating the cognate response regulator Pho regulon: this process is negatively regulated by PhoU and the P-specific transporter PstSCAB (Wanner, 1993; Morohoshi et al., 2002). Although these regulatory cascades are administered based on the availability of the respective nutrient, interconnection between the individual signaling pathways is essential for the accurate coordination of all metabolic processes. However, the existence of such interconnections that facilitate crosstalk between regulatory cascades has remained unclear.
Because the FtsH1/3 complex has been shown to affect the expression of several selected genes belonging to the Fur regulon, we were interested in profiling its role at the whole-genome level in much greater detail. Therefore, in this study, we sought to identify regulons, operons, or single genes affected by changes in FtsH1 and/or FtsH3 levels in Synechocystis. We performed genome-wide expression analysis of a previously established strain of Synechocystis with conditionally suppressed FtsH3 expression, FtsH3down (Boehm et al., 2012; Krynická et al., 2014), under Fe-replete and -deplete conditions. Moreover, we performed a detailed proteomic analysis of a mutant with a suppressed level of FtsH1, called FtsH1down (Krynická et al., 2014). We applied whole-cell label-free quantitative proteomics and two-dimensional (2D) gel analysis consisting of clear native (CN) and SDS-PAGE under various nutrient stress conditions. The insufficient responses of mutants exposed to Fe, P, Ci, and N starvation indicate that FtsH1/3 is essential for mounting the full transcriptional response to these nutrient stress conditions.
RESULTS
FtsH Mutants Used to Study the Function of the FtsH1/3 Complex
To more deeply characterize the physiological role of the FtsH1/3 complex in nutrient stress responses, we used four previously characterized mutants: FtsH3down, FtsH1down, FtsH1over, and ΔFtsH2. FtsH3down and FtsH1down mutants exhibit knocked down expression of essential FtsH homologs ftsH3 and/or ftsH1 driven by regulatable promoters. In the FtsH3down mutant, the expression of ftsH3 is controlled by the nirA promoter, whose activity is downregulated by ammonium ions (NH4+). The presence of 13 mM NH4+ in the medium suppressed ftsH3 expression almost eightfold, leading to a significant decrease in the levels of both FtsH2/3 and FtsH1/3 protein complexes (Boehm et al., 2012). In FtsH1down, the expression of ftsH1 is controlled by the petJ promoter, which is downregulated in the presence of copper ions. The presence of 0.8 µM Cu2+ ions in the medium led to a sixfold decline in ftsH1 transcript levels compared with the wild type, resulting in a significant decrease in the level of the of FtsH1/3 complex (Krynická et al., 2014). By contrast, the third mutant, FtsH1over, contains eight times more ftsH1 transcript than the wild type, resulting in the accumulation of the FtsH1/3 complex. FtsH1 overexpression was achieved by inserting ftsH1 under the control of the psbA2 promoter (Krynická et al., 2014). In the FtsH2-less ΔFtsH2 mutant (Komenda et al., 2006), the FtsH2/3 complex is specifically knocked out.
FtsH3 Limitation Strongly Reduces the Transcriptional Response of Fur-Regulated Genes to Fe Deficiency
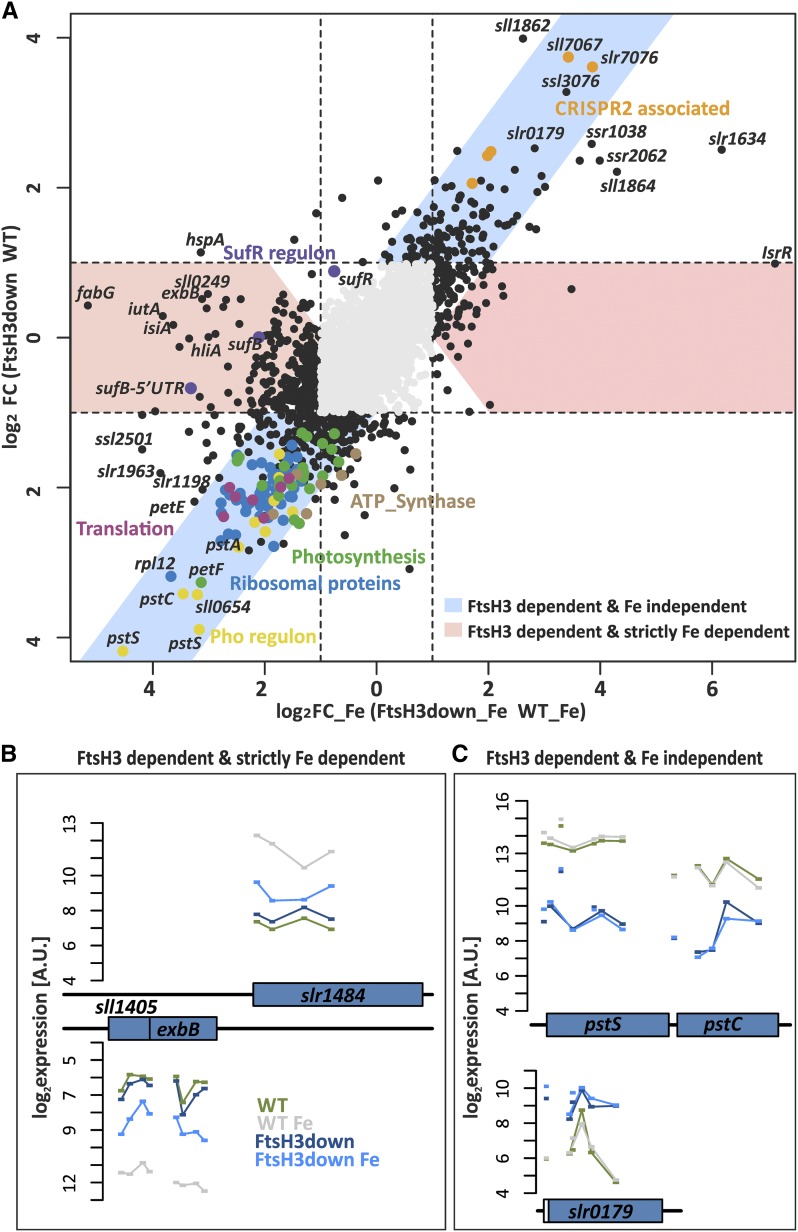
We previously reported that the FtsH1/3 protease affects the expression of two well-known Fur-repressed genes under Fe deficiency (Krynická et al., 2014). Unlike the wild-type control, mutants limited in FtsH1/3 accumulate the Fur protein under Fe depletion, resulting in the persistent repression of isiA (encoding an iron stress-induced chlorophyll binding protein) and futA1 (encoding an iron uptake protein A1; Krynická et al., 2014). To further characterize the impact of FtsH3 suppression on the expression of Fur-regulated genes, we compared the transcriptome profiles of the FtsH3down mutant and the wild type grown under Fe-deplete and -replete conditions (Figure 1; a genome-wide visualization of probe localization and signal intensities of mRNAs, noncoding RNAs, intragenic RNA elements, and 5′UTRs (untranslated regions) is shown in Supplemental Data Set 1). To distinguish the effects of FtsH3 limitation that depend on Fe depletion from those that were independent of Fe depletion, we defined two types of regulatory relationships. The first type of genes, classified as FtsH3 dependent and strictly Fe dependent, exhibited differential expression only under Fe depletion. For stringent classification, we used the concept of double differential expression based on the ∆∆ value: , where FC and FC Fe represent the extent of change (fold change) between FtsH3down and the wild type under Fe-replete and -deplete conditions, respectively. In particular, genes were considered to be FtsH3 dependent and strictly Fe dependent if they showed a significantly different response to FtsH limitation during Fe depletion () but no FtsH-specific response under standard conditions (), where the corresponding q values ≤ 0.05 (Figures 1A and 1B). The second group consists of genes differentially expressed in response to the reduction in FtsH3 level regardless of Fe abundance, where the corresponding q values ≤ 0.05 were classified as FtsH3 dependent and Fe independent (Figures 1A and 1C). Intermediate effects where the FtsH dependency was modified by Fe availability were not considered in this analysis.
Figure 1.
Identification of Genes Differentially Expressed in the FtsH3down Mutant.
(A) Scatterplot of the transcriptomic differences in log2 FC_Fe (x axis) and log2 FC (y axis), where FC and FC_Fe correspond to FCs between FtsH3down and the wild type (WT) under standard conditions and after Fe depletion, respectively. The threshold FC is indicated by dashed lines. Black or colored points represent transcriptional features with a significant FC (, q – value ≤ 0.05). Important functionally connected features are color coded.
(B) and (C) Part of a genome-wide overview of signal intensities. The mean normalized log2 expression values (scale on left; A.U., arbitrary units) of the four different microarray experiments are plotted for each probe as short horizontal bars that span the corresponding hybridization region. All probes of a single RNA feature are connected by lines. The following color coding was used: dark green, wild-type expression before Fe depletion (WT); light green, WT expression after Fe depletion (WT_Fe); dark blue, FtsH3down expression before Fe depletion (FtsH3); light blue, FtsH3down expression after Fe depletion (FtsH3_Fe). (B) FtsH3 dependent and strictly Fe dependent genes ( and ), as represented by slr1484 and sll1404. (C) Example of FtsH3 dependent and Fe independent genes. Genes downregulated independently of Fe (log2 FC_Fe ∼ log2 FC ≤ –1) are represented by sll0680 and sll0681 (belonging to the Pho regulon) and genes upregulated independently of Fe (log2 FC_Fe ∼ log2 FC ≤ –1) by sll0179 (encoding a hypothetical protein).
Our results confirmed that the limitation of FtsH3 caused strong repression of genes responsible for Fe homeostasis. The most strongly downregulated FtsH3-dependent and strictly Fe-dependent protein-coding genes are listed in Table 1. As expected, one of the strongest levels of downregulation (up to 14-fold difference) was observed for the isiA/B operon. Similar results were obtained for genes encoding different Fe transporter subunits, such as iutA (sll1206), futA1 (slr1295), futA2 (slr0513), and colocalized genes sll1404-sll1407, all encoding putative Fe-siderophore complex transporters (Table 1; Supplemental Data Set 2; Katoh et al., 2001). All of these genes were previously reported to be highly induced by Fe depletion (Hernandez-Prieto et al., 2012; Kopf et al., 2014), and some of their homologs were determined to be Fur targets in other cyanobacteria (González et al., 2014, 2016). Interestingly, the sufBCDS operon was also repressed in the FtsH3down strain specifically under Fe depletion (Figure 1A). The suf operon is regulated by the iron-sulfur cluster, containing transcriptional repressor SufR (Shen et al., 2007), and the Fur-dependent small RNA IsaR1 (Georg et al., 2017).
Table 1.
Most Strongly Downregulated FtsH3-Dependent and Strictly Fe-Dependent Protein-Coding Genes
| Functional Group Category | Gene Name(s) | TU No. | Protein Name(s) | ΔΔ |
|---|---|---|---|---|
| IsiA/B operon | sll0247–sll0249 | TU1556 | IsiA, IsiB, Slr0249 | −3.8 to −3.6 |
| ABC transport system proteins | sll1404–sll1406 | TU32 | ExbB, ExbD, FhuA, HP | −3.6 to −2 |
| Fec region | slr1316–slr1319 | TU23 | FecC, FecE, FecB | −1.9 to −1.3 |
| Other transporters | sll1206 | TU24 | IutA | −4.1 |
| slr1295 | TU288 | FutA1 | −2.6 | |
| slr1392 | TU689 | FeoB | −1.8 | |
| slr0513 | TU3085 | FutA2 | −1.8 | |
| sll1878 | TU1867 | ABC transporter | −1.7 | |
| slr1484–slr1488 | TU37 | TonB-type transport system | −2.9 to −1.8 | |
| SufR regulon | slr0074–slr0076 | TU3030 | SufB, C, D | −2.6 to −0.7 |
| sll0088 | TU3027 | SufR | −1.6 | |
| High light inducible proteins | ssl1633 | TU690 | HliC | −3.5 |
| ssl2542 | TU1176 | HliA | −2.6 | |
| Lipid metabolism | sll0330 | TU2474 | FabG | −5.6 |
| Phycobilisome degradation proteins | ssl0453, ssl0452 | TU1562 | NblA2, NblA1 | −2.4; −2.0 |
| Chaperonins | sll1541, sll0170 | TU445/2403 | Hsp17, DnaK | −3.15; −1.8 |
| Type II peroxiredoxin | sll1621 | TU1359 | AhpC/TSA family protein Sll1621 | −1.9 |
| Unknown proteins | ssl2501, ssl1762 | TU1144/2070 | −2.9; −2,5 | |
| sll0846 | TU1394 | −2.2 |
Differential expression in FtsH3down caused by Fe depletion was determined by the ΔΔ parameter; ΔΔ = log2 FC_Fe – log2FC = (FtsH3_Fe − WT_Fe) – (FtsH3 – WT), where wild type (WT), WT_Fe, FtsH3, and FtsH3_Fe correspond to log2 signal intensities of transcripts from the wild type before and after Fe depletion and the FtsH3down mutant before and after Fe depletion, respectively. All genes belonging to the same transcriptional unit (TU) are listed in one line. The organization and number of TUs were defined previously (Kopf et al., 2014). The P-values of all listed log2FC were ≤0.05.
FtsH3 Limitation Leads to the Repression of the Pho Regulon and RP Genes
Interestingly, FtsH3 suppression also affected the expression of a distinct set of individual genes and gene clusters under nutrient sufficiency. Of the 8394 mRNAs, noncoding RNAs, intragenic RNA elements, and 5′ untranslated regions identified, 1187 transcripts were detected as FtsH dependent and Fe independent (), 418 of which were mRNAs. Seventy-three percent of the differentially expressed genes were repressed by FtsH3 limitation in the mutant, whereas only 27% were upregulated (Figure 1A; Supplemental Data Set 2). The most strongly downregulated protein-coding genes are listed in Table 2. Surprisingly, suppression was most pronounced for genes belonging to the Pho regulon (Figures 1A and 1C; Table 2). The Pho regulon in Synechocystis encompasses genes encoding proteins related to P metabolism, such as the ABC-type transporter complex PstSACB, involved in phosphate import, and two additional genes, phoA and nucH (Su et al., 2007; Kopf et al., 2014). Remarkably, a major transcriptional response was observed for the promoter-proximal pstS1 gene (sll0680), for which we identified an almost 20-fold lower mRNA level in the mutant compared with the wild type. Genes encoding ribosomal proteins (RPs) constituted another set of genes downregulated by FtsH3 suppression under Fe sufficiency. The extended RP gene cluster sll1799–sll1822, as well as all other RP genes were, without exception, threefold to ninefold downregulated in the mutant (Table 2; Supplemental Data Set 2). The transcription of some genes encoding photosynthetic apparatus, including PSI, phycobilisomes, cytochromes, and ATP synthase, and that of the toxin-antitoxin system (ssl2245–sll1330), were also downregulated by FtsH3 suppression (Figure 1A; Table 2). Most upregulated genes associated with FtsH3 limitation encode hypothetical proteins with unknown function, some of which share homology with CRISPR2-associated proteins (Figure 1A).
Table 2.
Most Strongly Downregulated FtsH3-Dependent and Fe-Independent Protein-Coding Genes, log2FC ≤ −2
| Functional Category | Gene Name(s) | TU No. | Protein Name(s) | log2FC = (FtsH3 − WT) | |
|---|---|---|---|---|---|
| Pho regulon | sll0680–sll0684 | TU2785 | PstS1, PstC1, PstA1, PstB1, PstB1’ | −4.2 to −2.2 | |
| slr1247–slr1249 | TU1428 | PstS2, pstC2, PstA2 | −3.9 to −2.3 | ||
| sll0654, sll0656 | TU388 | Alkaline phosphatase, NucH | −3.4 to −2.4 | ||
| Translation (RP, EF) | sll1743–sll1746 | TU931 | RpL11, RpL1, RpL10, RpL12 RpL3, RpL4, | −3.2 to −2.1 | |
| sll1799–sll1807 | TU837 | RpL23, RpL2, RpS19, RpL22, RpS3, RpL16, RpL29, Rps17, RpL14, RpL24 | −2.8 to −2.1 | ||
| ssr0482 | TU2205 | RpS16 | −2.7 | ||
| sll1096–sll1101 | TU1986 | RpS12, RpS7, elongation factor EF-TU, Rps10 | −2.6 to −2.2 | ||
| ssr1398; ssr1399 | TU298 | RpL33; RpS18 | −2.6; −2.3 | ||
| sll1816–sll1822, | TU833 | RpS13; RpS11, Rps9 | −2.4; −2.1 | ||
| sll1426;sll0767 | TU2501 | Rpl35; Rpl20 | −2.2; −2.1 | ||
| slr0628 | TU2793 | RpS14 | −2.2 | ||
| ssl1784 | TU332 | RpS15 | −2.2 | ||
| ssl2233 | TU865 | RpS20 | −2.1 | ||
| slr1678 | TU2627 | RpL21 | −2.0 | ||
| slr1356 | TU1129 | RpS1 homolog A | −2.0 | ||
| sll1260 | TU1778 | RpS2 | −2.0 | ||
| sll1261 | TU1777 | Elongation factor Ts, EF-Ts | −2.2 | ||
| slr0083 | TU3046 | ATP-dependent RNA helicase CrhR | −2.1 | ||
| ssl3177 | TU1030 | Uncharacterized protein Ssl3177 | −2.0 | ||
| Photosynthesis, energy | ssl0020 | TU2593 | Ferredoxin-1, PetF | −3.3 | |
| sll0819 | TU1727 | PSI reaction center subunit III, PsaF | −2.1 | ||
| sll1317 | TU1200 | Apocytochrome f, PetA | −2.0 | ||
| ssl2615; sll1322 | TU166 | ATP synthases subunits AtpH; AtpI | −2.4; −2.3 | ||
| Toxin-antitoxin systems | ssl2245, sll1130 | TU1078 | Ssl2245 antitoxin, Sll1130 toxin | −2.8; −2.7 | |
| Unknown proteins | slr0287 | TU2207 | Slr0287 protein | −2.6 | |
| ssl2874 | TU3563 | UPF0296 protein Ssl2874 | −2.4 | ||
Genes were considered FtsH3-dependent and Fe-independent when log2 FC ≈ log2FC_Fe ≥ 1. FCs between the wild type (WT) and FtsH3down mutant were calculated by subtracting the corresponding wild-type value. The P-values of all listed log2FC were ≤0.05. All genes belonging to the same transcriptional unit (TU) are listed in one line, with the range in log2FC values shown at the end. TUs were defined previously (Kopf et al., 2014). EF, elongation factor.
Subdued Response to P Depletion: Relationship between Transcript and Protein Levels in Mutants Limited in FtsH1/3
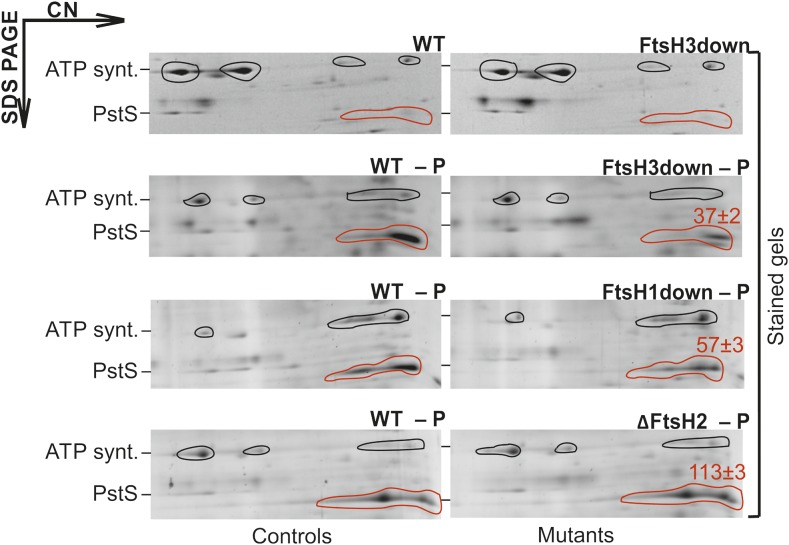
To evaluate the proteomic impact of reduced transcription of Pho regulon genes, we performed 2D gel protein analyses of mutant and wild-type cells to compare the levels of PstS1 (Sll0680), the most strongly downregulated representative of the Pho regulon. To test PstS1 inducibility, we exposed the wild type and the FtsH3down mutant to P limitation and analyzed the protein profiles after 72 h of P deficiency. The results confirmed that PstS1 accumulation in the wild type was clearly inducible and significantly higher than in the FtsH3down strain, whereas the signals of presumably noninvolved subunits of ATP synthase were comparable in both strains (Figure 2). FtsH3 is bound in two hetero-oligomeric complexes, FtsH1/3 and FtsH2/3, and both complexes are suppressed in FtsH3down (Krynická et al., 2014). To find out which complex is involved in controlling PstS1 accumulation, we also analyzed the protein composition of a P-depleted ΔFtsH2 mutant lacking the FtsH2/3 complex (Komenda et al., 2006) and the FtsH1down mutant limited specifically in the FtsH1/3 complex (Krynická et al., 2014). The lack of FtsH2/3 had no visible effect on PstS1 accumulation, whereas the reduction in FtsH1/3 levels significantly reduced PstS1 levels compared with the control (Figure 2). Thus, the transcriptional regulation of PstS1 is controlled by the FtsH1/3 complex.
Figure 2.
Differences in the Accumulation of PstS in FtsH Mutants under P Depletion.
Protein complexes isolated from membranes of FtsH mutants and their respective wild-type controls (WT) grown under nutrient sufficiency or after 72 h of P depletion (‒P) were analyzed by 2D CN/SDS-PAGE. The 2D gel was stained with Sypro Orange. Each loaded sample contained 5 μg of chlorophyll a. The overall PstS signal is indicated by a red outline, and the AtpA/B subunits of ATP synthase (ATP synt.) used for normalization by a black outline. The identity of ATP synt. and PstS proteins was verified by MS. Red numbers indicate the percentage of the PstS protein in the mutant related to the amount of PstS in the wild-type control, normalized to ATP synthase; means of three measurements ± sd. Quantification of bands was performed using ImageQuant TL software (GE Healthcare).
Detailed Proteomic Analysis of FtsH-Depleted Strains Exposed to Nutrient Stresses
Our transcriptomic and 2D gel analysis highlighted the roles of FtsH1/3 complexes in acclimation to P and Fe depletion. Since Ci and N are, together with P and Fe, the most limiting nutrients for cyanobacterial growth, we examined how the suppression of the FtsH1/3 complex also affects acclimation to N and C nutrient stress. To analyze the effect of missing FtsH1/3 on proteins important for nutrient stress acclimation in more detail, we performed label-free quantitative proteomic analysis of nutrient-limited cells of the wild type and the FtsH1down mutant that unlike FtsH3down, is limited specifically in the essential FtsH1/3 complex. We preferred this strain to FtsH3down in order to eliminate the possible additional effects of the missing FtsH2/3 complex. The number of proteins identified and quantified in the control and nutrient-depleted cells of the wild type and FtsH1down was in the range 1369 to 1701 (median, 1606) with inter-replicate correlation coefficients of 0.943 to 0.960 (Supplemental Figure 1). Primarily, we were interested in the level of stress-inducible proteins that belong to the Pho (P), NdhR (Ci), and NtcA (N) regulon (Figure 3; Supplemental Data Set 3). We also determined the abundance of FtsH homologs, with emphasis on FtsH1 and FtsH2, reflecting the quantity of FtsH1/3 and FtsH2/3, respectively (Supplemental Data Set 3). Notably, the mass spectrometry (MS) analysis did not identify the FtsH1 protease in the FtsH1down mutant, in contrast to the wild type (proteotypic peptides of FtsH1 were below the threshold for automatic selection for tandem mass spectrometry). Hence, FtsH1 repression in the mutant was very effective. On the other hand, the levels of FtsH2 in the mutant were comparable to those of the wild type (Supplemental Data Set 3). The results of quantitative proteomic analysis in response to Ci and N starvation were complemented with those of 2D gel analysis, as in the case of P depletion. Whenever possible, we also analyzed ΔFtsH2 and FtsH3down mutants.
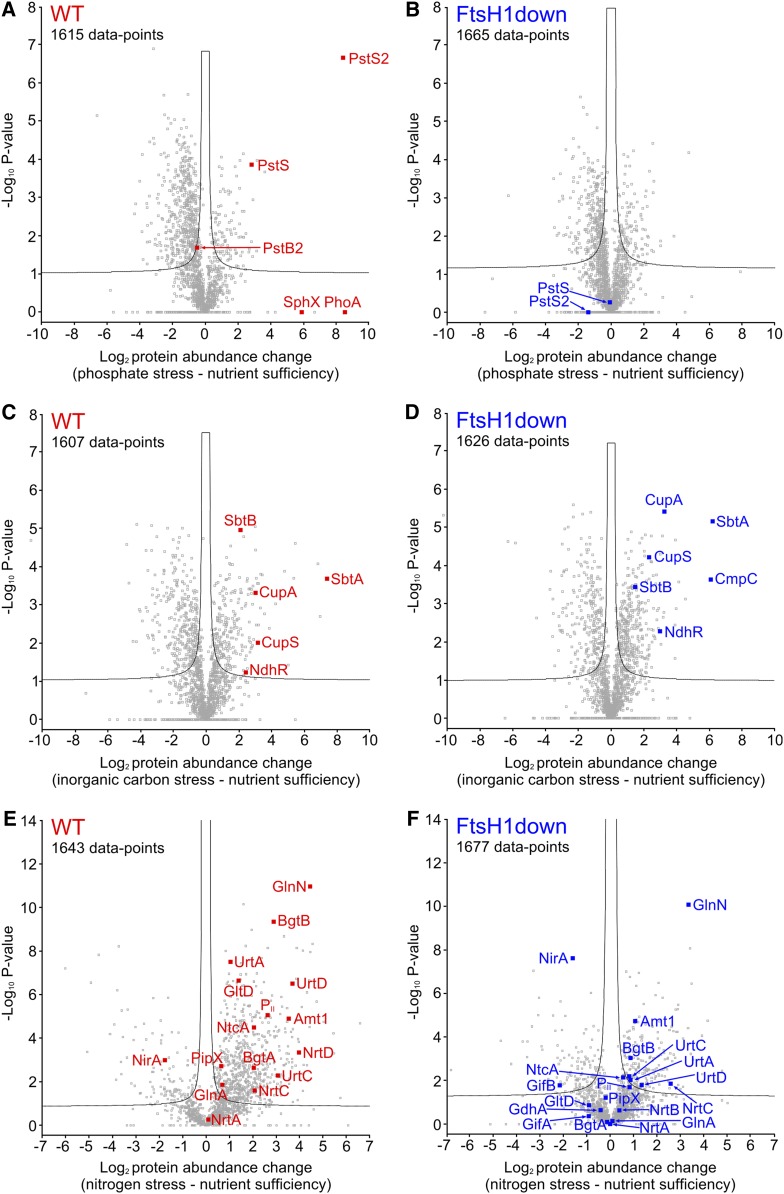
Figure 3.
Comparison of Proteomic Changes between the Wild-Type and FtsH1down Mutant Synechocystis Strains in Response to P, Ci, and N Stress.
(A) to (F) Volcano plots were constructed using Perseus (see Methods), with (A), (C), and (E) showing protein abundance changes in the wild-type control (WT) in response to P, Ci, and N stress, respectively. (B), (D), and (F) show corresponding changes in the FtsH1down mutant. P-values were calculated by a t test incorporating permutation-based false discovery rate with 250 randomizations of the complete data set for each comparison. The number of data points in each panel is indicated, and the significance threshold corresponding to a false discovery rate = 0.05 is shown by a black line. Proteins relevant to this study that were identified and therefore quantifiable by MaxQuant in at least one replicate are included in the statistical analysis and highlighted in red (wild type) or blue (FtsH1down). Other relevant proteins were not identified and therefore not quantifiable in either one or both of the nutrient sufficiency/stress analyses. Analyses from P and Ci stress were n = 3 (1× biological and 3× technical replicates) and analysis from N stress was n = 9 (3× biological and 1× technical replicates). Further details of both the highlighted proteins and those not quantified are given in Supplemental Data Set 3.
Global Proteomics
P Depletion
Similar to the FtsH3down mutant, the FtsH1down mutant exhibited downregulation of the complete Pho regulon. As shown in Figure 3A, the levels of several hundred proteins underwent significant changes in response to P stress in the wild-type strain. Focusing on proteins belonging to the Pho regulon (Su et al., 2007), the relative abundances of PstS, PstS2, SphX, and PhoA increased under P stress. In particular, PstS and PstS2 displayed increases of ∼4- and 170-fold, respectively. Only the level of PstB2 remained unchanged. Other Pho regulon proteins, PstB, PstB1’, PstC, and NucH, were below the detection threshold under control conditions, while under P starvation, these four proteins significantly exceeded the detection threshold (Supplemental Data Set 3). In clear contrast to the wild type, the FtsH1down strain did not display any significant change in the level of PstS or PstS2, and only SphX and PstB exceeded the detection threshold. The levels of all other proteins were either close to or below the threshold for their detection under both control and P stress conditions (Figure 3B; Supplemental Data Set 3).
Ci Limitation
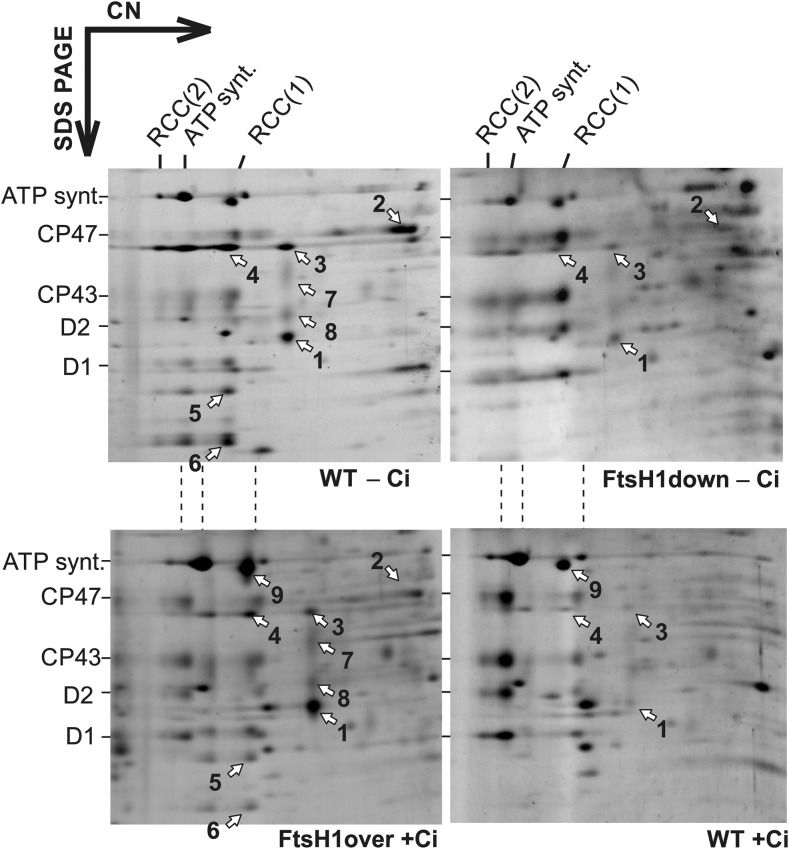
Our 2D gel analysis revealed no accumulation of NdhR-regulated proteins in our ΔFtsH2 and FtsH3down mutants after 48 h of Ci limitation (Supplemental Figure 2). This finding is in agreement with the role of FtsH2/3 in NdhR repressor regulation as proposed previously by Zhang et al. (2007). However, the FtsH1down mutant, which is limited specifically in FtsH1/3 but contains the wild-type levels of FtsH2/3, also exhibited a retarded response to Ci starvation. The 2D gel analysis revealed that after 48 h of Ci starvation, Ci transporters SbtA, NdhD3, NdhF3, and CupA, whose expression is repressed by NdhR (Klähn et al., 2015), accumulated to substantially lower levels in the mutant compared with the wild type (Figure 4). On the other hand, in the FtsH1over mutant, which overexpresses the FtsH1 protease, in addition to accumulating IsiA and FutA1 (belonging to Fur regulon; Krynická et al., 2014), a surplus of Ci-transporting proteins accumulated even under nutrient sufficiency (Figure 4; Supplemental Figure 3). Another sign of acclimation to low Ci is a reduction of the levels of both PSII dimer and monomer (Zhang et al., 2007). Interestingly, the FtsH1down mutant, unlike the wild-type strain, still maintained PSII monomer after 48 h of exposure to Ci depletion (Figure 4). Quantitative MS analysis of FtsH1down and the wild-type cells exposed to Ci limitation for 72 h identified only a few proteins belonging to the NdhR regulon, namely, SbtA/B, CupA/S, and NdhR. Other NdhR-regulated proteins were undetectable in all analyzed samples. Nevertheless, except for the NdhR transcription factor, the levels of all identified proteins were comparable in both the wild type and the mutant after 72 h of Ci depletion (Figures 3C and 3D; Supplemental Data Set 3). These results suggest that the response to Ci stress in the FtsH1down mutant is weaker compared with the wild type only at the earlier phase of the stress, whereas it later equalizes in both strains.
Figure 4.
Accumulation of Ci Acquisition Proteins in FtsH1 Mutants under Ci Replete/Deplete Conditions.
Protein complexes isolated from membrane fractions of FtsH1down and FtsH1over mutants and their respective wild-type controls (WT) grown under Ci-replete/-deplete conditions (+Ci/–Ci) were analyzed by 2D CN/SDS-PAGE. The 2D gel was stained with Sypro Orange. Each loaded sample contained 5 µg of chlorophyll a. Designation of complexes: RCC(2) and RCC(1), dimeric and monomeric PSII core complexes; ATP synt., ATP synthase; spots indicated by white arrows and numbers: 1, SbtA; 2, CmpA; 3, CupA; 4, NdhH; 5, NdhI; 6, NdhJ; 7, NdhF3; 8, NdhD3; 9, RbcL. Proteins were identified by MS (Supplemental Table) and according to Linhartová et al. (2014) and Zhang et al. (2007). Quantification of several protein spots is shown in Supplemental Figure 3.
N Depletion
We analyzed FtsH-dependent proteomic changes under N depletion by both quantitative MS analysis and 2D gel protein analysis of the FtsH1down and ΔFtsH2 mutants. The effect of N depletion on the FtsH3down mutant was not investigated due to its requirement for NH4+ for FtsH3 suppression. Only the ΔFtsH2 mutant, but not FtsH1down, was able to acclimate to N depletion via induction of the NtcA regulon, which comprises genes important for N assimilation (Giner-Lamia et al., 2017). 2D gel protein analysis confirmed the accumulation of the ammonium permease transporter Amt1 in the wild type as well as ΔFtsH2 after 24 h of stress, whereas the signal for Amt1 was almost undetectable in the FtsH1down mutant (Figure 5A; Supplemental Figure 4). The quantitative MS analysis also confirmed that the expression of all proteins within the NtcA regulon was affected in the FtsH1down mutant. The NtcA-regulated proteins either did not significantly change their levels or displayed smaller changes even after 72 h of N starvation compared with the wild type. Proteins induced in the wild type by NtcA with starvation, such as urea, ammonium, and nitrate transporters, remained downregulated in the FtsH1down mutant (Figures 3E and 3F; Supplemental Data Set 3). Phycobilin degradation, which normally provides N for the starving wild-type cells, was also slowed down more in FtsH1down than in the wild type, which was evident after both 24 and 72 h of N depletion (Figure 5B; Supplemental Data Set 3). These data show that the absence of FtsH1/3 caused diminished responses to N depletion, as was also observed for Fe, P, and Ci deficiency.
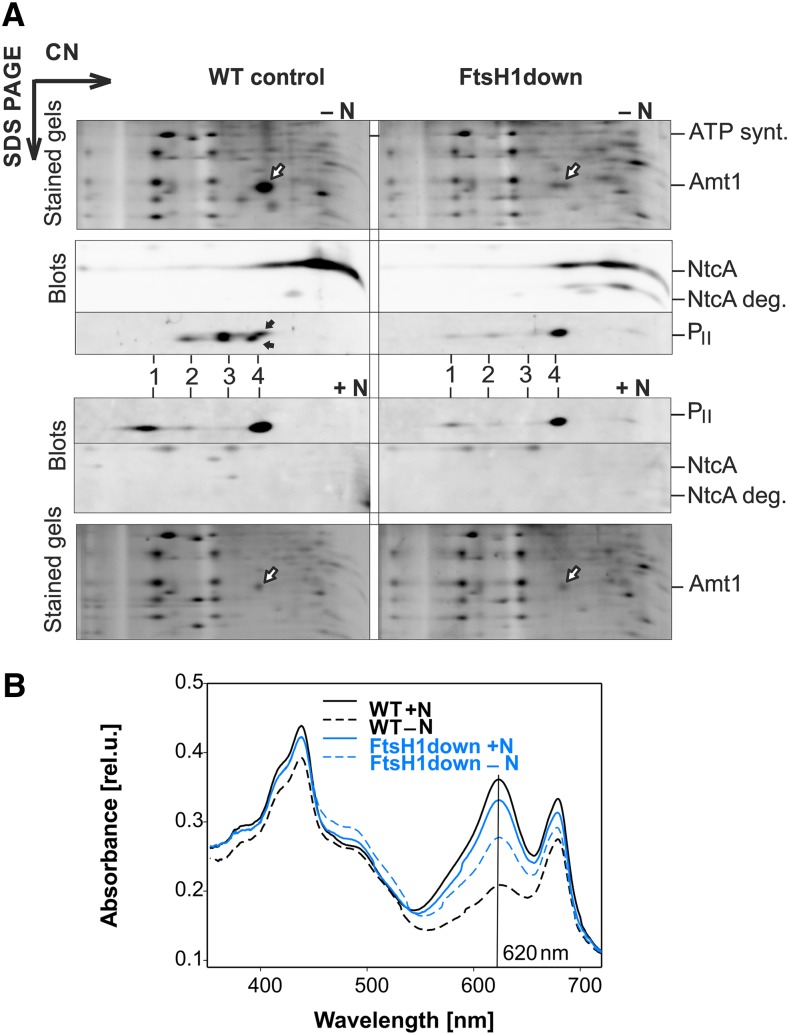
Figure 5.
Protein Distribution and Phycobilin Levels in FtsH1down under N Depletion.
Cells of FtsH1down and the respective wild-type control (WT) were exposed for 24 h to N-deplete conditions (‒N) or grown for another 24 h under N-replete conditions (+N).
(A) Protein complexes isolated from +N/‒N cells were analyzed by 2D CN/SDS-PAGE. The 2D gel was stained with Sypro Orange and used for immunodetection with specific antibodies against NctA and PII. White arrows indicate Amt1; the identity of Amt1 was verified by MS by Linhartová et al. (2014). Black arrows point to a shift in PII migration in the SDS gel, likely due to different phosphorylation states in the WT − N. NtcA was not detectable by immunodetection under N+. Other designations: ATP. synt., AtpA/B subunits of ATP synthase; NtcA deg., putative degradation product of NtcA; 1 to 4, different PII distribution in the CN direction. Each loaded membrane fraction sample contained 5 µg of chlorophyll a.
(B) Whole-cell absorption spectra were measured with cell suspensions of the same OD750 of 0.1. Absorbance at the phycobilin peak maximum (620 nm), indicated by the solid line, reflects the level of phycobiliproteins in the cell. rel.u., relative units.
Recombinant Fur (Sll0567) Is Not Digested by a Membrane Extract Lacking the FtsH1/3 Complex
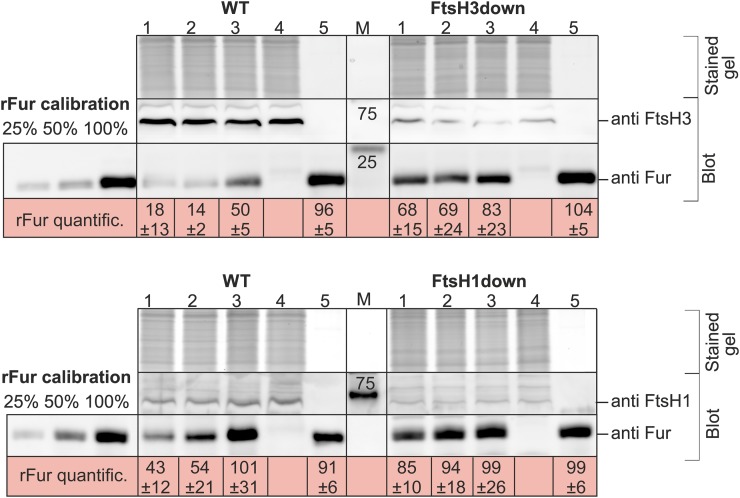
In our previous work (Krynická et al., 2014), we suggested that the diminished response to Fe stress in the FtsH1down or FtsH3down mutant was caused by the accumulation of the negative transcriptional regulator Fur. While the level of Fur decreased significantly in the wild type, it remained high in both FtsH1down and FtsH3down after 72 h of Fe stress. To determine whether the Fur protein might be a direct substrate for FtsH1/3 degradation, we performed an in vitro experiment using recombinant Fur (Sll0567) protein (hereafter rFur). Owing to a lack of in vitro systems for membrane proteases (Yang et al., 2018) and the low stability of purified FtsH1/3 complex, we used a membrane fraction isolated from the Fe-depleted wild-type cells enriched in FtsH1/3 complex (Krynická et al., 2014), and for comparisonwe used a membrane fraction isolated from Fe-depleted cells of the FtsH3down and/or FtsH1down mutant (Figure 6). After a 3-h incubation of rFur with the respective membrane fractions, we examined the level of remaining rFur in the reaction by immunoblotting. The level of rFur decreased significantly after incubation with the wild-type membranes but remained almost unchanged after incubation with FtsH1/3down membranes (Figure 6, sample 1). The addition of EDTA-free protease inhibitor cocktail did not affect the degradation of rFur in the wild-type sample (Figure 6, sample 2), implying that Ser proteases, such as Deg or Clp, were not responsible for Fur degradation. By contrast, the presence of EDTA inhibited the degradation of rFur in the wild-type sample, demonstrating that the degradation is caused by metalloproteases, such as FtsH (Figure 6, sample 3). These results support our hypothesis that FtsH1/3 directly degrades the Fur repressor, thereby regulating its action.
Figure 6.
rFur Degradation Assay in Vitro.
The rFur protein was incubated with the membrane fraction (MF) isolated from iron-depleted cells of FtsH3down and FtsH1down and their respective wild-type control (WT) as described in Methods. Description of reaction samples 1 to 5: 1, MF + rFur in buffer B; 2, MF + rFur + protease inhibitor cocktail (Sigma-Aldrich) in buffer B; 3, MF + rFur + protease inhibitor cocktail + EDTA in buffer B; 4, MF in buffer B; 5, rFur in buffer B; and M, marker. After a 3-h incubation, the samples were denatured by 1% (w/v) SDS and further analyzed by one-dimensional SDS-PAGE. The gel was stained with Sypro Orange for control of loading and used for immunodetection by antibodies specific for Fur, FtsH3, and/or FtsH1. Samples 1 to 4 contained MF with 20 μg of protein in total, before a 3-h incubation; samples 1, 2, 3, and 5 contained 0.5 μg of rFur corresponding to 100% in calibration. rFur quantification (rFur quantific.), percentage of rFur protein in the particular reaction sample related to the initial amount of rFur (100%) shown as means of three measurements of three biological replicates ± sd. Quantification of bands was performed using ImageQuant TL software (GE Healthcare).
Accumulation of Negative Transcriptional Regulators under P and Ci Depletion in the FtsH1down Mutant
To elucidate the role of FtsH in acclimation to other nutrient stresses, we explored the possibility that the diminished or retarded response to P, Ci, and N stress in FtsH1down is also related to the accumulation of transcriptional regulators, as in the case of Fur. Using quantitative proteomic data, we focused in particular on the levels of negative regulators because their accumulation leads to the regulon suppression observed in FtsH1down or FtsH3down mutants.
P Depletion (Pho Regulon)
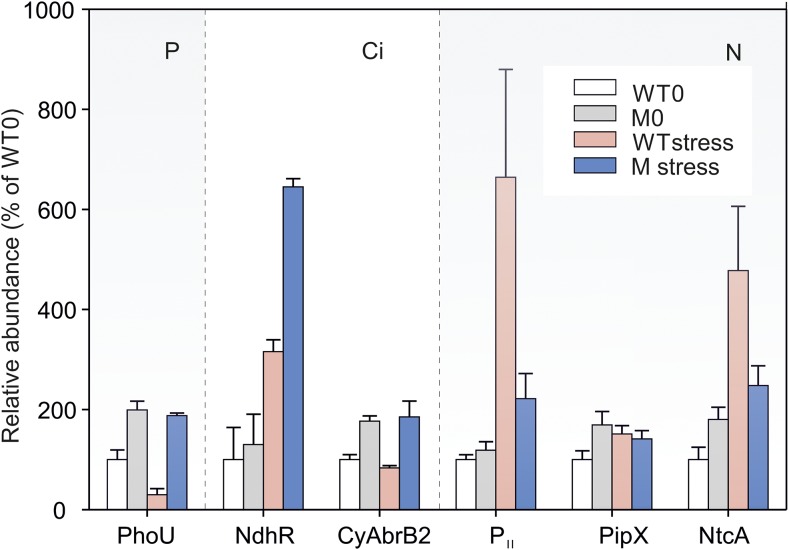
In cyanobacteria, transcription of the Pho regulon is negatively regulated both by PhoU (Slr0741) and the PstSCAB transporting complex (Wanner, 1993; Vuppada et al., 2018). Protein analyses have shown that the levels of Pst transporters were diminished in the FtsH1down mutant (Figures 2, 3A, and 3B). By contrast, the negative regulator PhoU (Slr0741) accumulated in the FtsH1down mutant under both P-replete and -deplete conditions. Under P sufficiency, the PhoU level in the FtsH1down mutant was twofold higher than in the wild type, but under P depletion it was 6.3-fold higher, indicating that PhoU levels rapidly decreased in the wild type (Figure 7). Similar to Fur, these results indicate that the depletion of FtsH1/3 leads to low-level PhoU degradation under P stress and consequent continuous repression of the regulon.
Figure 7.
Accumulation of Transcriptional Regulators under Nutrient Replete/Deplete Conditions.
Proteins extracted from cells of the FtsH1down mutant (M) and its respective wild-type control (WT) grown in nutrient sufficiency (0) or under nutrient depletion (stress) were quantified (except PhoU) as described in Methods. PhoU was quantified manually from extracted ion chromatograms (see Supplemental Data Set 3). PhoU was measured under P depletion, NdhR and CyAbrB2 under Ci depletion, and PII, PipX, and NtcA under N depletion. Relative abundances were expressed as percentage of wild type 0 (WT0) and were calculated from the values shown in the Supplemental Data Set 3. Mean ratio ± sd was n = 3 for PhoU, NdhR, and CyAbrB2, whereas n = 9 (3× biological and 3× technical replicates) for proteins under N stress. Although SDs for PII and NtcA were relatively high in wild type N stress, the difference between the wild type and mutant was statistically significant at P < 0.001 (Figures 3E and 3F; Supplemental Data Set 3).
Ci Depletion (NdhR Regulon)
The expression of Ci acquisition proteins is negatively regulated by NdhR, which represses gene expression under high Ci conditions (Burnap et al., 2015; Jiang et al., 2018). Under Ci-replete conditions, the level of NdhR determined by proteomic analysis was slightly higher in FtsH1down mutant compared with the wild type. After 72 h of Ci depletion, NdhR levels increased in both strains, and importantly they were twofold higher in FtsH1down compared with the wild type (Figure 7). Interestingly, we also detected the accumulation of the regulatory protein CyAbrB2 in FtsH1down under both Ci-replete and -deplete conditions (Figure 7). CyAbrB2 is involved in controlling the expression of several NdhR-regulated genes, such as sbtA/B and ndhF3/ndhD3/cupA. In contrast to NdhR, it activates their expression under low Ci (Orf et al., 2016). Thus, CyAbrB2 accumulation might have contributed to the increase in SbtA/B and CupA protein levels (to the wild-type levels) after 72 h of Ci starvation despite the NdhR accumulation in the FtsH1down mutant.
N Depletion (NtcA Regulon)
To date, no negative regulator of the NtcA regulon has been identified in cyanobacteria. The transcription factor NtcA, the signal transduction protein PII, and the PII-interacting protein PipX are involved in the induction of the NtcA regulon during nutrient stress. According to our quantitative proteomic analysis, the level of PipX in the FtsH1down mutant was comparable to the wild type (Figure 7). By contrast, the levels of both PII and NtcA were more than twofold lower in the N-limited FtsH1down mutant than in the wild type (Figure 7). This is consistent with the finding that both ntcA and glnB (encoding PII) are under the control of the NtcA regulon (Giner-Lamia et al., 2017), whose expression is suppressed in the FtsH1down mutant (Figures 3E and 3F). Notably, we observed signs of NtcA decay in the mutant after 24 h of N depletion (Figure 5A).
PII protein Phosphorylation Is Diminished in the FtsH1down Mutant under N Depletion
Under N-replete conditions, PII forms a complex with PipX (Forchhammer and Tandeau de Marsac, 1994; Spät et al., 2015). By contrast, under N deficiency, PII becomes phosphorylated, resulting in the dissociation of this complex. PipX subsequently binds to NtcA, and this hetero-complex functions as an activator of the NtcA regulon (Espinosa et al., 2014; Spät et al., 2015).
Since PII phosphorylation is required for the induction of the NtcA regulon during nutrient stress, we addressed possible differences between the wild type and the FtsH1down mutant in the distribution of PII complexes and their phosphorylation state in cells exposed to N starvation. 2D gel analysis revealed that under N-replete conditions, PII mainly migrated in the largest and smallest complexes, as found for both the wild type and FtsH1down (Figure 5A). After 24 h of N starvation, a fraction of PII in the wild type slightly increased its mobility in the direction of SDS gel separation (Figure 5A, from upper to lower black arrow), suggesting that it changed its phosphorylation state. The distribution of complexes also changed, with the largest one disappearing and the complexes of an intermediate size becoming more abundant (Figure 5A, band 2 and 3, respectively). Importantly, we observed no changes in PII complexes or PII phosphorylation in the FtsH1down mutant under N starvation compared with N-replete conditions. Comparative proteomic analysis of the mutant confirmed a defect in PII phosphorylation of residue Ser-49, as shown in Figure 8. Using MaxQuant software, we determined the ratio of the Ser-49 phosphorylated form of PII to total PII protein. Under N repletion, 4% of total PII was present in the phosphorylated form in the wild type and just 1.3% in the FtsH1down mutant. After 72 h of N depletion, more than 60% of total PII protein was phosphorylated in the wild type, whereas less than 17% was phosphorylated in the mutant. Consequently, we suggest that defective PII phosphorylation in the FtsH1down mutant results in persistent inhibition of the NtcA regulon. The ΔFtsH2 mutant did not exhibit compromised acclimation to N depletion, and PII distribution on a 2D gel was similar to the wild type (Supplemental Figure 4). Overall, the data point to the requirement of FtsH1/3 (but not FtsH2/3) for the correct acclimation of Synechocystis to N depletion.
Figure 8.
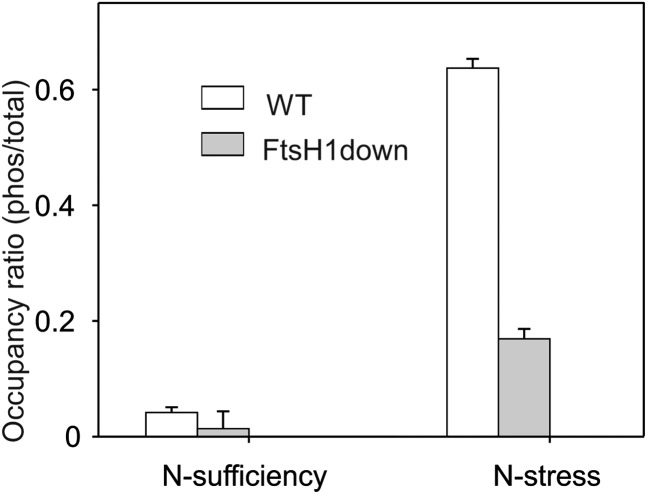
Nitrogen Stress-Induced Phosphorylation of the Ser-49 Residue of the PII Protein in the Wild Type and FtsH1down Mutant.
The tryptic peptides GSEYTVEFLQK and YRGSEYTVEFLQK (representing a missed cleavage site) from PII (Ssl0707), with the Ser-49 residue shown in bold, were detected by MS and database searching, as described in Methods. YRGSEYTVEFLQK was also identified as phosphorylated at Ser-49, as confirmed by product ion spectra shown in Supplemental Figure 6. Occupancy ratios are expressed as YRGpSEYTVEFLQK ion intensity (phos)/(GSEYTVEFLQK + YRGSEYTVEFLQK + YRGpSEYTVEFLQK) ion intensities. The phosphopeptide was only detected in two of the analyses in the FtsH1down mutant under nitrogen sufficiency; therefore, the mean ratio ± sd is n = 2. All other analyses were n = 9 (3× biological and 3× technical replicates).
DISCUSSION
Cyanobacteria have evolved the remarkable capability to rapidly acclimate to a wide range of environmental stimuli. The sophisticated responses of these bacteria to nutritional and environmental stress signals involve the activation or suppression of metabolically important genes. Three major modes of gene expression control function in bacteria: (1) signal transduction by two-component systems, with regulation occurring via response regulators and transcription factors; (2) the recruitment of alternative sigma factors (bacterial transcription initiation factors); and (3) posttranscriptional control via small regulatory RNAs (Hirani et al., 2001; Suzuki et al., 2004; Imamura and Asayama, 2009; Shimizu, 2013; Kopf and Hess, 2015). These regulatory cascades do not act independently of each other; crosstalk between individual regulatory pathways is required for maintaining and fine-tuning cellular metabolic homeostasis. Here, we show that FtsH proteases represent another key functional component of several regulatory pathways involved in stress responses in cyanobacteria. In Synechocystis, the essential FtsH1/3 complex has previously been shown to modulate the level of the transcription factor Fur during Fe deficiency. Here, we demonstrated that FtsH3 suppression leads to the downregulation and in some cases even abolishment of gene expression in the Fur regulon. In addition, under various nutrient stresses, FtsH1/3 affects the expression of other regulatory cascades in Synechocystis; we observed strong repression of the Pho regulon responsible for P metabolism, the NdhR regulon involved in Ci-dependent metabolic regulation, and the NtcA regulon responsible for N metabolism. The basis for these findings was a detailed examination of two mutants limited in FtsH1/3 hetero-complex formation using two different systems for the conditional downregulation of essential FtsH1 or FtsH3. The results were substantiated by both genome-wide transcriptomic and proteomic analyses. Evidence for the regulatory role of the FtsH1/3 complex is further supported by the observation that the excess of FtsH1/3 complex in the FtsH1over mutant led to the induction of the Fur and NdhR regulons, even under nutrient sufficiency (Krynická et al., 2014).
Phosphate Assimilation Is Regulated by FtsH1/3 via Proteolytic Control of the Pho Regulon
Transcriptomic analysis of the FtsH3down mutant indicated that the Pho and SufR regulons, RP genes, and several photosynthetic genes were suppressed due to the lack of FtsH1/3 and/or FtsH2/3 hetero-complexes. Considering that the FtsH2/3 complex is required for quality control of PSII in the thylakoid membrane of Synechocystis (Komenda et al., 2006; Nixon et al., 2010), it is likely that the lack of FtsH2/3 complex had a pleiotropic effect, with overall changes in the photosynthetic apparatus, including the observed downregulation of some photosynthetic genes in the mutant.
Suppression of the Pho regulon was further confirmed by proteomic analysis, and this suppression was proven to be linked exclusively with the absence of the FtsH1/3 complex (Figure 2). Our data suggest that Pho suppression is likely governed by the accumulation of the negative regulator PhoU (Figure 7). Since phoU transcript levels were unchanged under P depletion (Kopf et al., 2014), we suggest that the decreased PhoU levels in the wild type occur via regulation at the posttranscriptional level. Thus, it is tempting to speculate that PhoU abundance is modulated by FtsH1/3 complexes in a manner similar to that of Fur (Figure 6; Krynická et al., 2014). The diminished level of RP in FtsH3down could also be linked to permanent suppression of the Pho regulon, as the biosynthesis of nucleoside triphosphates and hence RNA depends on the availability of P (Tetu et al., 2009). Since we did not observe this phenomenon in the FtsH1down mutant under nutrient sufficiency, we speculate that the downregulation of RP was associated with an exhausted phosphate pool due to the addition of NH4+ for FtsH3 downregulation. NH4+ leads to enhanced phosphate consumption, normally resulting in the induction of the Pho regulon, as was observed here in the wild type (Figure 1). However, in the FtsH3down mutant, the addition of NH4+ led to P deprivation because P transport could not be de-repressed. This explanation is supported by the finding that RP deprivation was also detected in other analyzed strains only after exposure to P limitation (Supplemental Figure 5).
The suppression of SufR regulon genes involved in Fe-S cluster biogenesis appears to be analogous to the suppression of Fur-regulated genes, as their expression also responds to Fe availability and SufR functions as a repressor (Shen et al., 2007). Even though we do not have proteomic data demonstrating the accumulation of SufR protein in the mutant under Fe depletion, our transcriptomic results fit perfectly into the scheme of SufR as a repressor controlled by FtsH. Thus, hypothetically, SufR could be another candidate for FtsH1/3 control.
The Regulation of NdhR by Two FtsH Complexes Provides a Link between Nutrient Availability and Photosynthesis
Proteomic analyses have shown that both FtsH1/3 and FtsH2/3 complexes contribute to the cellular responses to Ci assimilation. Not only ΔFtsH2 (Zhang et al., 2007) but also FtsH3down and FtsH1down were unable to quickly induce the expression of the NdhR regulon. The quantitative MS data revealed that the NdhR transcription factor, functioning mainly as a regulon repressor, over-accumulated in the FtsH1down strain compared to the wild type (Figure 7). Since the ndhR gene is auto-regulated (Klähn et al., 2015) and hence its expression is suppressed in the FtsH1down mutant compared to the wild type, we suggest that the NdhR protein ectopically accumulates in this mutant, in a manner similar to that of Fur and PhoU, as a consequence of the lack of FtsH proteases. The requirements for both FtsH2/3 and FtsH1/3 complexes provide insights into the processes involved in the induction of Ci acquisition. Ci is an essential nutrient for photosynthesis. Thus, the FtsH2/3 complex, which is crucial for maintaining PSII activity, could facilitate the coordination between the light reaction and carbon fixation. In addition, the FtsH1/3 complex, which regulates the uptake of various nutrients, might provide further assistance to photosynthetic activities. This cooperation also points to the possibility that FtsH1/3 and FtsH2/3 can substitute for each other under certain circumstances.
Reduced Levels of the FtsH1/3 Complex Affect the NtcA Regulon via Its Effects on the Phosphorylation State of PII
We demonstrated that the FtsH1/3 complexes are also involved in N assimilation by regulating the expression of the NtcA regulon. However, the molecular mechanism of such regulation remains unclear. Unlike Fur, PhoU, and NdhR, NtcA functions as a regulon activator. Thus, in theory, NtcA is not a suitable candidate for FtsH digestion, since its degradation would lead to repression instead of induction of the NtcA regulon in the wild type under stress. In accordance with this theory, we did not observe NtcA accumulation in the FtsH1down mutant. Quite the opposite, owing to auto-regulation, its level was reduced due to the suppression of the NtcA regulon (Figure 7).
We showed that the absence of FtsH1/3 complexes in the FtsH1down mutant led to inefficient phosphorylation of PII, which may have resulted in the suppression of the NtcA regulon under N depletion (Figure 8). Nonphosphorylated PII prevents the formation of active NtcA-PipX complexes by itself binding to PipX in place of NtcA (Espinosa et al., 2014). Since FtsHs generally exhibit protease and chaperon, but not kinase, activity, we assume that they promote PII phosphorylation rather indirectly. However, the mechanism for this phenomenon remains unknown. Results from quantitative MS analysis refute the possibility that impaired PII phosphorylation is caused by the accumulation of PphA phosphatase (Sll1771), the enzyme responsible for PII dephosphorylation, under N-replete conditions (Kloft et al., 2005). Since the PphA level under N-depletion was significantly lower in the mutant compared with the wild type (Supplemental Data Set 3), we do not consider PphA to be a putative substrate for FtsH1/3 degradation. The observed block in PII phosphorylation could also be explained by low kinase levels/activity, but this question remains open, since the kinase responsible for PII phosphorylation has not yet been identified. Overall, the mechanism regulating PII and NtcA could be more complex than our current understanding. Nevertheless, FtsH1/3 is clearly a component of the regulatory pathway for N starvation.
The Essential FtsH Complex Might Function as an Epistatic Regulator of the Nutrient Stress Response
Our transcriptomic and proteomic analyses revealed that suppressing the FtsH1/3 complex affects the expression of several regulons associated with nutrient acquisition and that the diminished or retarded response of these regulons is related to their respective transcriptional regulators. Under nutrient deficiency, in mutants limited in FtsH1/3, the negative regulators remained at high levels instead of being degraded as in the wild type. In the case of Fur, our data support direct proteolytic degradation via FtsH1/3 (Figure 6). A similar regulatory mechanism might apply to P and Ci assimilation via FtsH1/3-mediated degradation of the PhoU and NdhR repressors. On the other hand, the NtcA regulatory cascade might function via FtsH1/3-mediated phosphorylation by an as-yet-unknown kinase. Based on our results, we propose a model describing how FtsH1/3 functions as an epistatic regulator of the transcriptional nutrient stress response in Synechocystis (Supplemental Figure 7). According to our model, transcriptional regulators can exist in an active form, which have high affinity for their targets (e.g., the Fur holo-enzyme with Fe as a cofactor targeted to promoters with a Fur-box) and an inactive form, with a lower but still relevant affinity for their targets (e.g., the Fur apo-protein). The ratio of the active to the inactive regulator is dependent on the nutrient state of the bacteria or general growth conditions. In our model, the inactive regulator is also strongly prone to degradation by FtsH, while the active regulator is a low-affinity target (Supplemental Figure 7A). The differential FtsH activity might be due to the protection of vulnerable sites of the active form by cofactor or DNA binding in a repressor complex. Inactive regulators might become accessible for degradation due to the loss of the relevant nutrient, which leads to their release from the repressor complex to the cytoplasm. Without FtsH, the inactive regulator can accumulate at relatively high concentrations and thus still bind to its target. In conclusion, the full response to nutrient stress requires not only the conversion of the regulator from the active to inactive form but also the degradation of the inactive form by FtsH. This model leads to the conclusion that the amplitude of the stress response is dependent not only on nutrient status but also on the concentrations of FtsH1/3 complexes (Supplemental Figure 7B).
FtsH might also regulate the levels of transcription factors indirectly by controlling other signal transducers superior to all regulatory pathways that operate between FtsH and individual regulons. Beyond this, FtsHs also maintain chaperone activity independent of their proteolytic activity (Li et al., 2013). Thus, we cannot exclude the possibility that the chaperone activity of these enzymes rather than their proteolytic activity might be important for the digestion of repressors, or perhaps the effects of FtsH1/3 could be seen as a combination of both functions.
Even though the mechanistic details have not yet been fully established, we demonstrated that the roles of FtsHs in regulating nutrient assimilation are more diverse than previously thought. In addition to the Fur regulon, the essential FtsH1/3 complex facilitates the full responses of the Pho, NdhR, and NtcA regulons (Figure 9). Since the FtsH1/3 complex functions upstream of individual transcription factors, it enables faster dynamics in tuning the cellular responses to stimuli. The FtsH-mediated regulation provides cyanobacteria—and possibly all FtsH-containing bacteria and cell organelles—with a pivotal mechanism to adjust the strength of transcriptional responses to stress conditions in a coordinated manner and thus to control the overall transcriptional plasticity of the organism.
Figure 9.
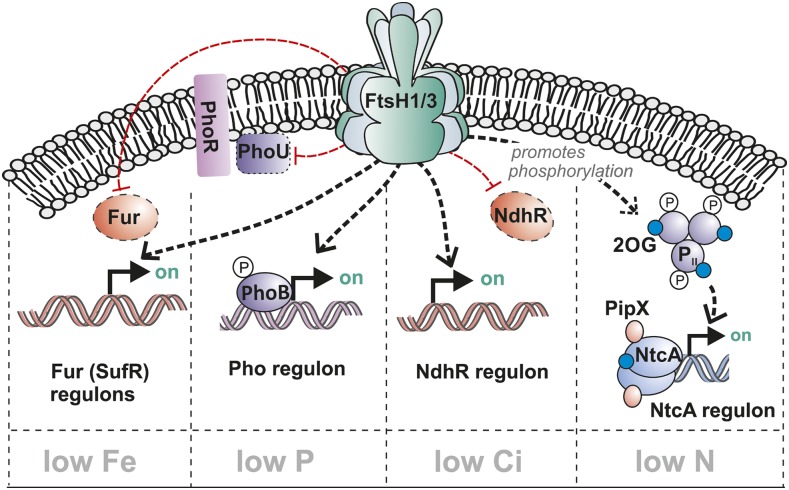
FtsH1/3 Complex Facilitates a Full Transcriptional Response to Fe, P, Ci, and N Stress Conditions.
Under nutrient limitation, the FtsH1/3 complex enhances the expression of the Fur, NdhR, and/or Pho regulon by reducing the levels of transcription factors Fur and NdhR and of PhoU, a negative effector of the two-component system PhoB/R. The FtsH1/3 complex also enhances the transcriptional response of the NtcA regulon: by an unknown mechanism, it promotes PII phosphorylation, enabling the formation of the PipX/NtcA complex, functioning as an activator of the NtcA regulon. These processes depend on the availability of the relevant nutrient. Being involved in several regulatory pathways, FtsH1/3 may function as a linker enabling the strength of transcriptional responses to be adjusted in response stress conditions in a coordinated manner. 2OG, 2-oxoglutarate.
METHODS
Strains and Cultivation
The previously described FtsH3down (originally termed SynFtsH3reg; Boehm et al., 2012), FtsH2-less (ΔFtsH2) (Komenda et al., 2006), FtsH1down (Krynická et al., 2014) Synechocystis sp PCC 6803 strains as well as the wild-type control were derived from the nonmotile Glc-tolerant Synechocystis strain obtained from the laboratory of Wim Vermaas (Tichý et al., 2016). All liquid cultures (50 mL) were grown in an orbital shaker in 250-mL conical flasks at 29°C, and 40 µmol photons m−2 s−1 of light was provided by cool-white fluorescence tubes. After initial growth, all cultures were maintained at OD750 nm of 0.3 to 0.6. The BG-11 growth medium for the suppression of FtsH3 and/or FtsH1, respectively, was modified as described in this section, and cells were grown in this medium for 5 d before the imposition of nutrient stress. This modification was also used for the duration of the nutrient stress. FtsH3down and the respective wild-type control were cultivated photo-autotrophically in the presence of 13 mM NH4+ to suppress the level of FtsH3 in FtsH3down as described by Boehm et al. (2012). The FtsH1down strain and the wild-type control were grown photoautotrophically in the presence of 0.8 μM copper to suppress the level of FtsH1 in FtsH1down as described by Krynická et al. (2014). The ΔFtsH2 strain and control (WTglc) were cultivated in the presence of 5 mM Glc. Samples prepared from the FtsH1down, FtsH3down, and ΔFtsH2 mutants were compared with their respective controls in all experiments. To deplete Fe, cells were transferred to an Fe-depleted medium (with a residual Fe concentration of ∼5 µM) supplemented with 10 μM deferoxamine B, an Fe chelator (Krynická et al., 2014). For P depletion, cells were transferred to BG-11 medium in which K2HPO4 (0.18 mM) had been replaced by an equimolar amount of KCl (total Cl− concentration was 0.31 mM; Aiba et al., 1993). For N depletion, cells were transferred to N-free medium as described by Schlebusch and Forchhammer, (2010). For Ci limitation, the access of the cells to air was limited using a Parafilm cover with a 2-mm-diameter hole. Growth curves of the FtsH1down mutant and its respective control under Ci and P depletion are presented in Supplemental Figure 8. Cells were analyzed at 24, 48, or 72 h after nutrient starvation. Unless indicated otherwise, experiments and measurements with cells were performed at least in duplicate (biological replicates).
RNA Extraction and Microarray Analysis
The wild type and FtsH3down cultures were harvested by rapid filtration on hydrophilic polyethersulfone filters (Pall Supor 800 Filter, 0.8 mm). The filter covered with cells was immediately immersed in 1 mL of PGTX (Pinto et al., 2009). RNA was extracted and analyzed by gel electrophoresis. The array design and the RNA processing and hybridization protocols are described by Hernandez-Prieto and Futschik (2012) and Hernandez-Prieto et al. (2012). Agilent Feature Extraction software version 10.7.3.1 with the protocol GE1_107_Sep09 was used to extract the raw signal intensities, which were analyzed with the limma package in R (Smyth, 2005). Data were preprocessed with normexp background subtraction and cyclicloess normalization. Control spots were omitted prior to the statistical evaluation of FCs. The P-values were corrected for multiple testing by the Benjamini–Hochberg method (Benjamini and Hochberg, 1995).
Analysis of Proteins and Their Complexes
Membrane protein complexes were analyzed by CN PAGE in combination with SDS-PAGE and immunoblot analysis as described by Komenda et al. (2012). The protein composition of the complexes was analyzed by electrophoresis in a denaturing 12 to 20% (w/v) linear gradient polyacrylamide gel containing 7 M urea. Complete lanes from the native gel were excised, incubated for 30 min in 25 mM Tris-HCl, pH 7.5, containing 1% SDS (w/v), and placed on top of the denaturing gel; two lanes were analyzed in a single denaturing gel. Proteins separated in the gel were stained with Coomassie Brilliant Blue G 250 and used for MS or with Sypro Orange and subsequently transferred onto a polyvinylidene difluoride membrane. Membranes were incubated with specific primary antibodies and then with secondary antibody-horseradish peroxidase conjugate (Sigma-Aldrich). The primary antibodies used in this study were anti-Fur (Krynická et al., 2014, dilution, 1:4000), anti-PII (Spät et al., 2015, dilution, 1:2000), and anti-NtcA (Agrisera, catalog no. AS12 1873, dilution, 1:1000). The Sypro Orange protein fluorescence and Coomassie blue protein stain in the gel, as well as chemiluminescence of the blot, were recorded using a LAS-4000 imaging system (Fuji), and quantification of bands was performed using ImageQuant TL software (GE Healthcare).
Tryptic In-Gel Digestion and Protein Identification by MS
The Coomassie blue–stained protein spots separated by 2D CN/SDS-PAGE were identified after tryptic digestion on a NanoAcquity UPLC (Waters) online coupled to an electrospray ionization Q-ToF Premier mass spectrometer (Waters) as described by Janouskovec et al., (2013).
Quantitative Proteomic Analysis by MS
Cell pellets from 30 mL of liquid culture were resuspended in 50 μL of 2% SDS (w/v) and 60 mM DTT and transferred to 1.5-mL Eppendorf tubes containing 50 μL of 0.1-mm silica/zirconia beads (BioSpec). The cells were lysed, and the proteins were solubilized by incubation at 95°C for 90 s and vortexing for 30 s. This sequence was repeated three more times before the addition of 50 μL of water. The samples were centrifuged briefly to pellet the beads and enable recovery of the cell extracts. Proteins were isolated from the extracts by precipitation using a 2D Clean-up kit (GE Healthcare) according to the manufacturer’s instructions and redissolved in 30 μL of 8 M urea and 100 mM Tris-HCl, pH 8.5. Aliquots containing 50 µg of protein (Nanodrop assay at 280 nm) were diluted to 10 μL with urea/Tris and subjected to reduction, S-carbamidomethylation, and endoproteinase Lys-C/trypsin digestion followed by analysis using nano-flow liquid chromatography on-line to a Q Exactive HF hybrid quadrupole-Orbitrap mass spectrometer according to Hitchcock et al. (2016). All samples were represented by three technical replicates (within a single experiment). N-stressed samples were also represented by three biological replicates (individual experiments), making a total of nine replicates. Proteins were identified by searching a Synechocystis proteome database (http://genome.microbedb.jp/cyanobase/) using MaxQuant (v. 1.6.0.16; Cox and Mann, 2008) with the intensity-based absolute quantification option (Schwanhäusser et al., 2011) selected. Phospho-Ser sites and their occupancy levels were determined by selecting this option as a variable modification. Protein identifications and associated intensity-based absolute quantification intensities were processed using Perseus (v. 1.6.0.7; Tyanova et al., 2016). The levels of AtpA and AtpC subunits of ATP synthase routinely used as a standard for immunoblot analysis (Rasala et al., 2013), and FtsH2 were shown as a control. None of these proteins is known to be a direct target of the regulatory pathways listed in the "Results" section.
Cloning, Expression, and Purification of Fur Protein
To obtain rFur protein (C-terminal His6 tag Sll0567), the Synechocystis sll0567 gene was cloned into the pET14b plasmid according to Adams et al. (2014) using 5′-CTAGAGCATATGTCCTACACCGCCGATTCCC-3′ (forward) and 5′-GAGGACGGATCCCTAGGCCAAGGAAATACTGCGG-3′ (reverse) primers and NdeI and BamHI restriction sites. The plasmid was transformed into Escherichia coli BL21 cells and grown in 500 mL of Lysogeny broth medium at 37°C to a D600 of 0.6. Expression of rFur protein was then induced with 0.4 mM isopropyl B-D-thiogalactopyranoside (IPTG), and the cultures were transferred to 20°C conditions overnight. rFur was purified according to Pellicer et al. (2010) using 5 mL of Ni-chelating Sepharose columns instead of Zn-iminodiacetate column. Bound protein was washed with 40 mM imidazole and eluted by 250 mM imidazole in buffer A (Pellicer et al., 2010). The purified protein was dialyzed against 10 mM acetic acid buffer at pH 4. Analysis of the resulting protein was performed by SDS-PAGE. The concentration of purified protein was assessed using Coomassie Plus (Bradford) Protein Assay Reagent (Thermo Fisher Scientific).
In Vitro rFur Digestion
Membranes (20 µg of total protein) isolated from the wild type, FtsH1down, and FtsH3down cells grown under iron depletion (as described in “Strains and Cultivation”) were incubated with 0.5 µg of rFur for 3 h at 37°C in buffer B (Figure 6, sample 1). We further performed the same experiment in the presence of EDTA-free protease inhibitor cocktail (Sigma-Aldrich; Figure 6, sample 2) or 5 mM EDTA (Figure 6, sample 3). As a control, we used a reaction mixture lacking membranes (Figure 6, sample 4) or rFur (Figure 6, sample 5). Composition of the reaction buffer B used for all reaction mixtures: MES buffer, pH 6.5, 10 mM MgCl2, 100 mM KCl, 0.4 mM ZnCl2, 3 mM ATP, and 5% (w/w) glycerol. Reaction volume was 30 µL. The reaction was stopped by denaturation using 1% (w/v) SDS, and 10 μL of each sample was analyzed by SDS-PAGE in combination with immunoblot analysis using the previously described primary antibody anti-Fur (Hernández et al., 2002; Krynická et al., 2014).
Accession Numbers
The microarray data can be accessed in the Gene Expression Omnibus database (GSE83303) and the CyanoEXpress database (http://cyanoexpress.sysbiolab.eu), which enables visualization and interactive inspection of the obtained expression patterns (Hernandez-Prieto and Futschik, 2012).
The MS proteomics data, including MaxQuant analysis txt files containing all information about identification scores, elution times, and so on, have been deposited in the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the data set identifier PXD013401.
Supplemental Data
Supplemental Figure 1. Correlation matrix for three replicate proteomic analyses of thylakoid membranes from wild-type (WT) Synechocystis cells grown under nutrient sufficiency.
Supplemental Figure 2. Accumulation of CCM and PSII in WT, FtsH3down, and ΔFtsH2 cells after 72 h of Ci limitation.
Supplemental Figure 3. Quantification of the proteins from the gel.
Supplemental Figure 4. Amt1 levels and PII distribution in ΔFtsH2 under nitrogen depletion.
Supplemental Figure 5. Accumulation of RP in FtsH mutants.
Supplemental Figure 6. Evidence for the phosphorylation of residue Ser49 of the nitrogen regulatory protein PII.
Supplemental Figure 7. Proposed working model for FtsH1/3 as an epistatic regulator.
Supplemental Figure 8. Growth curves of the WT and FtsH1down strains during Ci and P stress.
Supplemental Table. Identification of the protein composition of the three gel bands shown in Figure 5 by MS.
Supplemental Data Set 1. Genome wide overview of signal intensities.
Supplemental Data Set 2. Microarray-based gene expression analysis.
Supplemental Data Set 3. Whole cell comparative proteome analysis.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Peter Koník for the MS analyses of selected protein gel spots. This work was supported by the Germany Federal Ministry of Education and Research (de.STAIR grant 031L0106B to W.R.H.), by the Grant Agency of the Czech Republic (P501-12-G055 to V.K. and J.K.), and the Czech Ministry of Education (projects LO1416 to V.K. and J.K.). Funding was also provided by the Portuguese Fundação para a Ciência e a Tecnologia (Foundation for Science and Technology; PTDC/BIA-MIC/4418/2012, IF/00881/2013, and UID/Multi/04326/2013–The Centre of Marine Sciences to M.E.F.), by the United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC; BB/M012166/1 to M.J.D. and BB/M000265/1 to P.J.J. and C.N.H.), and by the European Research Council (Advanced Award 338895 to P.J.J. and C.N.H.).
AUTHOR CONTRIBUTIONS
V.K., W.R.H., and J.K. designed the research; V.K., J.G., and P.J.J. carried out the experiments; V.K., J.G.,.P.J.J., and M.E.F analyzed the data; V.K., M.E.F., J.G., and W.R.H. developed the theory; and V.K., J.G., P.J.J., M.J.D., C.N.H., M.E.F., W.R.H., and J.K. discussed the results and contributed to the final article.
References
- Adams N.B., Marklew C.J., Brindley A.A., Hunter C.N., Reid J.D. (2014). Characterization of the magnesium chelatase from Thermosynechococcus elongatus. Biochem. J. 457: 163–170. [DOI] [PubMed] [Google Scholar]
- Aiba H., Nagaya M., Mizuno T. (1993). Sensor and regulator proteins from the cyanobacterium Synechococcus species PCC7942 that belong to the bacterial signal-transduction protein families: Implication in the adaptive response to phosphate limitation. Mol. Microbiol. 8: 81–91. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Boehm M., Yu J., Krynicka V., Barker M., Tichy M., Komenda J., Nixon P.J., Nield J. (2012). Subunit organization of a synechocystis hetero-oligomeric thylakoid FtsH complex involved in photosystem II repair. Plant Cell 24: 3669–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnap R.L., Hagemann M., Kaplan A. (2015). Regulation of CO2 concentrating mechanism in Cyanobacteria. Life (Basel) 5: 348–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Crosa J.H. (1997). Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol. Mol. Biol. Rev. 61: 319–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian C., Vitale S., Leonard G.A., Bahlawane C., Fauquant C., Leduc D., Muller C., de Reuse H., Michaud-Soret I., Terradot L. (2011). The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol. Microbiol. 79: 1260–1275. [DOI] [PubMed] [Google Scholar]
- Escolar L., Pérez-Martín J., de Lorenzo V. (1999). Opening the iron box: Transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181: 6223–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa J., Rodríguez-Mateos F., Salinas P., Lanza V.F., Dixon R., de la Cruz F., Contreras A. (2014). PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc. Natl. Acad. Sci. USA 111: E2423–E2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K., Tandeau de Marsac N. (1994). The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J. Bacteriol. 176: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J., et al. (2017). Acclimation of oxygenic photosynthesis to iron starvation is controlled by the sRNA IsaR1. Curr. Biol. 27: 1425–1436. [DOI] [PubMed] [Google Scholar]
- Giner-Lamia J., Robles-Rengel R., Hernández-Prieto M.A., Muro-Pastor M.I., Florencio F.J., Futschik M.E. (2017). Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 45: 11800–11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Angarica V.E., Sancho J., Fillat M.F. (2014). The FurA regulon in Anabaena sp. PCC 7120: In silico prediction and experimental validation of novel target genes. Nucleic Acids Res. 42: 4833–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Bes M.T., Peleato M.L., Fillat M.F. (2016). Expanding the role of FurA as essential global regulator in Cyanobacteria. PLoS One 11: e0151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A., Bes M.T., Valladares A., Peleato M.L., Fillat M.F. (2012). FurA is the master regulator of iron homeostasis and modulates the expression of tetrapyrrole biosynthesis genes in Anabaena sp. PCC 7120. Environ. Microbiol. 14: 3175–3187. [DOI] [PubMed] [Google Scholar]
- Hernández J.A., Bes M.T., Fillat M.F., Neira J.L., Peleato M.L. (2002). Biochemical analysis of the recombinant Fur (ferric uptake regulator) protein from Anabaena PCC 7119: Factors affecting its oligomerization state. Biochem. J. 366: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Prieto M.A., Futschik M.E. (2012). CyanoEXpress: A web database for exploration and visualisation of the integrated transcriptome of cyanobacterium Synechocystis sp. PCC6803. Bioinformation 8: 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Prieto M.A., Schoen V., Georg J., Barreira L., Varela J., Hess W.R., Futschik M.E. (2012). Iron deprivation in synechocystis: Inference of pathways, non-coding RNAs, and regulatory elements from comprehensive expression profiling. G3 2: 1475–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirani T.A., Suzuki I., Murata N., Hayashi H., Eaton-Rye J.J. (2001). Characterization of a two-component signal transduction system involved in the induction of alkaline phosphatase under phosphate-limiting conditions in Synechocystis sp. PCC 6803. Plant Mol. Biol. 45: 133–144. [DOI] [PubMed] [Google Scholar]
- Hitchcock A., Jackson P.J., Chidgey J.W., Dickman M.J., Hunter C.N., Canniffe D.P. (2016). Biosynthesis of chlorophyll a in a purple bacterial phototroph and assembly into a plant chlorophyll-protein complex. ACS Synth. Biol. 5: 948–954. [DOI] [PubMed] [Google Scholar]
- Huergo L.F., Chandra G., Merrick M. (2013). P(II) signal transduction proteins: Nitrogen regulation and beyond. FEMS Microbiol. Rev. 37: 251–283. [DOI] [PubMed] [Google Scholar]
- Imamura S., Asayama M. (2009). Sigma factors for cyanobacterial transcription. Gene Regul. Syst. Bio. 3: 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskovec J., Sobotka R., Lai D.-H., Flegontov P., Koník P., Komenda J., Ali S., Prásil O., Pain A., Oborník M., Lukes J., Keeling P.J. (2013). Split photosystem protein, linear-mapping topology, and growth of structural complexity in the plastid genome of Chromera velia. Mol. Biol. Evol. 30: 2447–2462. [DOI] [PubMed] [Google Scholar]
- Jiang Y.L., et al. (2018). Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR. Proc. Natl. Acad. Sci. USA 115: 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H., Hagino N., Grossman A.R., Ogawa T. (2001). Genes essential to iron transport in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183: 2779–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klähn S., Orf I., Schwarz D., Matthiessen J.K., Kopka J., Hess W.R., Hagemann M. (2015). Integrated transcriptomic and metabolomic characterization of the low-carbon response using an ndhR mutant of Synechocystis sp. PCC 6803. Plant Physiol. 169: 1540–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloft N., Rasch G., Forchhammer K. (2005). Protein phosphatase PphA from Synechocystis sp. PCC 6803: The physiological framework of PII-P dephosphorylation. Microbiology 151: 1275–1283. [DOI] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C.W., Tichy M., Nixon P.J. (2006). The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281: 1145–1151. [DOI] [PubMed] [Google Scholar]
- Komenda J., Knoppová J., Kopečná J., Sobotka R., Halada P., Yu J., Nickelsen J., Boehm M., Nixon P.J. (2012). The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 158: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Hess W.R. (2015). Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol. Rev. 39: 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Klähn S., Scholz I., Matthiessen J.K.F., Hess W.R., Voß B. (2014). Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 21: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krynická V., Tichý M., Krafl J., Yu J., Kaňa R., Boehm M., Nixon P.J., Komenda J. (2014). Two essential FtsH proteases control the level of the Fur repressor during iron deficiency in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 94: 609–624. [DOI] [PubMed] [Google Scholar]
- Li W., Rao D.K., Kaur P. (2013). Dual role of the metalloprotease FtsH in biogenesis of the DrrAB drug transporter. J. Biol. Chem. 288: 11854–11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhartová M., Bučinská L., Halada P., Ječmen T., Setlík J., Komenda J., Sobotka R. (2014). Accumulation of the Type IV prepilin triggers degradation of SecY and YidC and inhibits synthesis of Photosystem II proteins in the cyanobacterium Synechocystis PCC 6803. Mol. Microbiol. 93: 1207–1223. [DOI] [PubMed] [Google Scholar]
- Mann N.H., Novac N., Mullineaux C.W., Newman J., Bailey S., Robinson C. (2000). Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479: 72–77. [DOI] [PubMed] [Google Scholar]
- Morohoshi T., Maruo T., Shirai Y., Kato J., Ikeda T., Takiguchi N., Ohtake H., Kuroda A. (2002). Accumulation of inorganic polyphosphate in phoU mutants of Escherichia coli and Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 68: 4107–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orf I., Schwarz D., Kaplan A., Kopka J., Hess W.R., Hagemann M., Klähn S. (2016). CyAbrB2 contributes to the transcriptional regulation of low CO2 acclimation in Synechocystis sp. PCC 6803. Plant Cell Physiol. 57: 2232–2243. [DOI] [PubMed] [Google Scholar]
- Pellicer S., Bes M.T., González A., Neira J.L., Peleato M.L., Fillat M.F. (2010). High-recovery one-step purification of the DNA-binding protein Fur by mild guanidinium chloride treatment. Process Biochem. 45: 292–296. [Google Scholar]
- Perez-Riverol Y., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1): D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto F.L., Thapper A., Sontheim W., Lindblad P. (2009). Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Barrera D.J., Ng J., Plucinak T.M., Rosenberg J.N., Weeks D.P., Oyler G.A., Peterson T.C., Haerizadeh F., Mayfield S.P. (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 74: 545–556. [DOI] [PubMed] [Google Scholar]
- Schlebusch M., Forchhammer K. (2010). Requirement of the nitrogen starvation-induced protein Sll0783 for polyhydroxybutyrate accumulation in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 76: 6101–6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G., Balasubramanian R., Wang T., Wu Y., Hoffart L.M., Krebs C., Bryant D.A., Golbeck J.H. (2007). SufR coordinates two [4Fe-4S]2+, 1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J. Biol. Chem. 282: 31909–31919. [DOI] [PubMed] [Google Scholar]
- Shimizu K. (2013). Regulation systems of bacteria such as Escherichia coli in response to nutrient limitation and environmental stresses. Metabolites 4: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Schwarz R., Forchhammer K. (2005). Acclimation of unicellular cyanobacteria to macronutrient deficiency: Emergence of a complex network of cellular responses. Microbiology 151: 2503–2514. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2005). Limma: linear models for microarray data In Bioinformatics and Computational Biology Solution Using R and Bioconductor, Gentleman R., Carey V., Dudoit S., Irizarry R., and Huber W., eds (New York: Springer; ), pp. 397–420. [Google Scholar]
- Spät P., Maček B., Forchhammer K. (2015). Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation. Front. Microbiol. 6: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Olman V., Xu Y. (2007). Computational prediction of Pho regulons in cyanobacteria. BMC Genomics 8: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Ferjani A., Suzuki I., Murata N. (2004). The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in synechocystis. J. Biol. Chem. 279: 13234–13240. [DOI] [PubMed] [Google Scholar]
- Tetu S.G., Brahamsha B., Johnson D.A., Tai V., Phillippy K., Palenik B., Paulsen I.T. (2009). Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J. 3: 835–849. [DOI] [PubMed] [Google Scholar]
- Tichý M., Bečková M., Kopečná J., Noda J., Sobotka R., Komenda J. (2016). Strain of Synechocystis PCC 6803 with aberrant assembly of photosystem II contains tandem duplication of a large chromosomal region. Front. Plant Sci. 7: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13: 731–740. [DOI] [PubMed] [Google Scholar]
- Urech C., Koby S., Oppenheim A.B., Münchbach M., Hennecke H., Narberhaus F. (2000). Differential degradation of Escherichia coli sigma32 and Bradyrhizobium japonicum RpoH factors by the FtsH protease. Eur. J. Biochem. 267: 4831–4839. [DOI] [PubMed] [Google Scholar]
- Vuppada R.K., Hansen C.R., Strickland K.A.P., Kelly K.M., McCleary W.R. (2018). Phosphate signaling through alternate conformations of the PstSCAB phosphate transporter. BMC Microbiol. 18: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B.L. (1993). Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51: 47–54. [DOI] [PubMed] [Google Scholar]
- Yang Y., Guo R., Gaffney K., Kim M., Muhammednazaar S., Tian W., Wang B., Liang J., Hong H. (2018). Folding-degradation relationship of a membrane protein mediated by the universally conserved ATP-dependent protease FtsH. J. Am. Chem. Soc. 140: 4656–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Fu A., Aluru M., Park S., Xu Y., Liu H., Liu X., Foudree A., Nambogga M., Rodermel S. (2007). Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 30: 350–365. [DOI] [PubMed] [Google Scholar]
- Zhang P., Sicora C.I., Vorontsova N., Allahverdiyeva Y., Battchikova N., Nixon P.J., Aro E.-M. (2007). FtsH protease is required for induction of inorganic carbon acquisition complexes in Synechocystis sp. PCC 6803. Mol. Microbiol. 65: 728–740. [DOI] [PubMed] [Google Scholar]