Plant cGMP-dependent protein kinase has both kinase and phosphatase activities and regulates the nucleocytoplasmic distribution of the GAMYB transcription factor, modulating gibberellin signaling.
Abstract
Cyclic GMP (cGMP) is an important regulator in eukaryotes, and cGMP-dependent protein kinase (PKG) plays a key role in perceiving cellular cGMP in diverse physiological processes in animals. However, the molecular identity, property, and function of PKG in plants remain elusive. In this study, we have identified PKG from plants and characterized its role in mediating the gibberellin (GA) response in rice (Oryza sativa). PKGs from plants are structurally unique with an additional type 2C protein phosphatase domain. Rice PKG possesses both protein kinase and phosphatase activities, and cGMP stimulates its kinase activity but inhibits its phosphatase activity. One of PKG’s targets is GAMYB, a transcription factor in GA signaling, and the dual activities of PKG catalyze the reversible phosphorylation of GAMYB at Ser6 and modulate the nucleocytoplasmic distribution of GAMYB in response to GA. Loss of PKG impeded the nuclear localization of GAMYB and abolished GAMYB function in the GA response, leading to defects in GA-induced seed germination, internode elongation, and pollen viability. In addition to GAMYB, PKG has multiple potential targets and thus has broad effects, particularly in the salt stress response.
INTRODUCTION
Cyclic GMP (cGMP) is an important secondary messenger involved in various signaling events in eukaryotic cells (Newton and Smith, 2004; Schmidt et al., 2009; Gehring and Turek, 2017). In mammalian cells, cGMP is produced from GTP by nitric oxide (NO)–triggered soluble guanylate cyclases (GCs) and degraded by cGMP phosphodiesterases (PDEs; Lucas et al., 2000; Friebe and Koesling, 2003; Francis et al., 2010). cGMP elevation resulting from guanylate cyclase activation and/or PDE inhibition usually leads to significant physiological effects. In plants, NO-triggered cGMP production is detectable in response to different stimuli including phytohormones and stresses in a variety of plant species (Penson et al., 1996; Pagnussat et al., 2003; Bastian et al., 2010; Dubovskaya et al., 2011; Isner and Maathuis, 2011; Nan et al., 2014). The role of cGMP is tightly linked with the NO signal cascade in diverse cellular and physiological processes, including gibberellin (GA)–induced α-amylase production and seed germination (Penson et al., 1996; Teng et al., 2010; Wu et al., 2013), pollen tube growth (Prado et al., 2004), auxin-induced adventitious root formation and growth (Pagnussat et al., 2003; Bai et al., 2012; Nan et al., 2014), abscisic acid (ABA)–induced stomatal closure (Garcia-Mata et al., 2003; Bright et al., 2006; Joudoi et al., 2013), photomorphogenesis (Bowler et al., 1994), and pathogen defense (Durner et al., 1998; Klessig et al., 2000).
GCs with high levels of amino acid sequence similarity to animal GCs have not been identified in higher plants (Gross and Durner, 2016). However, several plant-type GCs containing the conserved amino acid residues in the catalytic center have been identified and experimentally confirmed in Arabidopsis (Arabidopsis thaliana; Mulaudzi et al., 2011; Gehring and Turek, 2017). Of those, soluble NO-SENSITIVE GUANYLATE CYCLASE1 (AtNOGC1) that contains a heme-NO and oxygen binding domain has a higher affinity for NO than oxygen and is capable of catalyzing GTP to cGMP in a NO-dependent manner (Mulaudzi et al., 2011; Gehring and Turek, 2017). In addition, NO-independent GCs were identified in plants; they include soluble GUANYLATE CYCLASE1 (AtGC1; Ludidi and Gehring, 2003) and membrane-associated receptor kinases with the GC and kinase dual activities, such as brassinosteroid receptor (AtBRI1; Kwezi et al., 2007), PEP PEPTIDE RECEPTOR1 (AtPepR1; Qi et al., 2010), WALL ASSOCIATED KINASE-LIKE10 (AtWAKL10; Meier et al., 2010), PHYTOSULFOKINE RECEPTOR1 (AtPSKR1; Kwezi et al., 2011; Muleya et al., 2014), and PLANT NATRIURETIC PEPTIDE RECEPTOR1 (AtPNP-R1; Turek and Gehring, 2016). These results suggest that stimulus-triggered cGMP production occurs in plants, but how the intracellular cGMP is perceived and transduced into cellular and physiological responses is not well understood.
cGMP is perceived by three targets including cGMP-dependent protein kinases (PKGs), cyclic nucleotide-gated ion channels (CNGCs), and cGMP-regulated PDEs in animal cells (Zagotta and Siegelbaum, 1996; Bender and Beavo, 2006; Francis et al., 2010). Among those targets, PKGs are recognized as major players that decode the NO-cGMP signal and phosphorylate downstream proteins at Ser/Thr residues, leading to changes in activity, subcellular localization, or protein interaction in mammalian cells (Hofmann, 2005; Francis et al., 2010). In plants, one cGMP target identified is CNGCs that are responsible for cation translocation involved in growth, development, and stress responses (DeFalco et al., 2016; Demidchik et al., 2018). cGMP is required for AtCNGC18-mediated, voltage-independent currents in pollen tubes (Gao et al., 2016), whereas AtCNGC5 and AtCNGC6 regulate plasma membrane Ca2+-permeable channels in a cGMP-dependent, abscisic acid (ABA)–insensitive manner in guard cells (Wang et al., 2013). Recently, a cGMP-stimulated PDE1 (AtCN-PDE1) that was responsible for UV-A–induced reduction in cGMP level was identified in Arabidopsis and was found to be important for inhibiting stomatal opening in response to short-term UV-A (Isner et al., 2019). In addition, receptor kinase GCs, such as AtBRI and AtPSKR1, possess dual GC and kinase activities in which the GC activity is promoted by its kinase, while the kinase is inhibited by cGMP, the product of GC activity, exhibiting an autoregulatory signaling loop (Kwezi et al., 2007; Qi et al., 2010; Kwezi et al., 2011; Muleya et al., 2014). These dual cGMP-making and kinase activities are thought to function as a switch between cGMP signaling and its downstream feedback to modulate precisely physiological status (Freihat et al., 2014).
However, no cGMP-dependent kinases homologous to animal PKG have been identified in plants to date (Świezawska et al., 2018). An early study provided some biochemical evidence for the presence of a PKG-like enzyme in Pharbitis nil as a purified protein was recognized by the antibodies against animal PKG and had the activity of cGMP-dependent phosphorylation of histone H2B (Szmidt-Jaworska et al., 2003, 2009). Pharmacological studies using an inhibitor of animal PKG also implicated the presence of PKG that was involved in auxin-stimulated adventitious root formation in Arabidopsis (Nan et al., 2014). The Arabidopsis genome has predicted proteins that possess both a cyclic nucleotide binding domain (CNBD) and a protein kinase domain (Maathuis, 2006). Phosphoproteomic study revealed that cGMP treatments led to altered phosphorylation of numerous proteins in Arabidopsis (Isner et al., 2012; Marondedze et al., 2016). These observations suggested the presence of PKG-like activities in plants, but the molecular and genetic evidence of PKG in plants is still lacking. Therefore, this study was undertaken to elucidate the molecular identity and function of PKGs in higher plants.
RESULTS
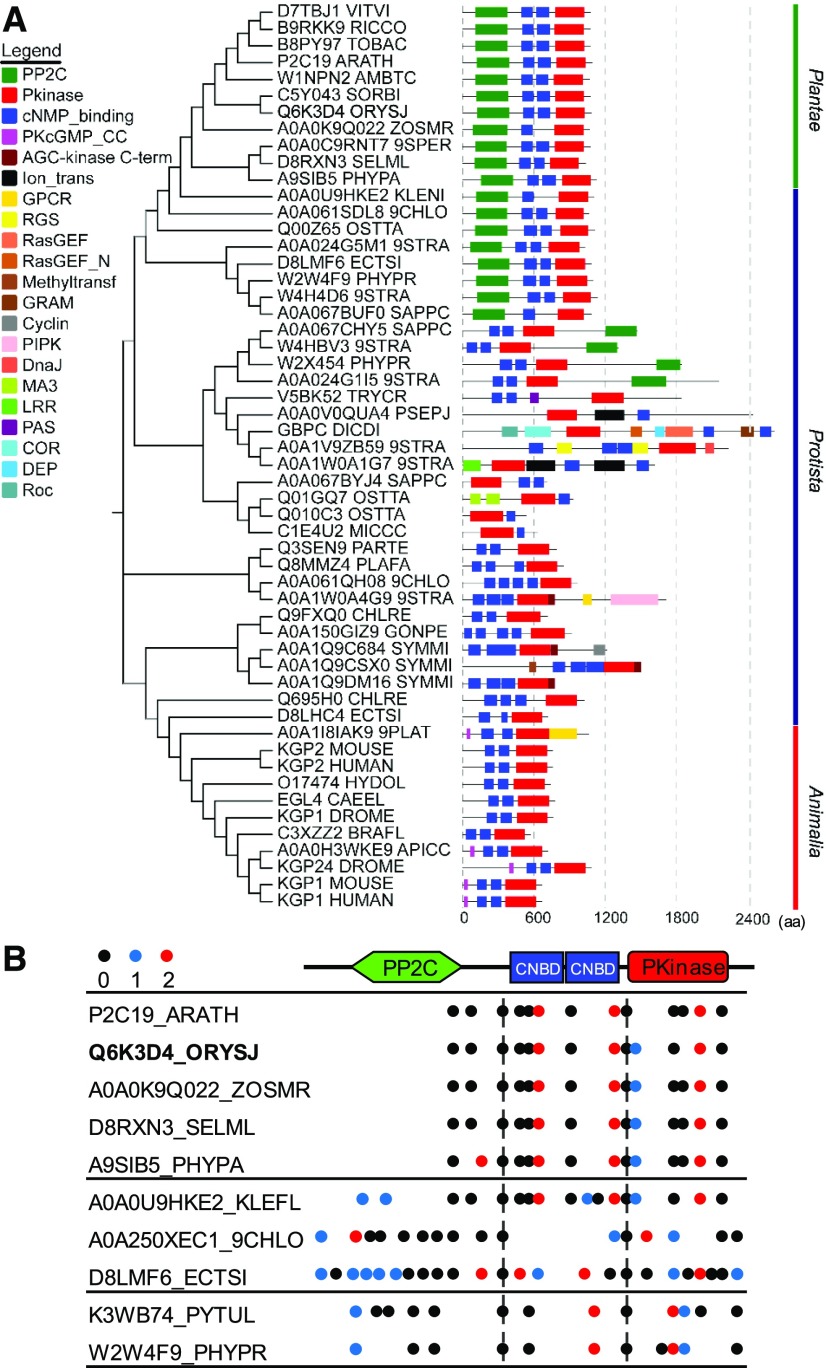
Plant PKGs Are Structurally Unique, Containing Both Protein Kinase and Phosphatase Domains
Animal PKGs contain two CNBDs in tandem arrangement at the N-terminal region and a catalytic protein kinase (Pkinase) domain at the C-terminal region (Wall et al., 2003; Hofmann, 2005). To identify PKG in plants, we used the conserved sequence of human PKGI (hPKGI) to query the InterPro database (http://www.ebi.ac.uk/interpro/) to search for proteins that contain both CNBD (IPR000595) and catalytic Pkinase domain (IPR000719) using the Domain architecture service http://www.ebi.ac.uk/interpro/search/ida/). This search identified PKG candidates across plant taxa (Figure 1A; Supplemental Data Set 1). The rice (Oryza sativa) and Arabidopsis genome each contain a single gene encoding a putative PKG. Like its counterparts in animals, plant PKGs possess two tandem CNBDs, CNBD-A and CNBD-B, followed by a catalytic Pkinase domain at the C-terminal region (Figure 1). The amino acid sequences of CNBD-CNBD-Pkinase domains (PKGcGK) of PKGs from rice and Arabidopsis share 49 and 47%, respectively, similarity to those of human PKGI (Supplemental Figure 1A). Both CNBD-A and CNBD-B of plant PKGs contain a conserved phosphate binding cassette motif (Supplemental Figure 1A) that captures the cyclic phosphate ribose moiety (Kim et al., 2016). The sequence in the CNBD-A domain of plant PKGs is more closely related to that of hPKGI than in CNBD-B (Supplemental Figure 1A). Plant PKGs harbor an activation loop conserved with a DFx(n)APE motif in the protein kinase domain (Supplemental Figure 1A) that is required for conformational changes for substrate binding and kinase activation (Taylor and Kornev, 2011). However, unique from animal PKGs is that plant PKGs contain a type 2C protein phosphatase (PP2C) domain with a (R/K)xx(M/N)(E/Q)D motif involved in divalent cation binding and a GxxDGHGxxG motif responsible for phosphatase catalytic activity at the N-terminal region (Figure 1; Supplemental Figure 1B).
Figure 1.
Identification and Evolutionary Analysis of Plant PKGs.
(A) Phylogenetic relationships and domain structures of PKGs across Protista, Plantae, and Animalia. Alignment and phylogenetic analyses were conducted using MAFFT (v7.452) and the neighbor-joining method in MEGA 5 for unrooted phylogeny tree construction. Domain architectures of PKG proteins were retrieved from Pfam. Boxes with different colors (left) indicate different domains of PKG homologs. Numbers at the nodes labeled with vertical gray dashed lines indicate the length of amino acid sequence. aa, amino acids.
(B) Intron phase analysis in plant-type PKG genes across Oomycota, algae, and plants. Intron positions of PKG genes and intron phase 0, 1, and 2 are indicated by black dots, blue dots, and red dots, respectively. Two conserved intron positions indicated by intron phase 0 are shown by vertical dashed lines.
To probe how the PP2C domain of plant PKG was derived evolutionarily, we compared PKG homologs across Protista, Plantae, and Animalia using the Uniprot database for phylogenetic and structural analysis. PKGs from Animalia contain a typical domain arrangement of CNBD-CNBD-Pkinase, which is similar to human PKGI. PKGs from Plantae are exclusively featured with the PP2C-CNBD-CNBD-Pkinase arrangement. PKGs from Protista are structurally diverse, possessing domain structures similar to either plant PKGs or animal PKGs, or containing additional domains, such as G protein–coupled receptors (GPCR), regulator of G protein signaling (RGS), Ras guanine nucleotide exchange factors (RasGEFs), and phosphatidylinositol phosphate kinase (PIPK) that are related to cGMP/PKG signaling (Figure 1A; Supplemental Figures 2A to 2C). Higher plant PKGs are structurally similar to algal PKGs, suggesting that higher plant PKGs originated from algal PKGs. Furthermore, the numbers of CNBD domains varied in PKGs across Protista taxa, as did the order of CNBD and Pkinase domains present, indicating that Pkinase-CNBD and its inversed form are the simplest structures and ancestral PKG (Figure 1A; Supplemental Figures 2A to 2C). Analysis of the exon/intron organization of PKGs across Plantae and Protista species reveals that the gene structures of plant-type PKGs contain two conserved intron positions indicated by intron phase 0 located in the borders of PP2C/CNBD and CNBD/Pkinase (Figure 1B). These results suggest that plant PKGs and animal PKGs originated from ancestral PKGs through shuffling and/or fusion of individual domains of CNBD, Pkinase, and/or PP2C during evolution.
Rice PKG Has cGMP-Dependent Protein Kinase but cGMP-Inhibited Phosphatase Activities
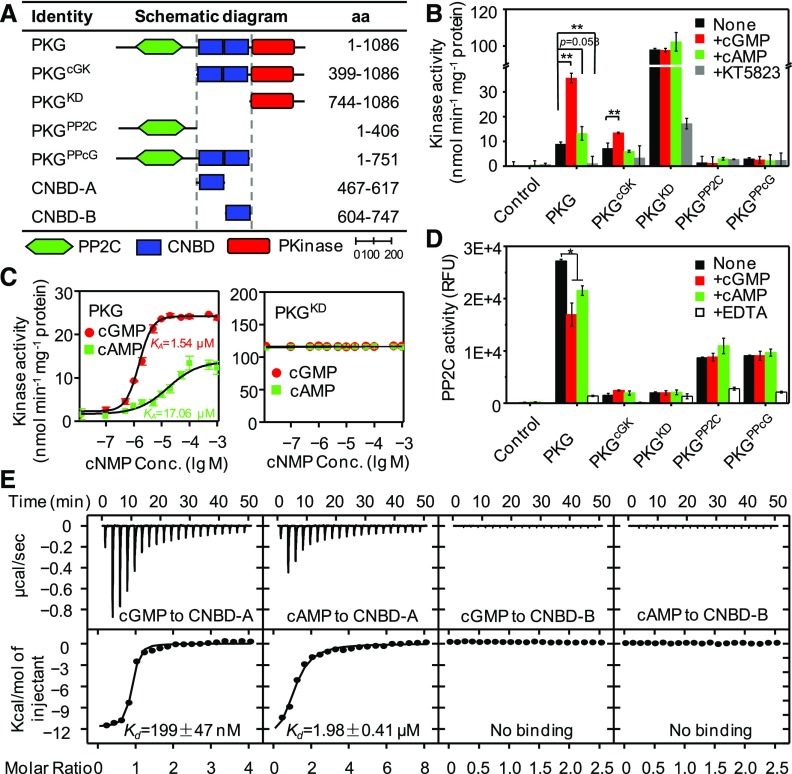
To test whether the PKG homologs in plants encode cGMP-dependent protein kinase, we focused on the PKG from the staple food crop rice. The full-length coding sequence (CDS) of PKG and its truncated mutants encoding CNBD-CNBD-Pkinase (PKGcGK, 76 kD), Pkinase domain (PKGKD, 38 kD), PP2C-CNBD-CNBD (PKGPPcG, 83 kD), and PP2C domain (PKGPP2C, 45 kD) were cloned from rice and expressed in and purified from Escherichia coli (Figure 2A; Supplemental Figures 3A and 3B) for an enzymatic activity assay. The protein kinase activity of full-length PKG was negligible in the absence of cyclic nucleotides but was greatly stimulated by cGMP with a fourfold increase (Figure 2B; Supplemental Figure 3C). The kinase activity of PKGcGK was dependent on cGMP but was lower than that of the full-length PKG (Figure 2B). However, the Pkinase domain PKGKD exhibited a constitutive activity in the absence and presence of cyclic nucleotides and had a much higher activity with a fivefold greater than Vmax value of the full-length PKG (Figures 2B and 2C; Supplemental Figure 3C). The addition of the PKG inhibitor KT5823 inhibited the protein kinase activity (Figure 2B). By contrast, the Pkinase-lacking fragments PKGPP2C and PKGPPcG had no kinase activity under the conditions tested (Figure 2B). The protein kinase activity of full-length PKG was increased in a cGMP concentration-dependent manner (Figure 2C). The Vmax value of PKG in the presence of cGMP is 24.22 ± 0.15 nmol min−1 mg−1 protein, which is 11-fold higher than that of the control without cyclic nucleotide supplementation. cAMP stimulated the PKG activity, but the magnitude was much lower than that of cGMP. The activation constant of cGMP was 1.54 μM, whereas activation constant for cAMP was 17.06 μM, which was 11-fold greater than that of cGMP (Figure 2C). These results suggest that rice PKG encodes a cGMP-dependent protein kinase and the PKinase domain of PKG is essential for its protein kinase activity.
Figure 2.
Molecular Characterization of Rice PKG.
(A) Domain structures of rice PKG and its truncated mutants. aa, amino acids.
(B) Protein kinase activity assay. The reaction mixture was supplemented without (none) or with 1 μM cGMP, 1 μM cAMP, or 1 μM KT5823 at 30°C for 15 min.
(C) Dose-dependent effects of cyclic nucleotides on protein kinase activity. KA, activation constant. Conc., concentration.
(D) Protein phosphatase activity assay. The reaction mixture supplemented without (none) or with 10 μM cGMP, 10 μM cAMP, or 50 mM EDTA at 30°C for 30 min. RFU, relative fluorescence unit.
(E) Binding affinity between cyclic nucleotides and CNBDs measured by ITC. µcal, microcalorie; Kcal, kilocalorie; Kd, dissociation constant.
Control, the reaction containing purified protein from E. coli carrying the empty vector. Values in (B), (C), and (D) are means ± sd (n = 3 independent experiments). Student’s t test: *P < 0.05, **P < 0.01.
To determine how cGMP regulates PKG activity, we assayed the interaction between cGMP and the two CNBD domains. PKG contains two CNBD domains, CNBD-A and CNBD-B, that are located in tandem upstream of the Pkinase catalytic domain (Figure 2A). Each CNBD contains eight β sheets and five α-helices (Supplemental Figure 4A). We used the isothermal titration calorimetry (ITC) assay to test whether PKG binds cGMP directly using purified CNBD-A and CNBD-B domains (Supplemental Figure 4B). cGMP and cAMP bound to CNBD-A at a 1:1 ratio, with a Kd value of 0.20 μM for cGMP and 1.98 μM for cAMP (Figure 2E). Thus, the CNBD-A affinity to cGMP is 10-fold higher than that to cAMP. Neither cGMP nor cAMP showed detectable binding to the CNBD-B domain (Figure 2E). These results show that the CNBD-A domain of PKG is responsible for cGMP binding and response.
To determine whether the PP2C domain at the N-terminal region of plant PKG has phosphatase activity, the full-length and truncated mutants of PKG expressed in and purified from E. coli were used for enzymatic assay. The full-length PKG showed phosphatase activity in the presence or absence of cyclic nucleotides (Figure 2D), as did the PP2C domain–containing fragments PKGPP2C and PKGPPcG, whereas the PP2C domain–lacking fragments PKGcGK and PKGKD had no phosphatase activity (Figure 2D). cGMP or cAMP did not promote the phosphatase activity of PKG; instead, they inhibited the phosphatase activity of the full-length PKG, but not the PP2C domain–containing mutants (Figure 2D). These data indicate that rice PKG has protein phosphatase activity that is inhibited by cyclic nucleotides and that the PP2C domain is responsible for the phosphatase activity. Taken together, these biochemical and mutational analyses indicate that rice PKG has dual activities with a cGMP-dependent protein kinase and a cGMP-inhibited protein phosphatase.
Loss of PKG Led to Defects in Seed Germination, Internode Elongation, and Pollen Viability in Rice
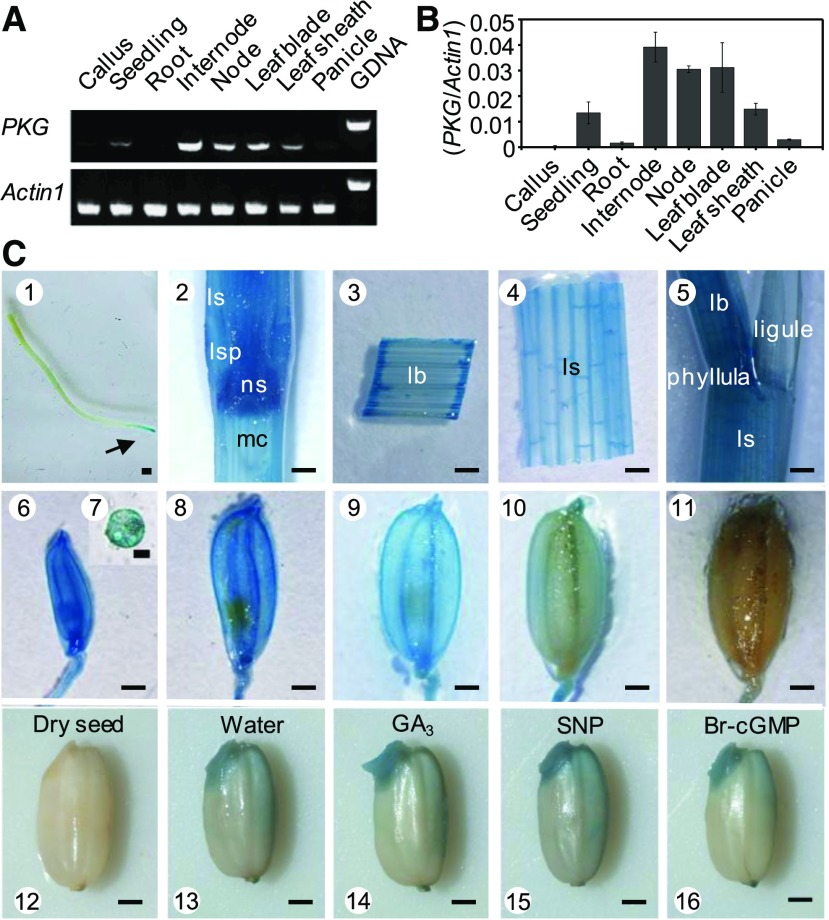
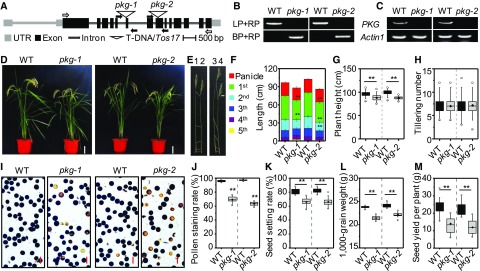
The results of RT-PCR analysis indicated that the expression of PKG was high in the rice internode, node, leaf blade, leaf sheath, and seedling but negligible in the whole root, panicle, and callus (Figures 3A and 3B). To assess the spatial distribution of PKG more precisely, the promoter region of PKG was fused with the β-glucuronidase (GUS) reporter gene and was expressed in rice plants. PKGpro:GUS activity was detected histochemically in various tissues, and strong expression was observed in the stem, leaf blade, leaf sheath, flower, pollen, developing seed, and germinating seed (Figure 3C). In addition, PKGpro:GUS activity was detected in the root tip, but not in other root tissues (Figure 3C). To investigate the physiological function of PKG, two independent mutants, pkg-1 and pkg-2, were isolated from rice cv Zhonghua 11 (Figures 4A and 4B). The full-length mRNA of PKG was detected in wild-type plants but absent in the pkg mutants (Figure 4C). The RT-PCR data are consistent with the possibility that the pkg alleles are nulls. The pkg mutants exhibited shorter internodes, in particular the first internode, with a 10% reduction in plant height at the flowering and mature stages relative to the wild-type plants grown in the field under normal conditions (Figures 4D to 4G). The pkg mutants also exhibited a lower pollen viability and seed setting rate than the wild type (Figures 4I to 4K). The grain weight of pkg mutants was also lower than that of the wild type (Figure 4L); consequently, these plants exhibited a 35% lower seed yield than the wild type due to the combined effects of reduced seed setting and seed weight (Figure 4M). In addition, the loss of PKG significantly reduced the seed germination rate under salt stress conditions (Supplemental Figure 5). These results suggest that rice PKG plays important roles in plant growth, reproduction, seed production, and seed germination.
Figure 3.
Expression Profile of PKG in Rice.
(A) and (B) Expression pattern of PKG in rice by RT-PCR (A) and by RT-qPCR (B). Total RNA was extracted from various tissues at different stages in rice. Actin1 was used as an internal standard control. Values are means ± sd (n = 3 separated samples). GDNA, genomic DNA.
(C) Histochemical analysis of GUS activity in PKGpro:GUS transgenic plants. 1, root; 2, stem; 3, leaf blade; 4, leaf sheath; 5, leaf; 6, spikelet at the pollen meiosis stage; 7, pollen grain; 8, spikelet at the vacuolated pollen stage; 9, mature spikelet; 10, seed at milk stage; 11, mature seed; 12, dry seed; 13, germinating seed; 14, seed soaked in 50 μM GA3 for 24 h; 15, seed soaked in 200 μM SNP for 24 h; 16, seed soaked in 1 μM Br-cGMP for 24 h. lb, leaf blade; ls, leaf sheath; lsp, leaf-sheath pulvinus; mc, medullary cavity; ns, nodal septum. Arrow indicates the root tip. Bar = 1 mm, except in pollen grain (7), where bar = 20 μm.
Figure 4.
Loss of PKG Resulted in Defects in Internode Elongation and Pollen Viability in Rice.
(A) T-DNA/Tos17 insertion in rice PKG. Two independent mutants, pkg-1 and pkg-2, contain the T-DNA insertion in the sixth exon and the Tos17 insertion in the eighth exon of rice PKG, respectively. Black boxes indicate exons in coding region, while gray boxes indicate exons in untranslated region (UTR). Black and gray lines between boxes indicate introns. Black arrows indicate the positions of primers used for PCR genotyping; white arrows indicate the positions of primers used for RT-PCR.
(B) PCR verification of homozygous pkg-1 and pkg-2 mutants. BP, border primer; LP, left primer; RP, right primer; WT, wild type.
(C) Loss of full-length PKG transcript in pkg-1 and pkg-2 mutants by RT-PCR. Actin1 was used as an internal standard control. WT, wild type.
(D) Growth phenotype of the pkg mutants and the wild-type plants grown in a paddy field at the mature stage. Bar = 20 cm. WT, wild type.
(E) and (F) Internode elongation of pkg mutants and the wild-type (WT) plants at the mature stage. The internode observation (E) and internode length (F) of pkg mutants and the wild type plants. The numbers at the top of (E) indicate different genotypes: 1, WT; 2, pkg-1; 3, WT; 4, pkg-2. The boxes with different colors labeled with the 1st to the 5th at the left panel of (F) indicate different internodes from top to bottom of plants (n = 10 plants).
(G) and (H) Plant height (G) and tiller number (H) of mature plants (n = 100 plants). WT, wild type.
(I) and (J) Viability of pollen grains stained with I2-KI solution (n = 6 independent experiments). Scale bar = 100 μm. WT, wild type.
(K) to (M) Seed setting rate (n = 24 plants) (K), 1000-grain weight (n = 20 plants) (L), and seed yield per plant (n = 24 plants) (M). WT, wild type.
Values are means ± sd. Student’s t test: **P < 0.01.
PKG Mediates NO-cGMP Signaling in Response to GA
To address how the loss of PKG altered rice growth and development, we probed the role of PKG in GA signaling because the reduced seed germination rate, shortened internode, and reduced pollen viability in pkg mutants are similar to the phenotypes resulting from mutations defective in GA responses (Kaneko et al., 2004; Ueguchi-Tanaka et al., 2005). In addition, it was previously shown that GA induced cGMP production in cereal aleurone cells during seed germination (Penson et al., 1996; Isner and Maathuis, 2011). These results indicate that PKG may be involved in GA signaling to modulate seed germination and growth in rice.
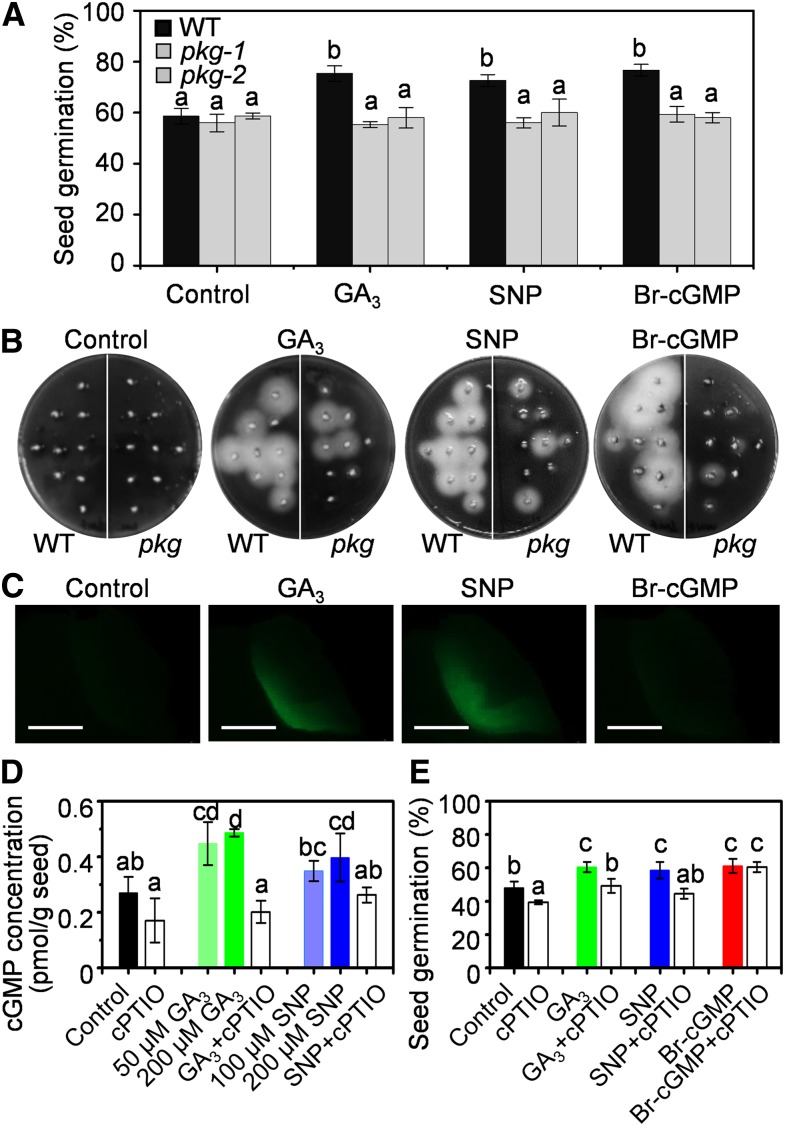
To test this possibility, pkg mutant and the wild-type seeds were germinated in GA-containing medium. GA promoted germination and amylase production of the wild-type seeds, but not pkg seeds. Supplementation with membrane-permeable 8-bromo-cGMP (8-Br-cGMP) or sodium nitroprusside (SNP), a NO donor, had a similar effect as did GA on the germination of the wild-type seeds, but this effect was impaired in pkg mutants (Figures 5A and 5B; Supplemental Figures 6A and 6C). On the fifth day after imbibition, the seed germination rate of both the wild-type and pkg mutant seeds was ∼58% under the control condition, but in the presence of gibberellin A3 (GA3), SNP, or Br-cGMP, the germination rate of the wild type was stimulated to 75% but that of the pkg mutants remained ∼58% (Figure 5A). As a control, 8-Br-cAMP treatment failed to promote seed germination in both the wild type and pkg mutants (Supplemental Figure 6B). These results suggest that PKG is required for mediating the effects of GA, NO, and cGMP on seed germination.
Figure 5.
NO-cGMP-PKG Cascade Mediated Seed Germination in Response to GA
(A) Seed germination under control conditions or supplemented with 50 μM GA3, 200 μM SNP, or 10 μM Br-cGMP on the fifth day. WT, wild type.
(B) Amylase activity assayed using embryoless half-seeds in starch-containing agar plates in the absence (control) or presence of 50 μM GA3, 200 μM SNP, or 1 μM Br-cGMP at 30°C for 4 d. WT, wild type.
(C) NO fluorescent signal monitored by 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate in germinating seeds as shown in (B). Bar = 1 mm.
(D) cGMP content in germinating seeds under the indicated conditions for 12 h by an ELISA immunoassay. cPTIO (50 μM) was applied.
(E) Effect of NO scavenger (cPTIO) on seed germination. The seed germination rate was measured on the fifth day after seeds were sowed in water (control) or supplemented with 50 μM GA3, 200 μM SNP, 10 μM Br-cGMP, and 100 μM cPTIO.
Values are means ± sd (n = 3 independent experiments). Different letters denote significant difference at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
To dissect the relationship between PKG and GA, NO, and cGMP in the signaling cascade, NO production was fluorescently monitored in germinating seeds. Supplementation of GA, but not cGMP, resulted in NO production in the embryo (Figure 5C; Supplemental Figure 7). cGMP accumulation in seeds was triggered by GA or NO, which was suppressed by 2-(4-carboxyphenyl)-4,5-dihydro-4,4,5,5-tetramethy-1H-imidazolyl-1-oxy-3-oxide (cPTIO), a NO scavenger (Figure 5D). Meanwhile, cPTIO also diminished GA- or NO-potentiated seed germination, but it did not suppress cGMP-enhanced seed germination (Figure 5E). These results suggest that NO functions downstream of GA to trigger cGMP production and subsequently activates PKG that upregulates amylase production and enhances seed germination. Thus, PKG is involved in the GA response via the GA→NO→cGMP→PKG signaling cascade during seed germination.
PKG Interacts with GAMYB and Catalyzes the Reversible Phosphorylation of GAMYB
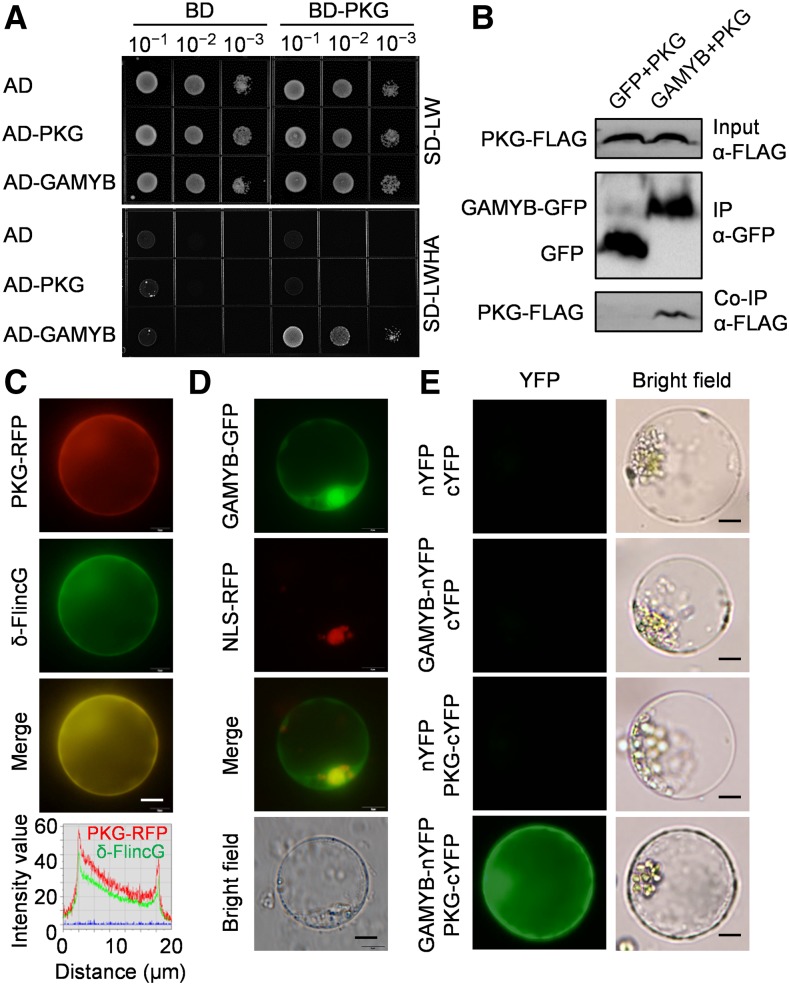
To determine how PKG transduces the cGMP signal to downstream targets for physiological effects, PKG was used as a bait to identify potential PKG-interacting proteins using a yeast two-hybrid (Y2H) system constructed using a rice cDNA library. Twenty-two candidate proteins were identified, including GAMYB (Supplemental Figure 8A; Supplemental Table 1), a major transcription factor in GA response involved in seed germination, reproduction, and internode elongation (Kaneko et al., 2004; Aya et al., 2009, 2011). The PKG-GAMYB interaction was confirmed by the cotransformation of pGBKT7-PKG and pGADT7-GAMYB in yeast (Figure 6A; Supplemental Figure 8B). This interaction was further verified by coimmunoprecipitation of PKG and GAMYB from Nicotiana benthamiana leaves (Figure 6B). In addition, PKG was found in the cytoplasm and colocalized with cGMP, which was monitored by δ-FlincG (Figure 6C; Supplemental Figure 9), whereas GAMYB was localized in both the cytoplasm and nucleus (Figure 6D). The PKG-GAMYB interaction occurred in the cytoplasm as verified by a bimolecular fluorescence complementation (BiFC) assay in rice protoplasts isolated from leaf sheath (Figure 6E).
Figure 6.
PKG Interacts with GAMYB Transcription Factor.
(A) Y2H testing of PKG interaction with GAMYB. Dilutions of saturated cultures were spotted onto a plate lacking Leu and Trp (SD-LW) or a plate lacking Leu, Trp, His, and adenine (SD-LWHA). AD, activation domain; BD, binding domain.
(B) Coimmunoprecipitation assay for the interaction between PKG and GAMYB. Total proteins from N. benthamiana leaves coexpressing PKG-FLAG and GAMYB-GFP were immunoprecipitated using anti-GFP antibody, and PKG was detected by immunoblotting using anti-FLAG antibody. IP, immunoprecipitation; Co-IP, co-immunoprecipitation.
(C) Colocalization of PKG and cGMP. PKG-RFP and δ-FlincG were transiently coexpressed in rice protoplasts, and fluorescent signals were observed after 12 h. cGMP was visualized by δ-FlincG. Intensity profiling of PKG-RFP and δ-FlincG signals across the merged image is shown, and the two channels showed identical distribution and similar intensity. Bars = 5 μm.
(D) Distribution of GAMYB in the nucleus and cytoplasm. GAMYB-GFP and NLS-RFP were transiently coexpressed in rice protoplasts, and fluorescent signals were observed after 12 h. NLS-RFP, nuclear marker. Bars = 5 μm.
(E) PKG interaction with GAMYB in rice cells in a BiFC assay. Bars = 5 μm. cYFP, C-terminal YFP; nYFP, N-terminal YFP.
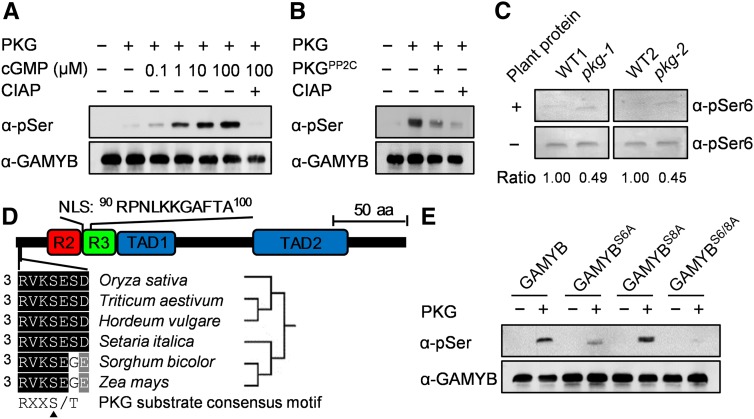
To investigate whether the PKG-GAMYB interaction affects the phosphorylation status of GAMYB, purified GAMYB was used as a substrate for PKG. The GAMYB phosphorylation level by PKG was negligible in the absence of cGMP but was greatly stimulated by cGMP in a cGMP concentration–dependent manner (Figure 7A). The phosphorylation level of GAMYB was decreased by 91% when the PP2C domain of PKG was added to the phosphorylated GAMYB compared with that without added PP2C domain (Figure 7B), suggesting that the PP2C phosphatase of PKG can dephosphorylate GAMYB. These results suggest that PKG phosphorylates GAMYB, whereas its PP2C phosphatase activity dephosphorylates the phosphorylated GAMYB.
Figure 7.
PKG Catalyzes Reversible Phosphorylation of GAMYB at Ser6.
(A) PKG phosphorylation of GAMYB was stimulated by cGMP. Purified GAMYB was used as a substrate. α-pSer, anti-phosphoserine; CIAP, calf intestinal alkaline phosphatase.
(B) Dephosphorylation of pGAMYB by the PP2C domain of PKG. Purified GAMYB was used as a substrate in the reaction mixtures supplemented with or without PKGPP2C or calf intestinal alkaline phosphatase (CIAP); α-pSer, anti-phosphoserine.
(C) pkg mutant has substantially reduced dephosphorylation activity toward phospho-GAMYB. Phosphorylated GAMYB by PKGKD was used as a substrate in the reaction mixture supplemented with or without plant proteins extracted from the wild-type and the pkg mutant plants. The reaction mixture containing 1 μM KT5823 was incubated at 25°C for 1 h. The relative ratio of dephosphorylation activity between the reaction with and without supplemented plant proteins was quantified by ImageJ software and is denoted at the bottom. α-pSer6, anti-phosphoGAMYBS6; WT, wild type.
(D) Conserved RXXS/T motif in cereal GAMYBs. NLS, nuclear localization sequence; R2 and R3, MYB repeat domains; TAD1 and TAD2, transcriptional activation domains. aa, amino acids.
(E) PKG phosphorylation of GAMYB at S6 indicated by site-directed mutagenesis. The purified GAMYB and its site-directed mutants (GAMYBS6A, GAMYBS8A, GAMYBS6/8A) were used as substrates in the reaction mixtures containing 100 μM cGMP.
In addition, the two rice pkg mutants with T-DNA/Tos17 inserted in the central region of PKG might express the N-terminal PP2C domain (Figure 4A), leading to a truncated version of PKG with constitutive PP2C activity. To test this possibility, we assayed PP2C activity in the pkg mutants. The results showed that the pkg-1 and pkg-2 mutants had only 49 and 45%, respectively, of wild-type phosphatase activity toward phospho-GAMYB (Figure 7C), suggesting that PP2C activity in the pkg mutants is markedly reduced relative to the wild type. Moreover, both kinase and phosphatase of PKG catalyze the reversible phosphorylation of GAMYB at the same site. Loss of kinase activity in the pkg mutants may lead to a defect in phosphorylation status of GAMYB, which may cause an unavailable substrate for PP2C action. Thus, the pkg mutants do not harbor a dominant-negative mutation that imparts constitutive PP2C activity.
The N-terminal region of GAMYB contains a Rxx(S/T) motif in which an Arg (R) residue is located at the −3 position around the phosphorylation site (S/T) for facilitating substrate affinity (Francis et al., 2010). Sequence analysis revealed that Ser6 (S6) in the motif is well conserved in GAMYBs from different cereal species (Figure 7D) and would be a potential phosphorylation site by PKG. To test this possibility, the S6 and S8 of GAMYB (GAMYBS6 and GAMYBS8) were mutated to Ala (A), yielding GAMYBS6A and GAMYBS8A, respectively. The phosphorylation level was greatly reduced at the GAMYBS6A mutation but to a lesser extent at the GAMYBS8A mutation. No phosphorylation was detected when both S6 and S8 of GAMYB were mutated (GAMYBS6/8A; Figure 7E). These results suggest that PKG predominantly phosphorylates GAMYB at the S6 residue.
PKG Phosphorylation Modulates the Nucleocytoplasmic Distribution of GAMYB in the GA Response
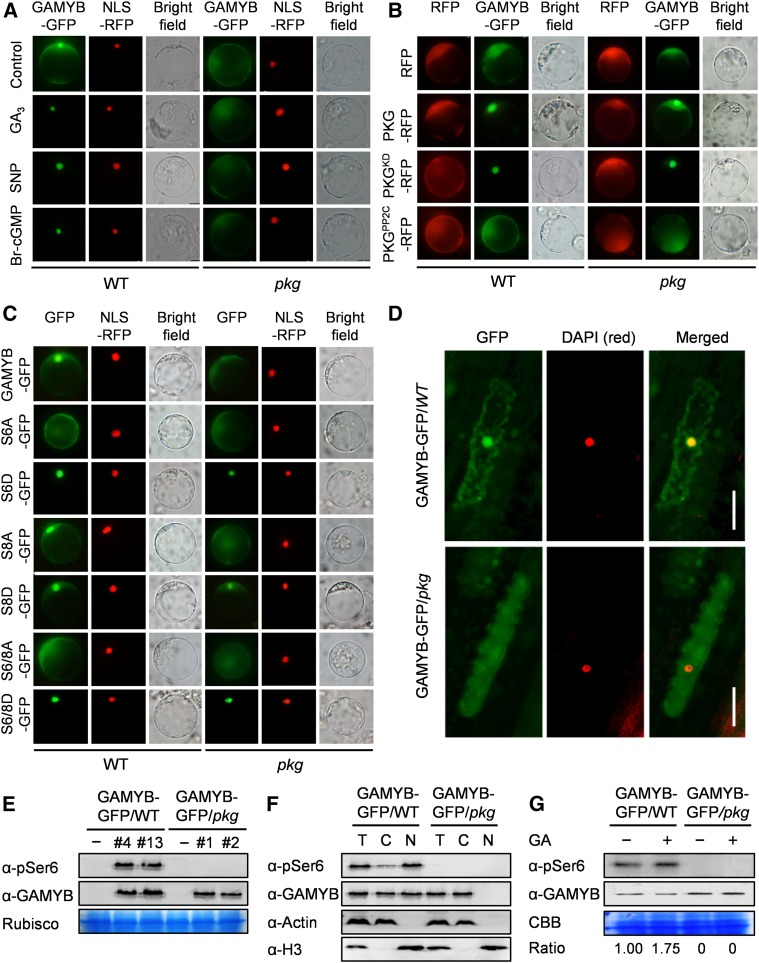
To determine the effect of PKG on GAMYB’s function, we first used an electrophoretic mobility shift assay to test whether the phosphorylation by PKG affected GAMYB’s binding to its target DNA. No such effect was observed (Supplemental Figure 10). However, the loss of PKG resulted in a loss of nuclear localization of GAMYB; GAMYB in pkg mutant cells was in the cytoplasm and undetectable in the nucleus. By comparison, GAMYB in the wild type was detectable in both the cytoplasm and nucleus (Figure 8A). Supplementation of GA, NO, or cGMP enhanced the nuclear localization of GAMYB in the wild-type cells, but not in pkg cells (Figure 8A). Introduction of PKG into pkg cells restored the GAMYB subcellular distribution to that of the wild type (Figure 8B). Moreover, expressing the Pkinase domain of PKG enhanced the GAMYB nuclear translocation, and GAMYB was completely localized to the nucleus in both the wild-type and pkg mutant cells. Conversely, expressing the PP2C domain of PKG reversed this process, and GAMYB was totally retained in the cytoplasm in both the wild-type and pkg cells (Figure 8B).
Figure 8.
PKG Modulates GAMYB Nucleocytoplasmic Distribution.
(A) PKG is required for GA-, NO-, and cGMP-triggered nuclear translocation of GAMYB. The protoplasts of the wild type and pkg mutant were transformed with plasmids carrying GAMYB-GFP for 12 h. Images were captured from protoplasts treated without (control) or with 50 μM GA3, 200 μM SNP, or 1 μM Br-cGMP for 4 h. NLS-RFP was used to label the nucleus. NLS, nuclear localization sequence; WT, wild type.
(B) Phosphorylation by protein kinase of PKG potentiated the nuclear translocation of GAMYB, while dephosphorylation by PP2C of PKG reversed this process. The protoplasts of the wild type and pkg mutant were cotransformed with plasmids carrying RFG (empty vector), PKG-RFP, PKGKD-RFP, and PKGPP2C-RFP, respectively, with GAMYB-GFP for 12 h. WT, wild type.
(C) Phosphorylation at S6 by PKG is critical for the nuclear localization of GAMYB. GAMYBS6, GAMYBS8, and GAMYBS6/8 were mutated to Ala (GAMYBS6A, GAMYBS8A, GAMYBS6/8A) and aspartate (GAMYBS6D, GAMYBS8D, GAMYBS6/8D), respectively. NLS-RFP was used to label the nucleus. NLS, nuclear localization sequence; WT, wild type.
(D) Effect of PKG on the subcellular localization of GAMYB observed in leaf sheath cells of the wild-type and pkg mutant plants expressing GAMYB-GFP. 4′,6-Diamidino-2-phenylindole (red) was used to label the nucleus. Bar, 20 μm. DAPI, 4′,6-diamidino-2-phenylindole; WT, wild type.
(E) Protein levels of total GAMYB and phospho-GAMYB (Ser6) in the wild-type and pkg mutant plants expressing GAMYB-GFP tested by immunoblotting using anti-GAMYB and anti-pSer6 antibodies, respectively. The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco, RbcL) was used as a loading control. WT, wild type.
(F) Effect of PKG on the subcellular localization and phosphorylation of GAMYB tested by cellular fractionation combined with immunoblotting using anti-pSer6 and anti-GAMYB antibodies. Actin and Histone H3 were used as a cytoplasmic marker and nuclear marker, respectively. C, cytoplasmic proteins; N, nuclear proteins; T, total proteins.
(G) Effect of PKG on phosphorylation status of GAMYB in germinating seeds. Proteins were extracted from germinating seeds in water or supplemented with 50 μM GA3 for 24 h. The relative ratio between phospho-GAMYB and total GAMYB is denoted at the bottom. CBB, Coomassie brilliant blue staining; WT, wild type.
Furthermore, the phosphorylation-deficient mutant GAMYBS6A blocked the nuclear localization (Figure 8C). Conversely, the phosphorylation mimic mutant GAMYBS6D, in which S6 is replaced with Asp, was localized to the nucleus in both the wild-type and pkg cells (Figure 8C). The subcellular distribution of the mutant GAMYBS8A remained similar to that of GAMYB, whereas GAMYBS8D in pkg cells promoted partial localization to the nucleus (Figure 8C). The subcellular distribution of the double mutant proteins GAMYBS6/8A and GAMYBS6/8D were similar to those of GAMYBS6A and GAMYBS6D, respectively (Figure 8C). These results suggest that PKG phosphorylation of GAMYB at S6 is required for GAMYB localization to the nucleus, whereas the phosphorylation of GAMYB at S8 is dispensable but enhances the nuclear translocation. Together, these results suggest that PKG plays an important role in regulating the nucleocytoplasmic distribution of GAMYB through reversible phosphorylation of GAMYB at the S6 residue in response to GA. Its kinase activity promotes the nuclear localization of GAMYB, whereas its phosphatase activity has an opposite effect, retaining GAMYB in the cytoplasm.
To verify the effect of PKG on GAMYB in planta, GAMYB-GFP was expressed in the wild-type and pkg rice plants (Figure 8E). GAMYB-GFP in the wild-type plants was localized predominantly in the nucleus with some in the cytoplasm, whereas GAMYB-GFP in the pkg mutant plants remained in the cytoplasm and was undetectable in the nucleus (Figures 8D and 8F). Furthermore, phospho-GAMYBS6 in the wild-type plants was mainly localized in the nucleus, whereas phospho-GAMYBS6 in the pkg mutant plants was undetectable in both the cytoplasm and nucleus (Figures 8E and 8F). Likewise, the effect of PKG on GAMYB phosphorylation in germinating seeds exhibited a similar behavior as shown in planta and in protoplasts. The addition of GA enhanced the phosphorylation level of GAMYB in the wild-type germinating seeds, but the GAMYB phosphorylation was absent in the pkg mutant seeds (Figure 8G). These results demonstrate that PKG is required for GAMYB phosphorylation and translocation to the nucleus.
PKG Is Required for GAMYB Action in GA Signaling
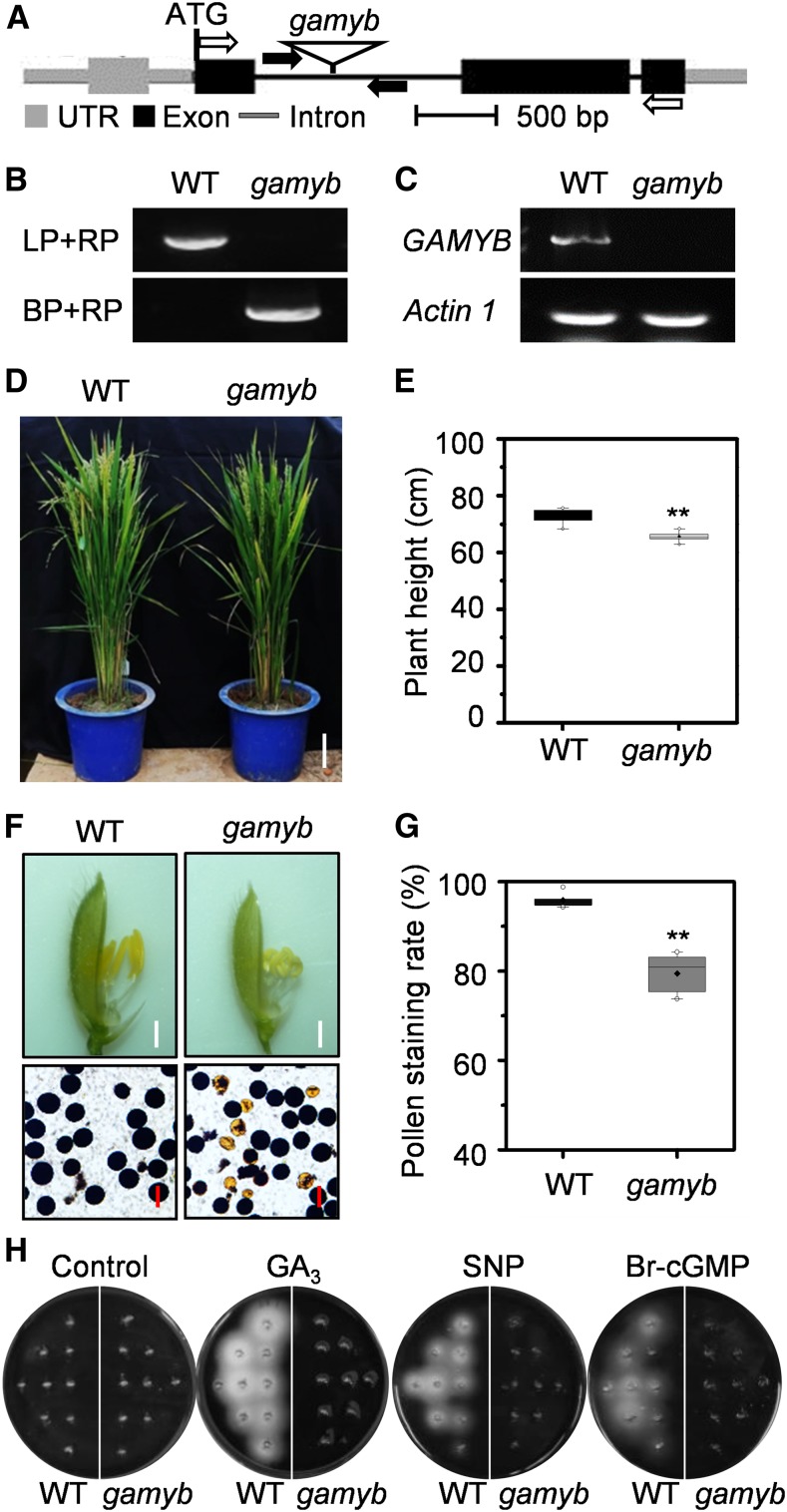
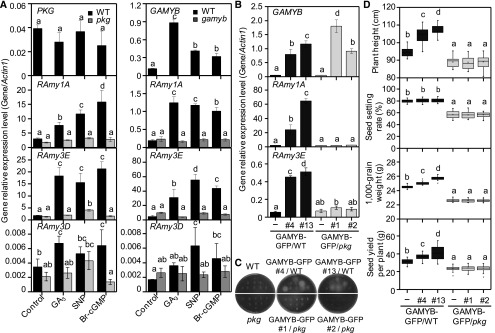
To investigate the functional relationship between PKG and GAMYB in planta, a rice GAMYB mutant gamyb caused by a T-DNA insertion was isolated from the Hwayoung genetic background (Figures 9A and 9B). RT-PCR showed that the full-length GAMYB transcript was detected in the wild type but absent in the gamyb mutant (Figure 9C), suggesting that gamyb is a null mutant. The gamyb mutant mimicked the pkg mutant phenotypes, exhibiting a defect in GA-induced seed germination, reduced pollen viability, and moderately shorter internodes, with a 9.7% reduction in plant height at the flowering stage, compared with the wild-type plants (Figures 9D to 9G; Kaneko et al., 2004; Aya et al., 2009). The GA-, NO-, and cGMP-induced amylase activity that occurred in germinating wild-type seeds was not observed in the gamyb seeds (Figure 9H), which was similar to that of the pkg mutants (Figure 5B). In response to GA, NO, and cGMP, the transcript level of RAmy1A and RAmy3E encoding amylase in the wild type was induced by 4- to 12-fold, but no such induction occurred in the pkg or gamyb mutant during seed germination. By comparison, the RAmy3D transcript level was relatively low and was much less induced by GA, NO, and cGMP. Unlike RAmy1A and RAmy3E, the RAmy3D expression in the pkg and gamyb mutants was less suppressed and exhibited an inconsistent pattern in response to GA, NO, and cGMP (Figure 10A), suggesting that RAmy3D is not coordinately regulated by PKG and GAMYB. Moreover, the transcript level of RAmy1A and RAmy3E was greatly upregulated by GAMYB overexpression (OE) in the wild-type seeds, but not in pkg seeds during germination (Figure 10B). GAMYB-OE in the wild-type seeds induced amylase production without added GA, NO, or cGMP, but no such auto-induction of amylase was observed in GAMYB-OE in pkg mutant seeds (Figure 10C). GAMYB-OE in the wild-type plants led to increased plant height, seed weight, and thus increased seed yield compared with the wild-type plants, whereas GAMYB-OE in pkg plants had no such promotion (Figure 10D). Collectively, these results further support that PKG is required for GAMYB action and that PKG mediates GA signaling through the GA→NO→cGMP→PKG→GAMYB cascade in modulating plant growth and development.
Figure 9.
Loss of GAMYB Led to Reduced Growth, Pollen Viability, and Impaired Response to GA-NO-cGMP Signaling.
(A) T-DNA insertion in rice GAMYB. The gamyb mutant is caused by a T-DNA insertion in the first intron of GAMYB. Black boxes indicate exons in coding region, while gray boxes indicate exons in untranslated region. Black and gray lines between boxes indicate introns. Black arrows indicate the positions of primers used for PCR genotyping; white arrows indicate the positions of primers used for RT-PCR. UTR, untranslated region.
(B) Identification of homozygous mutant gamyb by PCR. BP, border primer; LP, left primer; RP, right primer; WT, wild type.
(C) Loss of full-length GAMYB transcript in the gamyb mutant by RT-PCR. Actin1 was used as an internal standard control. WT, wild type.
(D) and (E) Loss of GAMYB exhibited reduced growth. The growth phenotype (D) and plant height (E) of the gamby mutant and wild type (WT). Plants were grown in a paddy field under natural conditions during summer and fall seasons. Values are means ± sd (n = 10). Bar = 10 cm.
(F) Loss of GAMYB exhibited defects in anther development and pollen viability. Pollen grains were stained with I2-KI solution. Bar = 1 mm in spikelets and 50 μm in pollens. WT, wild type.
(G) Pollen viability indicated by I2-KI staining rate (n = 7 biological replications). Student’s t test: **P < 0.01. WT, wild type.
(H) Amylase activity was detected using embryoless half-seeds in starch-containing agar plates in the absence (control) or presence of 50 μM GA3, 200 μM SNP, or 1 μM Br-cGMP at 30°C for 4 d. WT, wild type.
Figure 10.
PKG Integrates GA, NO and cGMP Signaling to Modulate GAMYB Action.
(A) Expression levels of PKG, GAMYB, and α-amylase genes in germinating seeds of pkg, gamyb, and the wild type by RT-qPCR. RNA was extracted from germinating seeds in agar plates in the absence (control) or presence of 50 μM GA3, 200 μM SNP, or 1 μM Br-cGMP at 30°C for 24 h. Actin1 was used as an internal standard. Values are means ± sd (n = 3 separated samples). WT, wild type.
(B) Upregulated RAmy1A and RAmy3E in germinating seeds by GAMYB OE in the wild-type plants, but not in pkg plants tested by RT-qPCR. Actin1 was used as an internal standard. Values are means ± sd (n = 3 separated samples). WT, wild type.
(C) Auto-induced amylase secretion by GAMYB-OE in the wild-type seeds, but not in pkg seeds. WT, wild type.
(D) Enhanced growth and seed production by GAMYB-OE in the wild-type plants but not in pkg plants (n = 20 plants). WT, wild type.
Different letters denote significant difference at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
PKG, but Not GAMYB, Is Involved in Salt Stress Response
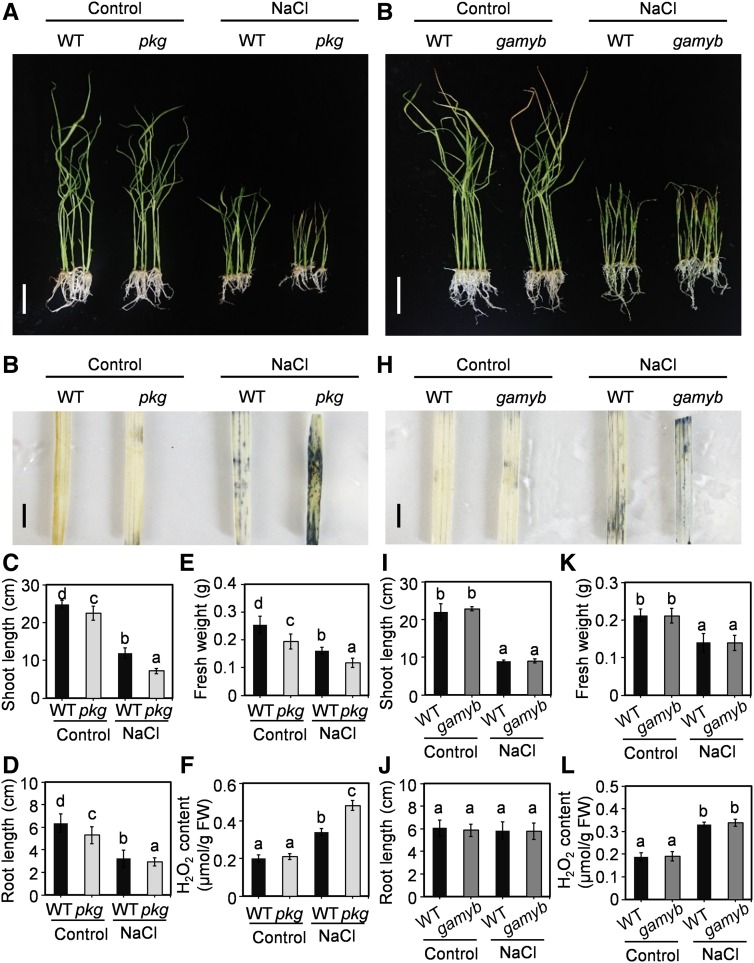
PKG mRNA exhibits a broader tissue distribution than that of GAMYB (Figure 3; Kaneko et al., 2004), suggesting that PKG may have other targets besides GAMYB. Our Y2H screen identified several other proteins potentially interacting with PKG, such as PEROXIDASE1 (LOC_Os05g41990) and CYTOCHROME B-C1 COMPLEX SUBUNIT8 (LOC_Os06g07969), which are involved in redox homeostasis; zeaxanthin epoxidase (ABA2; LOC_Os04g37619), which functions in ABA synthesis; and CML10-calmodulin–related calcium sensor protein (LOC_Os01g72100) and calreticulin precursor protein (LOC_Os04g32950), which are involved in Ca2+ signaling (Supplemental Figure 8A; Supplemental Table 1). The above-identified putative PKG-interacting proteins are involved in osmotic stress including salt stress responses, and salt stress was reported to induce cGMP production (Donaldson et al., 2004). These results could mean that GAMYB is not a sole target of PKG, and PKG function is not limited to GA signaling. To test this hypothesis, both pkg and gamyb mutants were subjected to salt stress treatments using their corresponding wild types as controls at the seedling stage. Loss of PKG resulted in growth arrest with shorter shoots and roots and reduced fresh weight under both the control and salt stress conditions at the seedling stage, with a more pronounced phenotype under salt stress, compared with the wild-type plants (Figures 11A to 11F). Moreover, the pkg mutant exhibited increased reactive oxygen species (ROS) accumulation under salt stress conditions (Figure 11B). H2O2 content in the pkg mutants was increased by 36% compared with the wild type when plants were treated with 125 mM NaCl for 14 d (Figure 11F). By comparison, the loss of GAMYB had no effect on the salt stress response relative to the wild type (Figures 11G to 11L). The results suggest that the PKG-affected response to salinity is not mediated through GAMYB, and thus PKG has targets in addition to GAMYB.
Figure 11.
pkg Mutant, but Not gamyb Mutant, Displayed Increased Sensitivity to Salt Stress.
(A) Rice growth phenotype of pkg mutant and the wild type under control and 125 mM NaCl treatment for 14 d. Bar = 5 cm. WT, wild type.
(B) ROS accumulation indicated by NBT staining in pkg mutant and the wild-type leaves with and without NaCl treatment. The plant growth conditions are shown in (A). Bars = 1 cm. WT, wild type.
(C) to (F) Shoot length (n = 7; see [C]), root length (n = 7; see [D]), fresh weight (n = 7; see [E]), and H2O2 content (n = 3; see [F]) of the wild type and pkg mutants under control and salt stress conditions. FW, fresh weight; WT, wild type.
(G) Rice growth phenotype of the gamyb mutant and the wild type under control and 125 mM NaCl treatment for 14 d. Bar = 5 cm. WT, wild type.
(H) ROS accumulation indicated by NBT-staining in leaves of gamyb mutant and the wild-type plants with and without NaCl treatment. Plant growth conditions are shown in (G). Bars = 1 cm. WT, wild type.
(I) to (L) Shoot length (n = 7; see [I]), root length (n = 7; see [J]), fresh weight (n = 7; see [K]), and H2O2 content (n = 3; see [L]) of the wild type and gamyb mutant under control and salt stress conditions. FW, fresh weight; WT, wild type.
Different letters denote significant difference at P < 0.05 according to ANOVA in combination with Duncan’s multiple range test.
DISCUSSION
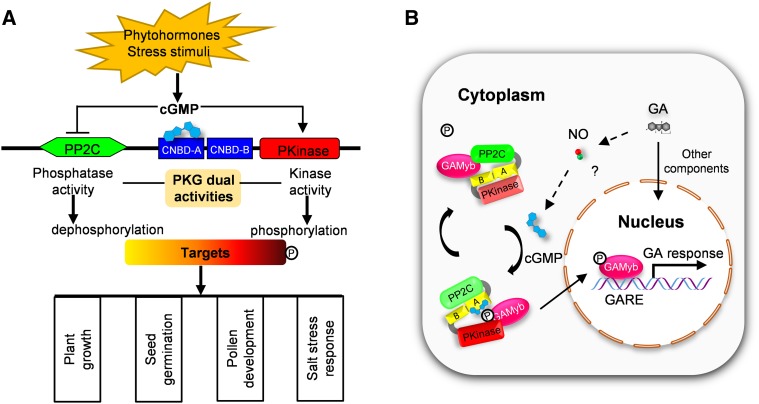
The presence and biological significance of PKG have long been suggested in plants (Szmidt-Jaworska et al., 2003; Nan et al., 2014; Marondedze et al., 2016), but the direct molecular and genetic evidence has been lacking. Here, we present molecular, genetic, biochemical, cellular, and physiological evidence for the presence of PKG in plants and have characterized in detail how PKG functions in rice in response to GA. Plant PKG is unique, possessing protein kinase and protein phosphatase activities. The dual activities of PKG fine-tune the reversible phosphorylation and nucleocytoplasmic distribution of GAMYB, an important transcription factor in GA signaling, through perceiving cGMP in response to GA, thus modulating multiple aspects of plant growth and development (Figure 12).
Figure 12.
Proposed Model for the Regulation of Dual Activities of Plant PKG and Its Coordination with GAMYB in GA Signaling.
(A) Dual activities of plant PKG are distinctly regulated by cGMP in rice and its roles in different biological processes. In response to environment stimuli, cGMP is produced and binds to the CNBD-A domain of PKG, subsequently promotes the kinase activity of PKG, while it inhibits PP2C phosphatase activity. Thus, PKG modulates the phosphorylation status of targets involved in plant growth, seed germination, pollen development, and salt stress response.
(B) PKG coordinates with GAMYB in response to GA. In response to GA, NO is produced and then triggers cGMP production. Subsequently, cGMP binds to and activates Pkinase of PKG, while it inhibits PP2C phosphatase of PKG. The Pkinase of PKG phosphorylates GAMYB, resulting in the nuclear translocation of GAMYB to upregulate the expression of GA-responsive genes. When cellular cGMP is low or absent, the Pkinase of PKG is less activated, whereas inhibited PP2C phosphatase is relieved. Thus, the PP2C phosphatase of PKG dephosphorylates pGAMYB, retaining GAMYB in the cytoplasm to downregulate the expression of GA-responsive genes. The letter P in circle indicates phophate group; the letters A and B in the boxes indicate CNBD-A and CNBD-B, respectively; the question mark (?) indicates the linkage among GA, NO and cGMP is still needed to be confirmed genetically. GARE, GA-responsive element.
Plant PKGs share similarities with animal PKGs but also display distinct properties. Current results indicate that most plant species have a single gene encoding PKG, unlike mammalian PKGs that are encoded by two PKG genes, PKGI and PKGII (Hofmann, 2005; Schmidt et al., 2009). The biochemical property of rice PKG is more similar to PKGI than to PKGII. For example, rice PKG is localized to the cytoplasm and so is mammalian PKGI, whereas PKGII is a membrane-associated protein (Chikuda et al., 2004; Hofmann, 2005). Mammalian PKGI has a higher cGMP binding affinity of CNBD-A than CNBD-B, whereas that of two CNBDs in PKGII exhibit the opposite behavior (Taylor and Uhler, 2000). The CNBD-A domain of rice PKG is completely responsible for cGMP binding, whereas the CNBD-B domain has no affinity for cGMP or cAMP, which is more closely related to that of mammalian PKGI. In addition, rice PKG binds cGMP and cAMP, but cGMP has a much higher binding affinity and stimulation of PKG kinase activity than that of cAMP, and this property is similar to that of mammalian PKGI (VanSchouwen et al., 2015). Mammalian PKGI is widely distributed in various tissues and plays key roles (Francis et al., 2010). Plant PKGs and mammalian PKGI are conserved in the CNBD-CNBD-Pkinase domain structures and cGMP-dependent protein kinase properties.
The most unique aspect of plant PKGs is that plant PKGs contain both protein kinase and protein phosphatase domains, whereas animal PKGs have no protein phosphatase domain or activity (Francis et al., 2010). Gene fusion and exon shuffling enhance the diversity of genes by rearranging the original functional domains or creating additional domains, and intron position is generally well conserved in orthologous genes over long evolutionary process (Long et al., 2013). Analysis of intron phase patterns of PKG genes across Protista, Plantae, and Animalia suggests that the PP2C domain in plant PKGs may have arisen from an ancient evolution event through shuffling or fusion of PP2C. In comparison, animal PKGs lack the PP2C domain that is encoded by a separated gene (Hofmann, 2005). Plant PP2C plays an important role in the stress response (Schweighofer et al., 2004). Plants are sessile and unable to escape from adverse environments. The combination of a kinase with a phosphatase allows one PKG to perform reversible phosphorylation that may offer an efficient regulatory strategy for plants to respond to an ever-changing environment.
Our data show that rice PKG has the dual activities of protein kinase and phosphatase. The full-length PKG displays a cGMP-dependent kinase activity. Deletion analyses indicate that the Pkinase domain is essential for its kinase activity, while the CNBD domain is required for cGMP binding and response. Furthermore, the rice Pkinase domain alone exhibited a constitutive and higher kinase activity than did the full-length PKG, suggesting that the Pkinase domain is sufficient for PKG kinase activity, whereas the N-terminal region of the full-length PKG has an inhibitory effect on kinase activity. In contrast to its kinase activity, rice PKG’s protein phosphatase activity is inhibited by cGMP or cAMP. The cyclic nucleotide inhibition of phosphatase occurs with the full-length PKG, but not with the Pkinase-deleted mutants, suggesting that the Pkinase domain is required for the inhibition. The inhibitory effect of cGMP and cAMP on phosphatase activity may be due to cGMP/cAMP binding resulting in an allosteric change to unfavored conformation for phosphatase catalytic activity. Thus, cGMP activates the protein kinase activity while inhibiting the phosphatase activity of PKG, presenting an efficient mechanism of regulation of PKG activity. These results suggest the cellular cGMP level distinctively regulates the protein phosphorylation and dephosphorylation activities of PKG. The cGMP concentration is dynamic and well controlled in cells (Lucas et al., 2000; Isner et al., 2012). Various stimuli, including hormones and osmotic stress affect the cellular cGMP level in plants (Durner et al., 1998; Pagnussat et al., 2003; Isner and Maathuis, 2011; Isner et al., 2012; Joudoi et al., 2013). Elevated cGMP stimulates PKG’s protein kinase activity and inhibits its protein phosphatase activity, enhancing the phosphorylation of target proteins.
Both NO and cGMP are widely involved in different biological processes in eukaryotic organisms (Francis et al., 2010; Domingos et al., 2015). In plants, NO is emerging as an important signaling molecule, and its roles are coupled with cGMP in many biological functions, including seed germination, pollen development, root formation, stomatal closure, and Ca2+ homeostasis (Garcia-Mata et al., 2003; Pagnussat et al., 2003; Bright et al., 2006; Bai et al., 2012; Joudoi et al., 2013; Wu et al., 2013; Nan et al., 2014). GA is an important hormone in seed germination, plant growth, and flower and pollen development in plants (Yamaguchi, 2008). Both GA and NO induced cGMP formation and seed germination (Penson et al., 1996; Teng et al., 2010; Isner and Maathuis, 2011; Wu et al., 2013). How cGMP is perceived in these processes remains to be elucidated. Our results show that PKG is a direct target of cGMP and plays a key role in the response to GA and NO in rice. In addition, the relationship among GA, NO, and cGMP in plant signal cascades remained undefined. Our data suggest that PKG mediates GA action through perceiving and transducing the NO-cGMP signals via the GA→NO→cGMP→PKG signal cascade. Arabidopsis AtNOGC1 catalyzes cGMP production in a NO-dependent manner (Mulaudzi et al., 2011; Gehring and Turek, 2017) and is important for stomatal movement in response to NO (Joudoi et al., 2013). Our search of the rice genome database (http://rice.plantbiology.msu.edu/) found several flavin-containing monooxygenase FMO GS-OX–like proteins, which share ∼50% identity to AtNOGC1. However, the residues in the heme-NO and oxygen binding motif and relaxed GC motif in rice NOGC1 homologs are less conserved than those in AtNOGC1. Thus, the GA→NO→cGMP signal cascade remains to be further confirmed genetically.
To determine how PKG mediates GA signaling to execute its physiological functions, we screened proteins potentially interacting with PKG and found that PKG interacted with and phosphorylated GAMYB, a positive regulator in GA signaling. GAMYB was first identified as a GA-responsive transcription factor through binding to GA-responsive element containing the TAACAA/CA element in the promoter region of genes encoding amylases involved in seed germination in cereals (Gubler et al., 1995; Kaneko et al., 2004). The function of GAMYB is not limited to seed germination; it is also involved in the male reproductive process, stem elongation, and flower and seed development (Gocal et al., 2001; Diaz et al., 2002; Kaneko et al., 2004; Aya et al., 2009, 2011), which are also found in PKG action. However, unlike the canonical dwarf phenotype caused by a severe defect in GA biosynthesis or GA signaling, such as GA-deficient GA2oxs-OE rice (Lo et al., 2008), a moderately reduced plant height in both pkg and gamyb mutants did not markedly affect the tiller number relative to the wild type. Little effect on tiller number is also found in the gamyb mutant reported previously (Kaneko et al., 2004). PKG coordinates with GAMYB to exert physiological functions in the GA response. One possible explanation is the site of effect of GAMYB, which is expressed in flowers, germinated seeds, shoot apical, and root tips (Kaneko et al., 2004). Transcriptome analysis revealed that GA-responsive element was enriched in the promoters of genes that were upregulated by GA and cGMP signaling (Bastian et al., 2010). All of these findings suggest that GAMYB plays ubiquitous roles in the GA response throughout the life cycle of plants. Our data showed that loss of PKG or GAMYB resulted in an impaired GA response, conferring plants with defects in GA-induced seed germination, internode elongation, and pollen development. Overexpressing GAMYB in the wild type led to increased plant height, seed weight, and seed yield and upregulated expression of amylases, but these promotions were not found in the pkg mutants, suggesting that PKG is required for GAMYB actions. The results suggest that PKG coordinates functionally with GAMYB in the GA signaling pathway.
GAMYB transcript is induced by GA and is suppressed by SLENDER1 (SLN1), a negative regulator in GA signaling in barley (Hordeum vulgare; Gubler et al., 2002). In addition, GA promotes the cytoplasmic-to-nuclear translocation of GAMYB in aleurone cells (Hong et al., 2012), but its mechanism for the nuclear translocation was unknown. Our data show that GAMYB is a target of PKG, that PKG phosphorylation of GAMYB modulates its cytoplasmic-to-nuclear localization, and that the PKG phosphorylation-mediated GAMYB nuclear localization is required for GAMYB action of the plant response to GA, NO, and cGMP. PKG catalyzes the reversible phosphorylation of GAMYB at S6; thus, the nucleocytoplasmic distribution of GAMYB is modulated tightly by PKG with its dual protein kinase and PP2C phosphatase activities, depending on cellular cGMP availability (Figure 12B).
It was shown that GAMYB-OE plants displayed aborted pollens (Murray et al., 2003; Chen et al., 2006), which is different from our observation that showed pollens from GAMYB-OE plants developing normally. Transgenic barley strongly overexpressing HvGAMYB was completely or partially male sterile, but some OE lines in which GAMYB-GFP with moderate expression level were phenotypically normal, exhibiting the higher level of GAMYB OE, the greater defect in male sterility (Murray et al., 2003; Chen et al., 2006). Our GAMYB-OE plants were under the control of 35S promoter, whereas those OE plants reported previously were driven by the Ubi promoter (Chen et al., 2006; Hong et al., 2012). In addition, we found that pollen defects in the pkg and gamyb mutants were temperature sensitive, with more severe defects under high temperature (>35°C) than at ∼25 to 30°C. Therefore, the level of OE, growth condition, and genetic background may be attributable to the difference.
It should be noted that GAMYB is one of the targets of PKG action. Consistent with this is our identification of multiple PKG-interacting proteins and broader effects of PKG than those of GAMYB. For example, the loss of PKG led to increased sensitivity to salt stress, which is not found in the gamyb mutant. Osmotic/salt stress induced cGMP production in Arabidopsis (Donaldson et al., 2004). Of identified PKG-interacting proteins from Y2H screening, several candidates are related to osmotic stress response such as Ca2+ signaling, ROS homeostasis, and ABA biosynthesis (Supplemental Table 2). The pleiotropic effects observed with pkg mutants likely stemmed from different signaling events in which PKG is involved. These results support the working model that plant PKG has broader effects than those of GAMYB (Figure 12). It would be of great interest to dissect how PKG decodes the salt and other stimuli to downstream effects in a future study.
METHODS
Plant Materials, Growth Conditions, and Chemical Supplementations
Rice (Oryza sativa) plants were grown in a paddy field at the experiment station of Huazhong Agricultural University (Wuhan, China) under natural growth conditions during summer and fall seasons. In some experiments, rice seedlings were grown on one-half-strength Murashige and Skoog medium with or without different chemical supplementations in growth chambers under 12-h light/12-h dark photoperiods (30°C/24°C), with white light intensity of ∼150 μmol m−2 s−1 and 60% humidity. For seed treatment with GA, germinating seeds were supplemented with GA3, with various concentrations ranging from 1 μM to 1 mM being tested. The concentrations of ∼50 to 200 μM GA3 have the most effect on promoting seed germination and were applied in most experiments in this study, based on our test and results from previous studies by Chen et al., (2006) and Gong and Bewley, (2008).
Mutant Isolation
The pkg mutants in the cv Zhonghua 11 background were obtained from the Rice Mutant Database (RMD; http://rmd.ncpgr.cn/). The pkg-1 mutants (RMD_04Z11JE75) and pkg-2 mutants (RMD_04Z11AH95) were generated by insertions of T-DNA and Tos17, respectively, in PKG. The gamyb mutant (PFG_1E-03950) caused by a T-DNA insertion in the first intron of GAMYB in the cv Hwayoung background was obtained from RISD_DB (http://cbi.khu.ac.kr/RISD_DB.html). The T-DNA/Tos17 insertion homozygous plants for each mutant were identified by PCR using gene-specific and T-DNA/Tos17–specific primer pairs. DNA sequences of the primers used are listed in Supplemental Table 2.
Measurement of Amylase Activity
For the starch plate assay, surface-sterilized embryoless seeds were placed on starch-containing agar plates (10 mM NaAc, pH 5.2, 5 mM CaCl2, 0.2% [w/v] starch, and 2% [w/v] agar) containing GA3, SNP, or 8-Br-cGMP and incubated at 30°C for 4 d, or amylase secretion was auto-induced at 30°C for 20 d. Subsequently, the amylase activity was visualized by staining with an iodine solution (0.1% [w/v] iodine and 1% [w/v] potassium iodide [I2-KI]). To quantify amylase activity, seed samples pretreated with water (control), GA3, SNP, or 8-Br-cGMP for 24 h were quickly frozen in liquid nitrogen. Samples were ground into fine powder and homogenized with extraction buffer (50 mM NaAc, pH 5.2, 10 mM CaCl2, 10% [v/v] glycerol, 1% [v/v] Triton X-100, and protease inhibitor cocktail) and then centrifuged at 14,000g at 4°C for 15 min. The supernatants were used for chromogenic reaction. α-Amylase activity was measured according to the 3,5-dinitrosalicylic acid colorimetric method (Bernfeld, 1955).
cDNA Synthesis and RT-qPCR
Total RNA was extracted from germinating seeds or various tissues with a modified CTAB reagent (100 mM Tris-HCl, pH 8.0, 2% [w/v] cetrimonium bromide, 2 mM NaCl, 2% [w/v] polyvinylpyrrolidone K-30, 5 mM EDTA, and 2% [v/v] 2-mercaptoethanol) and reverse transcribed by Moloney murine leukemia virus reverse transcriptase. RT-qPCR analyses were performed using a SYBR Green qPCR kit (TransGen Biotech) with a MyiQ system (Bio-Rad). Rice OsActin1 was used as an internal standard control. RT-qPCR conditions were as follows: 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s, for 50 cycles. Relative transcript abundance was calculated using the comparative threshold cycle method. The primers are listed in Supplemental Table 2.
OE of GAMYB in Rice Plants and GUS Activity Assay
It was reported that barley plants with strongly overexpressing HvGAMYB were completely or partially male sterile, but OE lines with moderate expression level were phenotypically normal (Murray et al., 2003). We found that gene OE driven by the Ubi promoter usually led to stronger expression than that of the 35S promoter in rice plants. To avoid the male sterile phenotype, GAMYB was overexpressed in rice plants under the control of 35S promoter instead of the Ubi promoter. The full-length CDS of GAMYB was amplified from rice cDNA by PCR and ligated into pCAMBIA1301s-GFP vector after digestion with KpnI/BamHI. The fidelity was verified by sequencing.
For the GUS activity assay, a 4.5-kb DNA fragment was amplified from the upstream region of the PKG transcriptional start site using rice genomic DNA as template and then cloned into the DX2181b vector. The resultant constructs were transformed into rice callus using Agrobacterium (EHA105) to generate plants with GAMYB OE or plants containing PKGpro:GUS. To observe GUS staining, rice plant tissues or seeds harboring PKGpro:GUS were infiltrated by vacuum and then stained overnight at 37°C with staining solution (50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM K3[Fe(CN)6], 0.5 mM K4[Fe(CN)6], 0.1% [v/v] Triton X-100, and 1 mM X-glucuronide). Photomicrographs were captured by stereoscopic microscope (SZX2-ILLK, Olympus).
Protein Expression and Purification
The full-length and truncated mutants of PKG CDS were cloned from rice cDNA by PCR and ligated into the pET28a vector in frame with 6×His tag. Likewise, full-length CDS of GAMYB was also cloned into the pET28a vector using the BamHI/XhoI cloning sites. The fidelity was verified by sequencing. Recombinant proteins were expressed in Escherichia coli Rosetta (DE3) strain by 0.4 mM isopropylthio-β-galactoside induction at 24°C for 8 h. Cells were harvested by centrifugation at 10,000g at 24°C for 30 min, resuspended in buffer A (25 mM Tris-HCl, pH 8.0, and 150 mM NaCl), and then lysed by a high-pressure cell disrupter (JNBIO). Supernatants were collected by centrifugation at 14,000g at 4°C for 1 h. His-tagged recombinant proteins were purified using the Ni-IDA resin (Sangon) according to the manufacturer’s protocol. The proteins were then separated by anion exchange chromatography (Source Q 10-100, GE Healthcare) using a linear NaCl gradient in buffer A. Proteins containing CNBD domain(s) were then dialyzed against buffer A overnight to remove endogenous cyclic nucleotides. Proteins were purified further by gel filtration on a Superdex-200 Increase 10/300 GL column (GE Healthcare) in buffer A. The purified proteins were used for enzymatic activity and binding assays after verification by SDS-PAGE stained with Coomassie Blue and by immunoblotting using His-tag antibody (1:3000 dilution; sc-8036, Santa Cruz).
Protein Kinase and Phosphatase Activity Assays
For protein kinase activity, purified full-length or truncated PKG (5 μg) was added to 50 μL of reaction mixture (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 μg/μL histones, 2 μCi of [γ-32P]ATP, and 50 μM cold ATP) supplied with or without 100 μM cGMP. After reactions were maintained at 30°C for 1 h, the proteins were separated by 15% SDS-PAGE and detected by autoradiography using the Typhoon imaging system (Typhoon FLA 9000, GE Healthcare). For quantification of PKG protein kinase activity, a Kinase-Glo plus luminescent kinase assay (V3771, Promega) was used according to the manufacturer’s protocol. Briefly, purified full-length or truncated PKG (1 μg) was added to a 50-μL reaction mixture (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 μg/μL histones, and 100 μM ATP) supplied with cGMP or cAMP using various concentrations ranging from 0.1 μM to 1 mM. The purified protein from cells harboring empty vector was used as a control. Reactions were performed at 30°C for 15 min and stopped by adding the Kinase-Glo reagent. After equilibrating the mixture at room temperature for 10 min, luminescence was monitored using an Infinite M200 reader (TECAN).
The protein phosphatase activity was measured using a ProFluor Ser/Thr PPase assay kit (V1260, Promega) according to the manufacturer’s protocol. Briefly, purified full-length or truncated PKG (1 μg) was added to 50 μL of reaction mixture supplemented with or without cGMP, cAMP, or EDTA. Fluorescence resulting from dephosphorylated substrate was monitored using an Infinite M200 reader (TECAN).
Isothermal Titration Calorimetry
The cyclic nucleotide binding was assayed by ITC at 25°C using Auto-iTC200 titration calorimetry (MicroCal). The purified protein (200 μL 50 μM in buffer A) was added in the sample cell, and then cyclic nucleotide (625 μM) was gradually injected into the sample cell with the first injection of 0.4 μL followed by 19 injections of 2 μL. cGMP or cAMP injected into buffer A in a reference cell was used as a negative control. The reference value was subtracted from the experimental curves before data analysis. The data were processed using ORIGIN software (MicroCal) with a manufacturer-supplied custom-addon ITC subroutine.
Subcellular Localization and BiFC Assay
For PKG and GAMYB subcellular localization, the full-length and different fragments of PKG CDS were amplified from rice cDNA by PCR and cloned into pGEM-T easy vector (Promega), and the fidelity was verified by sequencing. The full-length and different fragments of PKG CDS were released from pGEM-T easy vector by XbaI digestion and ligated with GFP in pM999-sunlight GFP or pM999-RFP vector to generate 35Spro:PKG-GFP or 35Spro:PKG-RFP. The 35Spro:GAMYB-GFP construct was generated by a similar strategy using XbaI digestions. For the BiFC assay, the CDSs of rice PKG and GAMYB were cloned into the pSPYNE and pSPYCE vectors, respectively, after digestion with XbaI/AscI. The resultant plasmids were transformed into protoplasts that were isolated from leaf sheaths of 14-d-old etiolated rice seedlings by a polyethylene glycol–mediated transient expression system (Yoo et al., 2007). The protoplasts were observed under a fluorescence microscope (BX53, Olympus) after transformation for 12 h.
Coimmunoprecipitation
The proteins were extracted from Nicotiana benthamiana leaves expressing 35Spro:PKG-FLAG/35Spro:GFP or 35Spro:PKG-FLAG/35Spro:GAMYB-GFP constructs using protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% [v/v] Nonidet P-40, and 0.1% [v/v] Triton X-100) and then incubated with anti-GFP agarose (sc-9996 AC, Santa Cruz) for 5 h at 4°C. After three washes with extraction buffer, the coimmunoprecipitated proteins were separated by SDS-PAGE and detected with anti-GFP (1:3000 dilution; sc-9996, Santa Cruz) and anti-FLAG (1:3000 dilution; sc-166384, Santa Cruz) antibodies, respectively.
Subcellular Fractionation
The nuclear fraction and cytoplasmic fraction were isolated from transgenic plants overexpressing GAMYB in the wild-type and pkg mutant backgrounds, respectively. Seedlings were sampled and ground into powder with liquid nitrogen and mixed with lysis buffer (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% [v/v] glycerol, 1% [v/v] Nonidet P-40, 2 mM Na3VO4, and 1 mM NaF) supplemented with a protease inhibitor cocktail. The homogenate was filtered through a double layer of Miracloth (no. 475855, Millipore). The extract was centrifuged at 1500g at 4°C for 10 min. The resultant pellet was the nuclear fraction as indicated by the nuclear marker H3, whereas the supernatant was further centrifuged at 10,000g at 4°C for 10 min to collect the soluble cytoplasmic fraction. The nuclear fraction was washed twice with 2 mL of lysis buffer and then resuspended with 500 μL of loading buffer (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% [v/v] glycerol, 1% [w/v] SDS, 2 mM Na3VO4, and 1 mM NaF). The separated fractions were detected by immunoblotting using protein-specific antibodies.
cGMP Content Measurement by ELISA
Seeds were weighed and germinated in water supplemented without (control) or with GA3, SNP, or cPTIO for 12 h. The germinating seeds were ground in 0.1 M HCl solution. The homogenate was centrifuged at 6000g at 25°C for 10 min, and the supernatant was transferred to a clear tube. After acetylation to increase the sensitivity, cGMP in the supernatant was detected by ELISA using a cGMP Enzyme Immunoassay kit (CG200, Sigma-Aldrich) according to the manufacturer’s protocol.
NO Detection
Seeds were germinated in water without (control) or with GA3, SNP, or cGMP supplementation at 30°C for 2 h and then transferred into 20 μM 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (NO-specific fluorescent probe, Sigma-Aldrich) solution in darkness for 1 h. After three washes with PBS (137 mM NaCl, 2.7 mM KCl, and 10 mM phosphate buffer, pH 7.5) to remove excess fluorophore, the seed sections were examined under a fluorescence microscope (BX53, Olympus) using a 488 nm excitation and 500 to 530 nm emission filter.
Y2H Assay
Y2H assay was performed according to the Matchmaker Gold Yeast Two-Hybrid System user’s manual (Clontech). The bait plasmid pGBKT7-PKG was cotransformed with the prey plasmids pGADT7-GAMYB and pGADT7-PKG, respectively, into Saccharomyces cerevisiae strain AH109. After growing in medium lacking Leu and Trp at 30°C overnight, different dilutions (10−1, 10−2, and 10−3) of saturated cultures were spotted onto a plate lacking Leu and Trp or a plate lacking Leu, Trp, His, and adenine for selection.
Measurement of ROS and H2O2 Contents
ROS was detected using nitrotetrazolium blue chloride (NBT; Sangon) based on a method described by Zhan et al., (2019). To visualize ROS, leaves were sampled and stained with 6 mM NBT in phosphate buffer (50 mM NaH2PO4, 50 mM Na2HPO4, 150 mM NaCl, and 10 mM KCl, pH 7.5) for 1 h after chlorophyll was removed with 70% (v/v) ethanol. H2O2 content was determined by peroxidase-based assay as described by Noctor et al., (2016).
Phylogenetic Analysis
Phylogenetic analysis was conducted using MAFFT (v7.452) and the neighbor-joining method in MEGA 5 for unrooted phylogeny tree construction supported by 1000 bootstrap reiterations. Alignments used to generate the phylogeny presented in Figure 1A are listed in Supplemental Data Set 1, and the tree file is provided in the Supplemental File.
Statistical Analysis
Statistical significance was analyzed by unpaired Student’s t test for two-group data or by ANOVA analysis in combination with Duncan’s multiple range test for multiple-group data. The details of statistical analyses are shown in Supplemental Data Set 2. The experimental repeat, sample size, and the significance level of P-values are described in figure legends.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: OsPKG (Os02g0281000), AtPKG (At2g20050), hPKGI (EF560730), OsGAMYB (Os01g0812000), OsRAmy1A (Os02g0765600), OsRAmy3E (Os08g0473600), OsRAmy3D (Os08g0473900), and OsActin1 (Os03g0718100).
SUPPLEMENTAL DATA
Supplemental Figure 1. Sequence analysis of plant PKGs compared with human PKGI (supports Figure 1).
Supplemental Figure 2. Phylogenetic analysis of PKGs in Eukaryota (supports Figure 1).
Supplemental Figure 3. Expression, purification and protein kinase activity assay of OsPKG and its different fragments (supports Figure 2).
Supplemental Figure 4. Secondary structures and the expression of CNBD domains (supports Figure 2).
Supplemental Figure 5. Loss of PKG led to reduced seed germination under salt stress (supports Figure 5).
Supplemental Figure 6. PKG mediates GA-, NO- and cGMP-induced seed germination (supports Figure 5).
Supplemental Figure 7. GA promoted NO production in rice cells (supports Figure 5).
Supplemental Figure 8. Screening of candidates that interact with PKG by yeast two-hybrid (supports Figure 6).
Supplemental Figure 9. Co-localization of PKG with cGMP in rice cell (supports Figure 6).
Supplemental Figure 10. PKG does not affect the DNA binding affinity of GAMYB (supports Figure 7).
Supplemental Table 1. Annotation of PKG-interacting proteins by Y2H screening.
Supplemental Table 2. Primers used in this study.
Supplemental Data Set 1. Alignments used to generate the phylogeny presented in Figure 1A.
Supplemental Data Set 2. Statistical analysis results.
Supplemental File. Phylogenetic tree of PKGs in Eukaryota.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ping Yin at the Center for Protein Research, Huazhong Agricultural University, for technical support with the ITC analysis. We thank Frans J. M. Maathuis (University of York) for kindly providing the δ-FlincG construct. This work was supported by the Ministry of Science and Technology of China (grants 2015CB150204 and 2012CB114200) and National Natural Science Foundation of China (grants 31271514 and 31470762).
AUTHOR CONTRIBUTIONS
Q.S. and Y.H. designed the experiments and analyzed data. Q.S. performed most of the experiments. X.Z. assisted with data analysis; P.Y. assisted with vector construction and agronomic trait measurements; J.L. assisted with protein expression and purification; J.C. helped with the generation and analysis of transgenic plants; B.T. assisted with Y2H library construction. Y.H. and Q.S. wrote the article; X.W. consulted on the study and revised the article.
Footnotes
Articles can be viewed without a subscription.
References
- Aya K., Hiwatashi Y., Kojima M., Sakakibara H., Ueguchi-Tanaka M., Hasebe M., Matsuoka M. (2011). The gibberellin perception system evolved to regulate a pre-existing GAMYB-mediated system during land plant evolution. Nat. Commun. 2: 544. [DOI] [PubMed] [Google Scholar]
- Aya K., Ueguchi-Tanaka M., Kondo M., Hamada K., Yano K., Nishimura M., Matsuoka M. (2009). Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Todd C.D., Desikan R., Yang Y., Hu X. (2012). N-3-oxo-decanoyl-L-homoserine-lactone activates auxin-induced adventitious root formation via hydrogen peroxide- and nitric oxide-dependent cyclic GMP signaling in mung bean. Plant Physiol. 158: 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian R., Dawe A., Meier S., Ludidi N., Bajic V.B., Gehring C. (2010). Gibberellic acid and cGMP-dependent transcriptional regulation in Arabidopsis thaliana. Plant Signal. Behav. 5: 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A.T., Beavo J.A. (2006). Cyclic nucleotide phosphodiesterases: Molecular regulation to clinical use. Pharmacol. Rev. 58: 488–520. [DOI] [PubMed] [Google Scholar]
- Bernfeld P. (1955). Amylases, α and β. Methods Enzymol. 1: 149–158. [Google Scholar]
- Bowler C., Neuhaus G., Yamagata H., Chua N.H. (1994). Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77: 73–81. [DOI] [PubMed] [Google Scholar]
- Bright J., Desikan R., Hancock J.T., Weir I.S., Neill S.J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45: 113–122. [DOI] [PubMed] [Google Scholar]
- Chen P.W., Chiang C.M., Tseng T.H., Yu S.M. (2006). Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. Plant Cell 18: 2326–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikuda H., et al. (2004). Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 18: 2418–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco T.A., Moeder W., Yoshioka K. (2016). Opening the gates: Insights into cyclic nucleotide-gated channel-mediated signaling. Trends Plant Sci. 21: 903–906. [DOI] [PubMed] [Google Scholar]
- Demidchik V., Shabala S., Isayenkov S., Cuin T.A., Pottosin I. (2018). Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 220: 49–69. [DOI] [PubMed] [Google Scholar]
- Diaz I., Vicente-Carbajosa J., Abraham Z., Martínez M., Isabel-La Moneda I., Carbonero P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29: 453–464. [DOI] [PubMed] [Google Scholar]
- Domingos P., Prado A.M., Wong A., Gehring C., Feijo J.A. (2015). Nitric oxide: A multitasked signaling gas in plants. Mol. Plant 8: 506–520. [DOI] [PubMed] [Google Scholar]
- Donaldson L., Ludidi N., Knight M.R., Gehring C., Denby K. (2004). Salt and osmotic stress cause rapid increases in Arabidopsis thaliana cGMP levels. FEBS Lett. 569: 317–320. [DOI] [PubMed] [Google Scholar]
- Dubovskaya L.V., Bakakina Y.S., Kolesneva E.V., Sodel D.L., McAinsh M.R., Hetherington A.M., Volotovski I.D. (2011). cGMP-dependent ABA-induced stomatal closure in the ABA-insensitive Arabidopsis mutant abi1-1. New Phytol. 191: 57–69. [DOI] [PubMed] [Google Scholar]
- Durner J., Wendehenne D., Klessig D.F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95: 10328–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis S.H., Busch J.L., Corbin J.D., Sibley D. (2010). cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol. Rev. 62: 525–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freihat L., Muleya V., Manallack D.T., Wheeler J.I., Irving H.R. (2014). Comparison of moonlighting guanylate cyclases: Roles in signal direction? Biochem. Soc. Trans. 42: 1773–1779. [DOI] [PubMed] [Google Scholar]
- Friebe A., Koesling D. (2003). Regulation of nitric oxide-sensitive guanylyl cyclase. Circ. Res. 93: 96–105. [DOI] [PubMed] [Google Scholar]
- Gao Q.F., Gu L.L., Wang H.Q., Fei C.F., Fang X., Hussain J., Sun S.J., Dong J.Y., Liu H., Wang Y.F. (2016). Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: 3096–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata C., Gay R., Sokolovski S., Hills A., Lamattina L., Blatt M.R. (2003). Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc. Natl. Acad. Sci. USA 100: 11116–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring C., Turek I.S. (2017). Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front. Plant Sci. 8: 1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal G.F., Sheldon C.C., Gubler F., Moritz T., Bagnall D.J., MacMillan C.P., Li S.F., Parish R.W., Dennis E.S., Weigel D., King R.W. (2001). GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 127: 1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Gong X., Bewley D.J. (2008). A GAMYB-like gene in tomato and its expression during seed germination. Planta 228: 563–572. [DOI] [PubMed] [Google Scholar]
- Gross I., Durner J. (2016). In search of enzymes with a role in 3′, 5′-cyclic guanosine monophosphate metabolism in plants. Front. Plant Sci. 7: 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Chandler P.M., White R.G., Llewellyn D.J., Jacobsen J.V. (2002). Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 129: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F., Kalla R., Roberts J.K., Jacobsen J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7: 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann F. (2005). The biology of cyclic GMP-dependent protein kinases. J. Biol. Chem. 280: 1–4. [DOI] [PubMed] [Google Scholar]
- Hong Y.F., Ho T.H., Wu C.F., Ho S.L., Yeh R.H., Lu C.A., Chen P.W., Yu L.C., Chao A., Yu S.M. (2012). Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant Cell 24: 2857–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isner J.C., Maathuis F.J. (2011). Measurement of cellular cGMP in plant cells and tissues using the endogenous fluorescent reporter FlincG. Plant J. 65: 329–334. [DOI] [PubMed] [Google Scholar]
- Isner J.C., Nühse T., Maathuis F.J. (2012). The cyclic nucleotide cGMP is involved in plant hormone signalling and alters phosphorylation of Arabidopsis thaliana root proteins. J. Exp. Bot. 63: 3199–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isner J.C., et al. (2019). Short- and long-term effects of UVA on Arabidopsis are mediated by a novel cGMP phosphodiesterase. Curr. Biol. 29: 2580–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joudoi T., Shichiri Y., Kamizono N., Akaike T., Sawa T., Yoshitake J., Yamada N., Iwai S. (2013). Nitrated cyclic GMP modulates guard cell signaling in Arabidopsis. Plant Cell 25: 558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Inukai Y., Ueguchi-Tanaka M., Itoh H., Izawa T., Kobayashi Y., Hattori T., Miyao A., Hirochika H., Ashikari M., Matsuoka M. (2004). Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell 16: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.J., Lorenz R., Arold S.T., Reger A.S., Sankaran B., Casteel D.E., Herberg F.W., Kim C. (2016). Crystal structure of PKG I:cGMP complex reveals a cGMP-mediated dimeric interface that facilitates cGMP-induced activation. Structure 24: 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig D.F., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 97: 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwezi L., Meier S., Mungur L., Ruzvidzo O., Irving H., Gehring C. (2007). The Arabidopsis thaliana brassinosteroid receptor (AtBRI1) contains a domain that functions as a guanylyl cyclase in vitro. PLoS One 2: e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwezi L., Ruzvidzo O., Wheeler J.I., Govender K., Iacuone S., Thompson P.E., Gehring C., Irving H.R. (2011). The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J. Biol. Chem. 286: 22580–22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.F., Yang S.Y., Chen K.T., Hsing Y.I., Zeevaart J.A., Chen L.J., Yu S.M. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20: 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M., VanKuren N.W., Chen S., Vibranovski M.D. (2013). New gene evolution: Little did we know. Annu. Rev. Genet. 47: 307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas K.A., Pitari G.M., Kazerounian S., Ruiz-Stewart I., Park J., Schulz S., Chepenik K.P., Waldman S.A. (2000). Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 52: 375–414. [PubMed] [Google Scholar]
- Ludidi N., Gehring C. (2003). Identification of a novel protein with guanylyl cyclase activity in Arabidopsis thaliana. J. Biol. Chem. 278: 6490–6494. [DOI] [PubMed] [Google Scholar]
- Maathuis F.J. (2006). cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 45: 700–711. [DOI] [PubMed] [Google Scholar]
- Marondedze C., Groen A.J., Thomas L., Lilley K.S., Gehring C. (2016). A quantitative phosphoproteome analysis of cGMP-dependent cellular responses in Arabidopsis thaliana. Mol. Plant 9: 621–623. [DOI] [PubMed] [Google Scholar]
- Meier S., Ruzvidzo O., Morse M., Donaldson L., Kwezi L., Gehring C. (2010). The Arabidopsis wall associated kinase-like 10 gene encodes a functional guanylyl cyclase and is co-expressed with pathogen defense related genes. PLoS One 5: e8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulaudzi T., Ludidi N., Ruzvidzo O., Morse M., Hendricks N., Iwuoha E., Gehring C. (2011). Identification of a novel Arabidopsis thaliana nitric oxide-binding molecule with guanylate cyclase activity in vitro. FEBS Lett. 585: 2693–2697. [DOI] [PubMed] [Google Scholar]
- Muleya V., Wheeler J.I., Ruzvidzo O., Freihat L., Manallack D.T., Gehring C., Irving H.R. (2014). Calcium is the switch in the moonlighting dual function of the ligand-activated receptor kinase phytosulfokine receptor 1. Cell Commun. Signal. 12: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray F., Kalla R., Jacobsen J., Gubler F. (2003). A role for HvGAMYB in anther development. Plant J. 33: 481–491. [DOI] [PubMed] [Google Scholar]
- Nan W., Wang X., Yang L., Hu Y., Wei Y., Liang X., Mao L., Bi Y. (2014). Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development. J. Exp. Bot. 65: 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R.P., Smith C.J. (2004). Cyclic nucleotides. Phytochemistry 65: 2423–2437. [DOI] [PubMed] [Google Scholar]
- Noctor G., Mhamdi A., Foyer C.H. (2016). Oxidative stress and antioxidative systems: Recipes for successful data collection and interpretation. Plant Cell Environ. 39: 1140–1160. [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Lanteri M.L., Lamattina L. (2003). Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol. 132: 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penson S.P., Schuurink R.C., Fath A., Gubler F., Jacobsen J.V., Jones R.L. (1996). cGMP is required for gibberellic acid-induced gene expression in barley aleurone. Plant Cell 8: 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado A.M., Porterfield D.M., Feijó J.A. (2004). Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714. [DOI] [PubMed] [Google Scholar]
- Qi Z., Verma R., Gehring C., Yamaguchi Y., Zhao Y., Ryan C.A., Berkowitz G.A. (2010). Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. USA 107: 21193–21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.H.H.W., Hofmann F., Stasch J.P. (2009). Handbook of Experimental Pharmacology 191. cGMP: Generators, effectors and therapeutic implications. Preface. Handb. Exp. Pharmacol. 191: v–vi. [PubMed] [Google Scholar]
- Schweighofer A., Hirt H., Meskiene I. (2004). Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 9: 236–243. [DOI] [PubMed] [Google Scholar]
- Świeżawska B., Duszyn M., Jaworski K., Szmidt-Jaworska A. (2018). Downstream targets of cyclic nucleotides in plants. Front. Plant Sci. 9: 1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmidt-Jaworska A., Jaworski K., Kopcewicz J. (2009). Cyclic GMP stimulates flower induction of Pharbitis nil via its influence on cGMP regulated protein kinase. Plant Growth Regul. 57: 115–126. [Google Scholar]
- Szmidt-Jaworska A., Jaworski K., Tretyn A., Kopcewicz J. (2003). Biochemical evidence for a cGMP-regulated protein kinase in Pharbitis nil. Phytochemistry 63: 635–642. [DOI] [PubMed] [Google Scholar]
- Taylor M.K., Uhler M.D. (2000). The amino-terminal cyclic nucleotide binding site of the type II cGMP-dependent protein kinase is essential for full cyclic nucleotide-dependent activation. J. Biol. Chem. 275: 28053–28062. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Kornev A.P. (2011). Protein kinases: Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Xu W., Ma M. (2010). cGMP is required for seed germination in Arabidopsis thaliana. J. Plant Physiol. 167: 885–889. [DOI] [PubMed] [Google Scholar]
- Turek I., Gehring C. (2016). The plant natriuretic peptide receptor is a guanylyl cyclase and enables cGMP-dependent signaling. Plant Mol. Biol. 91: 275–286. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.I., Kitano H., Yamaguchi I., Matsuoka M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- VanSchouwen B., Selvaratnam R., Giri R., Lorenz R., Herberg F.W., Kim C., Melacini G. (2015). Mechanism of cAMP partial agonism in protein kinase G (PKG). J. Biol. Chem. 290: 28631–28641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M.E., Francis S.H., Corbin J.D., Grimes K., Richie-Jannetta R., Kotera J., Macdonald B.A., Gibson R.R., Trewhella J. (2003). Mechanisms associated with cGMP binding and activation of cGMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 100: 2380–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.F., Munemasa S., Nishimura N., Ren H.M., Robert N., Han M., Puzõrjova I., Kollist H., Lee S., Mori I., Schroeder J.I. (2013). Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiol. 163: 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Wang F., Zhang C., Xie Y., Han B., Huang J., Shen W. (2013). Heme oxygenase-1 is involved in nitric oxide- and cGMP-induced α-Amy2/54 gene expression in GA-treated wheat aleurone layers. Plant Mol. Biol. 81: 27–40. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zagotta W.N., Siegelbaum S.A. (1996). Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci. 19: 235–263. [DOI] [PubMed] [Google Scholar]
- Zhan X., Shen Q., Chen J., Yang P., Wang X., Hong Y. (2019). Rice sulfoquinovosyltransferase SQD2.1 mediates flavonoid glycosylation and enhances tolerance to osmotic stress. Plant Cell Environ. 42: 2215–2230. [DOI] [PubMed] [Google Scholar]