Transcriptomic analysis of polyploid wheats and ancestral diploid grasses sheds light on the programming of embryo and grain development, gene activities, and their evolutionary implications.
Abstract
Modern wheat production comes from two polyploid species, Triticum aestivum and Triticum turgidum (var durum), which putatively arose from diploid ancestors Triticum urartu, Aegilops speltoides, and Aegilops tauschii. How gene expression during embryogenesis and grain development in wheats has been shaped by the differing contributions of diploid genomes through hybridization, polyploidization, and breeding selection is not well understood. This study describes the global landscape of gene activities during wheat embryogenesis and grain development. Using comprehensive transcriptomic analyses of two wheat cultivars and three diploid grasses, we investigated gene expression at seven stages of embryo development, two endosperm stages, and one pericarp stage. We identified transcriptional signatures and developmental similarities and differences among the five species, revealing the evolutionary divergence of gene expression programs and the contributions of A, B, and D subgenomes to grain development in polyploid wheats. The characterization of embryonic transcriptional programming in hexaploid wheat, tetraploid wheat, and diploid grass species provides insight into the landscape of gene expression in modern wheat and its ancestral species. This study presents a framework for understanding the evolution of domesticated wheat and the selective pressures placed on grain production, with important implications for future performance and yield improvements.
INTRODUCTION
Wheat is a global staple crop of the Poaceae family that is closely related to several wild and cultivated Triticum and Aegilops species. Given its global importance, improvements in wheat productivity are urgently needed to address the demands of a growing population (Avni et al., 2017; Appels et al., 2018; Ramírez-González et al., 2018). The majority of global wheat production comes from two species, Triticum aestivum and Triticum turgidum var durum. The hexaploid T. aestivum, or common/bread wheat, is used for bread making and accounts for 95% of global wheat production (Shewry, 2009). The remaining 5% of wheat production is contributed primarily from T. turgidum (durum or emmer), a tetraploid species with high protein and gluten content well suited to making pasta. These hexaploid (AABBDD) and tetraploid (AABB) genomes are the result of hybridization and polyploidization involving three putative diploid wild grass progenitor species, Triticum monococcum (AA), an Aegilops speltoides-related species (BB), and Aegilops tauschii (DD). Historical and recent reports suggest that the first polyploidization event between an A genome diploid (T. urartu) and a B genome diploid (Ae. speltoides relative) occurred 0.5 to 3.0 million years ago and created tetraploid T. turgidum (AABB), although substantial controversy surrounding the diploid progenitors exists (El Baidouri et al., 2017). Following a second polyploidization event, hexaploid wheat arose as little as 8000 years ago through hybridization of a tetraploid wheat (AABB) and a diploid grass (DD; Otto, 2007; Gegas et al., 2010; Matsuoka, 2011). Although bread wheat, durum wheat, and their putative ancestral grass species share many features, including being closely related in phylogeny and exhibiting high levels of sequence similarity (Luo et al., 2017; Ling et al., 2018), they differ in their genetic makeup and ploidy levels.
Grain and yield trait selection has shaped the characteristic features of cultivated wheats relative to their ancestral grass species. Yet, the mechanisms underpinning the regulation and reshaping of gene activities during grain production are not well understood. Uncovering gene expression differences and the evolutionary context associated with these changes has the potential to explain phenotypic alterations and the regulatory switches affecting gene expression, which could broaden genetic diversity and ultimately enable targeted crop yield improvements (Hofmann, 2013; Signor and Nuzhdin, 2018; Purugganan, 2019). In wheat breeding, the application of germplasm carrying new genes or allelic variations of adapted germplasm can dramatically increase grain yields, as evident in the Green Revolution, when the application of a dwarf gene in wheat and other crop species enabled significant grain yield improvements (Hedden, 2003; Saville et al., 2012). In contrast to polyploid wheat species, in which genetic diversity is often limited, the diploid ancestors have a large degree of genetic diversity within an accessible gene pool, which can be exploited for the development of new high-yielding varieties adapted to sustainable agricultural practices ((Salamini et al., 2002; )). Furthermore, as a result of polyploidization, most genes in tetraploid and hexaploid wheat species are present in multiple copies, referred to as homeologs from corresponding genes among A, B, and D subgenomes (Devos, 2010; Glover et al., 2016; Krasileva et al., 2017). Examining gene expression in the expanding ploidy levels of various wheat species and identifying and characterizing their corresponding homologs (referring to common ancestry) and homeologs (referring to corresponding genes from three subgenomes) in the diploid ancestral grass species has the potential to reveal their divergence and respective functionalities among wheat species subjected to agricultural selective pressures (Hills et al., 2007; Bansal et al., 2017; Ramírez-González et al., 2018). Despite the importance of hybridization and polyploidization in the history of wheat, our understanding of the gene expression changes and evolutionary divergence of embryogenesis from diploids to polyploids is limited.
The wheat grain consists of three major components: the diploid embryo, the triploid endosperm, and the pericarp (seed coat). The embryo and endosperm are produced by fertilization of the haploid egg cell and the diploid central cell by two sperm nuclei, respectively, while the pericarp is derived from maternal tissue of the female sporophyte. The wheat embryo, as the germline component of the grain, transmits expression programs and their genetic determinants between successive generations. The genetic crosstalk between embryo, endosperm, and pericarp tissues during development is highly complex and requires the cooperation of several biological processes (Xiang et al., 2011a). Gaining access to the transcriptomes of different components of the developing grain offers insight into the biological processes of grain formation beyond whole-tissue analyses, as evidenced by such investigations of the grains/seeds of valuable crops such as wheat, maize (Zea mays), and rapeseed (Brassica napus; Pfeifer et al., 2014; Yang et al., 2018; Yi et al., 2019; Ziegler et al., 2019). Furthermore, knowledge of the gene expression programs and associated regulatory networks in the developing embryo offers a valuable resource to identify and characterize the impact of hybridization, polyploidization, and breeding on gene activities during grain development and production in wheat.
Here, we present an atlas of global gene expression in the developing embryo in common and durum wheats and their three putative diploid ancestors. These comprehensive gene expression analyses of embryo development were combined with expression analysis of selected endosperm and pericarp tissues. The findings from this study provide valuable insights into the evolution of gene expression during embryogenesis and grain development in wheat.
RESULTS
Characteristics of Embryo and Grain Development in Various Wheat Species
Inspection of embryos and grain morphologies throughout their development revealed overlapping ontogenies and the same basic morphological progression of the embryo among the five species investigated, which allowed development to be grouped into the same discrete, sequential stages for all grass species examined (Guillon et al., 2012). We focused our study on the three main components of the grain: the embryo, endosperm, and pericarp (Figure 1A), and performed a detailed microscopic analysis of the isolated embryos, endosperm, and pericarp of developing grains of AC Barrie, a hexaploid cultivar grown in Canada (Figures 1B to 1U), focusing on 10 of the developmental stages and tissues (E1–E10) defined for all species in this study. Specifically, the developmental stages and tissues include the two-cell embryo (E1; Figure 1B), pre-embryo (E2; Figures 1C and 1E), transition (E3; Figures 1F and 1G), leaf early (E4; Figures 1H and 1I), leaf middle (E5; Figure 1J), leaf late (E6; Figure 1K), and mature embryo (E7; Figure 1L) stages; two stages of isolated endosperm including the transition stage endosperm (E8; Figures 1N and 1O) and leaf late stage endosperm (E9; Figures 1P and 1Q); and the leaf early stage pericarp (E10; Figures 1R and 1S). Additional detailed descriptions of the 10 developmental stages/tissues (E1–E10) and their corresponding reference time points reflecting hours/days after fertilization for the five wheat species are presented in Supplemental Table 1.
Figure 1.
Developmental Stages for Embryo, Endosperm, and Pericarp in the Hexaploid Wheat, AC Barrie.
Stages and tissues defined herein were used for the RNA-seq expression atlas for five different wheat and grass species. Light micrographs of the embryo ([B] to [G]), endosperm ([N] and [P]), pericarp (R), and grain ([T] and [U]); scanning electron micrographs of the grain (A) and embryo ([H] to [L]); and corresponding illustrations ([M], [O], [Q], and [S]) are shown.
(A) Longitudinally cut wheat grain in the leaf late stage of embryo development.
(B) Zygote.
(C) Quadrant.
(D) Octant.
(E) Dermatogen.
(F) and (G) Transition.
(H) and (I) Leaf early.
(J) Leaf middle.
(K) Leaf late.
(L) Mature embryo.
(M) Sequence of embryo stages used for RNA-seq analysis and bracketed groupings of embryo development with corresponding stages of development: E1, two-cell embryo; E2, pre-embryo; E3, transition embryo; E4, leaf early embryo; E5, leaf middle embryo; E6, leaf late embryo; E7, mature embryo.
(N) and (O) Transition stage endosperm (E8).
(P) and (Q) Leaf late stage endosperm (E9).
(R) and (S) Leaf early stage pericarp (E10).
(T) Leaf early stage grain.
(U) Mature grain.
Cp, coleoptile; Cr, coleorhiza; Em, embryo; En, endosperm; Epi, epiblast; LP, leaf primordia; Pc, pericarp; SAM, shoot apical meristem; Sc, scutellum; Sus, suspensor. Bars = 0.01 mm ([B]–[D], [N], and [P]), 0.05 mm ([E]–[L]), and 0.5 mm ([A], [R], [T], and [U]).
We visualized the developmental details of the early stage embryo by light microscopy (Figures 1B to 1G) and the surface features distinguishing later stages of embryo development by scanning electron microscopy (Figures 1H to 1L). Soon after fertilization, transverse division of the zygote formed the two-cell embryo (Figures 1B and 1M). The lower basal cell transformed into a large vesicular cell, while divisions of the upper apical cell gave rise to the quadrant component of the embryo (Figures 1C and 1M). Divisions of the middle radicle cell (situated beside the vesicular cell) produced the radicle initials and suspensor, while divisions of the middle lower embryo cell (situated between the radicle cell and the uppermost cells) formed the lower part of the embryo (Figures 1C and 1M). The quadrant continued to divide into an octant (Figures 1D and 1M), then dermatogen (Figures 1E and 1M), then transition stage embryo (Figures 1F, 1G, and 1M). A laterally placed dome marked the site of shoot apical meristem formation (Figures 1H and 1M) between the shield-shaped scutellum and the suspensor. Differentiation of coleorhiza cells was followed by the emergence of a bulging coleoptile and single leaf primordium (Figures 1H–1J and 1M), which characterize members of the monocotyledon clade. The leaf primordium and shoot apical meristem became engulfed by the developing coleoptile (Figures 1J, 1K, and 1M). After the emergence of the coleoptile and leaf primordium around the shoot apical meristem, the epiblast emerged (Figures 1J and 1M), expanded (Figures 1K and 1M), and formed a fan-like protrusion between the coleoptile and coleorhiza of mature embryos (Figures 1L and 1M). Significant expansion of the scutellum from its emergence in the leaf early stage of embryo development to the mature embryo stage was observed (Figures 1H to 1L and 1M), supporting its role in the transfer of nutrients from the endosperm to the embryo during germination.
In dicots, the endosperm cells are consumed to support embryo development, leaving a thin layer of endosperm on the inner wall of the maturing seed coat (Olsen, 2004). Conversely, in grass species such as wheat, the endosperm constitutes most of the grain and contains large amounts of carbohydrate and protein storage reserves. The endosperm progressively increased in size from endosperm initials that formed a cellularized endosperm by the transition stage (Figures 1N and 1O) to the leaf late embryo stage (Figures 1P and 1Q), where the endosperm occupied the majority of the grain space. The pericarp (exposed in Figure 1T) directly encapsulated the endosperm during grain development, beneath the external tissues of the mature grain (Figure 1U). Compared with polyploid wheat species, the ancestral diploid grass species had small grains, although the endosperm still occupied the majority of the grain. While the key stages and morphological features defining embryogenesis in tetraploid and hexaploid wheat and their putative diploid relatives largely followed the same progression, the developmental time required for embryo, endosperm, and pericarp maturation in the diploid species was reduced. The overlapping stages and morphological similarities across modern wheat and their diploid ancestors allowed us to select and isolate tissues to examine gene expression in the embryo, endosperm, and pericarp at comparable stages of embryogenesis.
Transcriptome Profiling of Developing Embryo, Endosperm, and Pericarp Tissue in Various Wheat Species
To obtain a global view of the transcriptomes during embryo, endosperm, and pericarp development in polyploid wheat cultivars and their putative diploid ancestors, we used the Illumina Hi-seq - platform for RNA sequencing (RNA-seq) analysis of transcripts from seven embryo stages, from the zygote to mature embryo (E1–E7), two endosperm stages (E8 and E9), and one pericarp stage (E10; Figure 1; Supplemental Data Sets 1 to 5). All samples and developmental stages were examined for each of the five species, including the diploids T. monococcum (AA, DV92-DV; a close relative of T. urartu), Ae. speltoides (BB, TA2780-SP), and Ae. tauschii (DD, TA101132-TA), tetraploid wheat (AABB, T. turgidum var durum, Canadian cultivar, Strongfield [SF]), and hexaploid wheat (AABBDD, T. aestivum, Canadian cultivar, AC Barrie [AC]), as summarized in Supplemental Table 1.
We assessed gene expression patterns using 98 RNA-seq samples comprising five species at seven stages of embryo development. On average, the mapping rate for the reads was 93.6%. Expressed genes with high confidence were selected based on the expression of more than five reads per 10 million in at least one of the seven stages of development, which identified 42,474 to 47,790, 31,383 to 35,747, 15,075 to 18,741, 17,883 to 21,613 and 18,639 to 25,008 genes expressed across all samples from AC, SF, DV, SP, and TA, respectively (Supplemental Table 2). These data sets were used to develop a gene expression atlas of embryo, endosperm, and pericarp tissues in bread wheat, durum wheat, and their putative diploid ancestors (see Methods; Supplemental Figure 1).
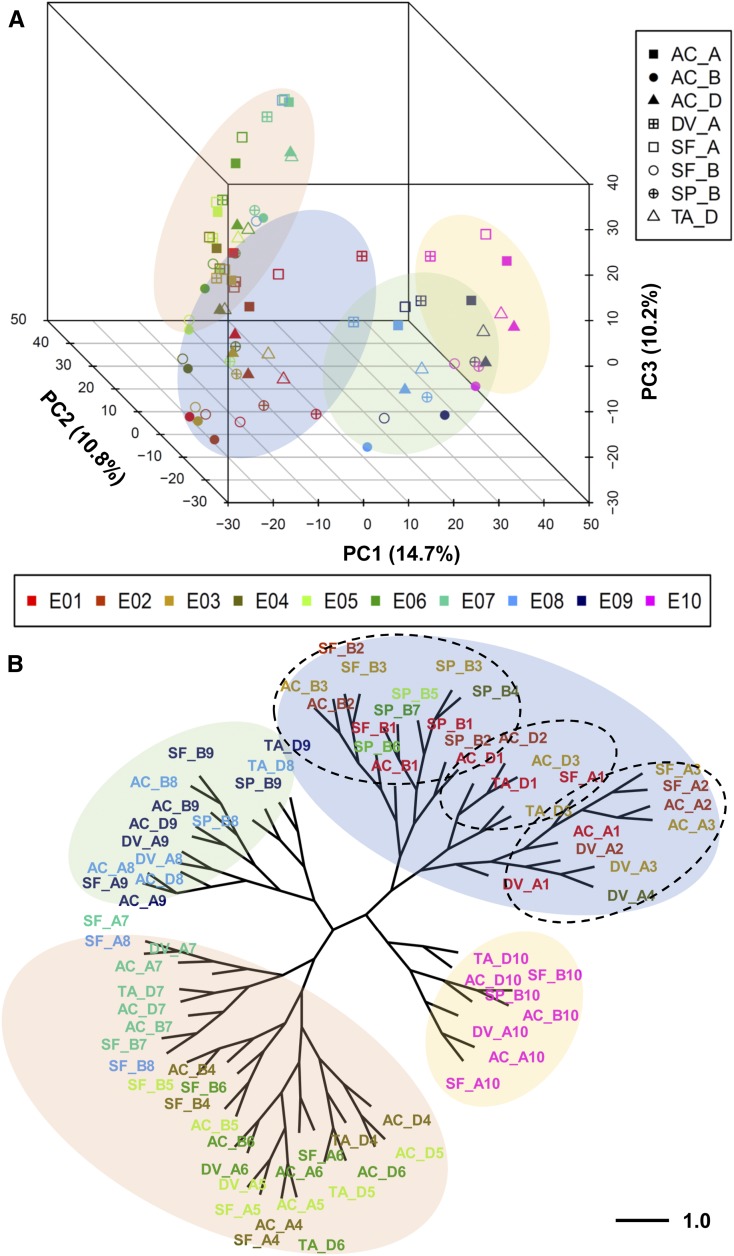
Changes in gene expression have been a driving force behind phenotypic divergence during the evolution of land plants (Wu and Sharp, 2013). To obtain an overview of gene expression patterns in developing embryo, endosperm, and pericarp tissues in wheat and its putative ancestors, we performed principal component analysis (PCA) using homeolog expression data (which included 20,702 identified homeologs; see Methods) for the aforementioned stages of embryo, endosperm, and pericarp development from a panel of eight subgenomes (AC_A, AC_B, AC_D, SF_A, SF_B, DV_A, SP_B, and TA_D). PCA variance was calculated for the data from these samples (Supplemental Figures 2A and 2B) and was found to explain 71.6% total variance from PC1 to PC10. We generated bi-plots for every two-component comparison (Supplemental Figure 3). The first component (PC1) separated samples based on tissue type and developmental stage, and the second component (PC2) showed clear separation among subgenomes (Figure 2A; Supplemental Figure 4). To further examine the association between species, we subjected the top 2000 homeologs with the greatest expression variance across eight subgenomes to 3D PCA analysis, and over 50% of variance was explained by PC1, PC2, and PC3 (Figure 2A; Supplemental Figure 2B). Cluster analysis showed the grouping of samples by developmental stage, tissue, and subgenome (Figure 2A). The four primary clusters represent early embryo development (E1–E3; denoted by blue shading), late embryo development (E4–E7; orange shading), endosperm (E8 and E9; green shading), and pericarp (E10; yellow shading; Figure 2A).
Figure 2.
Relationships of the Transcriptomes of Five Wheat and Grass Species from Different Stages of Grain Development, Tissues, and Subgenomes.
(A) 3D plot of PCA using the top 2000 variant homeologous and homologous genes in embryo, endosperm, and pericarp tissues for 80 individuals. x, y, and z axes indicate PC1, PC2, and PC3, respectively, and the proportion of variance for each principal component is shown in parentheses. The 10 stages of embryo (E01–E07), endosperm (E08 and E09), and pericarp (E10) development (defined in Figure 1) are labeled with different colors (see horizontal inset). Each species and subgenome is labeled with a different shaded shape, with A, B, and D subgenomes represented by squares, circles, and triangles, respectively (see vertical inset). Species names are abbreviated as follows: DV92 (DV), TA101132 (TA), TA2780 (SP), Strongfield (SF), and AC Barrie (AC; see Supplemental Table 1). Grouping of closely correlated individuals is indicated by color-shaded ovals, representing early embryo (E1–E3; denoted by blue shading), late embryo (E4–E7; orange shading), endosperm (E8 and E9; green shading), and pericarp (E10; yellow shading).
(B) Phylogenetic tree of homeolog expression in embryo, endosperm, and pericarp stages in five species. A, A genome; B, B genome; D, D genome. The 10 stages of development (E01–E10) are labeled with colors and the corresponding stage number. For example, AC-B1 represents AC Barrie B genome in stage E01 or the two-cell embryo stage. The scale bar for sample correlation distance is defined.
To provide an evolutionary context for gene expression in the germline during embryogenesis in diploid and polyploid wheat species, and to reveal the extent of evolutionary divergence, we constructed expression distance matrices (Supplemental Data Set 6) and a phylogenetic tree of homeolog expression (Figure 2B). In the early embryo development cluster (blue shaded oval), three subclusters based on subgenome type were observed (black dotted outlines). Similarly, in the late embryo development cluster (orange shaded oval) and endosperm (green oval) and pericarp (yellow oval) clusters, further clustering by subgenome A, B, or D was observed, with occasional separation of the D subgenome (Figure 2B). In the endosperm cluster, SP (B genome) and TA (D genome), AC_B (plus SF_B late endosperm stage) and AC_D, and AC_A (plus SF_A late endosperm stage) and DV (A genome) clustered into subgroups. In the pericarp cluster, subclusters were first distinguished by A, B, and D subgenomes, then by species (Figure 2B). Overall, based on the clustering of transcriptomes, it appears that tetraploid and hexaploid wheat species are more closely related than diploid species; the A and D genomes are more closely related than the B genome; and early stages of embryo development are more conserved than late stages. Thus, the results of two unsupervised clustering analyses suggest that the evolutionary divergence of gene expression is influenced by, in descending order, the tissue, general developmental phases (early, middle, or late embryogenesis), subgenome, species, and adjacent developmental stage of embryogenesis.
Cluster Analysis of DEGs during Wheat Grain Development
We identified 27,897, 22,208, 11,413, 11,129, and 17,239 differentially expressed genes (DEGs; see Methods) across all stages of embryo development from AC, SF, DV, SP, and TA, respectively. To identify differential expression patterns, we performed clustering analysis using the R package cutreeDynamic on the DEGs for each of the five species. A total of 32, 23, 28, 24, and 24 clusters were identified in AC, SF, DV, SP, and TA, respectively. Gene expression per cluster (also called “module”) was further condensed into module eigengene (ME) expression using the first principal component (Supplemental Figures 5A to 5E; Supplemental Data Set 7). To gain insight into the biological relevance and functional significance of modules, we performed Gene Ontology (GO) annotation and enrichment analysis for each module (Supplemental Data Set 7). GO term enrichment was largely conserved across all five species. Of the top 100 enriched terms from each species, 28 were conserved in all five species and 32 were conserved in four of the five species examined (Supplemental Data Set 7). Core biological processes such as DNA replication and histone methylation, cell division, and cell proliferation-associated GO terms were conserved across all five species (Supplemental Data Set 7). We identified differential expression patterns based on ME values. Although the gene expression data were derived from different species, high similarities in expression patterns during embryo development among clusters were identified by Pearson correlations between MEs (r > 0.8, P < 0.05; Supplemental Data Set 7). For example, genes from clusters AC_ME1, SP_ME4, and DV_ME4 displayed similar expression patterns, including low levels of expression during early stages of embryo development and high levels of expression during late stages. GO enrichment analysis suggested that these genes are involved in similar biological processes, such as the phosphorelay signal transduction system (e.g., TraesCS2A01G072100 and TraesCS2A01G072300 in AC and DV and TraesCS2B01G087100 and TraesCS4B01G010700 in AC and SP), glycogen biosynthetic process (e.g., TraesCS2A01G293400 and TraesCS2A01G310300 in AC and DV), and vesicle-mediated transport (e.g., TraesCS1A01G127000 and TraesCS1A01G363100 in AC and DV). Genes in clusters AC_ME10 and DV_ME6 were highly expressed during early stages but were rapidly downregulated during late stages of embryo development, while the remaining genes were highly expressed in developing endosperm. GO enrichment analysis revealed putative roles for the encoded gene products in cell fate specification, steroid metabolic processes, and storage protein synthesis.
Specific and Conserved Gene Expression during Grain Development
The identification of embryo- and endosperm-specific genes should provide a resource for understanding their respective functions and the crosstalk between these tissues. Moreover, fertilization signals appear to ensure synchronized development within an ovule and the underlying control of tissue or organ identity (Chevalier et al., 2011). To generate a comprehensive tissue-specific gene expression catalog, we compared the RNA-seq data from all five species across seven stages of embryo development plus endosperm and pericarp tissues. We classified these genes into six groups: embryo-specific, endosperm-specific, pericarp-specific, embryo-excluded, endosperm-excluded, and pericarp-excluded (Supplemental Data Set 8). In AC, SF, TA, DV, and SP, we identified 995, 285, 281, 320, and 271 embryo-specific genes, 146, 37, 5, 12, and 10 endosperm-specific genes, and 136, 96, 19, 16, and 6 pericarp-specific genes, respectively (Supplemental Data Set 8). These results indicate that the majority of embryo-specific genes were found in the two-cell and mature embryo stages.
To identify the conserved genes across subgenomes and species, we performed dynamic expression pattern analysis based on Pearson correlation coefficient, using all stages of embryo development. Conserved genes were defined as r > 0.8 based on comparisons of homeologs and homologs. Supplemental Figure 6 shows examples of conserved genes belonging to different categories. Seven conserved categories were identified, including conserved homologs in the A subgenome (CsA; AC_A, SF_A, and DV_A), B subgenome (CsB; AC_B, SF_B, and SP_B), and D subgenome (CsD; AC_D and TA_D); conserved homeolog triads in AC (CsAC; AC_A, AC_B, and AC_D); conserved homeolog diads in SF (CsSF, SF_A and SF_B); conserved homeologs in the three diploids (CsDP; DV_A, SP_B, and TA_D); and conserved homeologs across the five species, CsAll (AC-A, AC_B, AC_D, SF_A, SF_B, DV_A, SP_B, and TA_D). The conserved gene number and gene lists for each category are provided in Supplemental Data Set 9.
We performed GO annotation and enrichment analyses to investigate the putative functions associated with the seven gene categories identified. Supplemental Figure 7 shows the most significantly enriched GO terms in the Biological Process category. Enriched conserved genes involved in microtubule-based movement, cell division, cell cycle, and cytokinesis were found in A and D subgenomes, and enriched conserved genes involved in histone methylation and gene silencing were found in A and B subgenomes. Enriched conserved genes involved in DNA replication were common among the A, B, and D subgenomes (Supplemental Figure 7). The two polyploid wheat species were conserved in most genes in the Biological Process category, while diploid species were more divergent (Supplemental Figure 7). A total of 148 conserved genes were found across the five species and subgenomes analyzed, and the enriched GO terms were related to nucleosome assembly, DNA replication, and cell proliferation processes, supporting the conservation of these developmentally related essential processes among all subgenomes and species (Supplemental Data Set 9).
We used the expression patterns of the CsAll gene set from one subgenome (i.e., A genome of AC) to calculate gene correlation coefficients. Closely correlated genes were clustered, and a pair-wise comparison heatmap is shown in Supplemental Figure 8A. Two clusters (C1 and C2) clearly separated and exhibited contrasting expression patterns. Annotation of gene members in these two clusters revealed nine transcription factor (TF) genes in C1 but no TF genes in C2. The expression patterns of these nine TF genes are shown in Supplemental Figure 8B. All of these TF genes were highly expressed during early embryo development. The lack of conserved TF genes in C2 suggests that stringent transcriptional regulation is more evolutionarily conserved in early stages of embryo development than in late embryo development.
Coexpression Analysis of TF and Pathway Genes
The dynamic expression patterns of genes reflect their roles in development. Overlapping processes or common pathways can often be identified by finding sets of genes with distinct expression pattern changes. Similarly, grouping coexpressed TF genes into modules with an enrichment of tissue-specific genes may facilitate the identification of uncharacterized genes or processes in embryo and endosperm development. To identify such modules and expression shifts, we performed TF coexpression analysis to assess the dynamic reprogramming of the transcriptome and to identify spatial gene expression trends during embryo, endosperm, and pericarp development. In total, 3203 TF genes were identified, including 1028 genes from the A subgenome, 1182 genes from the B subgenome, and 993 genes from the D subgenome. The TF genes detected in the different developmental stages are shown in Supplemental Data Set 10. The expressed TF genes were significantly different during different embryo, endosperm, and pericarp developmental stages (36–75%), indicating that TF genes tended to be expressed in association with specific developmental stages and tissues (Supplemental Data Set 10). By examining the expressed TFs across all developmental stages and species, significantly enriched tissue-specific TF gene families were identified, which were consistent across different subgenomes and species. The expressed TF families B3, BHLH, bZIP, C2H2, G2-like, GRAS, DOF, ERF, MYB, NAC, WOX, WRKY, YABBY, and ZF-HD showed embryo-specific enrichment, whereas bZIP, ERF, MYB, NAC, and GRAS TFs were enriched in endosperm, and MADS, bZIP, MYB, NAC, BHLH, and C2H2 TFs were enriched in the pericarp (Supplemental Data Set 10).
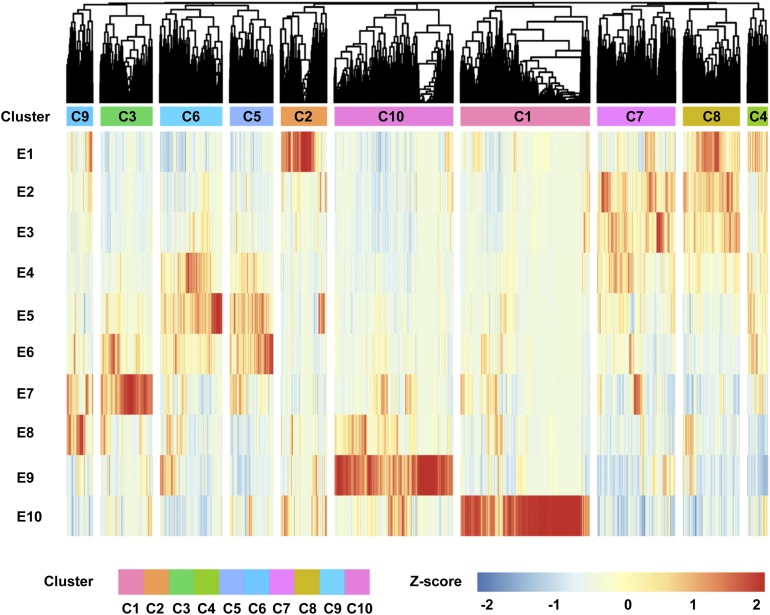
To identify transcriptional networks enriched in specific tissues, we compared gene distribution and expression patterns in the embryo, endosperm, and pericarp across the five species studied. Annotated genes associated with essential processes in embryo development, including carbohydrate metabolism, starch synthesis, and storage protein accumulation, were used to perform coexpression network analysis with TFs (named pathway categories). Querying Arabidopsis (Arabidopsis thaliana) embryo-defective mutant data sets, where loss-of-function mutations cause embryo defects in Arabidopsis (Xiang et al., 2011a), produced an essential embryo developmental gene list for wheat species. Coexpression analysis across the eight subgenomes (AC_A, SF_A, DV_A, AC_B, SF_B, SP_B, AC_D, and TA_D) revealed conservation among genes involved in embryo development across the five species examined. The essential embryo development gene list was characterized by medium to high expression levels and moderately dynamic expression patterns across embryo development (Supplemental Data Set 11). Furthermore, essential embryo development genes were coexpressed and clustered into five major groups, showing similarity across five species (Figure 3). Genes encoding storage proteins clustered in one major group, and genes associated with carbohydrates clustered in two major groups. Cluster 1 contained 672 TF genes enriched in the MYB, NAC, ERF, C2H2, and WRKY TF families, 173 embryo development essential genes, and 104 carbohydrate genes and showed coexpression patterns with storage protein, carbohydrate, and starch synthesis genes (Figure 3; Supplemental Data Set 11). Carbohydrate genes were also represented significantly in cluster 10, containing 136 carbohydrate-related genes, 322 embryo development essential genes, and 727 TF genes. The MYB, BHLH, NAC, WRKY, MADS, and bZIP TF genes were enriched in cluster 10 (Figure 3; Supplemental Data Set 11).
Figure 3.
Coexpression Analysis of TF and Pathway Genes in T. aestivum and T. turgidum Wheat Species and T. monococcum, Ae. speltoides, and Ae. tauschii Grass Species during Grain Development.
Using hierarchical clustering combined with a gene expression heatmap (Z-score normalized), 10 gene clusters (C1–C10) were identified from the five wheat or grass species’ gene homologs across the 10 stages of development (E1–E10) defined in Figure 1. Each cluster identifies genes that are dominant in different stages of embryo, endosperm, or pericarp development. Different clusters are indicated by different colors and the cluster number. The color scheme, from red through yellow to blue, indicates the level of normalized expression, from high to low.
To obtain further insight into the observed expression patterns, annotated processes, and respective contributions of homeologs and homologs to the polyploid genome of wheat, we analyzed the percentage of coexpressed genes in different clusters and categories. Ten clusters emerged for the same categories/pathway genes, with 41, 41, 46, and 35% homeologs in the storage protein, embryo essential, carbohydrate, and TF categories coexpressed, respectively. For homologs, the percentage of coexpressed categories/pathway genes differed among subgenomes. The storage proteins had the highest percentage of homologs (53–67%) coexpressed among the three subgenomes. In the A, B, and D subgenomes, 59, 47, 43, and 37%; 67, 32, 40, and 31%; and 53, 23, 45, and 29% homologs of storage protein, embryo essential, carbohydrate, and TF genes were coexpressed, respectively. These results indicate that the expression patterns of homeologs were more conserved than those of homologs during grain development in the evolution of the wheat lineage (Supplemental Data Set 11).
Homeolog Expression Divergence during Embryogenesis in Polyploid Wheat
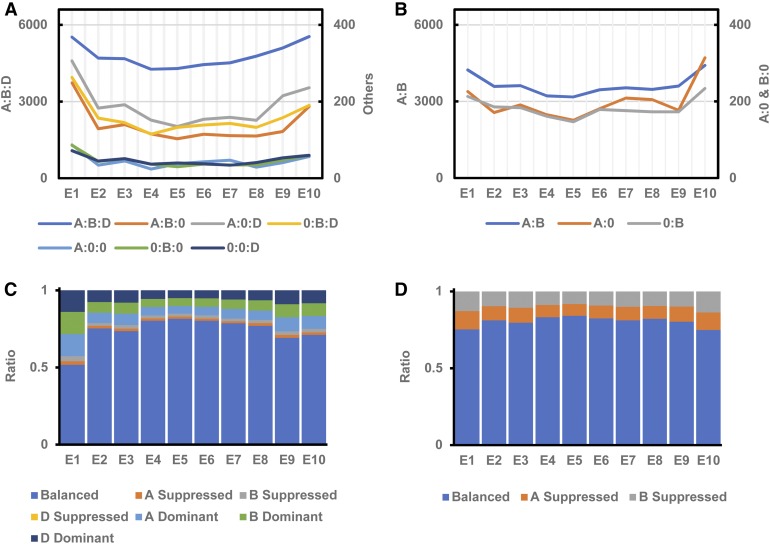
To systematically investigate genome-specific homeolog expression bias across embryogenesis in hexaploid and tetraploid wheat species, we identified triads and diads. To identify triads, or gene homeologs represented by each of the three subgenomes, 62,106 genes with a 1:1:1 correspondence ratio were analyzed across the A, B, and D subgenomes of AC. Similarly, to identify diads (also known as pairs), or gene homeologs represented in both of the subgenomes in SF, 41,404 genes across the A and B subgenomes were analyzed (see Methods; Supplemental Data Set 6). In AC, the triads with lowest and highest expression levels were observed in the two-cell embryo stage (46.2%) and leaf early stage pericarp (51.7%), respectively (Supplemental Data Set 6). In SF, the diads with lowest and highest expression levels were observed in the leaf late stage endosperm (53.6%) and two-cell embryo stage (60.9%), respectively (Supplemental Data Set 6). No strong similarities between the percentage of diads expressed in tetraploid wheat and triads expressed in hexaploid wheat across developmental stages were identified. However, homeolog groups with differential expression across all developmental stages were identified, revealing a higher number of differentially expressed homeologs in early stages of development in both AC and SF (Figures 4A and 4B) across all homeolog groups. Homeolog groups are dynamically expressed during early embryogenesis and become balanced before the leaf early embryo stages. The total percentage of expressed genes was lower in the middle stages of embryo development and higher in the two-cell embryo and leaf early stages of pericarp in tetraploid (71–73%) and hexaploid (71%) species (Figures 4A and 4B; Supplemental Data Set 6).
Figure 4.
Homeolog-Biased Expression in Polyploid Wheat Species T. aestivum and T. turgidum during Grain Development.
(A) Number of DEGs per homeologous group in hexaploid AC.
(B) Number of DEGs per homeologous group in tetraploid SF.
(C) Percentage of homeolog-biased expression across embryo, endosperm, and pericarp developmental stages in hexaploid AC.
(D) Percentage of homeolog-biased expression across embryo, endosperm, and pericarp developmental stages in tetraploid SF.
E1 to E10 represent the 10 stages of development defined in Figure 1.
We performed homeolog expression bias analysis across the 10 embryo developmental stages and tissues, focusing on the 20,702 triads and diads. Seven homeolog expression categories in hexaploid and three homeolog expression categories in tetraploid were used to perform the bias analysis (Figures 4C and 4D; Supplemental Data Set 12). In hexaploid wheat, 51.2, 75.2, and 73.5% of the expressed triads showed balanced expression in the two-cell, pre-embryo, and transition embryo stages, respectively. Approximately 14% of homeologs were predominantly expressed in the two-cell embryo stage, with only 2.3 and 3.4% suppressed in the A and B subgenomes, respectively. Both the suppressed and dominant homeologs showed decreased expression patterns until the transition embryo developmental stage in hexaploid wheat. Homeologs suppressed in subgenome D across all developmental stages and tissues were not identified (Figures 4C and 4D; Supplemental Data Set 12). In the tetraploid species, 75.2, 81.1, and 79.6% showed balanced expression, with 12.9, 9.6, and 10.7% dominantly expressed in the A subgenome and 11.8, 9.2, and 9.7% dominantly expressed in the B subgenome, in the two-cell, pre-embryo, and transition embryo stages, respectively (Figures 4C and 4D; Supplemental Data Set 12). More dominant or suppressed homeologs were detected in early embryo developmental stages in polyploid wheats than in diploid grasses (Figures 4C and 4D; Supplemental Data Set 12). Despite the dynamic changes in balanced, suppressed, and dominant gene expression across embryo developmental stages and tissues, the B subgenome had slightly more suppression of expressed genes compared with the A and D subgenomes (B > A > D); the D subgenome had the most dominantly expressed genes in hexaploid wheat (D > A > B); and the A subgenome had more dominantly expressed genes compared with the B subgenome in tetraploid wheat (Figures 4C and 4D; Supplemental Data Set 12). Similar to the homeolog group gene expression dynamics, the homeolog expression bias showed dynamic patterns of expression before the leaf early embryo stage and maintained the stability of expression patterns in subsequent stages.
Dynamic Expression of Subgenome Homologs in Polyploid Wheats and Diploid Ancestors
Considering the changes in subgenome homolog expression patterns in polyploids relative to their diploid ancestors, we propose that these expression changes result from the regulation of new divergent genomes. Our analyses provide a framework to describe the evolution of individual subgenome homolog expression patterns using five species across distinct embryo developmental stages. A detailed comparison of subgenome homologs in wheat polyploids and their diploid ancestors would provide a better understanding of the mechanisms determining the evolutionary events during polyploidization. To understand how subgenome homologs are coordinately expressed during embryogenesis, we performed DEG analysis using a differential expression feature extraction method (Pan et al., 2018) to identify differentially expressed subgenome homologs in the five species of interest. We separated the hexaploid and tetraploid wheat data sets into the five A, B, and D subgenome data sets and compared each data set with the A, B, and D subgenomes of the diploid species to generate the DEGs across embryogenesis (Supplemental Data Set 13). Seven sets of differential gene expression analyses were performed and four differential expression feature extraction (DEFE) pattern schemes were applied based on the design of three A subgenome comparisons: AC versus DV, SF versus DV, and AC versus SF; three B subgenome comparisons: AC versus SP, SF versus SP, and AC versus SF; and one D subgenome comparison: AC versus TA. DEGs were found in five species using pairwise stage comparisons (Figure 5; Supplemental Data Set 13). The number of DEGs during embryo, endosperm, and pericarp development are presented in Supplemental Data Set 13. In all three subgenome pairwise comparisons, the number of DEGs was significantly higher in the two-cell embryo, pre-embryo, transition stage endosperm, leaf late stage endosperm, and leaf early stage pericarp compared with the other stages (Figure 5; Supplemental Data Set 13). Hierarchical clustering revealed that both the A subgenome and B subgenome clustered in three corresponding major groups (Supplemental Figures 9A to 9C). In A subgenome comparisons, AC versus DV and SF versus DV were more closely related, whereas in B subgenome comparisons, AC versus SP and SF versus SP were more closely related than AC versus SF.
Figure 5.
Dynamic Expression of Homologs in Wheat Species Subgenomes.
Distribution of DEGs across embryo developmental stages was determined by subgenome pairwise comparisons. The numbers of upregulated genes (left panel) and downregulated genes (right panel) for each pairwise comparison are shown. E1 to E10 represent the 10 stages of development defined in Figure 1. Wheat and grass species are referred to with abbreviated names (AC, SF, DV, SP, and TA) as defined in Supplemental Table 1.
TFs play an important role in regulating development and metabolic pathway programs. The percentages of coexpressed subgenome homologs were 37, 31, and 29% in the A, B, and D subgenomes, respectively (Supplemental Data Set 11). However, 35% of the identified homeologous TF genes were coexpressed in polyploid wheat, suggesting that polyploidization may have altered the expression levels and/or roles of some subgenome TFs. To investigate -evolutionary dynamics, activation, and suppression of subgenome homologs after polyploidization in the wheat species, we compared the subgenome TF data sets from tetraploid and hexaploid species with those of their related diploid ancestors. Comparative analysis of subgenome TFs revealed that the activation and suppression of TF families from subgenomes are dynamic across development (Supplemental Figure 10). The majority of TF families derived from the A subgenome did not exhibit expression changes or activation in AC and SF, and fewer were suppressed in comparison with their diploid ancestors. The TFs derived from the A subgenome showed spatial activation of genes in the WRKY, TALE, MADS, LBD, HSF, HD-ZIP, GATA, G2-like, DOF, and AP2 families. By contrast, the majority of TFs derived from B and D subgenomes were constitutively suppressed in AC and SF compared with their relative expression in diploid ancestors, and fewer TF transcripts showed activated expression, including ZF-HD and WOX transcripts in the B subgenome and B3 in the D subgenome. The activation and suppression of TF families in the A and B subgenomes showed similarities across all embryo developmental stages but were significantly modulated in endosperm and pericarp tissues, with suppressed temporal and spatial expression in AC compared with SF (Supplemental Figure 10).
The Expression of Genes Involved in Endosperm Storage Protein and Carbohydrate Processes
Wheat grain endosperm contains storage proteins that influence dough elasticity and extensibility and the processing quality of a range of food products. Starch is the major carbohydrate component in wheat, which represents 65 to 70% of wheat flour and provides an excellent source of caloric energy. In order to explore the patterns and evolutionary divergence of gene expression associated with these important grain constituents in wheat and its diploid ancestors, we examined the expression of storage protein and carbohydrate genes in the A, B, and D subgenomes of diploid, tetraploid, and hexaploid species across embryo developmental stages and different grain tissues (Supplemental Figures 11A to 11E), respectively. Of the 94 storage protein genes (36 genes from the A subgenome, 30 genes from the B subgenome, and 28 genes from the D subgenome), 4, 6, 4, 1, and 2 were not expressed in any developmental stages or tissues examined in AC, SF, DV, SP, and TA, respectively. Sixty-three genes were categorized in expression modules with significantly high expression in the transition stage endosperm and leaf late stage endosperm (Supplemental Figures 11A to 11E; Supplemental Data Set 14). Among these 63 genes, 14 showed endosperm-specific expression. All of the 65 starch synthesis genes (20 genes from the A subgenome, 20 genes from the B subgenome, and 25 genes from the D subgenome) were expressed in the five species. By contrast, only 1 starch synthesis gene was specifically expressed in either endosperm or pericarp, whereas 19 genes were highly expressed in transition stage endosperm and leaf late stage endosperm. Unlike the storage protein synthesis genes, the dynamic changes in starch synthesis gene expression levels were smaller across embryo developmental stages and tissues. The remaining carbohydrate metabolic genes had expression patterns that were consistent with that of the starch synthesis genes. The carbohydrate metabolic pathway genes, including genes required for sucrose, glucose, fructose, and trehalose biosynthesis, exhibited a longer phase of expression after activation compared with storage protein genes (Supplemental Figures 11A to 11E; Supplemental Data Set 14-). These results suggest that the mechanisms regulating different carbohydrate metabolic pathway genes appear to be similar but may differ from those of storage protein synthesis genes.
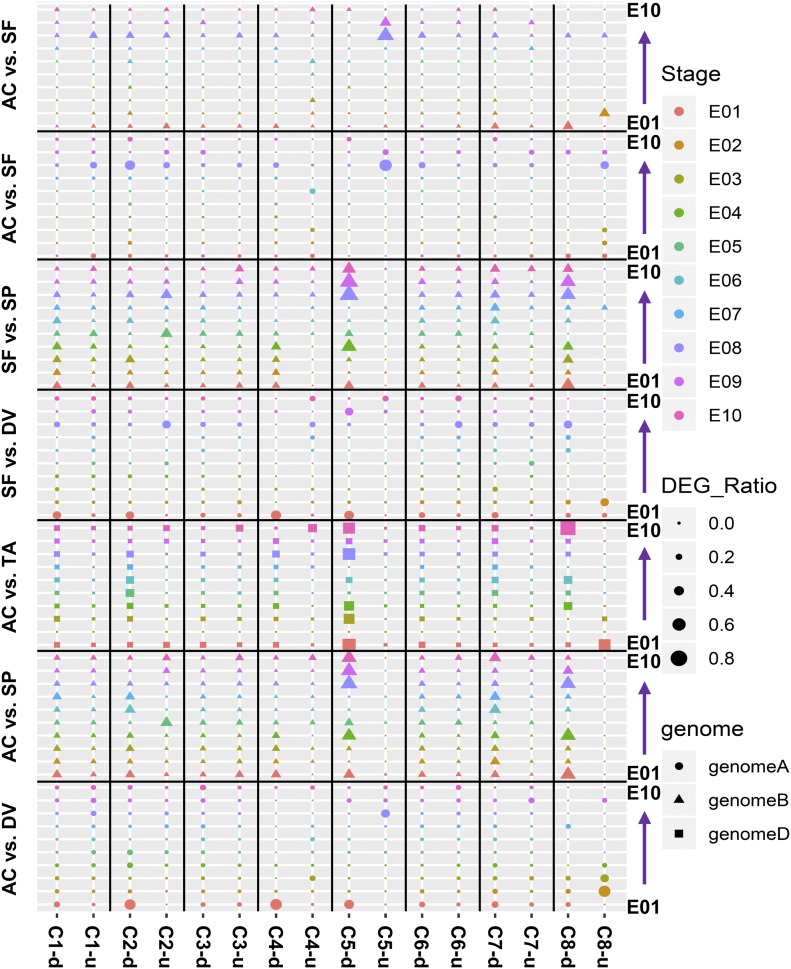
A hallmark of wheat embryo and endosperm tissues is their accumulation of storage reserves and secondary metabolites (Olsen, 2004). To gain biological insights into the gene expression associated with storage reserve metabolic networks and the contributing roles of subgenome homologs, we compared the ratio of homologs with differential expression in storage reserves and secondary metabolite-related processes (Figure 6). Pairwise species comparisons revealed that the activation of homologs related to embryo development was quite stable, and homologs related to storage reserves and secondary metabolites were dynamic in the A, B, and D subgenomes. More carbohydrate and storage protein-related genes derived from the B and D subgenomes were downregulated, whereas storage protein homologs derived from the A subgenome were upregulated, in the tetraploid and hexaploid wheat species compared with their diploid ancestors. Storage protein genes from the A and B subgenomes were upregulated in hexaploid species compared with tetraploid wheat (Figure 6).
Figure 6.
The Activation and Suppression of Homologs in Wheat and Grass Species Subgenomes.
The distributions of DEGs across embryo developmental stages are displayed by subgenome using pairwise comparisons between five species. Wheat and grass species are referred to with abbreviated names (AC, SF, DV, SP, and TA) as defined in Supplemental Table 1. Different stages of development (E01–E10, as defined in Figure 1) are indicated by different colors, with the progression of development indicated by arrows along the y axis. DEG ratios are indicated by different shape sizes. The three subgenomes are indicated by different shapes. Gene categories are represented by C1 to C8: C1, glucose genes; C2, starch genes; C3, embryo development essential genes; C4, fructose genes; C5, storage protein genes; C6, stress-related genes; C7, sucrose genes; C8, trehalose genes. d, downregulated genes; u, upregulated genes.
Phylotranscriptomic Hourglass Patterns during Embryogenesis in Various Wheat Species
To investigate ontogenetic divergence patterns in various wheat species, we performed phylotranscriptomic studies, which allow the average transcriptome age or transcriptome divergence for biological processes at each stage to be retrieved, using RNA-seq data derived from embryos in seven ontogenetic developmental stages from hexaploid wheat, tetraploid wheat, and diploid grass species. Fourteen phylostratum (clade of genes derived from the common ancestor) levels (PS1–PS14) were defined along the taxonomic lineage in accordance with the National Center for Biotechnology Information phylogeny leading to bread wheat with reference to 20 fully sequenced genomes (Figure 7A; Supplemental Data Set 15; Domazet-Lošo and Tautz, 2010; Quint et al., 2012; Drost et al., 2016). PS1 includes the evolutionarily oldest genes with homologous sequences in prokaryotes, while PS14 includes the evolutionarily youngest genes with no homologs beyond Triticum. The polyploid wheats and their diploid ancestors showed similarity in the percentage of conserved genes, with the majority of gene conservation in four levels, including 31.86 to 33.24%, 14.98 to 17.02%, 11.80 to 12.77%, and 6.99 to 8.50% at Embryophyta (PS4), Eukargota (PS2), Viridplantae (PS3), and Poaceae (PS9), respectively. A small number of genes (1.11–1.83%) were specific to the genus Triticum (Figure 7A; Supplemental Table 3).
Figure 7.
Evolutionary Age and Sequence Divergence of Various Wheat Species.
(A) Phylostratigraphic map of various wheat species.
(B) TAI difference between DV and AC.
(C) TAI difference between SF and AC.
(D) TAI difference between SP and AC.
(E) TAI difference between TA and AC.
The y axes in (B) to (E) represent TAI value difference at each developmental stage.
(F) PS level distribution across embryo developmental stages in wheat species.
(G) Transcriptome indices across AC embryogenesis.
(H) Transcriptome indices across SF embryogenesis.
For each species, we analyzed two different transcriptome indices, the transcriptome age index (TAI) and the transcriptome divergence index (TDI). The TAI is based on evolutionary age, whereas TDI is based on sequence divergence. A high TAI value represents a newly diverged (young) transcriptome, while a high TDI indicates a larger divergence. Using the most recent polyploidization event (forming hexaploid wheat [AC]) to compare TAI with other wheat species across developmental stages and PS levels (Figures 7B to 7F), the ancestral genes (PS1–PS5) had greater TAI variation across development than the younger genes (PS9–PS14). The patterns of TAI differences revealed similarities across the seven stages of embryo development in AC versus TA, AC versus SF, and AC versus SP comparisons (except in the two-cell embryo stage) and two patterns observed in AC versus DV (Figures 7B to 7F). We investigated the profiles of these two transcriptome indices across the seven stages of embryo development in the five species to determine if and to what degree they show an hourglass pattern of expression, predicting the divergent and conserved stages of embryogenesis. TAI and TDI supported hourglass expression patterns during embryogenesis for the five species (Figures 7G and 7H; Supplemental Figures 12A to 12C). However, the phylotypic (middle) stage, representing the embryonic stages of development with the oldest and the most conserved/least divergent transcriptome, differed slightly among these five species. The polyploid wheats had consistent TAI and TDI patterns across the seven embryo developmental stages (Figures 7G and 7H). In the diploid ancestors, DV and SP had similar patterns of TAI. We also calculated the average distance of TAI and TDI between adjacent stages of development (Supplemental Table 4). This identified the transition embryo stage as the phylotypic stage for SF and TA and the leaf early embryo stage as the phylotypic stage for AC, TA, and SP (Supplemental Table 4). Despite minor differences in the phylotypic stage of embryogenesis in the five species examined, the identification of adjacent transition and leaf early embryo developmental stages of embryogenesis, which are characterized by organ and primordia initiation and differentiation, supports the phylotranscriptomic hourglass pattern of evolutionary divergence during embryogenesis for wheat and its ancestral diploid species.
Droplet Digital PCR and in Situ Assays Validate RNA-Seq Results
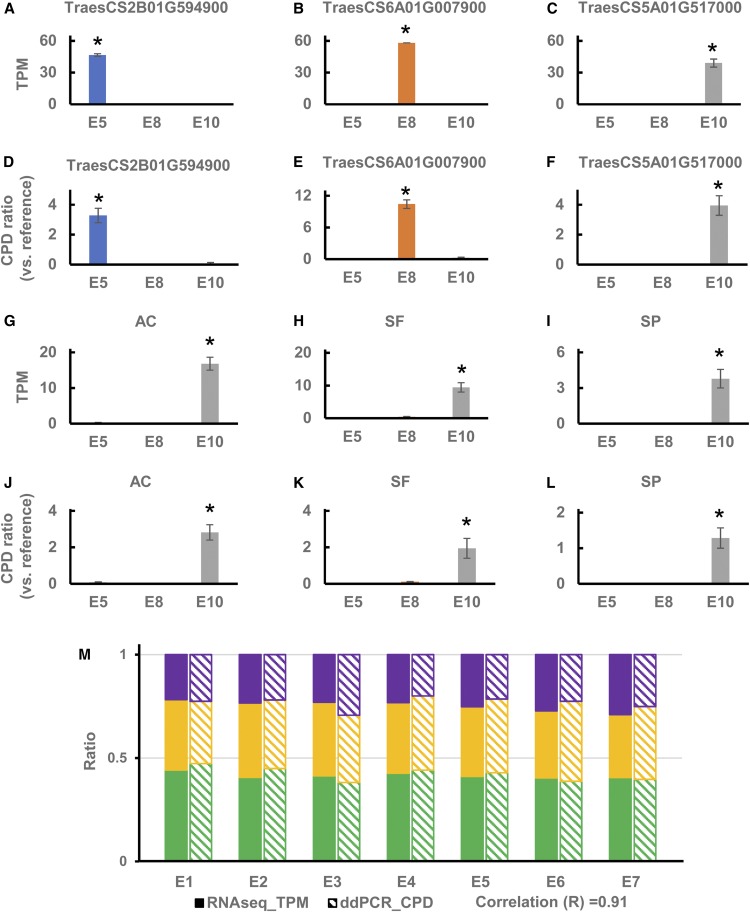
To confirm the expression of tissue-specific (Figures 8A to 8C), species-conserved (Figures 8G to 8I), and homeolog triad genes (Figure 8M) identified in the RNA-seq transcriptional data, we performed droplet digital PCR (ddPCR) assays for select genes representative of each category. We examined the expression of three genes identified for their tissue-specific expression patterns, TraesCS2B01G594900, TraesCS6A01G007900, and TraesCS5A01G517000, by ddPCR in the embryo, endosperm, and pericarp using gene-specific probes (Supplemental Table 5). Transcript copy per droplet (CPD) was calculated for each target, and normalization was performed based on an internal reference gene, TraesCS2B01G409000. The results from ddPCR analysis revealed embryo-, endosperm-, and pericarp-specific expression for TraesCS2B01G594900, TraesCS6A01G007900, and TraesCS5A01G517000, respectively (Figures 8D to 8F), which is consistent with and validates the corresponding RNA-seq results (Figures 8A to 8C).
Figure 8.
Expression of Select Tissue-Specific and Homeolog Triad Genes in the Developing Wheat Grain.
(A) to (F) Tissue-specific expression of embryo-specific (TraesCS2B01G594900), endosperm-specific (TraesCS6A01G007900), and pericarp-specific (TraesCS5A01G517000) genes, assayed by RNA-seq ([A]to [C]) and ddPCR ([D] to [F]) in AC Barrie.
(G) to (L) Conserved tissue-specific expression of a pericarp-specific gene (TraesCS6B01G331700) in hexaploid AC, tetraploid SF, and diploid SP species, assayed by RNA-seq ([G] to [I]) and ddPCR ([J] to [L]).
Expressed levels in the embryo (E5, leaf middle stage), endosperm (E8, transition stage), and pericarp (E10, leaf early stage) are shown as transcripts per million (TPM; [A] to[C] and [G]–[I]) or as the copies of target per droplet (CPD) relative to the copies of reference (TraesCS2B01G409000) per droplet in each sample ([D] to [F] and [J] to [L]). *, P < 0.05.
(M) Expression of a selected homeolog triad (composed of A, B, and D genome copies) in the embryo across the seven stages of embryogenesis (E1–E7), assayed by RNA-seq (solid filled bars) and ddPCR (line-filled bars) in AC Barrie. Expression levels are shown as a ratio of TPM (solid filled bars) or CPD (line-filled bars) for each homeolog in the A (green), B (yellow), and D (purple) subgenomes, relative to the sum of TPD or CPD for all three subgenomes, respectively. A, B, and D genome homeolog genes are TraesCS5A01G074900 (green), TraesCS5B01G081300 (yellow), and TraesCS5D01G088400 (purple), respectively. Transcript ratios (A:B:D) at each stage of embryo development for RNA-seq and ddPCR analyses had a Pearson correlation coefficient of r = 0.91.
To test the conservation of tissue-specific expression across species for genes identified by RNA-seq analyses (Figures 8G to 8I), we performed ddPCR assays to determine the expression pattern of a representative pericarp-specific gene, TraesCS6B01G331700, in the hexaploid (AC), tetraploid (SF), and diploid (SP) wheat and grass species. Consistent with the RNA-seq analysis, ddPCR analysis showed that the pericarp tissue specificity of TraesCS6B01G331700 was conserved in all three species, with varying levels of expression (Figures 8J to 8L).
To examine the accuracy of the RNA-seq analysis for identifying homeologous gene expression, we selected a representative expression triad with balanced expression among the three subgenomes (TraesCS5A01G074900, TraesCS5B01G081300, and TraesCS5D01G088400) for independent validation by ddPCR (Figure 8M) using three specific probes addressing single-nucleotide polymorphisms among the three homeologous genes (Supplemental Table 5). Clear separation in the detection of the three homeologs based on these specific probes was observed using the QX200 Droplet Reader. The transcript ratios (A:B:D) obtained from RNA-seq and ddPCR analyses were highly correlated (r = 0.91) for each stage of embryo development (E1–E7; Figure 8M), thus supporting the balanced expression pattern of the selected homeolog triad genes represented in the three subgenomes.
To further assess the tissue-specific gene expression in developing wheat grains, as identified by RNA-seq and validated by ddPCR, we employed in situ assays to spatially examine the localization of expression of two selected genes, TraesCS2B01G594900 (with embryo-specific expression) and TraesCS5A01G074900 (constitutively expressed in embryo, endosperm, and pericarp), within the compartments of developing AC Barrie wheat grain sections (Figure 9). The gene-specific primers used to assay the expression of these two genes by ddPCR were also utilized for in situ PCR (Supplemental Table 5). TraesCS2B01G594900 transcripts were localized to the embryo in developing grains (Figures 9A to 9C), while TraesCS5A01G074900 exhibited a broader expression pattern across the embryo, endosperm, and pericarp tissues in sections of developing grain (Figures 9D to 9F). These spatially resolved expression observations were further supported by the lack of expression signal in the negative controls, in which the reverse transcription step was omitted (Figures 9G to 9I). These results confirm the accuracy of the RNA-seq data analyses of this study and the value of this transcriptome database for identifying gene activities associated with embryo, endosperm, and grain development in wheat and its putative ancestral diploid species. Together, these validation studies provide independent confirmation and supporting evidence to RNA-seq-based global data sets and their analysis.
Figure 9.
Expression Patterns of Select Genes within the Tissues of the Developing AC Barrie Grain.
In situ PCR was used to localize TraesCS2B01G594900 and TraesCS5A01G074900 in the developing embryo, endosperm, and pericarp. Blue stain indicates the presence of in situ PCR-amplified target gene transcripts.
(A) to (C) Representative micrographs of longitudinal sections of grains for TraesCS2B01G594900, with embryo-specific expression.
(D) to (F) Representative micrographs of longitudinal sections of grains for TraesCS5A01G074900, with constitutive expression in the embryo, endosperm, and pericarp.
(G) to (I) Negative controls omitting the reverse transcription step.
Left panels show low-magnification overviews of the grain, sectioned longitudinally to show all three tissues of interest, including the embryo (Em), endosperm (En), and pericarp (Pc). Solid boxed regions highlight a region of the embryo and endosperm and are magnified in the middle panels. Dashed boxed regions highlight the pericarp and are magnified in the right panels. Bars = 5 mm ([A], [D], and [G]), 1 mm ([B], [E], and [H]), and 0.5 mm ([C], [F], and [I]).
DISCUSSION
In this study, we generated a high-resolution transcriptome atlas of grain development for two polyploid wheats and their putative diploid ancestors for seven stages of embryogenesis (from the two-cell to mature stage), two endosperm stages, and the pericarp. This comprehensive resource enabled gene expression programs to be defined in diploid grasses and during the evolution of polyploid wheat. The transcriptional signatures and developmental similarities among the five species identified herein suggest that the evolutionary divergence of expression is primarily affected by the tissue, followed in decreasing order by the general developmental phases (early, middle, or late embryogenesis), subgenome, species, and adjacent developmental stages of embryogenesis. Consistent with observations in other plant and animal species (Wang et al., 2010; Chen et al., 2014; Dylus et al., 2018), the evidence suggests that the developmental stage, rather than the genome of origin (subgenome), plays a major role in distinguishing gene expression profiles in grain tissues during wheat grain development (Figure 2). Coexpression and subgenome comparative analyses provided further insight into the dynamic reprogramming of the transcriptome by revealing functional transitions during embryogenesis in various wheat species.
Embryo and Endosperm Transcriptomes Are Complex and Overlapping
Many studies have examined grain development, gene expression, and storage reserve formation in important crops, including maize and wheat (Sekhon et al., 2011; Chen et al., 2014; Li et al., 2014; Pfeifer et al., 2014; Rangan et al., 2017). Here, we generated comprehensive data sets from fertilization to embryo maturity, capturing detailed gene expression, regulation, and evolutionary divergence in various wheat species. Although the data were derived from different developmental stages, tissues, and species, global gene expression patterns emerged. Embryo and endosperm tissues were distinguished by the expression of storage reserve genes and spatially expressed genes. Large sets of genes, including those putatively encoding TFs, showed endosperm-specific expression patterns, such as activation during transitions in endosperm development (Supplemental Data Set 8). Common expression patterns were observed across the five wheat and grass species examined, indicating that these genes likely play conserved roles in biological pathways. TFs that mediate crosstalk between the embryo and endosperm to coordinate development remain to be determined. Both starch and storage proteins serve as storage reserves, and this study suggested that storage protein-encoding genes are more specifically expressed in the endosperm, whereas starch/carbohydrate synthesis-related genes are coordinately expressed in the embryo and endosperm.
Since genes functioning in the same pathway tend to appear in the same or similar expression modules, the regulatory programs for synthesizing storage proteins and carbohydrates are expected to differ. Unlike most of the homeologous genes, storage and carbohydrate genes exhibited biased expression or expression shifts between the subgenomes, suggesting that genes derived from the A, B, and D subgenomes may play different roles in these pathways to produce qualitative and/or quantitative differences in diploid and polyploid species (Ramírez-González et al., 2018). These subgenome expression shifts have potential functional implications for some of the most significant changes in genes controlling important pathways in polyploid wheats, which could be associated with increases in grain size and production.
Transcriptional Reprogramming during Embryogenesis in Polyploid Wheats
Polyploidy may confer phenotypic plasticity through neospecialization, allowing some homeologs to be differentially expressed across development (Dubcovsky and Dvorak, 2007; Ramírez-González et al., 2018). Given the genome complexity of polyploid wheat, our results suggest that two types of transcriptional reprogramming likely shaped the evolution of polyploid wheats. In the case of dynamic biased expression of homeologs across embryogenesis, genes expressed in one particular stage were preferentially maintained in the subsequent stage (Supplemental Data Sets 6 and 12). The percentages of both suppressed and dominantly expressed genes were highest during early embryogenesis. Compared with suppressed genes, a higher percentage of genes were dominantly expressed. As embryo development progressed, biased homeologs gradually turned into balanced groups until the transition developmental stage and stabilized during subsequent embryo stages of development (Supplemental Data Sets 6 and 12). Furthermore, as shown in Supplemental Figure 10, unlike the biased expression patterns of homeologs, the subgenome homologs exhibited two major expression pattern trends, including spatial and constant expression in polyploid wheats. Most of the spatially upregulated TF genes were observed in the A subgenome homologs, whereas constantly downregulated TF genes were observed in B and D subgenome homologs. Our results demonstrate biased expression of homeologs and provide a dynamic overview of subgenome reprogramming, highlighting the fundamental transcriptional regulation and developmental phases present in embryo, endosperm, and pericarp during polyploid wheat embryogenesis and grain development. Our data provide a comprehensive transcriptome resource to facilitate hypothesis generation and the identification of functional processes, regulatory networks, and discovery of their associated flexibilities and constraints in the context of polyploid wheat grain development and breeding.
Ontogenetic Divergence and Evolution during Embryogenesis
Morphogenetic diversity during embryo development in plants and animals is known as the embryonic hourglass (Duboule, 1994; Raff, 1996), where the middle stage of embryogenesis is referred to as the phylotypic stage (Domazet-Lošo and Tautz, 2010; Quint et al., 2012). A phylotranscriptomic hourglass pattern has been used to predict organ, tissue, and gene evolution trends in animal and plant species, where the phylotypic stage was found to represent the oldest and most conserved transcriptome (Domazet-Lošo and Tautz, 2010; Quint et al., 2012; Drost et al., 2017). By applying phylotranscriptomic approaches based on TAI and TDI, we analyzed transcriptomic hourglass patterns during the embryogenesis of wheat and its progenitor grass species. Embryogenesis in plants can be divided into three major phases: asymmetric cell divisions to establish apical and basal polarity during the early stages, organ and primordia initiation and differentiation to establish the embryonic body plan during the middle stages, and the accumulation of storage reserves during the late stages of embryogenesis (Meyerowitz, 2002; Quint et al., 2012). Our data show that the middle embryo developmental stages (transition and leaf early stages) represent the phylotypic stages of embryogenesis in wheat species. In Arabidopsis, the torpedo stage marks the transition from morphogenesis to the maturation phase and is the phylotypic stage of embryogenesis (Quint et al., 2012). Based on our data, the phylotypic stage appears to occur slightly earlier in some wheats compared with Arabidopsis. The difference in the timing of primordium initiation and organ differentiation in monocots and dicots may explain this distinction. We also observed ontogenetic divergence and phylotypic stage differences among diploid ancestors and polyploid wheats, suggesting that embryo morphogenesis and maturation were reprogrammed after polyploidization. Convergent evolution of a phylotranscriptomic pattern in wheat suggests the operation of a fundamental developmental program controlling the expression of evolutionarily young or rapidly evolving genes across Poaceae species and during polyploidization. Thus, we speculate that the biased expression of homeologs and subgenome reprogramming may have been required to enable spatiotemporal organization and the evolution of polyploid wheat species.
The data generated in this study provide a comprehensive resource for the study of transcriptome dynamics over wheat embryogenesis and grain development. By characterizing the transcriptional programming of embryogenesis in tetraploid and hexaploid wheats and diploid ancestral grass species, this study provides insights into the evolution of gene expression in wheat and the selective pressures placed on grain production during domestication and breeding. As a comprehensive study of embryo transcriptomes, this research should guide and facilitate future investigations of wheat genomics and polyploid biology.
METHODS
Plant Material, Growth, and Vernalization Treatments
Polyploid (AC and SF) and diploid (DV) wheat plants were grown in growth chambers under long-day conditions of 16 h of light, 22°C and 8 h of dark, 20°C, with light intensity of 100 to 120 μmol m−2 s−1 (Philips high-output F54T5/835-841 bulbs) for the whole life cycle. SP and TA plants were initially grown in a growth chamber at 22°C under long days (16-h day/8-h night), then moved at the fifth leaf stage into a cold room with 4°C and the long-day photoperiod for 1 month (vernalization treatment), and returned to 16 h of light, 22°C and 8 h of dark, 20°C, with light intensity of 100 to 120 μmol m−2 s−1 to complete their life cycle.
Embryo, Endosperm, and Pericarp Isolation
Spikelets were emasculated and pollinated at the heading stage to ensure sufficient and developmentally coordinated grain production for embryo isolation. Embryo isolation was performed as described previously, with some modifications (Xiang et al., 2011a). For each embryo sample in early stages of development, ∼30 embryos were pooled in each biological replicate sample. For each sample in late embryo stages, a minimum of 10 embryos were pooled in each biological replicate sample. A minimum of 10 grains were used for pericarp and endosperm isolation in each biological replicate sample. A total of two biological replicates for each tissue at each stage of development were used. For transition stage endosperm isolation, a hole was punctured in the grain in a Petri dish containing isolation solution (4.8% Suc solution + 0.1% RNAlater [Ambion catalog no. AM7020]) to allow exposure of the inside of the grain to isolation solution and subsequent extraction of the endosperm by pipetting (Figures 1N and 1O). For leaf late stage endosperm isolation, when the endosperm occupies nearly the whole grain, the pericarp and embryo were removed from the grain and the remaining endosperm was kept for RNA isolation (Figures 1P and 1Q). For leaf early stage pericarp isolation, the embryo and endosperm were manually removed and the remaining pericarp was kept (Figures 1R and 1S). A major concern is the contamination of early stage embryo mRNA (in the two-cell, pre-embryo, and transition stages) by the pericarp and endosperm. To ensure that we had clean embryo samples for mRNA isolation, we performed the isolation of embryos from the ovules in Petri dishes containing 4.8% sucrose solution + 0.1% RNAlater (Ambion catalog no.AM7020). The isolation procedure involved making two precise incisions at the micropylar end with needles (Fine Science Tools catalog no. 10130-05) as described previously with some modifications (Xiang et al., 2011a). This resulted in the separation of the micropylar region that houses the early stage embryo, allowing dissection of the embryo with needles. Isolated embryos were moved away from the maternal ovule tissue and endosperm cells and transferred into Eppendorf tubes sitting on dry ice using fine drawn-out glass pipettes. To verify the clean collection of embryos, representative embryos were placed inside depressions made by a Mini PAP pen (Invitrogen catalog no.008877) on glass slides. After applying cover slips, we identified the embryo stages with a compound microscope (Leica DMR), captured images using a MicroFire camera (Optronics), and further confirmed no visible contamination from the ovule tissue or the early endosperm nuclei/cells. Since the ovule soon after fertilization contains very few endosperm cells/nuclei, the risk of endosperm contamination is greater after the leaf middle stages, when a dense endosperm sticks to the embryo. To ensure endosperm cell removal, embryos were carefully washed by repeated isolation solution exchanges.
RNA Isolation, Antisense RNA Amplification, and RNA-Seq Analysis
Total RNA was extracted from embryo, endosperm, and pericarp of different developmental stages following the protocol provided by the RNAqueous-Micro kit (Ambion catalog number 1927), with two replicates for each developmental stage. The quantity of RNA isolated from early stage embryos was insufficient for library preparation for RNA-seq experiments. Therefore, the mRNA from all stages was amplified and the antisense RNA (aRNA) was used for RNA-seq analysis. The mRNA amplification was conducted according to the protocol provided in the MessageAmp aRNA kit (Ambion catalog number 1750). For RNA-seq profile analysis, we prepared Illumina mRNA-seq libraries using the TruSeq RNA kit (version 1, rev A). Libraries were prepared with aRNA according to the manufacturer’s instructions. For HiSeq 2000 sequencing, four libraries were pooled per sequencing lane.
Microscopy
Wheat embryos were cleared in chloral hydrate solution (8:1:2, chloral hydrate:glycerol:water, w/v/v) and viewed with a Leica DMR compound microscope with Nomarski optics. Images were captured using a MagnaFire camera (Optronics) and were edited in Adobe Photoshop CS (Xiang et al., 2011b). Scanning electron microscopy was performed as described previously (Venglat et al., 2011) for isolated embryos. For the wheat grain (Figure 1A), longitudinal hand sections through the grain were made prior to submerging the samples in 25 mM PIPES, pH 7.0, containing 2% (v/v) glutaraldehyde for 2 h. After several washes, the samples were fixed in 2% OsO4 in 25 mM PIPES for 2 h, washed, and dehydrated in ethanol (30, 50, 70, 95, and three 100% exchanges).
After sample dehydration, substitution to amyl acetate was performed with increasing ratios of amyl acetate to ethanol (spanning 1:3 parts [v/v], 1:1 [v/v], 3:1 [v/v], then two pure amyl acetate exchanges). All solvent exchanges were separated by 15 min. Samples were critical-point dried with solvent-substituted liquid CO2 (Polaron E3000 Series II), mounted on aluminum specimen stubs with conductive carbon glue (Ted Pella), and rotary coated with 10 nm of gold (Edwards S150B sputter coater). Imaging was performed with a 3-kV accelerating voltage, 10-μA current, and 12.2-mm working distance on a Field Emission scanning electron microscope (Hitachi SU8010).
Mapping of RNA-Seq Read to Reference Genome, and Analysis of Expressed Genes
The IWGSC RefSeq v1.0 complete reference genome and corresponding annotation was used as a reference for the analysis of the RNA-seq data. Following the recommendation of the International Wheat Genome Sequencing Consortium, the chromosome-partitioned version (161010_Chinese_Spring_v1.0_pseudomolecules_parts. fasta) was used and the corresponding gff3 file was reformatted accordingly. The hexaploid AC Barrie (Triticum aestivum) was mapped to the entire wheat genome with 110,790 gene models; the tetraploid Strongfield (Triticum turgidum var durum) to the A and B subgenomes with 75,769 gene models; and the diploid Triticum monococcum (AA, DV92-DV), Aegilops speltoides (BB, TA2780-SP) and Aegilops tauschii (DD, TA101132-TA) to subgenomes A, B, and D with 39,031, 39,467, and 37,750 gene models, respectively. The RNA-seq reads were preprocessed by trimming the adaptor sequences, filtering low-quality reads (Phred Score ≤ 20), and eliminating short reads (length ≤ 20 bp) using the software package FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). After filtering, barcode, and adaptor removal, an average of 27.5 million RNA-seq reads per sample were retained for subsequent mapping. The cleaned RNA-seq reads in each sample were mapped against the reference genome using STAR v2.5.3a (Dobin et al., 2013) with default parameters to generate gene-level counts.
PCA
Relative relatedness and reproducibility among biological replicates were examined by PCA, and 2D plots using PC1 and PC2 were constructed with the built-in plotPCA function provided by the R DESeq2 package. Global comparisons of relative relatedness among 80 individuals including eight subgenomes (AC_A, AC_B, AC_D, SF_A, SF_B, DV_A, SP_B, and TA_D) and 10 developmental stages (E1–E10) from each subgenome were calculated by PCA using two different approaches. First, raw counts from all samples were normalized and logarithm transformed based on A, B, and D subgenomes (i.e., AC_A, SF_A, and DV_A subgenomes were grouped together, AC_B, SF_B, and SP_B were grouped together, and AC_D and TA_D were grouped together) using the Variance Stabilizing Transformation function in the R DESeq2 package. Then, based on the homeolog list generated, homeologs with expression values from A, B, and D genomes were extracted separately. Three homeologs from three subgenomes were given a new common name and treated as the same element (gene) in the downstream PCA and correlation analysis. Finally, a log-transformed expression value matrix of 80 (samples) × 20,702 (genes) was generated. Low-count genes (genes with >10 counts in less than two individuals) were filtered, and a matrix with 80 × 18,777 was regenerated. PCA was calculated with this matrix using the PCA function in the R FactoMineR package (http://factominer.free.fr). Variation explanation percentage for each principal component was calculated using the get_eigenvalue function in the R FactoMineR package. PC1 to PC10 plots were generated using the pairs function in the R Graphics package. To provide more than 50% variation explanation in the PCA 3D plots, the top 2000 genes with highly variable expression among 80 samples were selected using the rowVars function in the R metaMA package. PCA 3D plots were based on an 80 × 2000 expression matrix using the R scatterplot3d package.
Gene-Gene Correlation Coefficient Analysis
To investigate the correlations between homeologous and homologous genes, expression patterns across seven embryo developmental stages were used to calculate gene correlations. Eight matrices containing expression information from eight subgenomes were generated to achieve this target. Pairwise comparisons based on Pearson correlation coefficients of the same rows (genes) in different matrices were performed with the mapply function of the R base package. Correlation plots of r values of different comparisons from a single gene were generated using the R corrplot package. Venn diagrams among each homeologous and homologous genome were created, and a threshold of 0.8 was set to define evolutionarily conserved gene sets among different subgenomes. Pearson correlation coefficient values between different genes in the same gene set were calculated based on expression patterns of seven embryo developmental stages. P values were calculated using the corr.test function in the R psych package. Heatmaps were generated with the R pheatmap package Z-score transformation.
Functional Classification of Transcripts Based on GO and MapMan Pathway Enrichment
GO annotations of transcripts were compiled as described previously (Pan et al., 2018), and enrichments were performed using the R clusterProfiler package for diploid, tetraploid, and hexaploid species, respectively. GO enrichment analyses of DEGs in each species were performed separately using all genes in their genome as background. REViGO analysis (http://revigo.irb.hr) was used to slim the enriched GO terms based on the “medium similarity” parameter. The MapMan tool (Thimm et al., 2004) was used to facilitate the assignment of different gene sets into functional categories (bins). A MapMan mapping file that mapped the genes into bins via hierarchical ontologies through the searching of a variety of reference databases was generated using the Mercator tool (http://mapman.gabipd.org/web/guest/app/mercator).
Analyses of DEGs
Two sets of differential gene expression analyses were performed using DESeq2 (Love et al., 2014): (1) comparisons between each time point against the two-cell stage for each species, and (2) comparisons between each pair of species with the same subgenome. Taking subgenome A as an example of the second set of analyses, three pair-wise comparisons (AC versus SF, AC versus DV, and SF versus DV) were performed. Similarly, three pair-wise comparisons were performed for subgenome B (AC versus SF, AC versus SP, and SF versus SP) and one for subgenome D (AC versus TA). Genes with P < 0.01 (adjusting the false discovery rate using the Benjamini-Hochberg procedure) and log2 fold change ≥ 1 or ≤ −1 in at least one sampling time point were considered to be significantly DEGs. Genes with the highest read count less than 50 across all samples were considered to be expressed at low levels and were not included in subsequent analysis.
Cross-Species Comparison
Differential expression analyses were performed using DESeq2 (Love et al., 2014) based on the respective subgenomes. For example, the raw counts from subgenome A collected from AC, SF, and DV were combined and normalized together. Three pair-wise comparisons (AC versus SF, AC versus DV, and SF versus DV) were then performed using the criteria of log2 fold change ≥ 1, P ≤ 0.01, and max (pair of samples) ≥ 50.
Comparison of Expression Levels of Homologs among the Subgenomes in AC and SF
Through reciprocal best hit blast between the subgenomes, 20,702 homeolog triads were identified in AC and 24,339 homeolog diads in SF. For the purpose of comparison between AC and SF, only 20,702 homeolog diads in SF were used for subsequent analysis. This set of analyses consists of two parts. The first identifies similarity and difference in the expression level among the homeologs. A gene is considered expressed when its pseudo-read count after normalization is ≥5 and significantly expressed when it is ≥50. The categories of similarity and difference in expression of the homeologs among the subgenomes were defined for AC and SF (Supplemental Data Set 6). The second part identifies homeolog expression bias among the subgenome. For hexaploid wheat, the thresholds were defined as 33, 50, and 100% (Ramírez-González et al., 2018) for over 200 samples. In our analysis, we defined bias in AC and SF in Supplemental Data Set 12.
Data Reduction and Feature Pattern Identification
We applied the recently developed DEFE method (Pan et al., 2018, 2019) to identify DEGs. Four sets of differential gene expression analyses were performed using four DEFE pattern schemes.
There were nine comparisons between the later stage and the first two-cell stage; a DEFE feature pattern scheme was designed for this set of nine comparisons: T (E2/E1, E3/E1, E4/E1, E5/E1, E6/E1, E7/E1, E8/E1, E9/E1, E10/E1), where the prefix T stands for “time,” the numerical character 1 denotes “up” and 2 denotes “down” modulation between the time points.
The leaf early stage pericarp compared with other stages and a DEFE feature pattern scheme was designed for this set of nine comparisons: S (E10/E1, E10/E2, E10/E3, E10/E4, E10/E5, E10/E6, E10/E7, E10/E8, E10/E9), where, the prefix S stands for “leaf early stage pericarp”; here leaf early stage pericarp/two-cell is the same comparison as that in the first set of comparisons.
A DEFE feature pattern scheme was designed for the eight comparisons of late leaf stage endosperm with other stages: L (E9/E1, E9/E2, E9/E3, E9/E4, E9/E5, E9/E6, E9/E7, E9/E8), where the prefix L stands for “leaf late stage endosperm.”
A DEFE feature pattern scheme was designed for the seven comparisons of transition stage endosperm with embryo developmental stages: E (E8/E1, E8/E2, E8/E3, E8/E4, E8/E5, E8/E6, E8/E7), where the prefix E stands for “transition stage endosperm” and the acronym EE in each feature pattern stands for “transition stage endosperm.” The statistics of each feature pattern is available in the DEFE stats worksheet of each data file.
Determining TAI and TDI
Both TAI and TDI calculations were based on normalized gene expression data for different developmental stages, including the two-cell embryo, pre-embryo, transition embryo, leaf early embryo, leaf middle embryo, leaf late embryo, and mature embryo, for DV, SP, TA, SF, and AC. TAI for each developmental stage was computed based on the phylostratigraphic procedure (Domazet-Lošo and Tautz, 2010). The phylum level of each gene was determined using 20 genomes of the taxonomic lineage of wheat and its related species. The R code developed by Cheng et al. (2015) was adopted to calculate the phylum level of each gene. BLASTp was used to determine reciprocal best hits between wheat A, B, and D genomes and BP (i.e., A versus BP, B versus BP, and D versus BP). The ratios of gene conservation (divergent) were computed using MAFFT and PAL2NAL. The R package myTAI (Domazet-Lošo and Tautz, 2010; Quint et al., 2012; Drost et al., 2015) was used to compute TAI and TDI.
eFP Browser
The normalized RNA-seq expression data were used to generate an online portal providing a resource for wheat embryogenesis at http://bar.utoronto.ca/efp_wheat/cgi-bin/efpWeb.cgi?dataSource=Wheat_Embryogenesis and also included a custom image. These resources were used for the development and customization of the wheat eFP Browser; an output snapshot of one gene example is shown in Supplemental Figure 1.
ddPCR Assay
Primers and probes were designed to be specific to each selected tissue-specific gene and to exclude off-targets from homologous and homeologous genes. An intron-spanning feature was also included in primer design to eliminate off-target binding to potential genomic DNA contaminates. Probes were 5′ labeled with 6-carboxyfluorescein or 6‐carboxy‐2,4,4,5,7,7-hexachlorofluorescein succinimidyl ester as the reporter and 3′ labeled with ZEN and Iowa Black FQ as the double quenchers (Integrated DNA Technologies). Each homeolog-specific probe differed from its homeolog counterparts on the other two chromosomes by at least one single-nucleotide polymorphism. Primer and probe sequences for target and reference genes are provided in Supplemental Table 5.
The extracted RNA (as described above) was treated with DNase I (Thermo Fisher Scientific), and reverse transcription was performed using the SuperScript IV VILO system (Thermo Fisher Scientific) according to the manufacturer’s instructions. Transcript abundance was measured using the QX200 ddPCR System (Bio-Rad). In brief, each 20-μL 1× ddPCR SuperMix of probe reaction mixture (no dUTPs; Bio-Rad) containing cDNA templates, forward and reverse primers, and specific probes with optimized concentration was mixed with 70 μL of Droplet Generation Oil for Probes in a DG8 Cartridge (Bio-Rad). The cartridge was covered with a DG8 gasket and loaded into the QX200 Droplet Generator (Bio-Rad) to generate PCR droplets. From each droplet mixture, 40 μL was transferred to a 96-well PCR plate and sealed using a PX1 PCR plate sealer (Bio-Rad). PCR thermal cycling was optimized, and amplification signals were read using the QX200 Droplet Reader and analyzed using QuantaSoft software (Bio-Rad).
In Situ Assay
Grain samples for in situ PCR were prepared based on a combination of protocols described previously (Bagasra, 2007; Athman et al., 2014) with modifications detailed below. Leaf middle-stage grain samples from AC Barrie were fixed overnight in fresh 2.5% glutaraldehyde and 4% paraformaldehyde in 1× PBS. After PBS washes, dehydration in an ethanol series, and substitution with xylene, the grains were embedded in paraffin, longitudinally sectioned at 10 μm thickness using a histology microtome, and mounted on precleaned glass slides on a 45°C hotplate. Subsequent treatments, including deparaffinization, rehydration, postfixation, proteinase K treatment, DNase treatment, in situ reverse transcription, in situ PCR, and colorimetric detection of digoxin (DIG)-labeled PCR products, were performed on slides in Frame-Seal incubation chambers.
Specific in situ PCR primers for TraesCS2B01G594900 (embryo-specific) and TraesCS5A01G074900 (constitutively expressed in embryo, endosperm, and pericarp) were designed for the in situ PCR assay (Supplemental Table 5). In situ reverse transcription was performed on the DNase-treated grain sections using the SuperScript IV VILO system (Thermo Fisher Scientific catalog number 1176050). In situ PCR was performed using Phusion High-Fidelity DNA Polymerase (Invitrogen catalog number F530S) additionally containing 4 μM DIG-11-dUTP (Sigma-Aldrich catalog number 11093088910). Colorimetric detection of DIG-labeled PCR products was performed with Anti-DIG-AP (Sigma-Aldrich catalog number 11093274910), and sections were stained using BM-purple (Sigma-Aldrich catalog number 11442074001). Visualization was processed using Leica DMR equipped with a MicroFire camera (Optronics) under bright-field illumination. Negative controls were performed and analyzed using sections from the same grain samples processed as described above except that the in situ reverse transcription step was omitted.
Accession Numbers
All RNA-seq raw data generated from this study can be found in the Gene Expression Omnibus under accession number GSE129695.
Supplemental Data
Supplemental Figure 1. Snapshot of TraesCS1A01G005700 as an example displayed by the eFP Browser.
Supplemental Figure 2. Principal component analysis (PCA) of transcriptomes for seven embryo developmental stages, two endosperm stages, and one pericarp stage in five wheat and grass species.
Supplemental Figure 3. Principal component analysis (PCA) of transcriptomes for seven embryo developmental stages, two endosperm stages, and one pericarp stage in five wheat and grass species.
Supplemental Figure 4. Relationship of the transcriptomes of five wheat and grass species from different stages of grain development, tissues, and sub-genomes.
Supplemental Figure 5. Cluster analysis using DEGs derived from wheat and grass species.
Supplemental Figure 6. Examples of gene correlation coefficients among sub-genomes and species.
Supplemental Figure 7. Comparison of enriched GO (Biological Process) terms among conserved gene sets of polyploid wheats and diploid ancestral grass species.
Supplemental Figure 8. Analysis of conserved genes in the CsAll gene set.
Supplemental Figure 9. Hierarchical clustering analysis of sub-genomes across species.
Supplemental Figure 10. Dynamic expression of homologs in wheat and grass species sub-genomes.
Supplemental Figure 11. Hierarchical clustering analysis of transcription factor (TF) and selected pathway genes in five wheat and grass species.
Supplemental Figure 12. TAI and TDI in three diploid species.
Supplemental Table 1. Wheat species and sampling stages/tissues used in the study.
Supplemental Table 2. Number of expressed genes across 10 developmental stages/tissues.
Supplemental Table 3. Gene phylostratum (PS) level distribution in wheat grass species.
Supplemental Table 4. Distances between TAI and TDI of the adjacent points.
Supplemental Table 5. Primer used for ddPCR and in situ PCR validations.
Supplemental Data Set 1. Gene expression data for hexaploid wheat AC at different developmental stages.
Supplemental Data Set 2. Gene expression data for tetraploid wheat SF at different developmental stages.
Supplemental Data Set 3. Gene expression data for diploid wheat DV at different developmental stages.
Supplemental Data Set 4. Gene expression data for diploid wheat SP at different developmental stages.
Supplemental Data Set 5. Gene expression data for diploid wheat TA at different developmental stages.
Supplemental Data Set 6. Expression data for homeologs in AC and SF.
Supplemental Data Set 7. Gene Ontology (GO) annotation and enrichment analysis of MEs.
Supplemental Data Set 8. Genes specifically expressed during grain development in various wheat species.
Supplemental Data Set 9. Conserved genes involved in grain development in various wheat species.
Supplemental Data Set 10. Specifically expressed TF genes during grain development in various wheat species.
Supplemental Data Set 11. Co-expression analysis of TFs during grain development in various wheat species.
Supplemental Data Set 12. Biased expression of A, B and D homeologs during different developmental stages.
Supplemental Data Set 13. Dynamic expression of sub-genome homologs.
Supplemental Data Set 14. Expression of genes involved in storage protein and carbohydrate in different wheat species.
Supplemental Data Set 15. Gene model of hourglass construction in wheat embryogenesis.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
Assistance with sample preparations provided by the Western College of Veterinary Medicine Imaging Centre and Guosheng Liu at the Department of Biology, University of Saskatchewan, is gratefully acknowledged. We thank Wentao Zhang for reviewing the article and providing suggestions. This work was supported by the Wheat Flagship Program of Aquatic and Crop Resource Development Research Division of the National Research Council of Canada (ACRD manuscript number 56451).
AUTHOR CONTRIBUTIONS
D.X. and R.D. conceived and coordinated the study; D.X., P.G., and T.D.Q. performed experiments; D.X., T.D.Q., and P.G. performed data analysis, prepared figures, and wrote the article with R.D.; Z.L., P.G., Y.P., Q.L., E.W., P.V., K.T.N., Y.W., R.W., Z.Z., and Z.H. contributed to bioinformatic data analysis, imaging analysis, and drafting the figures and tables; T.D.Q., E.E., A.P., and N.J.P. created the eFP Browser for wheat grain development; Y.W., R.C., L.K., A.S., D.W., C.S.G., and C.P. contributed to materials, reagents, and article preparation; all authors read and approved the final article; R.D. secured funding.
Footnotes
Articles can be viewed without a subscription.
References
- Appels R., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Athman A., Tanz S.K., Conn V.M., Jordans C., Mayo G.M., Ng W.W., Burton R.A., Conn S.J., Gilliham M. (2014). Protocol: A fast and simple in situ PCR method for localising gene expression in plant tissue. Plant Methods 10: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. [DOI] [PubMed] [Google Scholar]
- Bagasra O. (2007). Protocols for the in situ PCR-amplification and detection of mRNA and DNA sequences. Nat. Protoc. 2: 2782–2795. [DOI] [PubMed] [Google Scholar]
- Bansal M., Kaur S., Dhaliwal H.S., Bains N.S., Bariana H.S., Chhuneja P., Bansal U.K. (2017). Mapping of Aegilops umbellulata-derived leaf rust and stripe rust resistance loci in wheat. Plant Pathol. 66: 38–44. [Google Scholar]
- Chen J., Zeng B., Zhang M., Xie S., Wang G., Hauck A., Lai J. (2014). Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Hui J.H., Lee Y.Y., Wan Law P.T., Kwan H.S. (2015). A “developmental hourglass” in fungi. Mol. Biol. Evol. 32: 1556–1566. [DOI] [PubMed] [Google Scholar]
- Chevalier É., Loubert-Hudon A., Zimmerman E.L., Matton D.P. (2011). Cell-cell communication and signalling pathways within the ovule: From its inception to fertilization. New Phytol. 192: 13–28. [DOI] [PubMed] [Google Scholar]
- Devos K.M. (2010). Grass genome organization and evolution. Curr. Opin. Plant Biol. 13: 139–145. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T., Tautz D. (2010). A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468: 815–818. [DOI] [PubMed] [Google Scholar]
- Drost H.G., et al. (2016). Post-embryonic hourglass patterns mark ontogenetic transitions in plant development. Mol. Biol. Evol. 33: 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost H.G., Gabel A., Grosse I., Quint M. (2015). Evidence for active maintenance of phylotranscriptomic hourglass patterns in animal and plant embryogenesis. Mol. Biol. Evol. 32: 1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost H.-G., Janitza P., Grosse I., Quint M. (2017). Cross-kingdom comparison of the developmental hourglass. Curr. Opin. Genet. Dev. 45: 69–75. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J., Dvorak J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. (1994). Temporal colinearity and the phylotypic progression: A basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994: 135–142. [PubMed] [Google Scholar]
- Dylus D.V., Czarkwiani A., Blowes L.M., Elphick M.R., Oliveri P. (2018). Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. Genome Biol. 19: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M., Murat F., Veyssiere M., Molinier M., Flores R., Burlot L., Alaux M., Quesneville H., Pont C., Salse J. (2017). Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytol. 213: 1477–1486. [DOI] [PubMed] [Google Scholar]
- Gegas V.C., Nazari A., Griffiths S., Simmonds J., Fish L., Orford S., Sayers L., Doonan J.H., Snape J.W. (2010). A genetic framework for grain size and shape variation in wheat. Plant Cell 22: 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover N.M., Redestig H., Dessimoz C. (2016). Homoeologs: What are they and how do we infer them? Trends Plant Sci. 21: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon F., Larré C., Petipas F., Berger A., Moussawi J., Rogniaux H., Santoni A., Saulnier L., Jamme F., Miquel M., Lepiniec L., Dubreucq B. (2012). A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. J. Exp. Bot. 63: 739–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. (2003). The genes of the Green Revolution. Trends Genet. 19: 5–9. [DOI] [PubMed] [Google Scholar]
- Hills M.J., Hall L.M., Messenger D.F., Graf R.J., Beres B.L., Eudes F. (2007). Evaluation of crossability between triticale (X Triticosecale Wittmack) and common wheat, durum wheat and rye. Environ. Biosafety Res. 6: 249–257. [DOI] [PubMed] [Google Scholar]
- Hofmann N.R. (2013). Getting there faster: Genome-wide association studies point the way to increasing nutritional values. Plant Cell 25: 4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K.V., et al. (2017). Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 114: E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., et al. (2014). Temporal patterns of gene expression in developing maize endosperm identified through transcriptome sequencing. Proc. Natl. Acad. Sci. USA 111: 7582–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H.-Q., et al. (2018). Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 557: 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.-C., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551: 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y. (2011). Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52: 750–764. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E.M. (2002). Plants compared to animals: The broadest comparative study of development. Science 295: 1482–1485. [DOI] [PubMed] [Google Scholar]
- Olsen O.-A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 (suppl.): S214–S227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P. (2007). The evolutionary consequences of polyploidy. Cell 131: 452–462. [DOI] [PubMed] [Google Scholar]
- Pan Y., Li Y., Liu Z., Surendra A., Wang L., Foroud N.A., Goyal R.K., Ouellet T., Fobert P.R. (2019). Differential expression feature extraction (DEFE) and its application in RNA-seq data analysis. bioRxiv 511188. [Google Scholar]
- Pan Y., Liu Z., Rocheleau H., Fauteux F., Wang Y., McCartney C., Ouellet T. (2018). Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genomics 19: 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M., Kugler K.G., Sandve S.R., Zhan B., Rudi H., Hvidsten T.R., Mayer K.F., Olsen O.A. (2014). Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345: 1250091. [DOI] [PubMed] [Google Scholar]
- Purugganan M.D. (2019). Evolutionary insights into the nature of plant domestication. Curr. Biol. 29: R705–R714. [DOI] [PubMed] [Google Scholar]
- Quint M., Drost H.G., Gabel A., Ullrich K.K., Bönn M., Grosse I. (2012). A transcriptomic hourglass in plant embryogenesis. Nature 490: 98–101. [DOI] [PubMed] [Google Scholar]
- Raff R.A. (1996). The Shape of Life: Genes, Development, and the Evolution of Animal Form.. (Chicago: University of Chicago Press; ). [Google Scholar]
- Ramírez-González R.H., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361: eaar6089. [DOI] [PubMed] [Google Scholar]
- Rangan P., Furtado A., Henry R.J. (2017). The transcriptome of the developing grain: A resource for understanding seed development and the molecular control of the functional and nutritional properties of wheat. BMC Genomics 18: 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamini F., Ozkan H., Brandolini A., Schäfer-Pregl R., Martin W. (2002). Genetics and geography of wild cereal domestication in the Near East. Nat. Rev. Genet. 3: 429–441. [DOI] [PubMed] [Google Scholar]
- Saville R.J., Gosman N., Burt C.J., Makepeace J., Steed A., Corbitt M., Chandler E., Brown J.K., Boulton M.I., Nicholson P. (2012). The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. J. Exp. Bot. 63: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563. [DOI] [PubMed] [Google Scholar]
- Shewry P.R. (2009). Wheat. J. Exp. Bot. 60: 1537–1553. [DOI] [PubMed] [Google Scholar]
- Signor S.A., Nuzhdin S.V. (2018). The evolution of gene expression in cis and trans. Trends Genet. 34: 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939. [DOI] [PubMed] [Google Scholar]
- Venglat P., et al. (2011). Gene expression analysis of flax seed development. BMC Plant Biol. 11: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xie W., Chen Y., Tang W., Yang J., Ye R., Liu L., Lin Y., Xu C., Xiao J., Zhang Q. (2010). A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 61: 752–766. [DOI] [PubMed] [Google Scholar]
- Wu X., Sharp P.A. (2013). Divergent transcription: A driving force for new gene origination? Cell 155: 990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D., Venglat P., Tibiche C., Yang H., Risseeuw E., Cao Y., Babic V., Cloutier M., Keller W., Wang E., Selvaraj G., Datla R. (2011a). Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol. 156: 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D., et al. (2011b). POPCORN functions in the auxin pathway to regulate embryonic body plan and meristem organization in Arabidopsis. Plant Cell 23: 4348–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Liu Z., Gao L., Yu K., Feng M., Yao Y., Peng H., Hu Z., Sun Q., Ni Z., Xin M. (2018). Genomic imprinting was evolutionarily conserved during wheat polyploidization. Plant Cell 30: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., et al. (2019). High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31: 974–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler D.J., Khan D., Kalichuk J.L., Becker M.G., Belmonte M.F. (2019). Transcriptome landscape of the early Brassica napus seed. J. Integr. Plant Biol. 61: 639–650. [DOI] [PubMed] [Google Scholar]
- Cox, T.S. (1997). Deepening the wheat gene pool. J. Crop Production 1: 1–25.