A gene cluster encoding two promiscuous enzymes modulates volatile emissions from Arabidopsis thaliana flowers, formation of insect-deterring terpene oxides, and the composition of the floral microbiome.
Abstract
Flowers are essential but vulnerable plant organs, exposed to pollinators and florivores; however, flower chemical defenses are rarely investigated. We show here that two clustered terpene synthase and cytochrome P450 encoding genes (TPS11 and CYP706A3) on chromosome 5 of Arabidopsis (Arabidopsis thaliana) are tightly coexpressed in floral tissues, upon anthesis and during floral bud development. TPS11 was previously reported to generate a blend of sesquiterpenes. By heterologous coexpression of TPS11 and CYP706A3 in yeast (Saccharomyces cerevisiae) and Nicotiana benthamiana, we demonstrate that CYP706A3 is active on TPS11 products and also further oxidizes its own primary oxidation products. Analysis of headspace and soluble metabolites in cyp706a3 and 35S:CYP706A3 mutants indicate that CYP706A3-mediated metabolism largely suppresses sesquiterpene and most monoterpene emissions from opening flowers, and generates terpene oxides that are retained in floral tissues. In flower buds, the combined expression of TPS11 and CYP706A3 also suppresses volatile emissions and generates soluble sesquiterpene oxides. Florivory assays with the Brassicaceae specialist Plutella xylostella demonstrate that insect larvae avoid feeding on buds expressing CYP706A3 and accumulating terpene oxides. Composition of the floral microbiome appears also to be modulated by CYP706A3 expression. TPS11 and CYP706A3 simultaneously evolved within Brassicaceae and form the most versatile functional gene cluster described in higher plants so far.
INTRODUCTION
Flowers are essential reproductive organs of plants, but also comprise their most vulnerable tissues and thus require sophisticated defense strategies to preserve plant fertility. The “optimal defense theory” predicts that tissues within a plant are defended in proportion to their fitness value and risk of attack (McKey, 1979). Indeed, flowers are vehicles of Darwinian fitness due to their involvement in pollen and seed production, and at the same time are particularly exposed to attack by insects and pathogens (Li et al., 2017). In addition, pollinators are often florivores at another stage of their life cycle (Abdalsamee and Müller, 2015). Surprisingly, however, flower defense mechanisms have rarely been investigated, especially at the metabolic level (Junker, 2016). For example, it has been known since the Middle Ages that one of the most potent natural insect repellents (presently identified as pyrethrin) is generated mainly by the buds and flowers of some Asteraceae, in particular Tanacetum cinerariifolium (Katsuda, 2012). Although the metabolic pathway leading to pyrethrin has now been elucidated (Kikuta et al., 2012; Xu et al., 2018), its direct impact on plant defenses has not been reported. In Nicotiana attenuata, one of the best described models of flower chemical ecology, a complex interplay between several plant pathways, signaling, and insects or hummingbirds has been observed in field studies, revealing different plant strategies to attract pollinators, and to deter or attract florivores (Euler and Baldwin, 1996; Kessler et al., 2010, 2015; Li et al., 2017, 2018). In Brassicaceae and especially the Arabidopsis (Arabidopsis thaliana) model, such a multipartite interplay largely remains to be investigated.
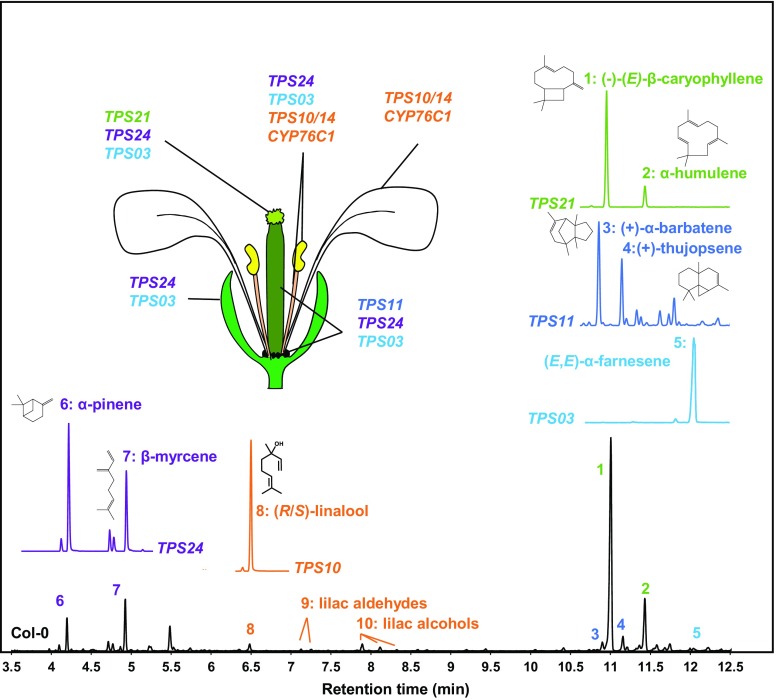
Arabidopsis is a selfing species, with flowers emitting an essentially terpenoid volatile bouquet that is complex but weak in intensity (Chen et al., 2003; Tholl and Lee, 2011). It is produced by six terpene synthases (Figure 1; Chen et al., 2003; Tholl and Lee, 2011) and is dominated by (E)-β-caryophyllene generated by TPS21 (Tholl et al., 2005). The next two most significant contributors to Arabidopsis floral emissions are TPS24, which generates monoterpenes such as α-pinene or β-myrcene, and TPS11, which produces a complex mixture of sesquiterpenes dominated by (+)-α-barbatene and (+)-thujopsene (Tholl et al., 2005). The only metabolic process so far reported to prevent excessive damage from pollinators and florivores in flowers from wild-type Arabidopsis is the oxidative metabolism of linalool, which generates both volatile and soluble repellent compounds (Boachon et al., 2015). The production of volatile linalool by the flower-expressed terpene synthases TPS10 and TPS14, which attract insects, is significantly reduced by P450-mediated oxidative metabolism depending on CYP76C1, leading to the production of repellent lilac aldehydes, lilac alcohols, and carboxylinalool. These oxygenated metabolites are largely converted to glucosides, which facilitates their storage in flower tissues. It thus seems reasonable to speculate that this oxidative metabolism constitutes an adaptation for fitness optimization and decreased risk of attack by consumers.
Figure 1.
Flower-Expressed Terpene Synthases in Col-0 Arabidopsis and Resulting Flower-Emitted Terpenoids.
Top: GC-MS chromatograms of products generated by different TPSs expressed in N. benthamiana. Expression in specific flower organs is indicated.
Bottom: Representative GC-MS chromatogram of headspace collected from Col-0 flowers focusing on major mono- and sesquiterpenes (m/z: 93) produced by flower-expressed TPSs.
A second mechanism for Arabidopsis flower protection is associated with the production of the sesquiterpene (E)-β-caryophyllene (Huang et al., 2012). In this case, (E)-β-caryophyllene was demonstrated to protect flowers from bacterial pathogens, although its impact on insect herbivory was not tested. The antibacterial activity of (E)-β-caryophyllene against Pseudomonas syringae pv tomato DC3000 (Pto DC300) was demonstrated in vitro, and tps21 mutants lacking (E)-β-caryophyllene showed enhanced growth of (Pto DC300) on their stigmas, and production of lighter seeds upon infection with the pathogen.
To uncover further metabolic processes associated with floral defense in Arabidopsis, in particular those relying on oxygenated nonvolatile compounds, we investigate here the function of a small gene cluster of two genes encoding a sesquiterpene synthase (TPS11, At5g44630) and a cytochrome P450 (CYP706A3, At5g44620), which are tightly coexpressed in Arabidopsis flowers and flower buds. We demonstrate that CYP706A3 efficiently oxidizes TPS11 products and the products of other flower-expressed mono- and sesquiterpene synthases, thereby suppressing floral volatile emissions. We show that expression of CYP706A3 impacts the composition of the floral microbiome, as well as the feeding behavior of Plutella xylostella on flower buds. Flower protection provided by the TPS11-CYP706A3 cluster starts well before anthesis and extends throughout floral bud development. This cluster of two physically and functionally linked genes is unusual, and CYP706A3 represents the most promiscuous plant P450 oxygenase reported to date.
RESULTS
CYP706A3 and TPS11 Are Clustered on Chromosome 5 and Tightly Coexpressed in Flowers
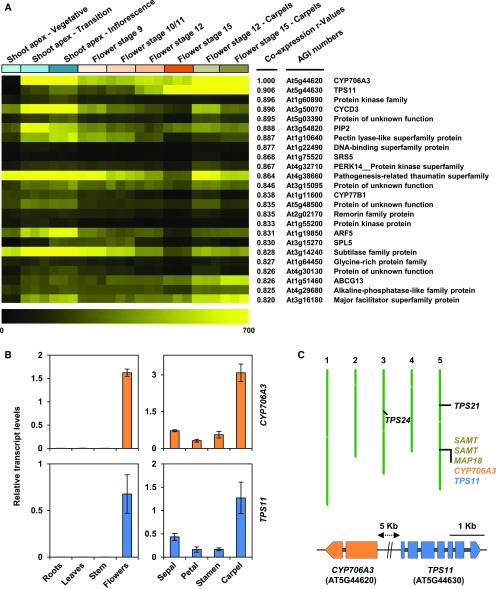
Coexpression of CYP706A3 and TPS11 in Arabidopsis flowers was first evidenced by an in silico analysis aimed at identifying P450s involved in specific metabolic pathways. CYP706A3 and TPS11 emerged as among the most tightly coexpressed terpene synthase and cytochrome P450 genes (correlation coefficient r = 0.94) in the plant organ data set (Ehlting et al., 2008; http://www-ibmp.u-strasbg.fr/∼CYPedia/CYP706A3/CoExpr_CYP706A3_Organs.html). This result was confirmed and refined using BioAanalyticResource Expression Angler (Toufighi et al., 2005) with CYP706A3 as the bait, which indicated that TPS11 was the gene most tightly coregulated with CYP706A3 when using the “Extended Tissue Compendium” data set (Figure 2A; Supplemental Data Set 1). Conversely, CYP706A3 was the P450 gene the most tightly coregulated with TPS11. Both genes were spatially and developmentally coexpressed, with the highest levels detected in carpels from flower stage 12 to 15, and in samples from the shoot apical meristem and inflorescence apex. Coexpression in flowers and especially carpels at anthesis was further validated by RT-qPCR (Figure 2B). Although suggesting that both genes might contribute to reproductive fitness, their expression patterns also provided a first hint that CYP706A3 might be acting on TPS11 products.
Figure 2.
CYP706A3 and TPS11 Are Coexpressed and Physically Clustered on Chromosome 5 in Arabidopsis.
(A) Expression heatmap of the 25 genes most coregulated with CYP706A3. Analysis was performed using the BAR Expression Angler tool and the AtGenExpress extended Tissue Compendium data set (Toufighi et al., 2005). The heatmap shows expression levels in selected flowers’ tissues: vegetative shoot apex; transition and inflorescence tissues; flower stages 9, 10/11, 12, 15; and carpels of flower stages 12 and 15. Pearson correlation coefficients (r values) of each gene coexpressed with CYP706A3, AGI numbers of the genes, and gene annotations from The Arabidopsis Information Resource are shown.
(B) Relative transcript levels of CYP706A3 and TPS11 in plant (left) and flower (right) organs. Relative transcript levels were determined by RT-qPCR (details in Methods). Results represent means ± se of four biological replicates (pooled tissues from individual plants).
(C) Map of the CYP706A3 and TPS11 gene cluster on chromosome 5 (top) and representation of introns/exons and genomic distances between both genes (bottom). The genomic localizations of TPS24, TPS21, and cluster-neighboring genes are also indicated. Numbers 1 to 5 indicate chromosomes 1 to 5. Line represents chromosome 5 on which are located the exons (CYP706A3: orange-filed, TPS11: blue-filled boxes) and introns (gray-filled boxes).
Genomic organization further supported this hypothesis: CYP706A3 and TPS11 are two adjacent genes on chromosome 5 of Arabidopsis, separated only by 5 kb of noncoding sequence and transcribed in opposite directions (Figure 2C). As this hinted that these two genes might form the core of a larger metabolic cluster (Nützmann and Osbourn, 2014; Nützmann et al., 2016), neighboring genes were also inspected. Two genes coding for S-adenosyl methyltransferases, and both predicted to be located in plastids, showed expression profiles not matching those of TPS11 and CYP706A3. Other genes were unlikely to contribute to a terpenoid-tailoring pathway (Figure 2C; Supplemental Figure 1). Moreover, no relevant metabolic gene(s) closely associated with the TPS11/CYP706A3 locus was part of the list of most coexpressed genes in BAR Expression Angler. It thus seems likely that TPS11 and CYP706A3 form a minimal metabolic cluster of two cotranscribed genes. Both proteins are located in the cytoplasmic compartment, with CYP706A3 anchored to the endoplasmic reticulum (Supplemental Figure 2).
CYP706A3 Oxidizes Major TPS11 Products in Yeast
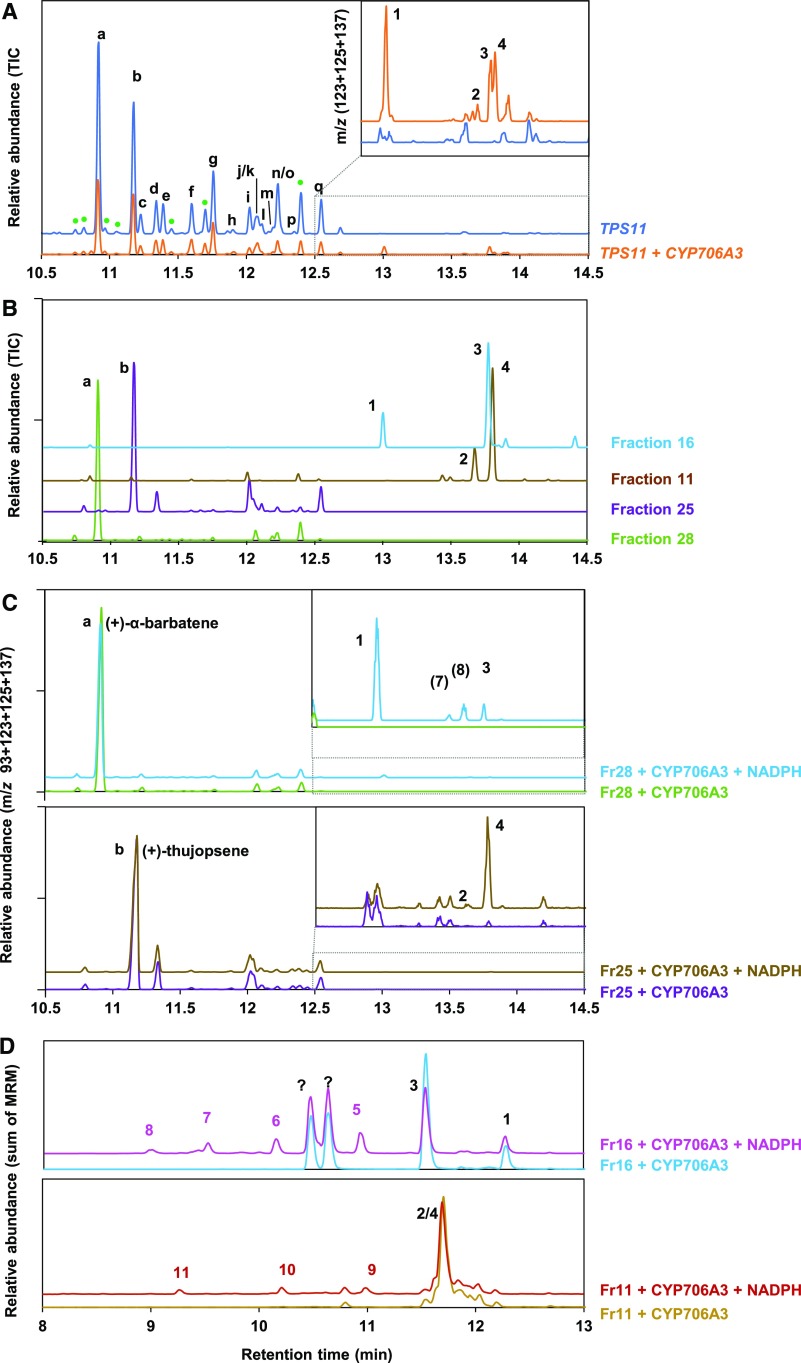
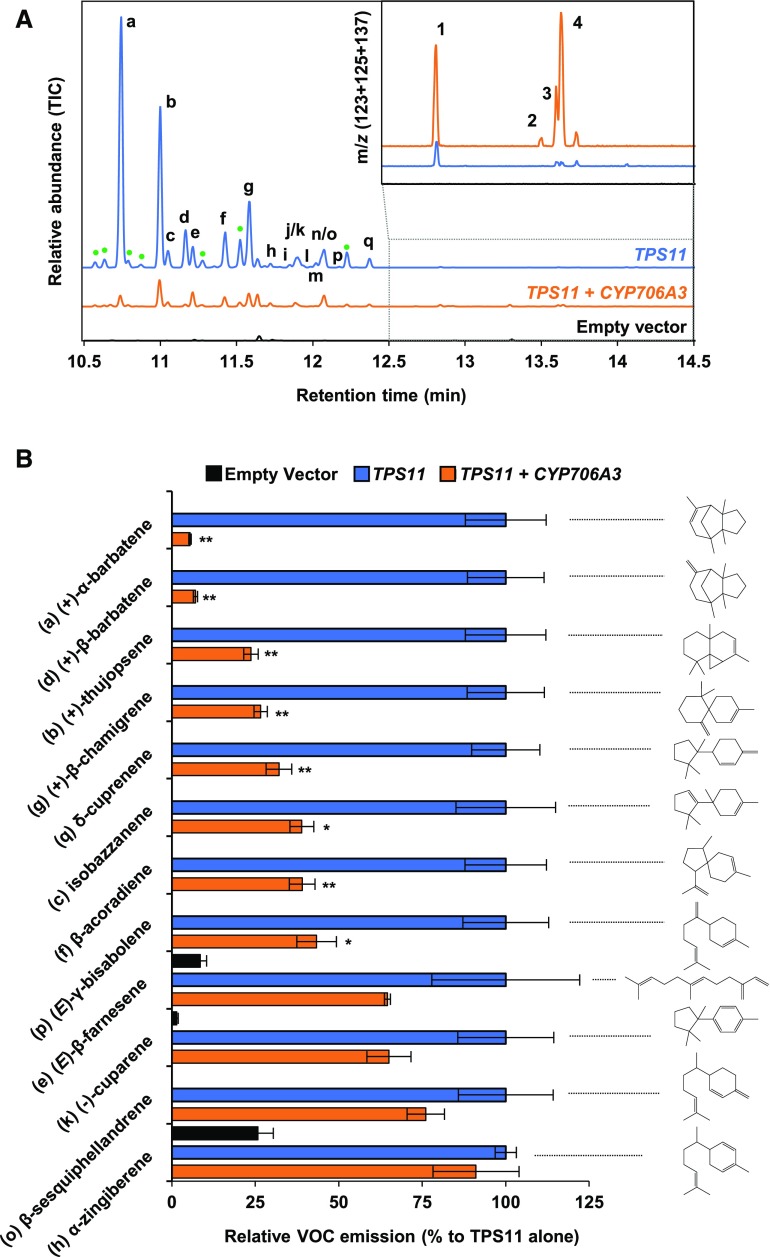
TPS11 was previously reported to generate a complex blend of more than sixteen sesquiterpenes, including (+)-α-barbatene and (+)-thujopsene as major components, when expressed in E. coli (Tholl et al., 2005). To test the hypothesis of a functional link between the two clustered genes, we tested the activity of CYP706A3 on TPS11 products in yeast, a more suitable host for expression of P450 enzymes (Supplemental Figure 3). The full coding sequences of TPS11 and CYP706A3 were independently expressed in yeast. Headspace, collected from yeast cultures expressing TPS11 alone and analyzed by gas chromatography coupled to mass spectrometry (GC-MS), featured the expected mixture of volatile compounds, with (+)-α-barbatene (a) and (+)-thujopsene (b) as major products (Figure 3A). Headspace collected from mixtures of cocultivated yeasts expressing TPS11 and CYP706A3 revealed up to 22 additional products, likely resulting from CYP706-dependent oxidation of TPS11 products (Figure 3A; Supplemental Figure 3D). None of the major products 1, 2, 3 and 4 shown in Figure 3A could be identified by comparing their mass spectra (Supplemental Figure 4) with those of compounds present in the National Institute of Standards and Technology (NIST) library. Thus, headspace from scaled-up cultures of yeasts expressing TPS11 and CYP706A3 was collected for 8 d (Supplemental Figure 3B). Up to 0.7 mg/L of major TPS11 products (+)-α-barbatene (a) and (+)-thujopsene (b), and 0.3 mg/L of major CYP706A3 product 1 were collected at the peak of production. Pooled compounds were analyzed and separated on semi-preparative high-pressure liquid chromatography coupled to mass spectrometry (LC-MS). A total of 28 fractions were recovered (Supplemental Figure 5) and subsequently checked for purity by GC-MS. Only four fractions were further analyzed, two of them containing the major TPS11 products (+)-α-barbatene (a = fraction 28) and (+)-thujopsene (b = fraction 25), and two others containing the major CYP706A3 oxidized products (1 and 3 in fraction 16, and 2 and 4 in fraction 11; Figure 3B; Supplemental Figure 5).
Figure 3.
Yeast-Expressed CYP706A3 Oxidizes TPS11 Products.
(A) GC-MS chromatograms of headspace collected from cultures of yeast expressing TPS11 or from mixtures of yeast expressing TPS11 or CYP706A3. Chromatograms show the relative abundance of total ion current (TIC) and the sum of extracted ion current as insets (m/z 123 + 125 + 137). See Methods for detailed protocol.
(B) GC-MS chromatograms of fractions purified from (A) and containing TPS11 and CYP706A3 products.
(C) GC-MS chromatograms of ethyl acetate extracts from incubations of purified TPS11 products [fraction 28 containing (+)-α-barbatene (top) or fraction 25 containing (+)-thujopsene (bottom)] with microsomal membranes prepared from yeast expressing CYP706A3 in the presence or absence (negative control) of NADPH. Chromatograms show the sum of extracted ion current (m/z 93 + 123 + 125 + 137). Insets show expanded chromatogram scale. CYP706A3-dependent metabolism of TPS11 products is probably underestimated due to low solubility of substrates in the incubation buffer.
(D) LC-MS/MS chromatograms of methanol extracts from incubations of purified CYP706A3 primary products [fraction 16 containing barbatene oxides (1) and (3; top) or fraction 11 containing thujopsene oxides (2) and (4; bottom)] with microsomal membranes prepared from yeast expressing CYP706A3 in the presence or absence (negative control) of NADPH. Chromatograms show the sum of different multiple reaction monitoring (MRM) listed in Supplemental Table 6.
TPS11 products are indicated with letters: (a) (+)-α-barbatene, (b) (+)-thujopsene, (c) isobazzanene, (d) (+)-β-barbatene, (e) (E)-β-farnesene, (f) β-acoradiene, (g) (+)-β-chamigrene, (h) α-zingiberene, (i) α-cuprenene, (j) α-chamigrene, (k) (-)-cuparene, (l) 1,2-dihydrocuparene, (m) (-)-zingiberene, (n) 1,2-dihydrocuparene, (o) β-sesquiphellandrene, (p) (E)-γ-bisabolene, and (q) δ-cuprenene. The products were identified based on a comparison of their MS and RT with libraries and published work (Tholl et al., 2005). CYP706A3 products are indicated by numbers and defined in Figure 4. Peaks labeled with question marks could not be identified, but their mass suggests that they are oxygenated sesquiterpenoids. These peaks appeared only after storage. See Supplemental Figures 3 and 4 for more extensive data.
To identify the compounds serving as substrates in formation of the major CYP706A3 products, fractions containing (+)-α-barbatene and (+)-thujopsene (fractions 28 and 25, respectively) were incubated with microsomal membranes prepared from yeast expressing CYP706A3 (Figure 3C; Supplemental Figure 6). (+)-α-Barbatene was converted by CYP706A3 into products 1 and 3 of purified fraction 16, and into two other products (7) and (8) previously detected in headspace of mixed yeast cultures (numbering according to Supplemental Figure 3). (+)-Thujopsene (Figure 3C) as well as pure (-)-thujopsene (as the only commercially available product) were both transformed into products 2 and 4 (Supplemental Figures 6B to D). Retention times and mass spectra of these products were compared with those of corresponding compounds present in fractions 11 and 16 and in headspace from mixed yeast cultures (Supplemental Figure 4). Products 1 and 3, derived from (+)-α-barbatene, yielded distinct fragmentation spectra, suggesting different structures. Products 2 and 4, derived from (-/+)-thujopsene, had similar fragmentation spectra and were thus most likely stereoisomers.
Nuclear Magnetic Resonance (NMR) spectroscopy enabled identification of the major CYP706A3 products as 2 diastereoisomers of (+)-1-oxo-thujopsene (products 2 and 4 in fraction 11), and as a mix of 2 diastereoisomers of (+)-6-oxo-α-barbatene and (+)-6-OH-α-barbatene (products 3 and 1 in fraction 16, respectively; Figure 3; Supplemental Figure 7).
Samples from yeast headspace, purified fractions, and in vitro incubations were analyzed and compared by LC-MS and LC-tandem MS (LC-MS/MS; Supplemental Figure 8). Mass spectra of compounds 1, 2, 3, and 4 in fractions 11 and 16 confirmed GC-MS and NMR structure assignments (Supplemental Figure 9). The purified fractions 11 and 16, respectively containing CYP706A3-generated thujopsene oxides and barbatene oxides, were then incubated with microsomal membranes isolated from yeast expressing CYP706A3. LC-MS/MS analysis of the resulting reaction medium extracts revealed activity of CYP706A3 on both thujopsene and barbatene oxides. The primary products of (+)-α-barbatene oxidation (fraction 16) were further converted by CYP706A3 into products 5, 6, 7, and 8, whereas those of (+)-thujopsene (fraction 11) were converted into products 9, 10, and 11 (Figure 3D; Supplemental Figure 8C). Retention times and mass spectra of these secondary products indicated that most of them were further oxygenated compounds (Supplemental Figure 10).
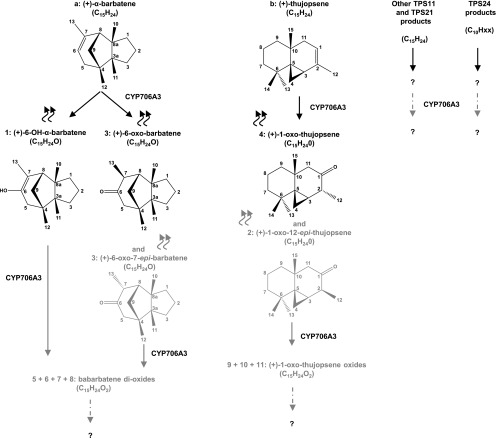
Taken together, these data suggest a scenario involving a sequential activity of CYP706A3 on (+)-α-barbatene, (+)-thujopsene, and their respective oxides, as summarized in Figure 4.
Figure 4.
Reactions Catalyzed by CYP706A3 on Floral TPS Products.
Wavy arrows indicate volatile compounds. Black arrows indicate primary and NADPH-dependent activities of CYP706A3 on TPS11 [(+)-α-barbatene and (+)-thujopsene], TPS21, and TPS24 products confirmed in vitro. Primary products 1, 2, 3 and 4 (in black and gray) were validated by NMR spectroscopy and represent the major CYP706A3 products emitted from Arabidopsis flowers. Gray arrows indicate secondary NADPH-dependent activities of CYP706A3 on primary products derived from (+)-α-barbatene and (+)-thujopsene obtained in vitro. Raw formulae of secondary products in gray were deduced from LC-MS spectra. Dashed-dotted gray arrows indicate CYP706A3 putative activities deduced from flower metabolic profiling. Unidentified compounds are indicated by question marks.
CYP706A3 Expression in Nicotiana benthamiana Reveals More Extensive Promiscuous Activity on TPS11 Products
To confirm the activity of CYP706A3 on TPS11 products in a plant environment, the corresponding genes were transiently coexpressed in leaves of Nicotiana benthamiana. The same set of sesquiterpenes emitted by TPS11-transformed yeast, and in similar proportions, was detected in the headspace of N. benthamiana leaves expressing TPS11 alone (Figure 5A). Coexpression of CYP706A3 along with TPS11 in N. benthamiana leaves resulted in the emission of the major CYP706A3 products 1, 2, 3, and 4 previously detected in yeast headspace, with concomitant decreased emission of all TPS11 products, although to different extents (Figures 5A and 5B). Terpene structures had a major impact on emission decrease that was more pronounced for multi-cyclic compounds such as barbatene, thujopsene, chamigrene, cuprenene, isobazzanene, and acoradiene (the emission of which decreased by more than 50%) than for monocyclic and acyclic compounds such as cuparene, sesquiphellandrene, zingiberene, and farnesene (Figure 5B; Supplemental Table 1).
Figure 5.
Transient Coexpression in N. benthamiana Reveals CYP706A3 Activity on Multiple TPS11 Products.
Headspace was collected on four leaves for 24 h, 3 d after agroinfiltration (see detailed protocol in Methods).
(A) Representative GC-MS chromatograms of headspace from N. benthamiana transiently expressing the empty vector, TPS11 alone, or coexpressing TPS11 and CYP706A3. Chromatograms show the relative abundance of total ion current (TIC) and the sum of extracted ion current in the inset (m/z 123 + 125 + 137).
(B) Relative quantification of emitted TPS11 products in the headspace of N. benthamiana expressing TPS11 alone and coexpressing TPS11 and CYP706A3 (only compounds for which quantification was not biased by low emission or coelution with compounds having similar mass spectra are shown). Histograms represent relative emission levels from 3 biological replicates (pooled leaves from individual transformed plants) ±se compared with levels of the same sesquiterpene emitted by N. benthamiana leaves expressing TPS11 alone (set at 100%). Statistically significant differences between leaves expressing TPS11 alone and leaves coexpressing TPS11 and CYP706A3 are indicated (two-tailed Student’s t test: *P < 0.05; **P < 0.01). Compounds are sorted from the most to the least transformed by CYP706A3. Chemical structure of each compound is shown on the right. Absolute quantifications and additional data are provided in Supplemental Table 1. Statistics can be found in Supplemental Data Set 2.
Decreased emission of TPS11 products was not due to the reduced production of TPS11 potentially resulting from its coexpression with another protein, because coexpression of TPS11 and CYP76C1 (the endogenous Arabidopsis linalool hydroxylase P450 inactive on TPS11 products) did not result in decreased emissions of TPS11 products (Supplemental Table 1).
To summarize, TPS11 and CYP706A3 coexpression in N. benthamiana confirmed CYP706A3-dependent formation of barbatene and thujopsene oxides in planta, and also revealed the promiscuity of CYP706A3 allowing oxidation of most TPS11 products, with a preference for more bulky multi-cyclic sesquiterpenoid structures.
CYP706A3 Suppresses Sesquiterpene and Monoterpene Emissions in Arabidopsis Flowers
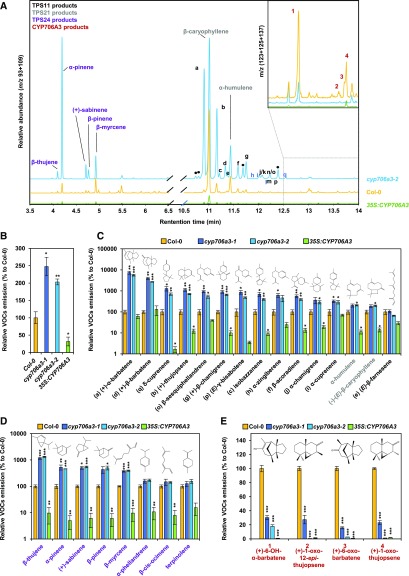
The functional relevance of the TPS11 and CYP706A3 cluster and role in Arabidopsis flowers was then investigated using cyp706a3 and 35S:CYP706A3 mutants. Two independent T-DNA insertion lines in CYP706A3, one in the promoter (cyp706a3-1) and one in the coding sequence (cyp706a3-2), resulted in a total absence of CYP706A3 transcripts in floral tissues. The ectopic overexpressor line was generated by transformation of Col-0 with a cauliflower mosaic virus 35S:CYP706A3 construct. Null mutant validation and selection of the overexpressing line are documented in Supplemental Figure 11. Comparison of volatiles collected from mutant inflorescences with that of the wild-type Col-0 provided clear evidence that CYP706A3 suppression leads to a dramatic increase in the emission of TPS11 products (Figure 6A). More surprisingly, this suppression also increased emission of other sesquiterpenes and monoterpenes generated by flower-expressed mono- and sesquiterpene synthases such as TPS21 or TPS24. Overall, the amounts of total flower-emitted volatile organic compounds (VOCs) more than doubled in the insertion mutants (Figure 6B). Conversely, in the CYP706A3 overexpressing line, total emission of flower VOCs was strongly decreased. A more precise quantification of the different compounds identified in the mutant headspace confirmed the data obtained in N. benthamiana for TPS11 and CYP706A3 coexpression, indicating that all volatile terpenes were not oxidized by CYP706A3 with the same efficiency (Figures 6C and 6D). Furthermore, emission of the barbatene oxides 1 and 3 and of the thujopsene oxides 2 and 4 (primary products of CYP706A3-dependent barbatene and thujopsene oxidation) was drastically reduced from cyp706a3 flowers relative to the wild-type (Figure 6E). These primary oxidized products were nevertheless not increased in the overexpressing line, in agreement with their further oxidation observed in the in vitro experiments.
Figure 6.
Flower-emitted VOCs in CYP706A3 Mutants.
(A) GC-MS chromatograms of headspace collected from flowers of Col-0, cyp706a3, and CYP706A3 overexpressing lines. Chromatograms show the relative abundance of the sum of extracted ion current (m/z 93 + 109) to display the major mono- and sesquiterpenes. Chromatograms in the inset show the relative abundance of the sum of extracted ion current (m/z 123 + 125 + 137) to highlight the major CYP706A3 primary and volatiles products.
(B) to (E) GC-MS relative quantification of total VOCs emissions (B), TPS11 and TPS21 products (C), TPS24 products (D), and CYP706A3 oxygenated products 1, 2, 3, and 4 (E). VOCs emitted by flowers of different lines were compared with those emitted by Col-0 (100%). Note that the y axis in (C) and (E) are in logarithmic scale. Compounds in (C) and (D) are sorted from the most to least increased in insertion mutants. Data are means ± se of three biological replicates (pooled inflorescences from individual plants). Statistically significant differences relative to Col-0 are indicated (two-tailed Student’s t test: *p < 0.05, **p < 0.01, ***p < 0.001). Statistics can be found in Supplemental Data Set 2.
Analyses of the headspace collected from the flowers of the CYP706A3 mutants (Figure 6A) further suggested an unexpected promiscuous activity of CYP706A3 on TPS21 (sesquiterpene) and TPS24 (monoterpene) products. CYP706A3 activity was thus further investigated by coexpression with TPS21 or TPS24 in yeast cultures and N. benthamiana leaves. Mixing yeasts producing terpene synthases and CYP706A3 and coexpressing TPS21 or TPS24 with CYP706A3 in N. benthamiana largely reduced terpene emissions compared with levels emitted by yeast or N. benthamiana leaves producing only terpene synthases (Supplemental Figures 12 and 13). CYP706A3 activity on TPS21 products resulted in the emission of oxidized derivatives in yeast and N. benthamiana (Supplemental Figures 12A and 12B). Some of these oxidized compounds were also generated by the incubation of (-)-E-β-caryophyllene (major TPS21 product) with the microsomal fraction of CYP706A3-transformed yeast. Among those, some were found at decreased levels in the headspace of cyp706a3 flowers as compared with the wild type (Supplemental Figures 12C to 12E). By contrast, no volatile oxidation products were detected in yeast or N. benthamiana headspace when both TPS24 and CYP706A3 were expressed (Supplemental Figures 13A and 13B). CYP706A3 activity on TPS24 monoterpene products was nevertheless observed following incubation of four pure standards [α-pinene, (+)-sabinene, β-pinene, and α-phellandrene] with a microsomal fraction of CYP706A3-transformed yeast, revealing the formation of several unknown oxygenated products (Supplemental Figure 13C). Taken together, these results confirm the promiscuous activity of CYP706A3 on a large subset of sesquiterpenes and monoterpenes generated by Arabidopsis flowers, still with a preference for cyclic compounds as substrates.
CYP706A3-Dependent VOC Metabolism Results in Accumulation of Oxygenated Terpenoids in Floral Tissues
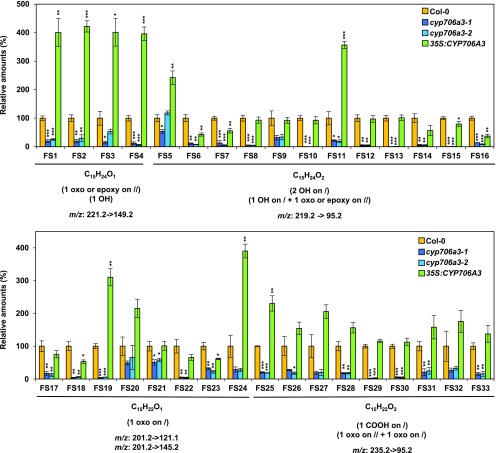
Only minor amounts of oxygenated terpenoid derivatives were detected in the headspace of wild-type and overexpressing 35S:CYP706A3 flowers, whereas CYP706-dependent conversion of primary oxygenated metabolites was observed in vitro. This raised the question of the fate of CYP706A3 products in flowers. We hypothesized that they might be retained in the floral tissues. Flowers were thus extracted with methanol after headspace collection, and the extracts were analyzed by targeted LC-MS/MS (Figure 7; Supplemental Figure 14), focusing on potential oxygenated sesquiterpene products. Up to 33 potential hydroxy-, oxo-, dihydroxy-, and carboxy- sesquiterpene derivatives were detected (FS1 to FS33: for flower soluble compounds), most of them essentially reduced in the flowers of cyp706a3 insertion mutants compared with the wild type. Conversely, sixteen of them were increased in the overexpressing line. None of the primary and secondary barbatene and thujopsene oxides (products 1 to 11) were detected in the methanol extracts of wild-type or mutant flowers, suggesting they were further metabolized. The same trend was observed for oxidation products accumulating in leaf tissues of N. benthamiana coinfiltrated with TPS11 and CYP706A3 (NbS1 to NbS21: for N. benthamiana soluble compounds; Supplemental Figure 15). β–Glycosidase treatment did not reveal new metabolites or increased amounts of detected oxidation products. This led us to conclude that most CYP706A3 products are sequentially oxidized, subjected to downstream modification, and become soluble enough to be stored in plant tissues.
Figure 7.
Soluble Products of CYP706A3-Dependent Sesquiterpene Oxygenation Detected in Flower Tissues.
Sesquiterpene oxides from wild-type and CYP706A3 mutant plants were quantified in flower methanol extracts. LC-MS/MS using multiple reaction monitoring of specific MS/MS transitions was targeted to identify sesquiterpene oxides resulting from single or multiple oxidations. For each compound differentially detected in mutant lines, the specific MS/MS transitions used are shown, as well as the expected raw formula. Data are given as means ± se of three biological replicates (pooled inflorescences from individual plants) and expressed relative to Col-0 set at 100%. Statistically significant differences relative to Col-0 are indicated (two-tailed Student’s t test: *P < 0.05, **P< 0.01, ***P < 0.001). Statistics can be found in Supplemental Data Set 2. Representative chromatograms and identification of numbered sesquiterpene oxides are shown in Supplemental Figure 14. FS, flower soluble compounds.
CYP706A3 Activity Is High in Developing Floral Buds
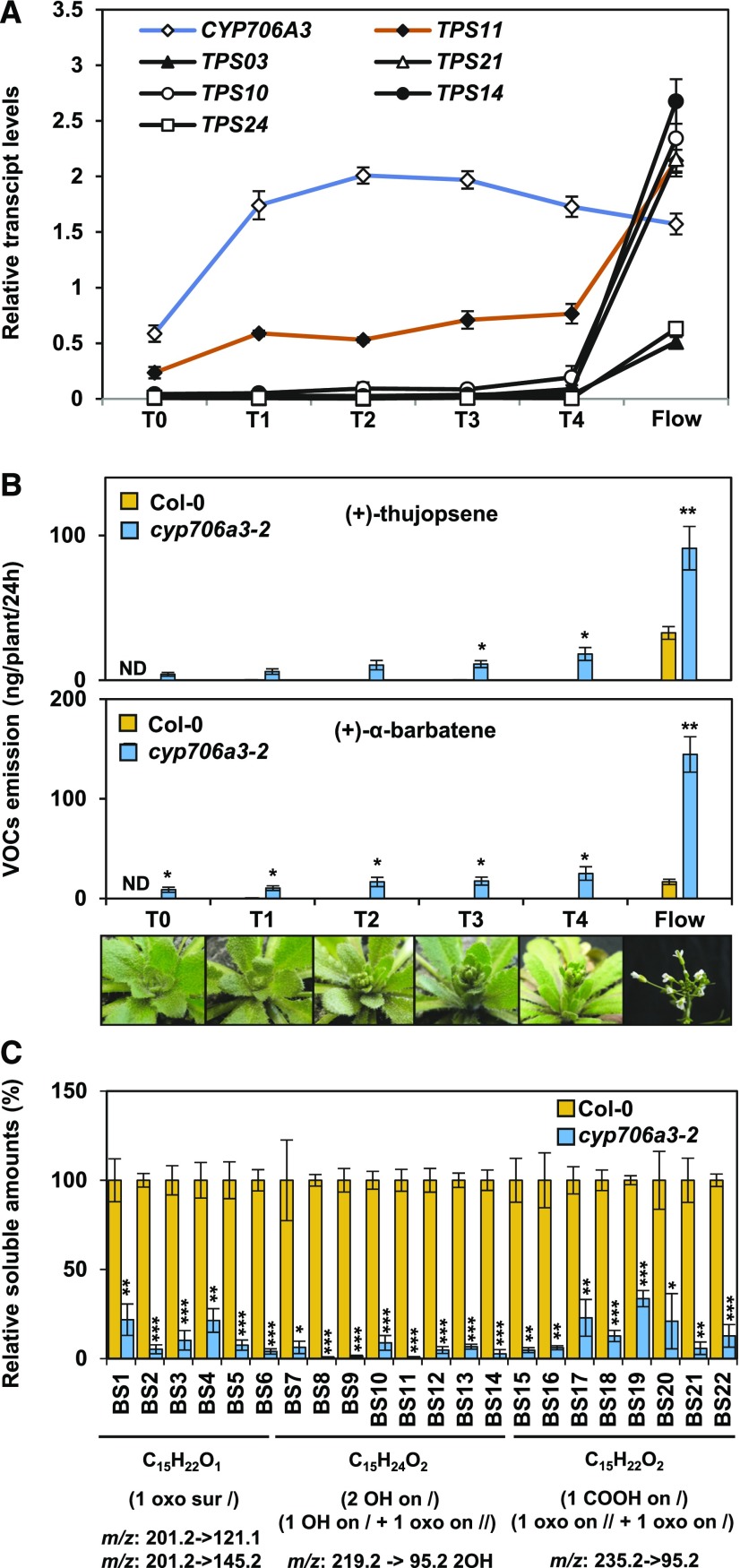
In silico transcriptome analysis shown in Figure 2 pointed to an intriguing expression pattern of CYP706A3 and TPS11 in the shoot apex, starting at the floral transition. In an attempt to determine the role of such gene expression during the early stages of inflorescence development, VOC emission and expression of terpene synthases and CYP706A3 were further evaluated (Figure 8). Unlike other floral terpene synthases, TPS11 was expressed upon floral transition and in the developing inflorescence, with concomitant, higher expression of CYP706A3. In agreement with these expression profiles, traces of (+)-α-barbatene and (+)-thujopsene were found in the headspace of bolting wild-type plants, and a dramatic increase was observed for cyp706a3 insertion mutants (Figure 8B). Conversely, accumulation of sesquiterpene oxides was detected in Col-0 flower buds (BS1 to BS22: for bud soluble compounds) and suppressed in cyp706a3 mutants (Figure 8C; Supplemental Figure 16). The observed inversion of relative gene expression at anthesis is interesting, suggesting that CYP706A3 almost completely prevents TPS11 product emissions in developing inflorescences to generate oxidation products (Figures 8A and 8B). The TPS11/CYP706A3 cluster and associated formation of sesquiterpene oxidation products are thus expected to play an important role during the early stages of flower development.
Figure 8.
The TPS11/CYP706A3 Cluster Is Active in Developing Floral Buds.
(A) Relative expression of CYP706A3 and of major flower-expressed terpene synthase genes (Figure 1) during floral transition and inflorescence development evaluated by RT-qPCR.
(B) Quantification of (+)-thujopsene (top) and (+)-α-barbatene (bottom) emission from wild-type and cyp706a3-2 mutant plants at the same development stages as in (A). VOCs were collected for 24 h from four plants per sample at the six stages of flower development shown in the photographs below from floral transition (T0) to opened inflorescence (Flow). T indicates the different transition phases.
(C) LC-MS/MS quantification of soluble sesquiterpene oxides identified in methanol extracts of stage T4 buds from wild-type and cyp706a3-2 mutant plants (as described in Figure 7). For each compound, specific MS/MS transitions used are indicated, as well as expected raw formulae. Representative chromatograms and identification of the numbered sesquiterpene oxides are shown in Supplemental Figure 16. BS, bud soluble compounds. Data in (B) are given as means ± se (n = 4 individual plants). Data in (A) and (C) are given as means ± se of three biological replicates (pooled inflorescences from individual plants). Statistically significant differences relative to Col-0 are indicated (two-tailed Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001). Statistics can be found in Supplemental Data Set 2.
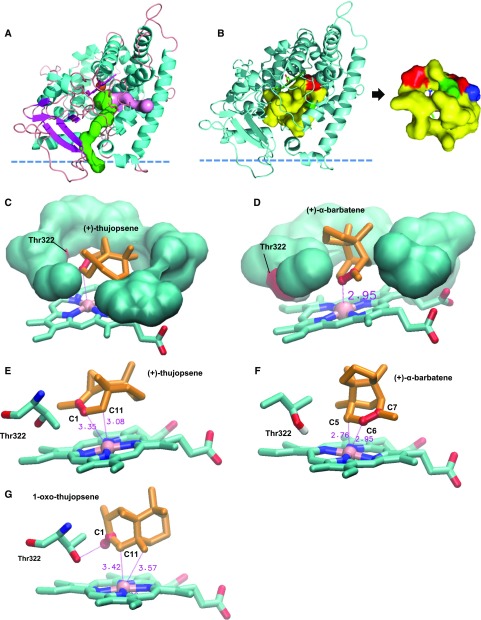
Structural Basis of CYP706A3 Activity
When compared with most other P450 enzymes, CYP706A3 shows an unusual promiscuity. It acts upon both sesqui- and monoterpenes and, in addition, catalyzes successive oxidation reactions on the same substrates. In an effort to understand the structural basis of this promiscuity, a three-dimensional (3D) homology model of the enzyme was generated using a multiple template approach, with an iterative search of templates based on sequence identity and on the search of distant homologs by HMM profiles (see Methods). CYP706A3 models rebuilt from various combinations of the detected templates (listed in Supplemental Table 2) were assessed and compared. The best homology model, yielding the highest QMEAN4 score (Benkert et al., 2008), was obtained from a three-template construction (Protein Data Bank [PDB] codes 3czh, 5irq, and 2 hi4, alignment displayed in Supplemental Figure 17). This structure exhibited two main access channels as computed by Mole 2.5 (Figure 9A; Supplemental Figure 18; Pravda et al., 2018), one markedly hydrophobic channel connected to the membrane, the other less hydrophobic pointing toward the membrane–water interface, corresponding respectively to channels 2f and 2c of Cojocaru et al., 2007. The CYP706A3 heme pocket revealed a rather small, constrained and essentially hydrophobic cavity (Figure 9B) composed of mainly apolar residues (Ile130, Ala383, Val382, Val387, Gly318, Leu386), with the exception of the only polar residue Thr322 in the first crown above the heme plane (Figures 9B and 9C). The upper part of the cavity was also predominantly hydrophobic (Val497, Ile496, Trp131, Leu317, and Leu229), with a single charged residue (Asp321). Together, these properties make the active site well-adapted to terpenoids such as sesquiterpenes, as exemplified by the good modeled fit of (+)-thujposene and (+)-α-barbatene in the heme vicinity (Figures 9C and 9D).
Figure 9.
3D Structural Model of the CYP706A3 Active Site and Comparative Docking of Relevant Substrates.
Structural model of AtCYP706A3 (segment 38-519) showing potential channels for substrate access, active site topology/characteristics, and docking positions for (+)-α-barbatene, (+)-thujopsene, and (+)-1-oxo-thujopsene determined by Autodock4.
(A) Overall structure displaying the two main channels (represented in surface mode in pink and green) with bottleneck values of 1.4 Å, positive hydropathy indexes, and logP values higher than 1. The predicted position of the membrane surface calculated by the PPM server of the OPM database (https://opm.phar.umich.edu/ppm_server) is schematized by the horizontal dashed line. The most hydrophobic channel (green) connects directly to the membrane bilayer while the other channel (pink) connects the active site to the interfacial medium.
(B) Display of the small internal cavity mostly delineated by hydrophobic residues (in yellow, surface mode). Two apertures are visible, corresponding to egress channels. Polar (green) and charged (red) residues correspond to Thr322 and Asp314, respectively. Thr322 is the only nonhydrophobic residue in contact with the active site cavity.
(C) View of (+)-thujopsene buried in the small hydrophobic pocket. Only the first crown of interacting residues is displayed. The only polar residue Thr322 of the cavity is shown on the left (red surface). The thujopsene double bond is indicated in red.
(D) View of (+)-α-barbatene in the small hydrophobic pocket. Only the first crown of residues is displayed. The only polar residue Thr322 of the cavity is shown on the left (red surface). The barbatene double bond is indicated in red. Distance (≈3Å) between the oxidation site and heme iron is indicated.
(E) Predominant docking pose of (+)-thujopsene, with high affinity energy binding of –9.2 kcal.mol–1. The two atoms closest to the heme iron are C1 and C11, in full agreement with oxidation site. Distances to heme iron are indicated in red with dotted lines, and double bond C1-C2 is shown in red.
(F) Predominant docking pose of (+)-α-barbatene, with high affinity energy binding of –9.39 kcal.mol–1. Two atoms (red ball-and-stick) closest to heme iron (pink) C5 and C6 are at equivalent distances. The polar side chain of Thr322 is displayed.
(G) Predominant docking pose of (+)-1-oxo-thujopsene, with high affinity energy binding of –9.8 kcal.mol–1. The keto group (C1=O), shown in red, is oriented on the same side as the polar side chain of Thr322.
Docking of different sesquiterpenes in this structural model yielded strikingly homogeneous distributions (Supplemental Figure 19), with a major or unique best-ranked cluster of poses corresponding in most cases to the observed metabolism. (+)-Thujopsene was found stacked to the heme porphyrin ring by its double bond, with the shortest iron–carbon distances observed for allylic (C11, Figure 9E) and vinyl positions (C1), in agreement with the primary oxidation product 1-oxo-thujopsene. Similarly, the best affinity histogram cluster for docked (+)-α-barbatene contained poses fully consistent with observed primary products (+)-6-OH- and (+)-6-oxo-α-barbatene, with the shortest iron–carbon distances observed for allylic (C5) and vinyl (C6) carbons (Figure 9F). The model is also consistent with re-uptake and further conversion of primary oxidation product 1-oxo-thujopsene (Figure 9G), with the best docking pose displaying a favorable heme–ligand distance (shortest distances observed for positions C11 and C15), with a slightly better binding score for (+)-thujopsene. This conformation is likely favored by the stabilizing presence of the polar Thr322 in the heme pocket. The low binding energies for docking of (E)-β caryophyllene, (E)-β-farnesene, α-pinene, and (+)-sabinene all indicate overall good affinities for the active site (Supplemental Table 3), with a preference for cyclic sesquiterpenes, fully supporting the CYP706A3-dependent oxidative conversions observed in vitro and in vivo, and the experimentally uncovered promiscuity of CYP706A3.
CYP706A3 Activity on TPS11 Products Results in Flower Protection against Insects
Oxidation of volatile floral terpenes can affect the behavior of flower visiting insects, as previously reported for the attractant linalool metabolized by CYP76C1 into repellent lilac compounds (Boachon et al., 2015). Thus, a potential role of TPS11/CYP706A3 in flower defense was investigated on the pollinator hoverflies that naturally visit Arabidopsis flowers (Jones, 1971; Snape and Lawrence, 1971; Hoffmann et al., 2003). Their preference for the blends of VOCs generated by yeast expressing TPS11 alone, or by mixed yeasts expressing TPS11 and CYP706A3 was tested in an olfactometer. Purified fractions of CYP706A3 products were also investigated (Supplemental Figure 20). Tests comparing TPS11 and CYP706A3 products as a blend or isolated fractions over the neutral field (control without scents) did not yield significant differences, but results all converged to suggest that nonoxidized sesquiterpenes tend to attract hoverflies, whereas oxidized sesquiterpenes repel them. In line with this trend, hoverflies significantly preferred TPS11 products over the mix of TPS11 and CYP706A3 products, thus confirming the repellent activity of the oxidized products.
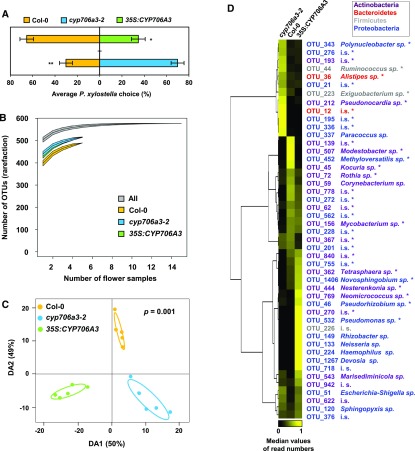
TPS11 and CYP706A3 were shown to be coexpressed during floral transition, suggesting a defensive role of the accumulated sesquiterpene oxides in developing buds (Figure 8). Thus, we tested the feeding behavior of the Brassicaceae specialist herbivore P. xylostella with dual-choice tests on T4 stage flower buds (as defined in Figure 8) of cyp706a3 and 35S:CYP706A3 compared with wild-type plants (Figure 10A). P. xylostella was previously shown to feed on Arabidopsis flowers (Boachon et al., 2015) and is a natural florivore on wild Brassicaceae (Knauer et al., 2018). Caterpillars clearly avoided wild-type buds, and favored foraging on buds of the cyp706a3-2 mutant lacking sesquiterpene oxides. Conversely, caterpillars preferred feeding on wild-type rather than 35S:CYP706A3 buds with increased sesquiterpene oxide content. Together, our data thus suggest that the CYP706A3 products can alter the preference of P. xylostella for developing buds and modify the behavior of insects visiting open flowers.
Figure 10.
The Cluster Oxidation Products Influence Florivore Behavior and Floral Microbial Populations.
(A) Feeding preference of larvae of Plutella xylostella for buds from Col-0, cyp706a3-2, and 35S:CYP706A3 in dual-choice test. Data represent the average proportion of consumed buds for 30 individual insects (±se). Statistically significant differences are indicated (Wilcoxon: *p = 0.0265, **p = 0.0019).
(B) Diversity of OTUs present on Col-0 flowers was lower than those of cyp706a3-2 and 35S:CYP706A3 when analyzed using a Chao rarefaction test. Graph shows OTUs accumulation curves for each flower line when increasing the number of flower samples analyzed (rarefaction). Data are means ± se from 5 biological replicates (pooled inflorescences from 5 individual plants): Col-0 = 538 ± 13, cyp706a3-2 = 565 ± 13, and 35S:CYP706A3 = 571 ± 15. Curve thickness represents ± se.
(C) Abundance of OTUs (counts) detected on the flowers of Col-0, cyp706a3-2, and 35S:CYP706A3 was analyzed by a PLS-DA using the three lines as discriminant factor. Graph shows the difference in overall bacterial communities for each of five replicates from the three different lines. A cross-validation test with 999 permutations confirms the significant difference between bacterial communities among lines (P = 0.001).
(D) Differential flower-associated bacterial populations between Col-0, cyp706a3-2, and 35S:CYP706A3 flowers. Heatmap represents the relative abundance of specific OTUs being significantly different between a pair of lines based on a Wilcoxon rank sum test (n = 5, P < 0.05). OTUs that significantly differed between all lines are marked with an asterisk (Kruskal-Wallis rank sum test, n = 5, P < 0.05). OTUs are hierarchically clustered based on the average number of reads per flower line with uncentered correlation. Each row gives the median proportional number of reads in each of the flower lines, that is, each row sums up to 1 to facilitate comparison between lines and OTUs despite differences in total read counts. Each color represents a phylum (Actinobacteria, Bacteroidetes, Firmicutes, or Proteobacteria) as indicated in the box. Taxonomic designation was based on the percentage of identity to reference sequences in the SILVA database and indicated at the genus level unless unknown (i.s., incertae sedis). Detailed taxonomic identification is presented in Supplemental Table 4. Statistics can be found in Supplemental Data Set 2.
CYP706A3 Expression Affects Flower-Associated Bacterial Communities
There is growing evidence for a close interplay between flower metabolites and the associated microbiome (Junker and Tholl, 2013; Boachon et al., 2019). Thus, we analyzed the effect of CYP706A3 expression on the floral microbiome by a nontargeted approach using commonly accepted methods (Bringel and Couée, 2015; Junker and Keller, 2015). DNA from bacterial communities colonizing wild-type, cyp706a3-2, and 35S:CYP706A3 flowers was extracted, and the 16S rRNA gene was used as a marker and sequenced to identify bacterial operational taxonomic units (OTU) based on DNA sequence similarities. OTU identities and counts were used to compare richness, diversity, and composition of the microbiomes present on the three flower types (Figures 10B to 10D). The average number of the different OTUs detected did not significantly differ between flower types (Col-0 = 337 ± 28 OTUs [mean ± sd], cyp706a3-2 =342 ± 41, and 35S:CYP706A3 = 360 ± 20 OTUs; Kruskal-Wallis rank sum test: Chi22 = 1.00, P = 0.61). Still, a Chao rarefaction analysis, which estimates the maximum number of OTUs to be expected after additional sampling, suggested a lower expected number of OTUs on wild-type flowers compared with cyp706a3-2 and 35S:CYP706A3 (Figure 10B). The lower rarefaction curve of OTU richness from wild-type flowers did not overlap with those of the cyp706a3-2 and 35S:CYP706A3. The abundance of the OTUs detected on each flower line was then further investigated with a multivariate analysis using the Partial Least Square Discriminant test (PLS-DA; Hervé et al., 2018). Clearly, the five biological replicates of each of the wild-type, cyp706a3-2, and 35S:CYP706A3 flowers were significantly separated in three different clusters, each corresponding to a respective genotype (Figure 10C). The two first discriminant axes of the PLS-DA explained 99% of the variance between the flower lines, and the permutation test indicated a significant difference between the composition of the microbial communities among the three flower lines (P = 0.001).
To identify the OTUs specifically colonizing each of the flower lines, a univariate analysis was then performed (Figure 10D; Supplemental Table 4). It revealed that 47 OTUs had significantly different colonization patterns on the flowers of the three lines. Specifically, 20 and 25 OTUs were significantly enriched in the microbiome colonizing cyp706a3-2 and 35S:CYP706A3 flowers relative to wild-type flowers, out of which 20 were barely detectable or absent on wild-type. By contrast, 19 and 19 OTUs found in wild-type flowers were significantly depleted from cyp706a3-2 and 35S:CYP706A3 flowers, respectively, out of which 14 and 6 were barely detectable or absent.
Such data suggest that the CYP706A3-generated metabolites play a role in the assembly of specific bacterial taxa colonizing Arabidopsis flowers.
TheTPS11/CYP706A3 Cluster Is Specific to Brassicaceae
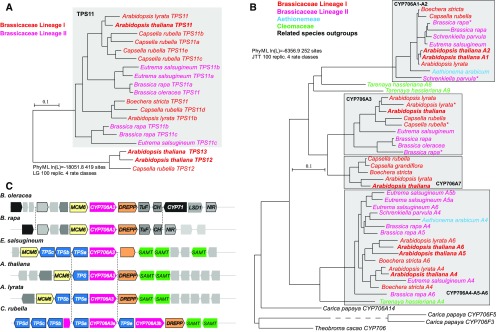
To evaluate the significance and conservation of the TPS11/CYP706A3 cluster, the emergence, phylogeny, and association of its constituents were examined, with a primary focus on Brassicales and related plants. TPS11 homologs were detected in all Brassicaceae from both lineages I and II (with one to four duplications in Arabidopsis lyrata, Eutrema salsugineum, Brassica rapa, and Capsella rubella), but not in earlier diverging Brassicales (Figure 11A). The TPS11 clade thus appears specific to Brassicaceae. Phylogenetic analysis also revealed that the CYP706A subfamily is specific to Brassicales, but that the CYP706A3 clade is represented only in Brassicaceae where it seems to be fixed, because CYP706A3 homologs (with a single duplication in A. lyrata and B. rapa) are present in all species of sequenced genomes (Figure 11B).
Figure 11.
Evolution of the TPS11/CYP706A3 gene cluster in Brassicaceae.
TPS11 (A) and CYP706A (B) phylogeny in Brassicales. Protein sequences were retrieved using a homology search with Arabidopsis TPS11 or CYP706A3 sequences as a template, from the Phytozome website (http://phytozome.jgi.doe.gov/), the BrassicaDB page (http://brassicadb.org/brad), or David Nelson’s website (http://drnelson.uthsc.edu/CytochromeP450.html), and used to build the tree using PhyML on Gblocks sets defined on the multiple alignment of protein sequences using Seaview (http://doua.prabi.fr/software/seaview). Alignments and global trees used to generate the figure are available in Supplemental Data Sets 4 to 7.
(A) Phylogeny of TPS11. TPS sequences closest to TPS11 in Capsella and Arabidopsis are used as an outgroup.
(B) Phylogeny of the available sequences assigned to the CYP706A subfamily of Brassicales and some closest homologs were used as an outgroup. If not specifically named yet, sequences were referred to according to their position in the different branches of the tree. Bootstrap values for the different branches are available in Supplemental Data Sets 5 and 7.
(C) Structure of the TPS11/CYP706A3 locus. Species possessing a CYP706A3 homolog were aligned at the genome level to show potential gene clusters. TPS11 and homologs were given different names corresponding to their position in the phylogenetic tree below. Conserved features and neighboring genes were highlighted by similar box shape, color, or putative annotation. Shown are CYP706A3 and duplicates (magenta); TPS11 and duplicates (blue); SAMT (green): S-adenosyl methyltransferase domain; MCM6 (yellow): DNA replication licensing factor; DREPP (orange): PF05558 DREPP plasma membrane polypeptide; TuF (gray): elongation factor Tu; CH (gray): charged multivesicular body protein 2A; LSD1 (gray); NiR: unnamed nitrate dehydrogenase. Light gray boxes: no annotation available. Scale bar = substitutions per site.
TPS11/CYP706A3 clustering is observed in all Brassicaceae belonging to lineage I and in Eutrema salgineum belonging to lineage II (Figure 11C). TPS11 and CYP706A3 clustering thus occurred early in the evolution of Brassicaceae, before the split between sister lineages around 45 million years ago. The cluster is duplicated in C. rubella, whereas tandem duplications of TPS11 are observed in several wild species. In domesticated members of the lineage II Brassica genus (thus submitted to a less stringent environment), TPS11 and CYP706A3 are still present, but no longer clustered (Figure 11C).
DISCUSSION
A Minimal and Versatile Functional Cluster Evolving within Brassicaceae
Functional gene clusters have been defined as groups of at least three nonhomologous biosynthetic genes for a distinct biosynthetic pathway adjacent to one another in the genome (Nützmann and Osbourn, 2014; Nützmann et al., 2016). We show here that a combination of just two physically clustered and coregulated genes can form an efficient functional unit, generating a whole range of terpenoid oxides that protect the plants against florivorous insects and selectively alter the flower microbiome. This gene combination still matches the criteria defined for metabolic gene clusters, since it contains the first committed step of the pathway generating initial chemical scaffolds (a terpene synthase) and a subsequent P450 tailoring. The originality of this uncommon cluster associated with a sesquiterpenoid-derived pathway is that both the terpene synthase and cytochrome P450 enzymes yield several products and/or transform a large set of substrates. CYP706A3, alone, catalyzes not only the oxidation of a broad range of sesqui- and monoterpenes, but also a cascade of reactions on the same original substrate. Our data suggest that both TPS11 and CYP706A3 evolved early within Brassicaceae, and became clustered before the split of Brassicaceae lineages I and II. This minimal functional cluster is possibly representative of an early stage of the formation of more complex metabolic gene clusters, such as those reported in angiosperms. It may also be representative of a more flexible and transient organization suited to the inherent promiscuity of enzymes involved in small isoprenoid (mono- and sesquiterpenes) metabolism for swift plant adaptation.
CYP706A3 Generates a Versatile Defense Arsenal Exapted from Flower Scent
Both TPS11 and CYP706A3 seem to be fixed in Brassicaceae, and their clustering is maintained in wild species. This might result from a dual selective advantage for such a structural arrangement. One is reduced emission and enhanced detoxification of membrane-damaging terpenes abundantly formed in flower tissues essential for reproduction. Another is the formation of defense molecules that protect flower organs from pathogens and herbivores. Our data demonstrate that CYP706A3 not only acts upon products of TPS11, but also on mono- and sesquiterpenes generated by other flower-expressed TPS enzymes, such as (E)-β-caryophyllene or α-pinene. Remarkably, primary oxidation products are further transformed by CYP706A3. The reason for this unusual activity of CYP706A3 with a broad range of olefins and oxygenated compounds is found in its predicted active site properties (Figure 9; Supplemental Table 3), and in the orientations of the enzyme on the membrane and of the two main access channels. Orientations of the two access channels, together with their physicochemical properties (Figure 9; Supplemental Figure 18), suggest that CYP706A3 has been evolutionarily adapted to facilitate uptake of small and hydrophobic/nonpolar substrates from the membrane, and release of polar products into the membrane–water interface. This configuration would also be compatible with the reported activities of three previously functionally characterized CYP706 enzymes, catalyzing oxidation of (+)-valencene in Alaska cedar (Callitropsis nootkatensis; Cankar et al., 2014), (+)‐δ‐cadinene in cotton (Luo et al., 2001), and phenylacetaldoxime in eucalyptus (Eucalyptus cladocalyx; Hansen et al., 2018), respectively. In addition, the CYP706A3 structural characteristics enable re-uptake of moderately polar metabolites from the membrane–water interface for further oxidation. As a result, CYP706A3-dependent metabolism of flower-emitted terpenes leads to a large variety of oxygenated products.
The production of such a large diversity of oxygenated terpenoids may provide the plant with a selective advantage and also bears potential for development of synergistic effects in support of flower defense. Arabidopsis is a selfing species and thus not dependent on pollinators. Our results support the hypothesis that the flowers of this species benefit from reducing the number and frequency of flower visitors, including taxa otherwise known to be pollinators and florivores. Interestingly, duplications of TPS11 or of the whole cluster are observed in several Brassicaceae species. Such duplications are expected to generate even broader chemical diversity of VOCs, possibly for adaptation to specific ecological niches, or to face increased ecological challenges. In favor of the latter hypothesis, C. rubella and A. lyrata show whole cluster and/or TPS11 duplications (Figure 11C). The C. rubella ancestor C. grandiflora and A. lyrata are both outcrossing and emit strong scents dominated by benzenoid compounds (Abel et al., 2009; Raguso, 2016). The selfing C. rubella only recently evolved from outcrossing C. grandiflora with concomitant loss in benzaldehyde emission, but keeps emitting significant amounts of benzoic acid (Sas et al., 2016). TPS11 and CYP706A3 duplications may thus have been required to reinforce the defense arsenal of floral tissues against visiting insects in outcrossing species. Terpene oxides have been shown earlier to serve as protections against florivores (Boachon et al., 2015). This is supported in the current study. Although we could not measure the development of P. xylostella larvae by feeding them with flower buds, nor test pure CYP706A3 products, our choice-test experiments show that flower buds containing lower amounts of oxidized sesquiterpenes were clearly preferred by insect larvae over buds with higher concentrations of these compounds. Scent, like outbreeding, is considered as an ancestral trait in angiosperms. A shift from outbreeding to selfing is one of the most common evolutionary transitions in flowering plants (Thien et al., 2009; Sicard et al., 2011; Doubleday et al., 2013) and is well-documented for Brassicaceae (Vekemans et al., 2014; Sicard and Lenhard, 2018). Following emergence of TPS11 and CYP706A3, their clustering and duplication might thus represent ancestral events in Brassicaceae. The multiple instances of gene or cluster duplications and loss observed in different taxa were potentially favored by the presence of transposable elements on both sides of the cluster (Boutanaev and Osbourn, 2018), as reported in Arabidopsis (http://signal.salk.edu/cgi-bin/tdnaexpress?GENE=At5g44620andFUNCTION=andTDNA=).
So far, flower defense and interactions have essentially been discussed with respect to volatile compounds emitted at anthesis, and their impact on insects or microbial pathogens. Our results support the notion that soluble oxygenated metabolites, diverted from flower-emitted volatiles and retained in flower tissues, also contribute to a versatile and complex defense line against insects (Figure 10A; Supplemental Figure 20). Together with our previous demonstration of the conversion of linalool generated by Arabidopsis flowers into soluble defensive compounds (Boachon et al., 2015), our results contribute to solving the long-standing puzzle of the modest residual amount of scent emitted by selfing plants (Raguso, 2016), by revealing the exaptation of floral volatile mono- and sesquiterpenes to raise antifeedant flower defense.
Flower Defense Starts with the Onset of Floral Bud Development
Previous investigations on floral defense focused on anthesis without considering the earlier stages of flower development. Worthy of note, TPS11 and CYP706A3 expression starts with floral transition, resulting in an accumulation of oxygenated sesquiterpenes already in young flower buds. Our data thus associate initiation of the defense of reproductive organs to early stages of flower development. In young buds, moreover, the ratio of TPS11 to CYP706A3 expression is inverted compared with open flowers, preventing emission of insect-attracting volatile terpenes and ensuring their complete conversion into soluble defense compounds. The observed ratio inversion also indicates that although both genes are essentially coregulated, independent developmental modulation of their respective expression is also possible.
CYP706A3 most likely Shapes Flower-Associated Bacterial Communities
Our data indicate a selective impact of CYP706A3-dependent terpenoid oxidative metabolism on the flower microbiome. Analyses of bacterial abundance, diversity, and composition suggest distinct microbial community characteristics for each flower genotype (Figure 10C). Flowers of both the cyp706a3 and 35S:CYP706A3 lines host OTU-enriched bacterial communities compared with the wild type (Figure 10B), which would be in agreement with a contribution of terpenoid oxidative metabolism to reduce bacterial colonization and thus diversity in wild-type flowers. Visual inspection of the ordination plot (PLS-DA) enables a clear separation of the communities associated with the flower genotypes. Such differences in community composition of the genotypes raise the possibility that all OTUs are equally not affected. Accordingly, we found plant line-specific effects on individual OTUs. For example, among OTUs found in higher abundance on the 35S:CYP706A3 plant, OTU_532 affiliated with Pseudomonas was detected (Figure 10D). In line with this observation, an OTU affiliated with Pseudomonas was found to be enriched in Petunia stigmas impaired in the accumulation of sesquiterpenes (Boachon et al., 2019). Strains from this genus were also recently described to stimulate conversion of terpenoid hydrocarbons to oxygenated sesquiterpenoids in the plant Atractylodes lancea (Zhou et al., 2018). Conversely, we found that specific OTUs colonizing wild-type flowers were depleted on cyp706a3 and the 35S:CYP706A3 flowers.
Only 47 out of 1485 OTUs detected significantly differed between the flower lines. This potentially reflects the selective effects of CYP706A3-dependent metabolic alterations in the different genotypes. Analysis from single mutant lines might cast doubt on the reliability of our data. The latter are nevertheless supported by the good reproducibility among replicates. Our sampling procedure might, in addition, conceal larger differences. CYP706A3 expression is mainly detected in the flower stigma, but due to the small size of Arabidopsis flowers, we analyzed the microbiome from whole inflorescences. Bacterial communities were previously shown to differ between flower organs (Junker and Keller; 2015). Pooling floral organs is thus expected to reduce visibility of microbiota modifications specifically associated with stigma biochemistry.
Altogether our results suggest that the TPS11 and CYP706A3 cluster may contribute to modulate bacterial communities on Arabidopsis flowers. This would be in concordance with earlier findings that volatile organic compounds shape bacterial community diversity and composition (Junker and Tholl, 2013; Burdon et al., 2018; Boachon et al., 2019). Recent studies demonstrated that removal of the floral microbiome resulted in a decrease of floral terpene emission (Peñuelas et al. 2014) and modified the floral metabolome (Gargallo-Garriga et al. 2016). In addition, epiphytic bacteria altered floral scent emissions (Helletsgruber et al. 2017). Conversely, floral volatiles can also affect microbes colonizing flowers, such as (E)-β-caryophyllene generated by TPS21 in Arabidopsis flowers that was shown to confer protection against the bacterial pathogen Pseudomonas syringae (Huang et al., 2012). The organ-specific ecological role of bacterial sub-populations and their advantage for flower fitness thus remain to be determined. Furthermore, the selective effects of CYP706A3 substrates and products on relevant microbial species merit further investigations. More complex levels of multi-organism interactions may also be considered to link the flower-associated microbiome and flower visitors.
METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) and Nicotiana benthamiana plants were cultivated in growth chambers under standard conditions as described previously by Boachon et al. (2015). Seeds were sown on a standard soil compost mixture in 7-cm diameter pots and cultivated in growth chambers under white fluorescent lamps with a light intensity of 100–150 µmol.m−2.s−1 at 22°C during the 12-h d period and 19°C during the 12-h night period for Arabidopsis, and at 24°C during the 16-h d period and at 20°C during the 8-h night period for N. benthamiana.
Gene Coexpression Analysis and Quantification of Gene Expression
CYP706A3 and TPS11 coexpression patterns were investigated using the ‘Expression Angler’ tool (Toufighi et al., 2005). Quantification of expression of CYP706A3, TPS03, TPS10, TPS11, TPS14, TPS21, and TPS24 genes was performed by RT-qPCR as previously described by Boachon et al. (2015). Primers used for each gene are listed in Supplemental Table 5. Relative transcript levels were calculated using the EΔCt method (Pfaffl, 2001), taking the specific amplification efficiency of each primer pair into account, and were normalized with four reference genes whose stable expression is validated (Czechowski et al., 2005). Four biological replicates (pooled organs collected from individual plants) were used for plant organ tissues and flower organs, and three for floral transition analysis.
Generation of Vector Constructs
To express CYP706A3 and terpene synthase genes in plant and yeast, plasmids were constructed as previously described by Höfer et al. (2014) and Boachon et al. (2015) using the USER cloning system (New England Biolabs) according to Nour-Eldin et al., 2006. Coding sequences (CDS) were inserted into the yeast expression plasmid pYeDP60u2 and into the plant expression vector pCAMBIA3300u. To study subcellular localization, CYP706A3 and TPS11 CDSs with modified 3′ ends were inserted in Gateway binary vectors 5′ to the sequence of G3GFP in pGWB451 and of RFP in pGWB461, respectively (Nakagawa et al., 2007). Plants and yeasts were transformed as previously described by Ginglinger et al. (2013), Höfer et al. (2014), and Boachon et al. (2015). Primers used for cloning are listed in Supplemental Table 5.
Confocal Microscopy of CYP706A3 and TPS11 in N. benthamiana
Fluorescent protein fusion constructs were transformed into the hypervirulent Agrobacterium tumefaciens strain LBA4404. The leaves of 3-week-old N. benthamiana plants were cotransformed by agro-infiltration with cultures of equal density of agrobacteria harboring the genes of interest or the p19 gene (silencing suppressor; Voinnet et al., 2003) in a ratio of 1/1 (v/v) for transient expression as described in Bassard et al., 2012. At 4 d after infiltration, leaf discs were excised for observation by laser scanning confocal microscopy. Cell imaging was performed using the A1R confocal system microscope (Nikon). Images were recorded with a 40× water immersion Apochromat Long Working Distance objective lens and a 1.15 numerical Aperture objective (Nikon). Excitation/emission wavelengths were 488/500–550 nm for G3GFP and CYP706A3:G3GFP constructs, and 561/570-620 nm for the TPS11:mRFP construct, respectively. Z-stack image series were sequentially acquired using the A1R confocal system with NIS-Element software (Nikon). Images were processed via contrasts and brightness corrections with ImageJ software version 1.51n (National Institutes of Health; http://rsb.info.nih.gov/ij).
Heterologous Expression in Yeast, VOCs Collection, and Purification
To test the activity of CYP706A3 on products formed by terpene synthases, yeast expression plasmids carrying CYP706A3, TPS11, and TPS21 CDSs were each transformed into the WAT11 yeast strain (Höfer et al., 2014; Boachon et al., 2015), whereas expression plasmid carrying TPS24 CDS was transformed into the K197G yeast strain as previously described by Fischer et al. (2011) and Ginglinger et al. (2013)). Individual colonies were verified by PCR and cultivation was performed in the appropriate media for WAT11 as previously described by Höfer et al. (2014) and Boachon et al. (2015) and K197G yeast strains described by Fischer et al. (2011) and Ginglinger et al. (2013). Individual transformed colonies were grown in minimum selection medium for at least 24 h, and then 10 times diluted in complete medium and grown for 30 h. Yeast culture (250 mL) of each transformant in coculture with 250 mL of yeast culture of the corresponding strain transformed with the empty vector, or cocultures of 250 mL of yeast transformed with CYP706A3 CDS mixed with 250 mL of yeast culture transformed with plasmids carrying either TPS11, TPS21, or TPS24 CDSs, were then induced overnight with Gal (20 g/L). Cultures were poured in 1L glass bottles equipped with a lid with 2 entries (in and out) on which was inserted a TEFLON tube inserted in the culture at the inlet and a glass cartridge containing 500 mg of Poropak Q (80–100 mesh; Sigma-Aldrich) at the outlet. Cultures were stirred with a magnetic bar, and a vacuum pump was used to pump out VOCs through the Poropak Q cartridges, enabling oxygenation of the cultures by bubbling the air flux through the inlet tube as shown on the scheme in Supplemental Figure 3. VOCs were collected from the yeast cultures for 2 d, and cartridges were eluted every 24 h with 1 mL dichloromethane spiked with nonyl acetate as internal standard before analysis by GC-MS. To produce and purify TPS11 products further converted by CYP706A3, yeast cultures were grown under similar conditions, but with optimization and upscaling. Briefly, 100 mL of a culture of yeast cotransformed with both TPS11 and CYP706A3 expression plasmids was mixed, at the start of Gal induction, with 900 mL of a culture of yeast transformed with only CYP706A3 expression plasmid. Up to six 1-L cultures were grown at the same time in 2-L glass bottles for 8 d, and VOCs were collected as described above. Cartridges were eluted every day with 1 mL dichloromethane; eluates were then pooled and concentrated under a gentle stream of argon. After analysis of diluted samples by GC-MS to check for products, concentrated samples were diluted in methanol and run on a semi-preparative High Performance Liquid Chromatography system (e2695 separation module Waters) coupled to an ACQUITY QDa Mass Spectrometer system (Waters). The semi-preparative system was equipped with a Kinetex 5 µm C18 100 Å Column 250 × 10.0 mm, Ea (Phenomenex), a diode array detector, and a fraction collector. Chromatography was run with water (A) and methanol (B) as mobile phase, both containing 0.1% (v/v) formic acid and starting for 1 min at 85% (v/v) B. A linear gradient was applied to reach 100% B in 12 min, followed by 100% B for 10 min. Return to initial conditions was achieved in 1 min followed by 22 min of column conditioning with 85% (v/v) B for a total run time of 45 min. The column was operated at 35°C with a flow rate of 3 mL/min, injecting 200 μL sample per run. Following separation on the column, 10% of the products were directed to the Q-da mass spectrometer while the remaining 90% was directed to the diode array detector and fraction collector through a split line. The Q-da mass spectrometer was set to ionize in positive mode, and the cone voltage was set at 10 V. Products were identified based on their putative oxygenated mass, and fractions collected accordingly. Each collected fraction was enriched through several runs, and then diluted with pure water so as to reduce the concentration of eluting solvent methanol to less than 5%. Sesquiterpene products in each fraction were subsequently concentrated by solid phase extraction on Oasis HLB extraction cartridges (Waters) as previously described by Höfer et al. (2014). Products were eluted with CDCl3 before GC-MS analysis to verify the purity of each fraction. Selected fractions were submitted to NMR analysis for product identification.
NMR Characterization of Products
Selected fractions containing substrates or products of CYP706A3 were analyzed on a 500 MHz Bruker Avance spectrometer equipped with a 5-mm DCH dual cryoprobe with z-gradient operating at 500.13 MHz for 1H and 125.758 MHz for 13C. 1D 1H, 1H -1H COSY, edited 1H -13C HSQC, and 1H -13C HMBC were recorded for each sample, adding 1H -1H NOESY and 1D 13C as required.
In Vitro Activity of CYP706A3 on Purified Sesquiterpenes
Yeast transformed with CYP706A3 expression plasmid was grown, expression induced, microsomal fraction extracted, and P450 expression quantified as previously described by Höfer et al. (2014). To analyze CYP706A3 activity on different substrates, assays were performed in 300 μL of 20 mM sodium phosphate buffer (pH 7.4) containing 100 µM substrates, 1 mM NADPH, and an adjusted amount of CYP706A3, as yeast microsomal fraction. To improve dissolution of sesqui- and monoterpenes, the latter were added directly to microsomal fractions before dilution with buffer containing up to 5% (v/v) DMSO. Samples were incubated at 28°C for 1 h and subsequently extracted with 600 μL ethyl acetate spiked with 10 µM nonyl acetate. After vortexing and short centrifugation at 4000 g for 2 min at room temperature, the ethyl acetate phase was recovered, dried on anhydrous Na2SO4 (Sigma-Aldrich), and analyzed by GC-MS.
For LC-MS/MS analysis of products, similar reactions were stopped by addition of 250 μL of methanol, vortexed for 10 s, and centrifuged at 5500 g for 5 min at room temperature. A volume of 400 μL of the supernatant was transferred into LC vials for analysis.
Isolation of Insertion Mutant and Overexpression Lines
Arabidopsis insertion lines cyp706a3-1 (SALK_057031) and cyp706a3-2 (SALKseq_052540) were identified using the T-DNA Express tool from SALK (Alonso et al., 2003), and requested from the Nottingham Arabidopsis Stock Center. Homozygous mutant lines were selected for absence of transcripts in insertion lines by RT-PCR amplifying the full CDS, and RT-qPCR in flower tissues as described above. Overexpressed lines (35S:CYP706A3) were generated by transforming the plant expression plasmid pCAMBIA3300 carrying the CYP706A3 CDS into the Agrobacterium GV3101 strain, before transformation of Col-0 plants by floral dip. Transformants in the T1 progeny were selected by germination on phosphinothricin (BASTA) at 10 ug/mL, and resistant lines were screened by RT-qPCR for CYP706A3 expression in flower tissues. Selected T1 lines showing highest expression were brought to T3 stable progeny by germination on BASTA. CYP706A3 overexpression was analyzed on T3 lines by RT-qPCR in flower tissues as described above.
VOC Collection from the Headspace of Arabidopsis Flowers and Transformed N. benthamiana Leaves
VOCs emitted from Arabidopsis flowers were collected as previously described by Boachon et al. (2015), with minor changes. About 50 to 60 inflorescences from each line were used for each sample collection, with at least three biological replicates. Detached inflorescences were placed in 12-mL glass tubes filled with water and placed in 1 liter glass jars equipped with an inlet and an outlet. Volatiles were pumped out from the jar with a vacuum pump at ∼100 mL min−1 and trapped on a cartridge filled with 30 mg Porapak Q (80–100 mesh, Grace scientific) at the outlet. A similar cartridge was placed at the inlet to ensure purification of the incoming air. VOCs were sampled for 24 h. After volatile collection, flowers were cut from inflorescences and weighed before further analysis of soluble compounds. Porapak Q cartridges were eluted with 200 μL dichloromethane spiked with 10 µM nonyl acetate, and VOC analysis and quantification were performed by GC-MS.
To measure VOCs emitted from Arabidopsis plants during floral transition, plants were grown individually on Jiffy-7 peat pellets (Jiffy) under standard conditions, until the appearance of the first inflorescence. Four plants were used per sample and placed in 1-L glass jars, and VOCs were collected as described above. Porapak Q cartridges were eluted every day until the first inflorescence opened, and samples were analyzed by GC-MS.
For headspace analysis of VOCs emitted from transformed N. benthamiana leaves, plant expression constructs carrying the CDS of CYP706A3, CYP76C1, TPS11, TPS21, or TPS24 were transformed into hypervirulent Agrobacterium LBA4404, which was used to infiltrate the N. benthamiana leaves as described previously (Ginglinger et al., 2013). After 2 d, infiltrated leaves were detached, and their petioles dipped into a 12-mL glass vial filled with water that was placed in a 1-L glass jar. VOCs emitted from the headspace were collected from 3 to 4 infiltrated leaves per sample for 24 h following the same procedure as described above for Arabidopsis flower VOCs and subsequently analyzed by GC-MS.
GC-MS Analysis
Capillary gas chromatography was performed as described previously (Boachon et al., 2015), on a PerkinElmer Clarus 680 gas chromatograph coupled to a PerkinElmer Clarus 600T mass spectrometer (PerkinElmer), using a HP-5ms column (30 m, 0.25 mm, 0.25 µm; Agilent technologies). Samples were injected by splitless injection, at 250°C injector temperature, using a program consisting of 0.5 min at 50°C, followed by 20°C min−1 to 320°C, then 5 min at 320°C, with a flow of 1.2 mL min−1 of He as carrier gas. Products were identified based on their retention times and electron ionization mass spectra (70 eV, m/z 50-300), and compared to those present in the NIST and WILEY libraries and to previously published Arabidopsis flower VOCs (Tholl et al., 2005).
Extraction of Soluble Compounds
Following collection of VOCs from Arabidopsis flowers and N. benthamiana transformed leaves, plant material was extracted with methanol for analysis of the soluble compounds. The extraction procedure was as previously described by Boachon et al. (2015), and samples were analyzed by LC-MS/MS.
LC-MS/MS Analysis of Soluble Compounds
LC-MS/MS analyses were essentially performed as described previously (Boachon et al., 2015). Quantitative analyses were performed using several multiple reaction monitoring channels, each specific for a potential oxidized sesquiterpene mass (based on soluble products from yeast-expressed cultures), with specific MS/MS transition and tunes as listed in Supplemental Table 6.
Insect Behavior
We tested florivory with the herbivore Plutella xylostella, because it is a Brassicaceae specialist that had been shown to feed on floral tissues of Arabidopsis in previous experiments (Boachon et al., 2015) and on Brassicaceae flowers in nature (Knauer et al., 2018). Preference of the P. xylostella L3 larvae for flowers from wild-type, cyp706a3, or 35S:CYP706A3 mutants was tested using a dual-choice feeding test, essentially as described previously (Boachon et al., 2015), except that closed flowers at stage T3 (see Figure 8) were used. Briefly, five Col-0 and five cyp706A3-2 or 35S:CYP706A3 closed buds were set in 1% (w/v) agarose on opposite sides of a Petri dish. One insect was placed in the center of the Petri dish. The number of flowers (or flower parts) consumed by the insect was recorded after 3 h.
The behavior of adult hoverflies (Episyrphus balteus) was tested with headspace collected from yeast cultures expressing CYP706A3 alone, together with TPS11, or with purified fractions, in a star-shaped olfactometer described previously (Junker et al., 2010). Experiments were performed as described previously (Boachon et al., 2015). The field of the olfactometer defined as substance field was supplied with an air stream from a desiccator in which 100 µg of compounds dissolved in methanol was placed on a filter. The field of the olfactometer defined as neutral was supplied with an air stream from a control desiccator in which the same amount of methanol was placed on a filter. Hoverflies were placed one by one for 4 min in the olfactometer and time spent in the neutral or substance fields was measured. Substances in the desiccator were replaced every 20 min.
Phylogenetic Analysis
Sequences of CYP706 and TPS11 from different available Brassicales genomes were retrieved using a Blast homology search with AtTPS11 and AtCYP706A3 as bait on the Phytozome website (http://phytozome.jgi.doe.gov/), the BrassicaDB page (http://brassicadb.org), or Dr. David Nelson’s website (http://drnelson.uthsc.edu/CytochromeP450.html). Homologous sequences were then aligned with other representative TPS (AtTPS12, 13, 23) and CYP76 family members, and more distantly related sequences as outgroup (CYP76C1, CYP76G1, CYP71A12). Sequences were discarded if not grouping with TPS11 or CYP706 in alignments including these outgroup sequences. Short incomplete sequences were manually curated and removed, and remaining sequences were then aligned using MUSCLE and default settings with Seaview (Edgar, 2004; Gouy et al., 2010), and Gblocks sets defined on protein sequence alignments. Phylogenetic trees were then built using PhyML (Guindon et al., 2010), using the LG model for TPS, and JTT model for CYP706, and consistency was tested by performing 100 bootstrap iterations. If not specifically named to date, sequences were referred to according to their position in the different branches of the tree.
Microbiome analysis
For microbial DNA extraction, micro-organisms were collected from inflorescences of 5- to 6-week- old Arabidopsis plants. For each sample, 5 fully opened inflorescences were harvested from one plant using sterile forceps. Each biological replicate was collected from a different plant. Inflorescences were placed in BashingBeads Lysis tubes from the Xpeditio Fungal/Bacterial DNA Miniprep kit (Zymo Research) containing lysis solution. Lysis tubes were shaken for 5 min at 20 Hz using TissueLyser II (Qiagen) to extract DNA following the manufacturer’s guidelines (Zymo Research). After DNA extraction, the 16S rRNA gene V5-V7 hypervariable region was PCR amplified using primers 799F/1193r as previously described (Supplemental Table 5; Bodenhausen et al., 2013). Amplicon DNA libraries were constructed by the Illumina TruSeq DNA library preparation protocol. Sequencing was performed at MR DNA (www.mrdnalab.com) by MiSeq (2×300bp) following the manufacturer’s guidelines. Sequence data were processed using the MR DNA analysis pipeline (MR DNA). In summary, sequences were joined, barcodes removed, and sequences <150 bp or with ambiguous base calls were discarded. Sequences were denoised, OTUs generated, and chimeras removed. OTUs were defined by clustering at 3% divergence (97% similarity). Final OTUs were taxonomically classified using BLASTn against a curated database derived from GreenGenes, RDPII, and NCBI (www.ncbi.nlm.nih.gov; http://greengenes.lbl.gov/Download/; http://rdp.cme.msu.edu). The resulting OTU table was used to determine taxonomic relative abundances and subsequent statistical analyses of alpha- and beta-diversity using the R package vegan (Dixon, 2003; Supplemental Table 4). The Kruskal-Wallis rank sum test was performed to probe differences in the OTU richness in wild-type, cyp706a3 mutant, and complemented flowers. Chao rarefaction analysis was used to estimate the maximal number of OTUs associated with the flowers of specific lines given a complete sampling. PLS-DA was performed to discriminate the entire bacterial community composition between the flowers of the three Arabidopsis lines. The PLS framework is suited to analyzed data when the number of variables is equal to or greater than samples, which was the case in our data with 15 samples and 1485 variables (number of OTUs; Hervé et al., 2018). Furthermore, this framework can be applied to quantitative data without any assumption on their distribution. PLS-DA tests whether lines have different bacterial communities. To validate the PLS-DA model, we used a permutation test to evaluate significant differences between groups (Hervé et al., 2018). Finally, a Kruskal-Wallis rank sum test was performed to identify OTUs that significantly differed between all lines, and a Wilcoxon rank sum test was performed to identify OTUs being significantly different between a pair of lines.
Construction of 3D Models of CYP706A3
Modeller9v14 and Autodock4 calculations were performed using the computing facilities of the CEA-DRF-Joliot (cluster Gabriel) at Saclay. GROMACS (2016.1) calculations were performed on the I2BC (CNRS/CEA-Gif) cluster.
The 3D models of Arabidopsis CYP706A3 (519 amino acids) were rebuilt without the N-terminal membrane segment (catalytic domain 38-519) using Modeller9v14 (Webb and Sali, 2016) and the crystal structures of PDB selected as templates. Choice of templates was guided by an iterative process that optimized both the multiple sequence alignment (MSA) between the CYP706A3 sequence and templates, and the quality score of models implied by each alignment. The protocol started with six templates selected on the basis of the best similarity according to Blast-PDB, HMMER (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) and HHPRED (https://toolkit.tuebingen.mpg.de/#/tools/hhblits), summarized in Supplemental Table 2, and on the basis of other criteria such as highest alignment length, best atomic resolution, and highest quantitative model energy analysis (QMEAN) scores. All suitable templates proved to be human isoforms, and exhibited rather low sequence identities with AtCYP706A3 (24 to 25%). However, corresponding 3D structures were well-aligned by structure alignment algorithms MUSTANG (Konagurthu et al., 2006) and PROMALS3D (Konagurthu et al., 2006; Pei et al., 2008). Resulting MSA were consistent with those obtained with the MAFFT-L-INS-I algorithm (Pei et al., 2008; Katoh and Standley, 2013). As a result of the iterative process, final construction giving rise to the best model involved the three templates CYP2R1 (pdb 3czh), CYP17A1 (pdb 5irq), and CYP1A2 (pdb 2 hi4; see features in Supplemental Table 2). The validated MSA used as input for MODELER is shown in Supplemental Figure 17.
Several runs of 100 models each were performed; generated models sorted by the MODELER objective function were also evaluated by their discrete optimized protein energy and GA341 scores calculated by MODELER. The best models having the lowest discrete optimized protein energy score or best objective function issued from each run were pooled and submitted to the online metaserver structural analysis and verification server (http://services.mbi.ucla.edu/SAVES), and finally to the QMEAN scoring function (Benkert et al., 2008) server (Benkert et al., 2009) for model quality assessment. The final model selected for docking studies was the best one according to a good compromise between the scores calculated by the structural analysis and verification server scoring programs and the QMEAN4 value and Z-score. As a result, the selected AtCYP706A3 model had a QMEAN4 value and Z-score equal to 0.71 and −2.2, respectively, which are reasonably good scores when compared with the QMEAN scores of individual PDB templates (Supplemental Table 2). The Ramachandran plot exhibited only one residue in disallowed regions (Ile55), located in the disordered N-terminal segment, upstream of A helix. Root Mean Square Deviations (RMSDs) calculated between a-carbons of each template and of the model ranged between 0.61 and 0.88 Å.
Preparation of AtCYP706A3 Structures for Docking Studies
Protein structures generated by Modeler were first stripped of all hydrogen atoms, and then atom charges and hydrogen atoms were added to the protein with the University of California San Francisco (UCSF) Chimera package (www.cgl.ucsf.edu/chimera) UCSF chimera 2004 (Pettersen et al., 2004) using AMBER ff14SB parameters. Parameters (geometry and atom charges) applied for the heme in the initial step of the catalytic cycle (defined as FeIII protoporphyrin IX) were taken from an AMBER-compatible heme model developed by Shahrokh et al., 2012, and in some docking experiments with (+)-thujopsene the heme cofactor was also defined as the highly reactive intermediate compound I (FeIV=O)+.. The atom charges of the proximal cysteine thiolate were taken from the same work.
Docking Procedure of Sesquiterpenes
Molecular docking experiments with sesquiterpene molecules into the CYP706A3 active site were performed using AutoDock 4 (release 4.2.6) in the semi-flexible mode, and prepared with AutoDock Tools (Morris et al., 2009). Ligand molecules were parameterized under the UCSF Chimera molecular modeling suite using AM1-BCC charges model, and the structure files saved under MOL2 format as input files for AutoDock. Usually, the semi-flexible mode in Autodock means that the ligand is fully flexible and the receptor kept rigid. In multi-cyclic sesquiterpenoid structures, however, there is no rotatable bond detected by Autodock in the torsion tree. For handling unsaturated ring flexibility, several conformations of the nonaromatic rings were generated by short molecular dynamics (MD) simulations and minimization steps performed under Chimera, and used as a starting point for docking to allow better conformational sampling of the ligands under Autodock 4. The MOL2 format files created in Chimera for the receptor and ligands were converted into PDBQT format with merging all nonpolar hydrogen atoms, and with original charges (AMBER 14ff and AM1-BCC) maintained. The receptor was kept either rigid or flexible. The modeled structure of AtCYP706A3 was also optimized by short 30 ns MD runs (GROMACS 2016) to generate various relaxed conformations of the protein, in addition to the initial MODELER structure. As a whole, in most cases, the best docking results were obtained using different conformations of multi-cyclic compounds docked in the rigid receptor mode, onto the initial MODELER structure, with the heme in the FeIII state.
The docking box, in which grid maps were computed using the program AutoGrid (Morris et al., 2009), included the active site with the iron-protoporphyrin group on one edge, and the whole distal moiety of the enzyme to allow a large sampling of potential poses. A “blind” docking test was first done with a grid built by AutoGrid with 64, 88, and 80 points in the x, y, and z directions and a grid spacing of 0.37Å to allow for a good compromise between the resolution of the explored volume and the size of the binding area. In most cases and remarkably, the docking experiments revealed a unique pose in the active site, close to the heme iron (Supplemental Figures 19A and 19B). The docking box size was then decreased to 52×54×54 points (x,y,z) with a grid spacing of 0.27Å to allow for a more detailed view of conformations docked in the active site (Supplemental Figure 19C as example). For each sesquiterpene conformer, 100 independent runs were performed using the Lamarckian genetic algorithm (Fuhrmann et al., 2010). The default settings were applied for all other parameters.
The resulting poses were assigned a score calculated by Autodock that can be considered as an estimated free energy of ligand binding (indicative of binding affinity), then clustered as a function of the closeness of their positions and conformations with RMSD set at 2.0 Å or 1.0 Å, and finally ranked by their binding score (for the best pose in the cluster). The resulting histograms of 100 poses of (+)-thujopsene docked into AtCYP706A3 shown in Supplemental Figure 19 revealed solutions markedly focused in the first cluster, sometimes gathered in a unique cluster within 2Å of RMSD as for conformer 2 of (+)-thujopsene (Supplemental Figure 19A). To resolve slightly different positions of the ligand in the vicinity of the heme, it proved to be more informative to cluster at 1Å of RMSD (Supplemental Figures 19B and 19C).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CYP706A3 (AT5G44620), TPS11 (AT5G44630), TPS21 (AT5G23960), TPS24 (AT3G25810), CYP76C1 (At2g45560), TPS03 (AT4G16740), TPS10 (At2g24210), and TPS14 (AT1G61680). The microbiome SRA accession number is PRJNA562511.
Supplemental Data
Supplemental Figure 1. CYP706A3-TPS11 cluster on chromosome 5 of Arabidopsis.
Supplemental Figure 2. CYP706A3 and TPS11 subcellular localizations.
Supplemental Figure 3. Activity of yeast-expressed CYP706A3 on TPS11 products detected by GC-MS.
Supplemental Figure 4. Mass spectra confirm the identity of compounds detected and further used in our experiments.
Supplemental Figure 5. Identification, separation and enrichment of TPS11 and CYP706A3 products from yeast headspace by semi-preparative LC-Qda.
Supplemental Figure 6. Activity of yeast-expressed CYP706A3 on (-)-thujopsene.
Supplemental Figure 7. Structure of sesquiterpene oxide products formed by CYP706A3 according to NMR spectra.
Supplemental Figure 8. Sequential activity of yeast-expressed CYP706A3 on TPS11 products analyzed by LC-MS/MS.
Supplemental Figure 9. LC-MS of CYP706A3 primary products.
Supplemental Figure 10. LC-MS of CYP706A3 secondary products
Supplemental Figure 11. Genotyping of CYP706A3 mutants.
Supplemental Figure 12. Activity of CYP706A3 on TPS21 products.
Supplemental Figure 13. Activity of CYP706A3 on TPS24 products.
Supplemental Figure 14. Targeted LC-MS/MS profiling of the sesquiterpene oxide metabolites present in the flowers of wild-type Arabidopsis and CYP706A3 mutants.
Supplemental Figure 15. Targeted LC-MS/MS profiling of the sesquiterpene oxide metabolites in N. benthamiana leaves transiently co-expressing TPS11 and CYP706A3.
Supplemental Figure 16. Targeted LC-MS/MS profiling of sesquiterpene oxide metabolites in buds of wild-type Arabidopsis and cyp706a3-2 insertion mutant.
Supplemental Figure 17. Multiple sequence alignment of templates used for homology modeling.
Supplemental Figure 18. Channel profiles of physico-chemical properties calculated by Mole2.5.
Supplemental Figure 19. Examples of clustering of 100 docking runs of two different initial conformations of (+)-thujopsene performed by Autodock 4.2.6 on the rebuilt 3D model of AtCYP706A3.
Supplemental Figure 20. Ecological impact of CYP706A3 products on hoverflies.
Supplemental Table 1. Quantification of sesquiterpene emissions upon heterologous expression in Nicotiana benthamiana.
Supplemental Table 2. List of potential templates for AtCYP706A3 modeling detected by remote homology methods (based on HMM profiles, or structure prediction by HMM-HMM comparison).
Supplemental Table 3. Binding energy scores obtained by docking the mentioned sesqui- and monoterpene substrates with the AtCYP706A3 model.
Supplemental Table 4. Detailed taxonomic designation for bacterial OTUs that are significantly different between a pair of Arabidopsis lines (Col-0, cyp706a3-2, and 35S:CYP706A3).
Supplemental Table 5. PCR primer list.
Supplemental Table 6. MS/MS conditions and set-up for the LC analysis in multiple reaction monitoring (MRM) of putative sesquiterpene oxides.
Supplemental Data Set 1. Raw and complete data for the fifty most co-expressed genes identified using BAR expression Angler (Toufighi et al., 2011) and CYP706A3 as bait.
Supplemental Data Set 2. Flower microbiome raw data.
Supplemental Data Set 3. Statistical analysis.
Supplemental Data Set 4. Protein sequence alignment used to generate TPS11 phylogeny.
Supplemental Data Set 5. Tree resulting from alignment in Data set 4.
Supplemental Data Set 6. Protein sequence alignment used to generate CYP706A3 phylogeny.
Supplemental Data Set 7. Tree resulting from Data set 6.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We acknowledge the use of the facilities of the Bindley Bioscience Center, a core facility of the National Institutes of Health–funded Indiana Clinical and Translational Sciences Institute. We thank Todd Blevins for contributing to the editing of the article. This work was supported by the European Fund for Regional Development in the program Interreg IVA Broad Region EU invests in your future to the SaarLorBiotech project (to B.B., Y.B., and D.W.-R.)
Author Contributions
B.B. designed and performed experiments, and interpreted results; Y.B. performed experiments and analyzed data; B.B., Y.B., R.R.J., I.K., and N.D. designed the insect behavior experiment and analyzed data; B.B. and D.W.-R. wrote the article; R.R. and F.A. performed modeling and docking studies; R.R.J. and X.-Y.C. generated and provided mutants; B.V., L.A., and L.M. performed RMN analysis and interpretation; S.M. constructed vectors and mutants; R.R.J., F.N., F.B., and S.V. performed microbiome analysis; A.L. performed yeast expression; L.H. and J.E.B. performed confocal microscopy; N.N. performed phylogenomic analysis; D.W.-R. received funding and designed work; all authors read and edited the article.
Footnotes
Articles can be viewed without a subscription.
References
- Abdalsamee M.K., Müller C. (2015). Uncovering different parameters influencing florivory in a specialist herbivore. Ecol. Entomol. 40: 258–268. [Google Scholar]
- Abel C., Clauss M., Schaub A., Gershenzon J., Tholl D. (2009). Floral and insect-induced volatile formation in Arabidopsis lyrata ssp. petraea, a perennial, outcrossing relative of A. thaliana. Planta 230: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Bassard J.-E., et al. (2012). Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24: 4465–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P., Künzli M., Schwede T. (2009). QMEAN server for protein model quality estimation. Nucleic Acids Res. 37: W510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P., Tosatto S.C.E., Schomburg D. (2008). QMEAN: A comprehensive scoring function for model quality assessment. Proteins 71: 261–277. [DOI] [PubMed] [Google Scholar]
- Boachon B., et al. (2015). CYP76C1 (cytochrome P450)-mediated linalool metabolism and the formation of volatile and soluble linalool oxides in Arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell 27: 2972–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boachon B., Lynch J.H., Ray S., Yuan J., Caldo K.M.P., Junker R.R., Kessler S.A., Morgan J.A., Dudareva N. (2019). Natural fumigation as a mechanism for volatile transport between flower organs. Nat. Chem. Biol. 15: 583–588. [DOI] [PubMed] [Google Scholar]
- Bodenhausen N., Horton M.W., Bergelson J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One 8: e56329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev A.M., Osbourn A.E. (2018). Multigenome analysis implicates miniature inverted-repeat transposable elements (MITEs) in metabolic diversification in eudicots. Proc. Natl. Acad. Sci. USA 115: E6650–E6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringel F., Couée I. (2015). Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 6: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon R.C.F., Junker R.R., Scofield D.G., Parachnowitsch A.L. (2018). Bacteria colonising Penstemon digitalis show volatile and tissue-specific responses to a natural concentration range of the floral volatile linalool. Chemoecology 28: 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cankar K., van Houwelingen A., Goedbloed M., Renirie R., de Jong R.M., Bouwmeester H., Bosch D., Sonke T., Beekwilder J. (2014). Valencene oxidase CYP706M1 from Alaska cedar (Callitropsis nootkatensis). FEBS Lett. 588: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Chen F., Tholl D., D’Auria J.C., Farooq A., Pichersky E., Gershenzon J. (2003). Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru V., Winn P.J., Wade R.C. (2007). The ins and outs of cytochrome P450s. Biochim. Biophys. Acta 1770: 390–401. [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14: 927–930. [Google Scholar]
- Doubleday L.A.D., Raguso R.A., Eckert C.G. (2013). Dramatic vestigialization of floral fragrance across a transition from outcrossing to selfing in Abronia umbellata (Nyctaginaceae). Am. J. Bot. 100: 2280–2292. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: Multiple sequence alignment with improved accuracy and speed. In Proceedings. 2004 IEEE Computational Systems Bioinformatics Conference. [Google Scholar]
- Ehlting J., Sauveplane V., Olry A., Ginglinger J.-F., Provart N.J., Werck-Reichhart D. (2008). An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler M., Baldwin I.T. (1996). The chemistry of defense and apparency in the corollas of Nicotiana attenuata. Oecologia 107: 102–112. [DOI] [PubMed] [Google Scholar]
- Fischer M.J.C., Meyer S., Claudel P., Bergdoll M., Karst F. (2011). Metabolic engineering of monoterpene synthesis in yeast. Biotechnol. Bioeng. 108: 1883–1892. [DOI] [PubMed] [Google Scholar]
- Fuhrmann J., Rurainski A., Lenhof H.-P., Neumann D. (2010). A new Lamarckian genetic algorithm for flexible ligand-receptor docking. J. Comput. Chem. 31: 1911–1918. [DOI] [PubMed] [Google Scholar]
- Gargallo-Garriga A., Sardans J., Pérez-Trujillo M., Guenther A., Llusià J., Rico L., Terradas J., Farré-Armengol G., Filella I., Parella T., Peñuelas J. (2016). Shifts in plant foliar and floral metabolomes in response to the suppression of the associated microbiota. BMC Plant Biol. 16: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginglinger J.-F., et al. (2013). Gene coexpression analysis reveals complex metabolism of the monoterpene alcohol linalool in Arabidopsis flowers. Plant Cell 25: 4640–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hansen C.C., Sørensen M., Veiga T.A.M., Zibrandtsen J.F.S., Heskes A.M., Olsen C.E., Boughton B.A., Møller B.L., Neilson E.H.J. (2018). Reconfigured cyanogenic glucoside biosynthesis in involves a cytochrome P450 CYP706C55. Plant Physiol. 178: 1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helletsgruber C., Dötterl S., Ruprecht U., Junker R.R. (2017). Epiphytic bacteria alter floral scent emissions. J. Chem. Ecol. 43: 1073–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé M.R., Nicolè F., Lê Cao K.A. (2018). Multivariate analysis of multiple datasets: A practical guide for chemical ecology. J. Chem. Ecol. 44: 215–234. [DOI] [PubMed] [Google Scholar]
- Höfer R., Boachon B., Renault H., Gavira C., Miesch L., Iglesias J., Ginglinger J.-F., Allouche L., Miesch M., Grec S., Larbat R., Werck-Reichhart D. (2014). Dual function of the cytochrome P450 CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol. 166: 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.H., Bremer M., Schneider K., Burger F., Stolle E., Moritz G. (2003). Flower visitors in a natural population of Arabidopsis thaliana. Plant Biol. 5: 491–494. [Google Scholar]
- Huang M., Sanchez-Moreiras A.M., Abel C., Sohrabi R., Lee S., Gershenzon J., Tholl D. (2012). The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 193: 997–1008. [DOI] [PubMed] [Google Scholar]
- Jones M.E. (1971). The population genetics of Arabidopsis thaliana I. The breeding system. Heredity 27: 39–50. [Google Scholar]
- Junker R.R. (2016). Multifunctional and diverse floral scents mediate biotic interactions embedded in communities In Deciphering Chemical Language of Plant Communication, Blande J., and Glinwood R., eds (Cham: Springer; ), pp. 257–282. [Google Scholar]
- Junker R.R., Heidinger I.M.M., Blüthgen N. (2010). Floral scent terpenoids deter the facultative florivore Metrioptera bicolor (Ensifera, Tettigoniidae, Decticinae). J. Orthoptera Res. 19: 69–74. [Google Scholar]
- Junker R.R., Keller A. (2015). Microhabitat heterogeneity across leaves and flower organs promotes bacterial diversity. FEMS Microbiol. Ecol. 91: fiv097. [DOI] [PubMed] [Google Scholar]
- Junker R.R., Tholl D. (2013). Volatile organic compound mediated interactions at the plant-microbe interface. J. Chem. Ecol. 39: 810–825. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuda Y. (2012). Progress and future of pyrethroids. Top. Curr. Chem. 314: 1–30. [DOI] [PubMed] [Google Scholar]
- Kessler D., Diezel C., Baldwin I.T. (2010). Changing pollinators as a means of escaping herbivores. Curr. Biol. 20: 237–242. [DOI] [PubMed] [Google Scholar]
- Kessler D., Kallenbach M., Diezel C., Rothe E., Murdock M., Baldwin I.T. (2015). How scent and nectar influence floral antagonists and mutualists. eLife 4: e07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta Y., Ueda H., Takahashi M., Mitsumori T., Yamada G., Sakamori K., Takeda K., Furutani S., Nakayama K., Katsuda Y., Hatanaka A., Matsuda K. (2012). Identification and characterization of a GDSL lipase-like protein that catalyzes the ester-forming reaction for pyrethrin biosynthesis in Tanacetum cinerariifolium–a new target for plant protection. Plant J. 71: 183–193. [DOI] [PubMed] [Google Scholar]
- Knauer A.C., Bakhtiari M., Schiestl F.P. (2018). Crab spiders impact floral-signal evolution indirectly through removal of florivores. Nat. Commun. 9: 1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagurthu A.S., Whisstock J.C., Stuckey P.J., Lesk A.M. (2006). MUSTANG: A multiple structural alignment algorithm. Proteins 64: 559–574. [DOI] [PubMed] [Google Scholar]
- Li R., Schuman M.C., Wang Y., Llorca L.C., Bing J., Bennion A., Halitschke R., Baldwin I.T. (2018). Jasmonate signaling makes flowers attractive to pollinators and repellant to florivores in nature. J. Integr. Plant Biol. 60: 190–194. [DOI] [PubMed] [Google Scholar]
- Li R., Wang M., Wang Y., Schuman M.C., Weinhold A., Schäfer M., Jiménez-Alemán G.H., Barthel A., Baldwin I.T. (2017). Flower-specific jasmonate signaling regulates constitutive floral defenses in wild tobacco. Proc. Natl. Acad. Sci. USA 114: E7205–E7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Wang Y.H., Wang G.D., Essenberg M., Chen X.Y. (2001). Molecular cloning and functional identification of (+)-delta-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 28: 95–104. [DOI] [PubMed] [Google Scholar]
- McKey D. (1979). The distribution of secondary compounds within plants In Herbivores: Their Interaction with Secondary Plant Metabolites. (New York: Academic; ), pp. 55–133. [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30: 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H.H., Nørholm M.H.H., Halkier B.A. (2006). Screening for plant transporter function by expressing a normalized Arabidopsis full-length cDNA library in Xenopus oocytes. Plant Methods 2: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H.-W., Huang A., Osbourn A. (2016). Plant metabolic clusters - from genetics to genomics. New Phytol. 211: 771–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H.-W., Osbourn A. (2014). Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 26: 91–99. [DOI] [PubMed] [Google Scholar]
- Pei J., Tang M., Grishin N.V. (2008). PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 36: W30-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñuelas J., Farré-Armengol G., Llusia J., Gargallo-Garriga A., Rico L., Sardans J., Terradas J., Filella I. (2014). Removal of floral microbiota reduces floral terpene emissions. Sci. Rep. 4: 6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravda L., Sehnal D., Toušek D., Navrátilová V., Bazgier V., Berka K., Svobodová Vareková R., Koca J., Otyepka M. (2018). MOLEonline: A web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46 (W1): W368–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raguso R.A. (2016). Plant evolution: Repeated loss of floral scent - A path of least resistance? Curr. Biol. 26: R1282–R1285. [DOI] [PubMed] [Google Scholar]
- Sas C., Müller F., Kappel C., Kent T.V., Wright S.I., Hilker M., Lenhard M. (2016). Repeated inactivation of the first committed enzyme underlies the loss of benzaldehyde emission after the selfing transition in Capsella. Curr. Biol. 26: 3313–3319. [DOI] [PubMed] [Google Scholar]
- Shahrokh K., Orendt A., Yost G.S., Cheatham T.E. III (2012). Quantum mechanically derived AMBER-compatible heme parameters for various states of the cytochrome P450 catalytic cycle. J. Comput. Chem. 33: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A., Lenhard M. (2018). Capsella. Curr. Biol. 28: R920–R921. [DOI] [PubMed] [Google Scholar]
- Sicard A., Stacey N., Hermann K., Dessoly J., Neuffer B., Bäurle I., Lenhard M. (2011). Genetics, evolution, and adaptive significance of the selfing syndrome in the genus Capsella. Plant Cell 23: 3156–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape J.W., Lawrence M.J. (1971). The breeding system of Arabidopsis thaliana. Heredity 27: 299–302. [Google Scholar]
- Thien L.B., Bernhardt P., Devall M.S., Chen Z.-D., Luo Y.-B., Fan J.-H., Yuan L.-C., Williams J.H. (2009). Pollination biology of basal angiosperms (ANITA grade). Am. J. Bot. 96: 166–182. [DOI] [PubMed] [Google Scholar]
- Tholl D., Chen F., Petri J., Gershenzon J., Pichersky E. (2005). Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 42: 757–771. [DOI] [PubMed] [Google Scholar]
- Tholl D., Lee S. (2011). Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K., Brady S.M., Austin R., Ly E., Provart N.J. (2005). The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J. 43: 153–163. [DOI] [PubMed] [Google Scholar]
- Vekemans X., Poux C., Goubet P.M., Castric V. (2014). The evolution of selfing from outcrossing ancestors in Brassicaceae: What have we learned from variation at the S-locus? J. Evol. Biol. 27: 1372–1385. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Webb B., Sali A. (2016). Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47: 5.6.1-5.6.37. [DOI] [PubMed] [Google Scholar]
- Xu H., Moghe G.D., Wiegert-Rininger K., Schilmiller A.L., Barry C.S., Last R.L., Pichersky E. (2018). Coexpression analysis identifies two oxidoreductases involved in the biosynthesis of the monoterpene acid moiety of natural pyrethrin insecticides in Tanacetum cinerariifolium. Plant Physiol. 176: 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-Y., Sun K., Chen F., Yuan J., Li X., Dai C.-C. (2018). Endophytic Pseudomonas induces metabolic flux changes that enhance medicinal sesquiterpenoid accumulation in Atractylodes lancea. Plant Physiol. Biochem. 130: 473–481. [DOI] [PubMed] [Google Scholar]