Abstract
Chronological age is an important predictor of morbidity and mortality; however, it is unable to account for heterogeneity in the decline of physiological function and health with advancing age. Several attempts have been made to instead define a “biological age” using multiple physiological parameters in order to account for variation in the trajectory of human aging; however, these methods require technical expertise and are likely too time-intensive and costly to be implemented into clinical practice. Accordingly, we sought to develop a metabolomic signature of biological aging that could predict changes in physiological function with the convenience of a blood sample. A weighted model of biological age was generated based on multiple clinical and physiological measures in a cohort of healthy adults and was then applied to a group of healthy older adults who were tracked longitudinally over a 5–10-year timeframe. Plasma metabolomic signatures were identified that were associated with biological age, including some that could predict whether individuals would age at a faster or slower rate. Metabolites most associated with the rate of biological aging included amino acid, fatty acid, acylcarnitine, sphingolipid, and nucleotide metabolites. These results not only have clinical implications by providing a simple blood-based assay of biological aging, but also provide insight into the molecular mechanisms underlying human healthspan.
Electronic supplementary material
The online version of this article (10.1007/s11357-019-00123-w) contains supplementary material, which is available to authorized users.
Keywords: Biological aging, Metabolomics, Healthspan, Precision medicine
Introduction
Advanced chronological age is one of the most important risk factors for many of the chronic diseases and disabilities affecting contemporary societies (Lunenfeld and Stratton 2013). The link between advancing age and increased chronic disease risk is predominately mediated by the progressive decline of multiple physiological systems (Franceschi and Campisi 2014; Kennedy et al. 2014). While aging itself is inescapable, the rate at which physiological functions decline with age is highly variable among individuals and is the combined result of genetic and non-genetic factors including lifestyle behaviors (e.g., diet and physical activity) and other environmental or occupational exposures (e.g., sun damage or proximity to volatile chemicals) (Brooks-Wilson 2013; Jiang et al. 2013). As such, individuals of the same chronological age may differ considerably with respect to their physiological function and overall health status, thus limiting the predictive capacity of chronological age alone in determining overall disease risk.
In order to address this issue, recent attempts have been made to define a “biological age” that is more reflective of the inherent heterogeneity of human aging than chronological age (Nakamura 1991; Cho et al. 2010; Levine 2013; Mitnitski et al. 2013; Belsky et al. 2015; Sebastiani et al. 2017). Central to this approach is the integration of multiple age-related “biomarkers” that are modifiable by lifestyle behaviors and other environmental factors and therefore more reflective of overall health than chronological age alone. In this regard, biological age, when determined using clinical and physiological parameters, predicts morbidity and mortality better than chronological age (Levine 2013); however, the clinical utility of this approach is limited by the substantial cost, time, specialized equipment, and training required to accurately assess multiple physiological functions. As such, the development of a surrogate blood-based measure of biological age would eliminate the burden of making multiple clinical and physiological assessments and more rapidly identify individuals at risk for faster aging. Moreover, such biomarkers may offer insight into the underlying mechanisms of aging and provide new targets for therapies aimed at improving human healthspan.
The metabolome, defined as all small molecules characterizing a biological system, is altered with age and reflective of age-related changes in physiological function (Lawton et al. 2008; Houtkooper et al. 2011; Mapstone et al. 2014; Johnson et al. 2018). In the present study, we sought to identify plasma metabolomic signatures associated with biological aging in healthy adults. To do this, we trained a model of biological age based on clinical and physiological measures in a large cohort of healthy adults and tested it in a smaller cohort of mostly healthy middle-aged and older adults who were tracked longitudinally over a 5–10-year time frame. We identified small molecule signatures present at baseline and/or follow-up in the longitudinal cohort that are associated with biological aging, including metabolites that are predictive of faster vs. slower aging.
Methods
Study design and subjects
All study procedures were reviewed and approved by the University of Colorado Boulder Institutional Review Board. Clinical and physiological measurements were performed at the University of Colorado Boulder Clinical Translational Research Center (CTRC). All study participants provided written informed consent after the nature, benefits, and risks of the study were explained. Subjects from the longitudinal cohort were re-contacted after at least 5 years and provided an option to re-enroll. All subjects were non-smokers, determined to be free of clinical disease as assessed by medical history, physical examination, blood chemistries, and resting and exercise ECG. To control for their menstrual cycle, premenopausal women were studied during the early follicular phase, while older women were postmenopausal for at least 1 year. All subjects followed a 12-h fast and 24-h abstention from alcohol, exercise, and prescription medication prior to testing.
Model of biological age
Biological age was calculated using the basic Klemera-Doubal equation without chronological age as a marker to avoid biasing or overfitting the model and to ensure that we capture the true heterogeneity of human aging (Klemera and Doubal 2006). Parameters for the 13 clinical and physiological measures used in the model were estimated from cross-sectional data in 355 men and 249 women (Table 1), generating separate equations for biological age in males and females due to basic differences in physiology. Specifically, a linear relationship with chronological age was estimated in men and women for each measure by individual linear regressions. Biological age (BAE) for an individual with measurements (xj) was calculated by the Klemera-Doubal equation with m = 13:
where kj is the slope, qj the intercept, and sj the standard deviation of residuals for the corresponding regression. Once the model was trained on our initial cohort of 604 individuals, the equation was applied to a longitudinal cohort of 31 individuals to determine rates of aging, calculated as the ratio of Δ biological age:Δ chronological age. ageDiff was calculated by assessing the difference in calculated biological age from chronological age at baseline.
Table 1.
Training cohort subject characteristics
| Subject characteristics | Male | Female |
|---|---|---|
| Subjects (n) | 355 | 249 |
| Avg. age (range) | 52 (18–79) | 55 (18–80) |
| Body mass index (kg/m2) | 26 ± 0.21 | 24 ± 0.25 |
| Waist-to-hip ratio | 0.89 ± 0.003 | 0.76 ± 0.004 |
| Body fat (%) | 24 ± 1 | 34 ± 1 |
| Bone mineral density (g/cm2) | 1.27 ± 0.01 | 1.14 ± 0.01 |
| Systolic blood pressure (mmHg) | 122 ± 1 | 116 ± 1 |
| Diastolic blood pressure (mmHg) | 74 ± 1 | 69 ± 1 |
| Glucose (mg/dL) | 90 ± 1 | 87 ± 1 |
| Total cholesterol (mg/dL) | 189 ± 2 | 200 ± 2 |
| LDL-C (mg/dL) | 116 ± 2 | 116 ± 2 |
| HDL-C (mg/dL) | 50 ± 1 | 66 ± 1 |
| eGFR (mL/min/1.73 m2) | 78 ± 1 | 75 ± 1 |
| Max heart rate (bpm) | 173 ± 1 | 168 ± 1 |
| VO2max (mL/kg/min) | 37.9 ± 0.5 | 30.7 ± 0.5 |
Data are mean ± SEM. LDL-C, low-density lipoprotein cholesterol; VLDL-C, very low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; eGFR, estimated glomerular filtration rate; VO2max, maximal oxygen consumption
Dietary analysis
Each participant was instructed to complete a 3-day food record (two weekdays and one weekend day) at each time point. Participants were provided with verbal and written instructions for reporting food intake including providing accurate descriptions of the foods consumed as well as estimated portion sizes. Upon completion, food records were checked for accuracy. Food Processor SQL Nutrition and Fitness Program (ESHA Research, Salem, OR, USA) was used for analysis. Percent calories from fat, protein, and carbohydrates were calculated for each individual, and differences in the distribution of these dietary variables were assessed using paired t tests and deemed significant at P < 0.05.
Metabolomics analysis
Sample preparation
Plasma was isolated from subjects and stored at − 80 °C until analysis. Prior to LC-MS analysis, samples were diluted 1:10 (v/v) with methanol:acetonitrile:water (5:3:2, v:v). Suspensions were vortexed continuously for 30 min at 4 °C. Insoluble material was removed by centrifugation at 10,000g for 10 min at 4 °C, and supernatants were isolated for metabolomics analysis by UHPLC-MS.
UHPLC-MS analysis
Analyses were performed as previously published (Nemkov et al. 2017). Briefly, the analytical platform employs a Vanquish UHPLC system (Thermo Fisher Scientific, San Jose, CA, USA) coupled online to a Q Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). Plasma extracts (10 μL) were resolved over a Kinetex C18 column, 2.1 × 150 mm, 1.7-μm particle size (Phenomenex, Torrance, CA, USA) equipped with a guard column (SecurityGuard™ Ultracartridge—UHPLC C18 for 2.1 mm ID Columns—AJO-8782—Phenomenex, Torrance, CA, USA) using an aqueous phase (A) of water and 0.1% formic acid and a mobile phase (B) of acetonitrile and 0.1% formic acid. Samples were eluted from the column using either an isocratic elution of 5% B flowed at 250 μL/min and 25 °C or a gradient from 5 to 95% B over 1 min, followed by an isocratic hold at 95% B for 2 min, flowed at 400 μL/min and 30 °C. The Q Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) was operated independently in positive or negative ion mode, scanning in Full MS mode (2 μscans) from 60 to 900 m/z at 70,000 resolution, with 4-kV spray voltage, 15 sheath gas, and 5 auxiliary gas. Calibration was performed prior to analysis using the Pierce™ Positive and Negative Ion Calibration Solutions (Thermo Fisher Scientific). Acquired data was then converted from .raw to .mzXML file format using Mass Matrix (Cleveland, OH, USA). Samples were analyzed in randomized order with a technical mixture injected after every 15 samples to qualify instrument performance. Metabolite assignments, isotopologue distributions, and correction for expected natural abundances of deuterium, 13C, and 15N isotopes were performed using MAVEN (Princeton, NJ, USA) (Clasquin et al. 2012). Metabolic pathway analysis was performed using the MetaboAnalyst 3.0 package (www.metaboanalyst.com) (Xia and Wishart 2016).
Statistics
The relation between chronological age and biological age in our initial training cohort was established using Pearson Correlation analysis. Additional comparisons between measures made at baseline and follow-up in the longitudinal cohort were performed using paired t tests and deemed significant at P < 0.05. For metabolomics analysis, features between the two time points in the longitudinal cohort with raw p values < 0.05 resulting from a two-tailed t test and a false-discovery rate (FDR) < 0.1 were classified as significant. To identify metabolites related with rate of aging, differences in metabolite concentrations were calculated and associated with rate of biological aging while including chronological age as a covariate. Subsequently independent linear regressions were employed to understand the relation between metabolite abundances, future rate of aging, and biological age while using percent calories from total fat, protein, and carbohydrate as covariates. Because three individuals did not complete all necessary dietary records, they were excluded and 28 individuals were included in the regression analyses. Removal of 3 individuals with missing dietary values did not change overall subject characteristics (Table S1).
Results
Selection of healthspan indicators and calculation of biological age
Thirteen (13) clinical and physiological indicators of human healthspan were used to generate a model biological age (Table 1) using a similar approach as previously described (Klemera and Doubal 2006; Belsky et al. 2015). Briefly, select clinical and physiological measures were evaluated for their relation to chronological age using individual linear regressions and necessary components of these regressions were incorporated into a weighted equation to calculate biological age (see “Methods” for complete description) (Klemera and Doubal 2006). The selection of healthspan indicators was based on their availability in our datasets, their association with chronological age in an independent training cohort (Figure S1) and/or their relevance to age-related disease risk as reported in the literature (Calle et al. 1999). Importantly, to avoid biasing or overfitting our model of biological age, chronological age was not a selected variable. The training cohort consisted of 604 healthy adults (aged 18–80 years), who had previously undergone testing in our laboratory between 2003 and 2017. Importantly, all subjects were free of clinical disease and disability at the time of initial testing as confirmed by a medical history and physical examination, and all healthspan indicators were within normal healthy ranges, providing a unique opportunity to address the underlying mechanisms of primary aging without the confounding and uncontrolled impact that various disease states may impart upon an aging cohort (Table 1). Due to inherent sex-related differences of several biomarkers included in our model such as body composition and maximal aerobic capacity (Fleg et al. 2005; Wells 2007), sex-specific models of biological age were created from our training cohort allowing us to compare men and women on the same scale for all subsequent analyses.
Biological age was significantly correlated with chronological age (Fig. 1, R2 = 0.68, P < 0.0001; both sexes combined); however, variability among subjects of similar chronological age was observed and reflects the expected heterogeneity of biological aging. To explore this heterogeneity further, we calculated each individual’s “ageDiff”—the mathematical difference (in years) between biological age and chronological age (Horvath 2013; Kim et al. 2017). A positive ageDiff implies that an individual is biologically older than their chronological age would suggest. In this regard, we observed a normal distribution of ageDiff across our training cohort (Fig. 2), suggesting that our model is sensitive to detect a wide range of differences in biological age.
Fig. 1.

Biological age and chronological age are significantly correlated in our training cohort of 604 healthy adults (R2 = 0.68, P value < 0.0001)
Fig. 2.
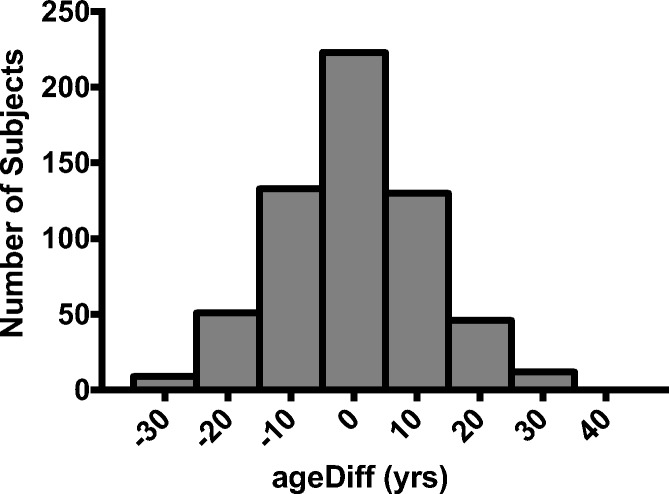
ageDiff was calculated, and although most individual’s biological age is within 5 years of their chronological age, many demonstrate greater differences between biological age and chronological age
Rate of biological aging in an independent longitudinal cohort
Using the sex-specific regression coefficients derived from our training models, we calculated biological age in a separate set of 31 healthy middle-aged and older adult men and postmenopausal women at two separate time points (Table 2). Subjects included individuals who had undergone testing in our laboratory between 5 and 10 years prior and agreed to return to the laboratory for follow-up assessments. All subjects were free of chronic disease or disability at the time of initial enrollment and remained mostly healthy over the time to follow-up, which averaged 8.6 years. Only late middle-aged and older adults were included in this longitudinal cohort to maximize our ability to detect changes in biological age within the relatively short follow-up period.
Table 2.
Subject characteristics of our longitudinal cohort
| Subject characteristics | Baseline | Follow-up |
|---|---|---|
| Sex (M/F) | 20/11 | |
| Age (years) | 59 ± 1 | 68 ± 1* |
| Body mass index (kg/m2) | 25 ± 1 | 25 ± 1 |
| Waist-to-hip ratio | 0.86 ± 0.02 | 0.86 ± 0.02 |
| Body fat (%) | 26 ± 2 | 27 ± 2 |
| Bone mineral density (g/cm2) | 1.23 ± 0.02 | 1.21 ± 0.02* |
| Systolic blood pressure (mmHg) | 119 ± 2 | 123 ± 2 |
| Diastolic blood pressure (mmHg) | 73 ± 2 | 73 ± 1 |
| Glucose (mg/dL) | 89 ± 2 | 85 ± 1* |
| Total cholesterol (mg/dL) | 204 ± 5 | 172 ± 5* |
| LDL-C (mg/dL) | 124 ± 4 | 100 ± 4* |
| HDL-C (mg/dL) | 57 ± 3 | 54 ± 3* |
| eGFR (mL/min/1.73 m2) | 71 ± 2 | 78 ± 3 |
| Max heart rate (bpm) | 168 ± 2 | 156 ± 3* |
| VO2max (mL/kg/min) | 36.1 ± 1.7 | 31.5 ± 1.5* |
Data are mean ± SEM. LDL-C, low-density lipoprotein cholesterol; VLDL-C, very low-density lipoprotein cholesterol; HDL-C, high-density lipoprotein cholesterol; eGFR, estimated glomerular filtration rate; VO2max, maximal oxygen consumption. *P < 0.05 vs. baseline
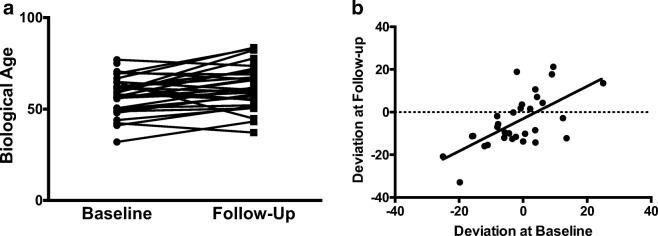
Overall, a significant increase in mean biological age was calculated in our longitudinal cohort from baseline to follow-up, which were free of any differences associated with sex, demonstrating the balance of our sex-specific algorithms. A total increase of approximately six biological aging years was observed, suggesting that as a group, the longitudinal cohort aged slower than expected over the time to follow-up (Fig. 3a). This may have been due in part to a slight decrease in LDL and total cholesterol (Table 2), which was most likely due to initiation of cholesterol-lowering medications in several of the subjects. Interestingly, ageDiff at baseline was significantly associated with ageDiff at follow-up, suggesting that individuals who were biologically older than their chronological age at baseline were likely to remain relatively older than their chronological age at follow-up, and vice versa (Fig. 3b).
Fig. 3.
a Change in biological age, calculated from clinical and physiological measures, in our longitudinal cohort. Although the change in biological age is significant (P < 0.01), the trajectories of aging are highly variable. b Significant relation between ageDiff at baseline and follow-up (P < 0.001)
Finally, to determine the rate of biological aging over the follow-up period, we normalized changes in biological age to changes in chronological age. This resulted in a ratio in which a value above or below one (1) is indicative of faster vs. slower biological aging, respectively. We observed a continuous distribution in the rate of biological aging across our longitudinal cohort, with approximately half of all subjects exhibiting faster biological aging and other half exhibiting slower biological aging (Fig. 4a).
Fig. 4.

a Rate of biological aging for each individual in the longitudinal cohort. A ratio smaller than one indicates slower aging, while a ratio greater than one indicates faster aging. b Heatmap of the change in abundance of 28 metabolites significantly associated with rate of biological aging. Individuals are aligned in columns, metabolites in rows. (See Table S2 for list of metabolites and associated pathways).
The plasma metabolome as a predictor of biological aging
Because the determination of biological age based off of clinical and physiological indicators is time-consuming and technically challenging, we sought to identify potential circulating metabolites that may serve as novel biomarkers of biological aging. To do this we measured the abundance of 360 individual metabolites in the plasma of our longitudinal subjects at both time points using ultra-high pressure liquid chromatography and mass spectrometry (UHPLC-MS). To account for any transient changes in the metabolome within an individual due to modifications to dietary consumption, macronutrient compositions were included as covariates in our baseline statistical models. The percent of each macronutrient in the diet showed no differences from baseline to follow-up, suggesting stability in diet over time. Therefore, dietary intakes were not included in the longitudinal analyses.
In total, eighty-one (81) metabolites were significantly altered from baseline to follow-up in the longitudinal cohort, confirming that age-related changes are detectable in the plasma metabolome over a relatively short period of time (Figure S2). To determine if any of these changes were also indicative of biological aging, we compared changes in the metabolome (normalized to follow-up time) to the rate of biological aging and identified 28 metabolites that were significantly associated with the rate of biological aging (Fig. 4b) and Table S2. Interestingly, more robust changes in metabolite abundance in either direction, as indicated by a higher or lower Z-score, appeared to occur in those individuals who exhibited the fastest or slowest biological aging, respectively (Fig. 4a). The metabolites most associated with the rate of biological aging included amino acid, fatty acid, acylcarnitine, sphingolipid, and nucleotide metabolites.
Although determining an individual’s actual rate of biological aging holds clinical value (e.g., precision medicine), it is technically challenging and requires repeat assessments of multiple clinical and physiological parameters, making it difficult to implement into clinical practice. Therefore, our ultimate goals were to (a) identify baseline metabolomic signatures indicative of one’s biological age at a single time point and (b) to predict one’s future rate of biological aging (i.e., if someone is at risk for faster vs. slower aging).
Metabolites associated with biological age
To understand if the plasma metabolome is reflective of one’s biological age, we explored whether metabolite abundances measured at baseline are associated with an individual’s biological age at the same time point, while controlling for chronological age and dietary macronutrient content in the analysis. Three individuals did not complete all necessary dietary records and were therefore excluded from this regression analysis due to missing values, although the elimination of these subjects did not change group characteristics (Table S1). In total, 21 metabolites were related to biological age, 16 of which were endogenous or secondary metabolites of microbe metabolism (Table 3). Greater abundances of metabolites associated with folate (6-lactoyl-5-6-7-8 tetrahydropterin), fatty acyl (phaseolic acid), ethanolamine (anandamide), and carboxylic acid (l-homocitrulline) metabolism were observed in individuals who were biologically older. Alternatively, greater concentrations of metabolites from carnitine and fatty acid metabolism (10-hydroxydecanoic acid, O-dodecanoyl-carnitine, tetradecenoyl carnitine, O-decanoyl-l-carnitine, acyl-C18:2-OH), TCA cycle (citrate), polyamine (putrescine), inositol (inositol 1-2-3-5-6-pentakisphosphate), sterol (pregna-4,9(11)-diene-3,20-dione), serine (phosphoserine), indole (indole-3-acetate), and ubiquinone (4-hydroxybenzoate) pathways were associated with a lower biological age.
Table 3.
Association of metabolite concentrations with biological age at baseline. Non-endogenous are metabolites notated in italic text
| Metabolite | Estimate | Error | P value |
|---|---|---|---|
| Greater values indicative of greater biological age | |||
| 6-Lactoyl-5-6-7-8-tetrahydropterin | 9.45E-06 | 4.07E-06 | 0.03 |
| Phaseolic acid | 1.23E-05 | 5.23E-06 | 0.03 |
| Anandamide | 4.56E-04 | 1.82E-04 | 0.02 |
| l-Homocitrulline | 2.61E-03 | 1.07E-03 | 0.02 |
| 4-Nitroaniline | 2.63E-06 | 8.61E-07 | 0.006 |
| Felbamate | 4.40E-06 | 1.82E-06 | 0.02 |
| Greater values indicative of lower biological age | |||
| 10-Hydroxydecanoic acid | − 1.17E-04 | 4.35E-05 | 0.01 |
| Inositol 1-2-3-5-6-pentakisphosphate | − 3.95E-04 | 1.52E-04 | 0.02 |
| Citrate | − 1.27E-06 | 5.15E-07 | 0.02 |
| Acyl-C18:2-OH | − 3.00E-04 | 1.22E-04 | 0.02 |
| Phosphoserine | − 1.57E-05 | 6.56E-06 | 0.03 |
| 1stgna-4-9(11)-diene-3-20-dione | − 1.54E-04 | 6.56E-05 | 0.03 |
| Indole-3-acetate | − 6.64E-06 | 2.87E-06 | 0.03 |
| 4-Hydroxybenzoate | − 1.26E-07 | 5.56E-08 | 0.03 |
| Tetradecenoyl carnitine | − 5.57E-06 | 2.47E-06 | 0.03 |
| O-Dodecanoyl-carnitine | − 5.90E-06 | 2.69E-06 | 0.04 |
| O-Decanoyl-l-carnitine | − 3.40E-06 | 1.60E-06 | 0.045 |
| Putrescine | − 7.86E-04 | 3.76E-04 | 0.048 |
| 6-Thioxanthine 5-monophosphate | -5.87E-05 | 1.88E-05 | 0.004 |
| Theogallin | -5.53E-06 | 1.67E-07 | 0.003 |
| Pantetheine | -5.80E-06 | 2.21E-06 | 0.02 |
Predicting faster vs. slower aging
Finally, after determining that the plasma metabolome is associated with biological age, we sought to determine if the concentrations of specific metabolites among individuals at baseline could predict whether an individual would experience faster or slower biological aging over the time to follow-up. While controlling for age and diet, twelve (12) metabolites measured at baseline predicted the rate of biological aging in our longitudinal cohort (Table 4). Of these, all four metabolites associated with faster aging were linked to fatty acid/TCA cycle metabolism (acyl-C5-OH, oxaloacetate, N-(heptadecanoyl)-ethanolamine, and oxalosuccinate), whereas eight metabolites related to glycolysis (2-phospho-d-glycerate), nucleotide (phosphate and 3′5′-cyclic IMP), glutathione (ascorbate), caffeine metabolism (5-acetylamino-6-formylamino-3-methyluracil), amino acid metabolism (selenohomocystine and pantetheine), and one exogenous metabolite (octylamine) were positively associated with a slower aging phenotype.
Table 4.
Metabolite concentrations at baseline are significantly associated with future faster or slower rate of aging. Non-endogenous metabolites are notated in italic text
| Metabolite | Estimate | Error | P value |
|---|---|---|---|
| Greater values indicative of future faster aging | |||
| Acyl-C5-OH | 9.47E-06 | 3.53E-06 | 0.01 |
| Oxaloacetate | 8.81E-06 | 3.38E-06 | 0.02 |
| N-(Heptadecanoyl)-ethanolamine | 9.74E-05 | 4.48E-05 | 0.04 |
| Oxalosuccinate | 2.13E-06 | 1.02E-06 | 0.048 |
| Greater values indicative of future slower aging | |||
| 2-Phospho-d-glycerate | − 2.11E-05 | 7.35E-06 | 0.01 |
| Ascorbate | − 2.50E-06 | 1.04E-06 | 0.02 |
| Selenohomocystine | − 9.00E-07 | 3.80E-07 | 0.03 |
| Pantetheine | − 3.00E-07 | 1.29E-07 | 0.03 |
| Phosphate | − 1.02E-07 | 4.50E-08 | 0.03 |
| 5-Acetylamino-6-formylamino-3-methyluracil | − 4.41E-07 | 1.98E-07 | 0.04 |
| 3′,5′-Cyclic IMP | − 4.11E-07 | 1.93E-07 | 0.045 |
| Octylamine | − 2.72E-07 | 1.25E-07 | 0.04 |
Discussion
Despite a growing body of literature, there is little consensus regarding the most appropriate method for determining biological age in humans. One popular approach has been to measure the levels of circulating biomarkers that are strongly correlated with chronological age such as the calculation of DNA methylation (DNAm) age using age-related CpG methylation sites within circulating leukocytes (Hannum et al. 2013; Horvath 2013). Although this approach is convenient and there is some agreement among methylation sites, it has been demonstrated that the ability of DNAm age to predict mortality is lost after adjusting for chronological age (Kim et al. 2017), suggesting that these circulating biomarkers may be more related to the passage of time than a true indicator of human healthspan. Although some analyses have associated molecular markers, such as DNAm age, with physiological measures, this approach only stands to validate against individual physiological risk factors instead of capturing changes in physiological function (Peters et al. 2015). An alternative approach is to define biological age using clinical and physiological parameters that are closely associated with age-related disease risk. Such models predict mortality better than those based only on chronological age-related molecular markers (Levine 2013; Belsky et al. 2015; Kim et al. 2017), but are more costly and time-consuming and require specialized training and equipment to measure, thus limiting their practicality in primary care or other clinical settings.
In the present study, we developed a hybrid approach in which we first developed a model of biological aging using clinical and physiological parameters indicative of risk for age-related disease and disability and then identified sets of circulating metabolites that are associated with, and/or predictive of biological age, thereby merging clinically relevant parameters with the simplicity of a blood sample. In general, we identified several metabolites associated with both baseline biological age and the future rate of aging that are known to be involved in the regulation of energy homeostasis (e.g., fatty acid metabolism). Cellular energy metabolism is tightly controlled, and its dysregulation has been implicated as a central mechanism, or “hallmark” of age-associated physiological declines (Finkel 2015; López-Otín et al. 2016); thus, our study may offer new indicators of aging trajectories that can suggest which individuals might be best targeted for early intervention. In our model, higher levels of carnitines (tetradecenoyl carnitine, O-dodecanoyl-carnitine, O-decanoyl-l-carnitine), necessary for fatty acid metabolism, were associated with lower biological ages at baseline. Elevated levels of carnitine metabolites may be indicative of increased capacity for fatty acid utilization, as reduced carnitine levels with age have been demonstrated to reduce mitochondrial performance (Noland et al. 2009) and support the association between biological aging and mitochondrial dysfunction. Additionally, lower biological age was also associated with higher levels of the TCA cycle intermediate citrate, indicating a greater capacity of the TCA cycle to produce ATP. Together, these findings indicate that lower biological age is associated with the potential for greater metabolic flux.
Metabolites associated with energy metabolism were also related to an individual’s future rate of biological aging. Elevated 2-phospho-d-glycerate (glycolysis) was associated with a slower rate of future aging, whereas higher concentrations of acyl-C5-OH, N-(heptadecanoyl)-ethanolamine, oxaloacetate, and oxalosuccinate were associated with a faster rate of biological aging in the future. Oxaloacetate and oxalosuccinate are key components of the citric acid cycle and indicate potential disruptions in this energy pathway may contribute to a pro-biological aging phenotype. Oxaloacetate is converted to citrate by the enzyme citrate synthase, while oxalosuccinate is converted from isocitrate to α-ketoglutarate. Considering that we observed higher levels of citrate in those with lower baseline biological age, these findings may suggest that diminished citrate synthase activity (resulting in an assumulation of oxaloacetate and oxalosuccinate accompanied by a decrease in citrate levels) could play a key role in the biological aging process. Indeed, citrate synthase activity has been shown to decline in human skeletal muscle with aging (Lanza et al. 2008), an effect that is protected by endurance exercise training, a known modulator of human healthspan.
Our analysis also highlighted amino acid metabolism in addition to energy homeostasis pathways. Several amino acids were also associated with an increased rate of biological aging, such as l-cysteine, l-lysine, glutamine, cystine, and l-methionine, further implicating changes in amino acid metabolism with aging (Houtkooper et al. 2011). Particularly, the positive association of circulating l-methionine levels and a faster rate of biological aging is intriguing, as previous findings have demonstrated that l-methionine restriction is associated with preserved physiological function in animal models across the lifespan (Sun et al. 2009; Hasek et al. 2010).
This analysis demonstrates that circulating plasma metabolomic profiles are associated with the rate at which changes in biological age occur, which is important in determining the molecular underpinnings of aging. Our novel set of blood-based markers of biological aging have potential applications in both biomedical research and clinical practice. In research settings, this approach provides an avenue to more efficiently test and validate novel interventions designed to target the basic mechanisms of human aging. Such trials will take years before their success can be confirmed; however, the concept of blood-based markers of biological age that could provide intermediate insight into the potential of an intervention to slow aging would be of great value considering the projected increase in the number of older adults over the next several decades.
Our unique blood-based markers of biological age could also be used in the primary care setting to more efficiently and cost-effectively track disease risk over time, and possibly screen candidates for more labor-intensive follow-up testing. For example, low cardiorespiratory fitness (usually assessed by measuring VO2max) is a known risk factor for cardiovascular and all-cause mortality in older adults (Blair et al. 1989; Laukkanen et al. 2004; Sui et al. 2007); however, it is not regularly assessed in the primary care setting due to costs and logistical challenges (Ross et al. 2016). Although non-exercise, regression-based prediction models of cardiorespiratory fitness currently exist (Jackson et al. 1990; Bradshaw et al. 2005; Cao et al. 2010; Nes et al. 2011), these models are typically based off of basic characteristics such as age, sex, and body composition (Ross et al. 2016) and may not capture more subtle differences among healthy, asymptomatic older adults. Because the plasma metabolome is thought to more closely reflect changes in physiological function (Soltow et al. 2010; Barallobre-Barreiro et al. 2013), our unique signatures of biological aging may be more capable of identifying subclinical changes indicative of worsening function (such as low cardiorespiratory fitness). While our approach is not meant to serve as a replacement for important clinical measures such as VO2max, the early and more frequent assessment of these novel biomarkers may trigger earlier follow-up testing and implementation of preventive measures.
In summary, our study identified unique metabolomic signatures that are associated with the rate of human biological aging and may be useful for the rapid detection of older adults at risk for physiological dysfunction. Although further analyses need to be performed in diverse patient populations to confirm and extend our observations, the present model of biological aging and subsequent molecular analyses has established an approach to investigate the molecular foundation of biological aging in humans.
Electronic supplementary material
(DOCX 1176 kb)
(DOCX 15.4 kb)
Author contributions
L.C.J. and C.R.M. conceived and developed the overall study design and collected data used in the biological aging model. K.P. established the mathematical models used to quantify biological age. K.P. and B.F.A. developed and packaged R scripts to execute the biological age algorithm. T.G.N. and A.D. conducted metabolomics analysis and contributed technical support. S.J. conducted dietary analyses. L.C.J. performed statistical analysis. L.C.J., C.R.M., and D.R.S. contributed critical input towards study design and manuscript development. All authors edited and approved the final version.
Funding information
This work was supported by National Institutes of Health awards T32 AG00027912 and Colorado CTSA UL1TR001082.
Compliance with ethical standards
All study procedures were reviewed and approved by the University of Colorado Boulder Institutional Review Board. Clinical and physiological measurements were performed at the University of Colorado Boulder Clinical Translational Research Center (CTRC). All study participants provided written informed consent after the nature, benefits, and risks of the study were explained.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Barallobre-Barreiro J, Chung Y-L, Mayr M. Proteomics and metabolomics for mechanistic insights and biomarker discovery in cardiovascular disease. Rev Esp Cardiol (Engl Ed) 2013;66:657–661. doi: 10.1016/j.recesp.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Belsky DW, Caspi A, Houts R, Cohen HJ, Corcoran DL, Danese A, Harrington H, Israel S, Levine ME, Schaefer JD, Sugden K, Williams B, Yashin AI, Poulton R, Moffitt TE. Quantification of biological aging in young adults. Proc Natl Acad Sci U S A. 2015;112:E4104–E4110. doi: 10.1073/pnas.1506264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair SN, Kohl H, Paffenbarger RS, Clark DG, Cooper KH, Gibbons LW. Physical fitness and all-cause mortality. JAMA. 1989;262:2395–2401. doi: 10.1001/jama.1989.03430170057028. [DOI] [PubMed] [Google Scholar]
- Bradshaw DI, George JD, Hyde A, LaMonte MJ, Vehrs PR, Hager RL, Yanowitz FG. An accurate VO2max nonexercise regression model for 18–65-year-old adults. Res Q Exerc Sport. 2005;76:426–432. doi: 10.1080/02701367.2005.10599315. [DOI] [PubMed] [Google Scholar]
- Brooks-Wilson AR. Genetics of healthy aging and longevity. Hum Genet. 2013;132:1323–1338. doi: 10.1007/s00439-013-1342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE, Thun MJ, Petrelli JM, Rodriguez C, Heath CW., Jr Body-mass index and mortality in a prospective cohort of U.S. adults. N Engl J Med. 1999;341:1097–1105. doi: 10.1056/NEJM199910073411501. [DOI] [PubMed] [Google Scholar]
- Cao Z-B, Miyatake N, Higuchi M, Miyachi M, Ishikawa-Takata K, Tabata I. Predicting VO2max with an objectively measured physical activity in Japanese women. Med Sci Sports Exerc. 2010;42:179–186. doi: 10.1249/MSS.0b013e3181af238d. [DOI] [PubMed] [Google Scholar]
- Cho IH, Park KS, Lim CJ. An empirical comparative study on biological age estimation algorithms with an application of Work Ability Index (WAI) Mech Ageing Dev. 2010;131:69–78. doi: 10.1016/j.mad.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Clasquin MF, Melamud E, Rabinowitz JD (2012) LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics. Chapter 14, Unit14 11 [DOI] [PMC free article] [PubMed]
- Finkel T. The metabolic regulation of aging. Nat Med. 2015;21:1416–1423. doi: 10.1038/nm.3998. [DOI] [PubMed] [Google Scholar]
- Fleg JL, Morrell CH, Bos AG, Brant LJ, Talbot LA, Wright JG, Lakatta EG. Accelerated longitudinal decline of aerobic capacity in healthy older adults. Circulation. 2005;112:674–682. doi: 10.1161/CIRCULATIONAHA.105.545459. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasek BE, Stewart LK, Henagan TM, Boudreau A, Lenard NR, Black C, Shin J, Huypens P, Malloy VL, Plaisance EP, Krajcik RA, Orentreich N, Gettys TW. Dietary methionine restriction enhances metabolic flexibility and increases uncoupled respiration in both fed and fasted states. Am J Phys Regul Integr Comp Phys. 2010;299:R728–R739. doi: 10.1152/ajpregu.00837.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Argmann C, Houten SM, Canto C, Jeninga EH, Andreux PA, Thomas C, Doenlen R, Schoonjans K, Auwerx J. The metabolic footprint of aging in mice. Sci Rep. 2011;1:134. doi: 10.1038/srep00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AS, Blair SN, Mahar MT, Wier LT, Ross RM, Stuteville JE. Prediction of functional aerobic capacity without exercise testing. Med Sci Sports Exerc. 1990;22:863–870. doi: 10.1249/00005768-199012000-00021. [DOI] [PubMed] [Google Scholar]
- Jiang T, Yu J-T, Tian Y, Tan L. Epidemiology and etiology of Alzheimer’s disease: from genetic to non-genetic factors. Curr Alzheimer Res. 2013;10:852–867. doi: 10.2174/15672050113109990155. [DOI] [PubMed] [Google Scholar]
- Johnson LC, Martens CR, Santos-Parker JR, Bassett CJ, Strahler TR, Cruickshank-Quinn C, Reisdorph N, McQueen MB, Seals DR. Amino acid and lipid associated plasma metabolomic patterns are related to healthspan indicators with ageing. Clin Sci. 2018;132:1765–1777. doi: 10.1042/CS20180409. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss-Coray T, Sierra F. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Myers L, Wyckoff J, Cherry KE, Jazwinski SM. The frailty index outperforms DNA methylation age and its derivatives as an indicator of biological age. Geroscience. 2017;39:83–92. doi: 10.1007/s11357-017-9960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemera P, Doubal S. A new approach to the concept and computation of biological age. Mech Ageing Dev. 2006;127:240–248. doi: 10.1016/j.mad.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–2942. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukkanen JA, Kurl S, Salonen R, Rauramaa R, Salonen JT. The predictive value of cardiorespiratory fitness for cardiovascular events in men with various risk profiles: a prospective population-based cohort study. Eur Heart J. 2004;25:1428–1437. doi: 10.1016/j.ehj.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA, Milburn MV. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- Levine ME. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J Gerontol A Biol Sci Med Sci. 2013;68:667–674. doi: 10.1093/gerona/gls233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Galluzzi L, Freije JM, Madeo F, Kroemer G. Metabolic control of longevity. Cell. 2016;166:802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- Lunenfeld B, Stratton P. The clinical consequences of an ageing world and preventive strategies. Best Pract Res Clin Obstet Gynaecol. 2013;27:643–659. doi: 10.1016/j.bpobgyn.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapstone M, Cheema AK, Fiandaca MS, Zhong X, Mhyre TR, MacArthur LH, Hall WJ, Fisher SG, Peterson DR, Haley JM (2014) Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med [DOI] [PMC free article] [PubMed]
- Mitnitski A, Song X, Rockwood K. Assessing biological aging: the origin of deficit accumulation. Biogerontology. 2013;14:709–717. doi: 10.1007/s10522-013-9446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura E. A study on the basic nature of human biological aging processes based upon a hierarchical factor solution of the age-related physiological variables. Mech Ageing Dev. 1991;60:153–170. doi: 10.1016/0047-6374(91)90128-M. [DOI] [PubMed] [Google Scholar]
- Nemkov T, Hansen KC, D’Alessandro A. A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun Mass Spectrom. 2017;31:663–673. doi: 10.1002/rcm.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes BM, Janszky I, Vatten LJ, Nilsen T, Aspenes ST, Wisloff U. Estimating VO2 peak from a nonexercise prediction model: the HUNT Study, Norway. Med Sci Sports Exerc. 2011;43:2024–2030. doi: 10.1249/MSS.0b013e31821d3f6f. [DOI] [PubMed] [Google Scholar]
- Noland RC, Koves TR, Seiler SE, Lum H, Lust RM, Ilkayeva O, Stevens RD, Hegardt FG, Muoio DM. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J Biol Chem. 2009;284:22840–22852. doi: 10.1074/jbc.M109.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R, Blair SN, Arena R, Church TS, Despres JP, Franklin BA, Haskell WL, Kaminsky LA, Levine BD, Lavie CJ, Myers J, Niebauer J, Sallis R, Sawada SS, Sui X, Wisloff U, American Heart Association Physical Activity Committee of the Council on L, Cardiometabolic H, Council on Clinical C, Council on E, Prevention, Council on C, Stroke N, Council on Functional G, Translational B, Stroke C Importance of assessing cardiorespiratory fitness in clinical practice: a case for fitness as a clinical vital sign: a scientific statement from the American Heart Association. Circulation. 2016;134:e653–e699. doi: 10.1161/CIR.0000000000000461. [DOI] [PubMed] [Google Scholar]
- Sebastiani P, Thyagarajan B, Sun F, Schupf N, Newman AB, Montano M, Perls TT. Biomarker signatures of aging. Aging Cell. 2017;16:329–338. doi: 10.1111/acel.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltow QA, Jones DP, Promislow DE. A network perspective on metabolism and aging. Integr Comp Biol. 2010;50:844–854. doi: 10.1093/icb/icq094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, LaMonte MJ, Blair SN. Cardiorespiratory fitness as a predictor of nonfatal cardiovascular events in asymptomatic women and men. Am J Epidemiol. 2007;165:1413–1423. doi: 10.1093/aje/kwm031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Sadighi Akha AA, Miller RA, Harper JM. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J Gerontol A Biol Sci Med Sci. 2009;64:711–722. doi: 10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JC. Sexual dimorphism of body composition. Best Pract Res Clin Endocrinol Metab. 2007;21:415–430. doi: 10.1016/j.beem.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics. 2016;55(14):10.1–14.10.91. doi: 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1176 kb)
(DOCX 15.4 kb)