Abstract
Age-related changes in human gut microbiota composition have been reported, and such changes might be influenced by the intake of nutrients or diets. To investigate the effects of aging on the gut microbiota independent of nutrient effects, we analyzed the gut microbiomes of 126 micro-pigs at a wide range of ages from newborns to 10 years old. The micro-pigs were reared in a constantly controlled environment. The diversity of the gut microbiome was found to continuously change with age. We also found associations between age and specific members and functions of the gut microbiome. Consistent with previous studies on the human gut microbiome, beneficial microbes including probiotic bacteria and short-chain fatty acid-producers decreased in older pigs, whereas Bacteroides increased with age. Based on the correlation network, Bacteroides seemed to have an important role in determining the relative abundances of other beneficial microbes. Our results suggest that maintaining beneficial gut microbes at a specific ratio corresponding to a certain age might contribute to a younger gut microbiome-age. Furthermore, due to similarities with the human system, micro-pigs are a useful animal model to elucidate the links between aging and the microbiome.
Electronic supplementary material
The online version of this article (10.1007/s11357-019-00121-y) contains supplementary material, which is available to authorized users.
Keywords: Micro-pig, Aging, Gut microbiome, 16S sequencing
Introduction
Over the past few decades, life expectancy around the world has increased dramatically. The challenge now is to extend not only lifespan but also health span and to reduce the burden of age-related chronic diseases. Human health span is known to be determined by complex interactions among biological processes such as metabolism and inflammation (Kennedy et al. 2014), which are under the influence of both genetic and environmental factors. Recently, the gut microbiota was recognized as an important environmental factor responsible for the development of metabolic and immune diseases (Clemente et al. 2012).
Microorganisms colonize the mammalian intestine immediately after birth. In humans, the adult-like configuration of the gut microbial composition is established over the first 3 years of life (Yatsunenko et al. 2012), and the gut microbiota remains largely stable thereafter (Faith et al. 2013; Lim et al. 2014), although it is influenced by various environmental factors including diet, lifestyle, and medication (Sommer and Backhed 2013). A recent study conducted on human subjects with a wide age range, specifically 0–104 years, showed that the gut microbiota composition changes sequentially with age, and that nutrients may play a key role in such changes (Odamaki et al. 2016). To gain more insights into the effects of aging on the gut microbiota independent of the effects of nutrients, it is important to characterize the gut microbiomes of animal models fed a consistent diet at each life stage.
Pigs are considered a primary model animal for studies on a wide range of human physiological functions and diseases, because of their similarity in size and physiology, as well as in organ development and disease progression (Lunney 2007). In a study comparing the gut microbiome gene catalogues of humans, mice, and pigs, it was shown that pig and human gut microbiomes are more similar to each other than to those of the mouse (Xiao et al. 2016). In addition, 96% of the functional pathways found in the human gut microbiome catalogue were also present in the pig gut microbiome catalogue, supporting the potential use of pigs for gut microbiota research.
Several studies have demonstrated the composition of the pig gut microbiome at a specific stage of life including early (Mach et al. 2015), growing (Metzler-Zebeli et al. 2015), and later stages (Pajarillo et al. 2015) or at multiple stages of life (Kim et al. 2015; Lu et al. 2018; Zhao et al. 2015). However, most of the previous studies were limited by small sample sizes or the fact that pigs tested at the later stage were under 1 year of age. Considering the average lifespan of domestic pigs is more than 10 years, a wider age range of pigs should be examined to better understand the characteristics of the gut microbiome in old-age pigs as a model for humans. Here, we investigated the micro-pig gut microbiomes using 126 fecal samples of micro-pigs ranging in age from newborn to 10 years and analyzed them for diversity, composition, and function.
Methods
Animals
The micro-pigs included in this study originated from crosses of miniatures (both naturally occurring and commercially-bred) and native pigs including Asian and South-East Asian native pigs, Yucatan, Pot-bellied Vietnamese, Meishan, and Pygmy pigs, which were then backcrossed with domestic swine breeds (White breeds). A total of 126 healthy micro-pigs of various ages (0 to 123 months) were randomly selected from both specific pathogen free (SPF) and non-SPF animal resource facilities (Supplemental Fig. S1). Among them, 25 micro-pigs aged 0 to 13 days were born by hysterectomy and were reared in an SPF zone, which was controlled by conducting regular screening for several viral, bacterial, parasitic, and fungal infections. They were fed with sterilized special milk replacer formula at approximately 2–3% of the body weight per day. The remaining 101 micro-pigs were born vaginally and were of 20 days to 123 months of age. They were maintained in specialized conventional housing and on PVC panel pens with steel bar flooring. They were maintained usually with their litter groups/mates to allow normal social interactions, except for breeding boars and pregnant sows nearing their terms. The conventional animal building was equipped with electronic devices for environmental control. These allowed the animal rooms (each equipped with a sensor probe) to be maintained at a specified target temperature (18–28 °C) or humidity (50 ± 10%) by controlling the operation of ventilation and heating equipment. The pigs reared in the conventional facility were fed based on life stage cycle/phase following NRC Nutrient Requirements for Swine (National Research Council 2012) as follows: pre-starter, pre-starter milk-based product with supplement for nursing stage/sucklings or 2–4-week-old piglets; starter, starter feed for pigs from as early as the 3rd week up to the 6th week of age; grower, grower mash in gradual transition for pigs from the 6th week to approximately the 12th week of age; breeder or finisher, breeder/adult rations for pigs from the 3rd month onwards, including mature/breeder micro-pigs, at a rate of up to 0.5 kg per head per day.
Sample collection
Fecal sampling was conducted in the morning before feeding. Each subject was restrained by manual handling or using a special restraining suspension cart to conduct fecal sampling via the anal/rectal access. If possible, each animal was briefly allowed to preliminarily excrete feces while restrained. Using sterile cotton swab sticks dipped in sterile normal saline solution, the animal’s anal canal was entered to the proximal portion of the rectum until some fresh fecal samples clung onto the cotton swab. Each cotton swab with adhering fecal sample was then placed in a sterile 50-mL conical tube with approximately 10–20 mL normal saline solution. Each tube was labeled accordingly, sealed using parafilm, and frozen at − 20 °C until microbiological analysis.
DNA extraction
DNA was extracted from the swabbed fecal samples using the QIAamp DNA Stool Mini Kit (Qiagen, Inc., Valencia, CA, USA) with the following modifications: each fecal swab in PBS was vigorously vortexed, and the bacterial cells were pelleted by centrifugation at 8000 rpm for 20 min at 4 °C. The swab was then removed, and the pellets were re-suspended in 500 μL ASL buffer. The suspension was transferred to a 2-mL tube containing 0.3 g of sterile 0.1-mm zirconia beads (BioSpec Products, Bartlesville, OK, USA) and homogenized with an additional 700 μL ASL buffer by vortexing for 2 min. The samples were then heated at 95 °C for 15 min, mechanically lysed twice by bead-beating for 1 min each, and centrifuged. The supernatant was transferred to a new tube. The remaining steps were performed according to the manufacturer’s instructions. The extracted DNA samples were stored at − 20 °C until use.
16S rRNA gene sequencing and analysis
The library preparation for the V3-V4 region of the 16S rRNA gene was conducted following the 16S Metagenomic Sequencing Library Preparation Illumina protocol (Part no. 15044223 Rev. B, Illumina, CA, USA). Sequencing was performed on an Illumina MiSeq platform using a 2 × 300 bp MiSeq Reagent Kit v3 (Illumina, San Diego, CA, USA). After quality filtering the raw sequences with Trimmomatic (Bolger et al. 2014), paired-end reads were assembled using PEAR (Zhang et al. 2014). Further removal of sequences shorter than 438 bp or longer than 469 bp was conducted to exclude inadequately paired reads from the downstream analysis. The sequences were further processed using QIIME version 1.9.0 (Caporaso et al. 2010). Chimeric sequences were eliminated using USEARCH and the non-chimeric sequences were clustered into operational taxonomic units (OTUs) at 97% identity using QIIME’s pick_closed_reference_otus.py. OTUs were classified using the Greengenes 13_8 reference database and grouped at different levels of classification. The microbial diversity of each sample was estimated by calculating Shannon, Chao 1, and observed OTUs. The distances between samples were calculated based on Bray-Curtis dissimilarity, and principle coordinate analysis (PCoA) was performed using the distance matrix. Functional abundances from 16S rRNA gene sequences were predicted with PICRUSt on the Galaxy platform (Langille et al. 2013).
Human and mouse gut microbiome dataset
To compare human, mouse, and micro-pig gut microbiomes, 16S rRNA sequence data of humans (Odamaki et al. 2016) and mice (Thevaranjan et al. 2017) were retrieved from the public databases. The human dataset consisted of 16S rRNA sequence data from the fecal samples of 367 healthy Japanese subjects between the ages of 0 and 104 years. In the mouse dataset, we only used 16S rRNA sequence data of 39 young wild-type SPF mice (10–14 weeks) and 39 aged wild-type SPF mice (18–22 months). Because the read lengths of the human and mouse datasets were different from those of our micro-pig dataset, we applied different filtering parameters to the human dataset (V3-V4, 399–426) and mouse dataset (V3, 174–199). The other steps were applied in the same way as those used for the micro-pig dataset.
Statistical analysis
Statistical analyses were carried out using R software and GraphPad Prism software packages. The Mann-Whitney test and Kruskal-Wallis test with Dunn’s correction were performed for comparisons of alpha diversity measurements. The strength and statistical significance of variables in determining variations in Bray-Curtis distances were determined using Adonis. Linear discriminant effect size analysis (LEfSe) was conducted to identify differentially-abundant taxa between sample groups (Segata et al. 2011). The association between taxonomic or functional abundances and age was determined through a multivariate algorithm while adjusting for sex with the MaAsLin package in R (Morgan et al. 2012).
The relationships among taxa (at the genus level) with higher than 0.5 prevalence were estimated by calculating the Spearman correlation. Networks of significantly correlated taxa were visualized using Cytoscape (Smoot et al. 2011). Pairs of nodes were linked by edges (correlation coefficients) only when their q values were < 0.05 and the coefficient values were > 0.5 or < − 0.5.
Results
Gut microbiome of hysterectomy-derived piglets
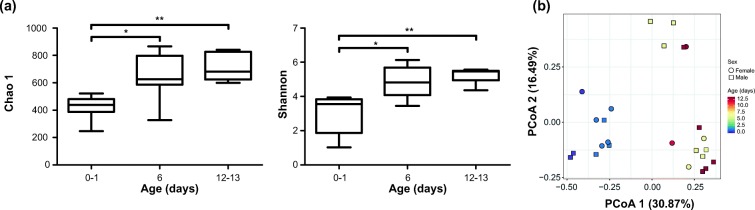
Among a total of 126 fecal samples from the micro-pigs, 25 were collected from hysterectomy-derived SPF piglets (from 0 to 13 days after birth), whereas the rest were from vaginally delivered pigs (from 20 days to 123 months after birth). Therefore, we first investigated the properties of early gut microbiomes using samples from the SPF piglets, separately from those of the vaginally delivered pigs. Regarding the alpha diversity, the Chao 1 and Shannon indexes increased with days of life (Fig. 1a). PCoA based on Bray–Curtis distance revealed that the overall gut microbial composition changed according to days of life, even before 2 weeks after birth (Fig. 1b). In particular, the gut microbiome of micro-pigs aged 0–1 days were clearly distinguished from those aged 6–13 days.
Fig. 1.
Gut microbial alpha and beta diversity of hysterectomy-derived specific pathogen-free (SPF) piglets from birth to 13 days of life. a Chao 1 (left) and Shannon (right) indexes. b Principle coordinate analysis (PCoA) based on Bray-Curtis distance (*p < 0.05; **p < 0.01; Kruskal-Wallis test with Dunn’s multiple comparisons test)
We next compared the relative abundances of microbial taxa between 0–1 and 6–13 days after birth using LEfSe (Supplemental Fig. S2). There were a large number of taxa showing significant differences in their relative abundances between these two groups. When only those taxa showing a log10 transformed LDA score > 3 were considered significant in this analysis, we found that Pseudomonas, Actinobacillus, and Clostridium were relatively enriched in the piglets at 0–1 days of life, whereas Akkermansia, Parabacteroides, and Lactobacillus were relatively enriched in the piglets at 6–13 days of life (Supplemental Fig. S2).
Age-related changes in gut microbial community structure
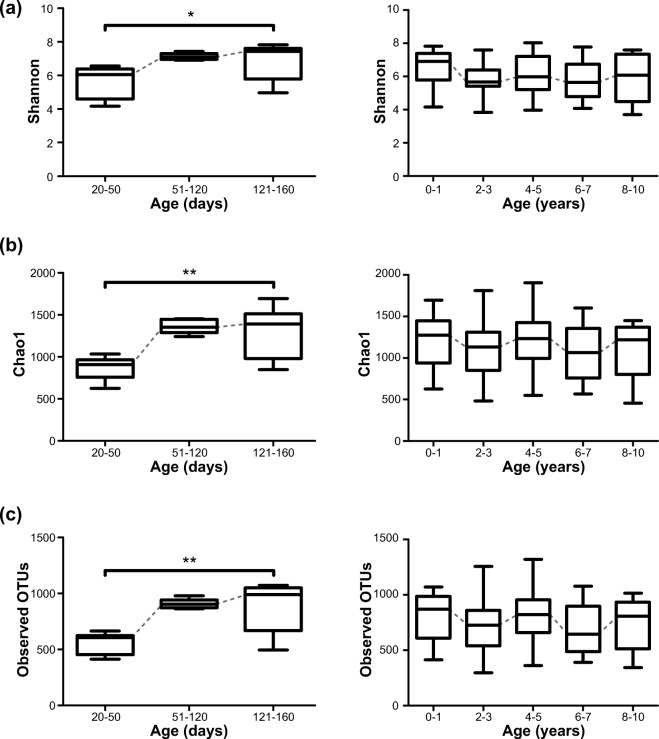
To investigate age-related changes in gut microbial community structure in the micro-pigs based on a wider range of ages, we next analyzed the microbiome data with only samples from vaginally delivered micro-pigs. The overall diversity scores sharply expanded with increasing age until approximately 20 weeks, and then slightly fluctuated throughout life (Fig. 2a–c). There were no significant differences in microbial diversity between males and females in terms of Shannon, Chao1, and observed OTUs (Supplemental Fig. S3).
Fig. 2.
Gut microbial diversity of vaginally-delivered pigs according to age. a Shannon index. b Chao 1 index. c Observed operational taxonomic units (OTUs). Left panel: microbial diversity of pigs at 20–160 days of life. Right panel: microbial diversity of pigs at 0–10 years of life (*p < 0.05; Kruskal-Wallis test with Dunn’s multiple comparisons test)
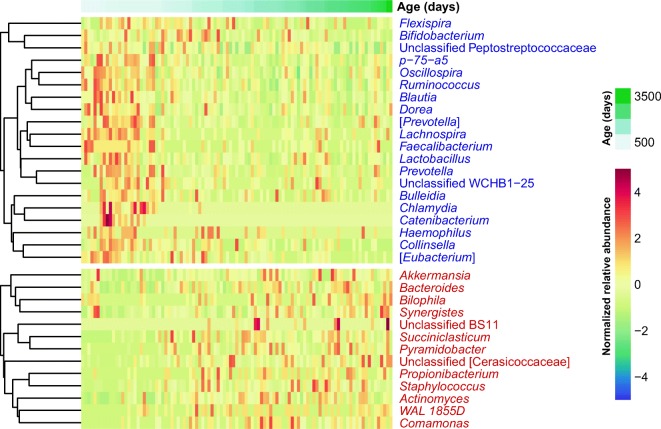
For the fecal samples from the vaginally delivered pigs, Adonis analysis based on the Bray-Curtis distance matrices indicated that age (days of life) explained 3.3% of variation in the gut microbiome (R2 = 0.033, p = 0.009) with sex contributing to a lesser extent (R2 = 0.023, p = 0.036) (Supplemental Fig. S4). This indicated that although there would be many other factors shaping the micro-pig gut microbiome, aging was a significant factor associated with composition distance. To identify the specific members of the gut microbial community that were significantly associated with aging after adjustments for sex, we performed multivariate analysis based on linear models. We found 20 negative associations and 13 positive associations between the days of life and microbial taxa (q < 0.1) (Fig. 3). For example, increasing age was inversely correlated with the relative abundances of Bifidobacterium, Lactobacillus, Blautia, and Ruminococcus and positively associated with those of Akkermansia and Bacteroides (Supplemental Fig. S5). We also found 19 significant associations between sex and microbial taxa. Among them, all taxa including Streptococcus, except for the unclassified Christensenellaceae, were more abundant in male samples (Supplemental Fig. S6).
Fig. 3.
Significantly changed taxa according to age (q value < 0.1) in micro-pig gut microbiomes. Red and blue letters indicate taxa positively and negatively associated with age, respectively
Additionally, we compared the gut microbiomes of micro-pigs with those of mice and humans obtained from the public databases (Odamaki et al. 2016; Thevaranjan et al. 2017). The entire dataset including both young and old individuals showed that gut microbiome compositions were significantly different according to species (Adonis R2 = 0.228, p < 0.001) (Supplemental Fig. S7a). When we compared only the microbiomes from aged individuals (mice: 18–22 months, micro-pigs: 7–10 years, humans: ≥ 60 years), the human microbiome was more similar to the micro-pig microbiome than to the mouse microbiome (Supplemental Fig. S7b, c).
Association between functional properties of the gut microbiome and age
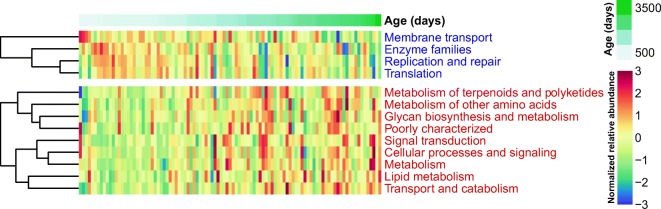
To investigate the functional potential of the gut microbiome associated with increasing age, we used PICRUSt to predict the metagenome based on 16S rRNA gene data. For KEGG pathway analysis (level 2), the relative abundances of metabolism-related pathways such as lipid metabolism, metabolism of terpenoids and polyketides, metabolism of other amino acids, and glycan biosynthesis and metabolism significantly increased with age. In contrast, the relative abundances of pathways involving genetic and environmental information processing such as translation, replication and repair, and membrane transport were negatively associated with aging (Fig. 4).
Fig. 4.
Significantly changed metabolic functions according to age (q value < 0.1) in micro-pig gut microbiomes. Red and blue letters indicate metabolic functions positively and negatively associated with age, respectively
Structure of co-occurrence relationships among the microbial taxa
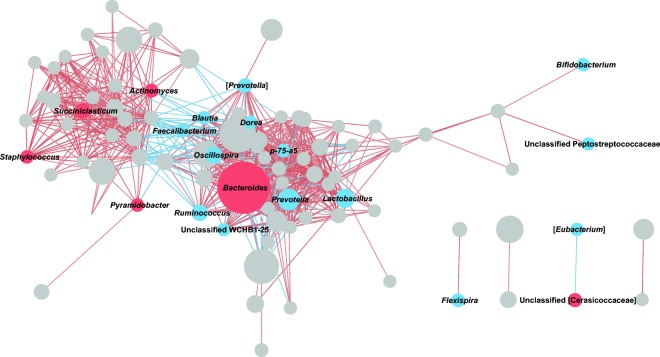
To predict potential relationships among the microbial taxa, we next constructed a bacterial co-occurrence network based on Spearman correlations between the relative abundances of genera. Among the 109 taxa with a prevalence > 0.5, we found that 91 had more than one significant correlation with other taxa, with a coefficient value > 0.5 or < − 0.5 (Fig. 5 and Supplemental Table S1). Among them, Coprococcus had the highest number of significant positive interactions. This genus co-occurred with members of Bacteroidetes including Parabacteroides and Prevotella, as well as members of Firmicutes including Dorea, Blautia, Ruminococcus, and Faecalibacterium. In contrast, the greatest number of significant negative correlations was observed for Bacteroides, which was positively associated with increasing age and exhibited the highest average relative abundance. Among a total of 27 significant correlations identified for this genus, 25 were inverse relationships with other taxa. For example, Bacteroides showed a strong negative correlation with Prevotella (Spearman’s correlation coefficient = − 0.706). In addition, Ruminococcus, Lactobacillus, Oscillospira, and Blautia, all of which decreased with age, were negatively correlated with Bacteroides.
Fig. 5.
Spearman correlation network of gut microbiome genera. Each node represents a genus, and node size indicates the average relative abundance of the genus. Red and blue nodes indicate genera positively and negatively associated with age, respectively. The entire list of correlations with FDRs < 0.1 and coefficient values > 0.5 or < − 0.5 is shown in Supplemental Table S1
Discussion
This study investigated the gut microbiome of micro-pigs at a wide range of ages. During the first 2 weeks of life, the gut microbiome of piglets became gradually diverse. This result is consistent with previous human infant gut microbiome studies (Bäckhed et al. 2015; Chernikova et al. 2018; Koenig et al. 2011), which indicates that like humans, newborn pigs are exposed to diverse bacteria from the environment starting from birth and that their gut microbiome changes according to days of life. Across different life stages, the diversity of the pig gut microbiome sharply increased after birth up to early breeder or finisher stages, and then fluctuated. In humans, a similar pattern was observed in US, Malawian, and Amerindian gut microbiomes, wherein bacterial diversity increased with age under 3 years old and then reached a relatively stable status (Yatsunenko et al. 2012). It is now widely accepted that considerable changes in the gut microbiome occur in the 3 years, after which a typical adult-like gut microbiota is established in humans. Based on our data, the maturation of the micro-pig gut microbiome might occur at the early stage of breeder or finisher.
Our results showed the sequential changes in gut microbiome composition and function that occur with age. The relative abundances of Bifidobacterium and Lactobacillus sequentially decreased with age; both of these are abundant in the infant gut microbiome (Bäckhed et al. 2015; Odamaki et al. 2016). Because Bifidobacterium has been shown to inhibit inflammatory responses in the gut epithelium (Khokhlova et al. 2012), the reduction in its relative abundance with increasing age might be associated with increased levels of chronic inflammation in the elderly. We also found that the relative abundances of butyrate-producing bacteria such as Blautia, Ruminococcus, and Faecalibacterium decreased with age. Although significant correlations between Bifidobacterium and these genera were not observed in the present study (correlation coefficient values > 0.5), it has been reported that acetate, which is one of the metabolites produced by Bifidobacterium, plays an important role in cross-feeding interactions between Bifidobacterium and butyrate-producing bacteria in the human gut (Falony et al. 2006). In addition, the relative abundances of Akkermansia and Bacteroides, which exist abundantly in the gut microbiome of human adults (Odamaki et al. 2016), increased with age in the pig gut microbiome. Most of our observations are thus consistent with the results from studies on the human gut microbiome. Therefore, the common age-related changes in the gut microbiome compositions of humans and pigs might be mainly affected by aging itself rather than other environmental conditions.
Gut dysbiosis has been proposed as one of the possible sources of chronic systemic inflammation, which is an aging hallmark (Sanada et al. 2018). The links between age-related gut dysbiosis and increases in intestinal permeability, the translocation of bacterial products to circulation, and the chronic systemic inflammation were previously studied in mice (Thevaranjan et al. 2017). More recently, differences in the compositions of serum microbiomes between young and older adults, as well as the associations between serum microbial members and age-related inflammatory markers, were reported (Buford et al. 2018). Due to the unavailability of blood biomarker data for micro-pigs, the association between age-related microbial changes and chronic systemic inflammation could not be determined, which is a limitation of the present study. However, micro-pigs exhibit similar age-related microbial changes to those observed in humans, and the human gut microbiome composition is more similar to the micro-pig microbiome than to the mouse microbiome (as shown in this study); moreover, they show similarities in physiology and disease progression. For these reasons, we suggest that micro-pigs are appropriate to investigate the mechanisms underlying interactions between the gut microbiome and the host with respect to aging and to test microbiome-based solutions to improve long-term human health.
In conclusion, this is the first study to examine the gut microbiome composition and function of micro-pigs over a wide range of ages, from newborns to 10 years old, reared under uniform conditions, to evaluate the effects of aging alone, independent of environmental factors. We observed that the diversity of the pig gut microbiome changes with age and that the relative abundances of specific gut microbes increase or decrease with age. Consistent with previous studies on the human gut microbiome, we found decreases in beneficial microbes including probiotic bacteria and short-chain fatty acid-producers in older pigs. In addition, the Bacteroides genus was positively associated with age and this genus was shown to have inverse relationships with other beneficial microbes. These results suggest that it might be important to maintain a certain proportion of beneficial microbes indicative of a younger gut microbiome age and that modulation of the relative abundance of Bacteroides in the elderly could be a target to promote healthy aging, although further studies are needed to understand the mechanism underlying the effects of the gut microbiome on aging. Further, in this respect, our study indicates that micro-pigs are a suitable animal model for studies on the links between aging and the microbiome.
Accession numbers
The sequences from this study have been deposited in the European Nucleotide Archive under the study accession number PRJEB33907.
Electronic supplementary material
(DOCX 710 kb)
(XLSX 41 kb)
Funding information
This research was supported by Main Research Program (E0170601-03) of the Korea Food Research Institute funded by the Ministry of Science and ICT.
Compliance with ethical standards
Procedures in this study were conducted in compliance with and approval of Korean laws and MK Institutional Animal Care and Use Committee (MK-IACUC) regarding the use of experimental animals.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buford TW, Carter CS, VanDerPol W, Chen D, Lefkowitz EJ, Eipers P, Morrow CD, Bamman MM. Composition and richness of the serum microbiome differ by age and link to systemic inflammation. Geroscience. 2018;40:257–268. doi: 10.1007/s11357-018-0026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova DA, Madan JC, Housman ML, Zain-Ul-Abideen M, Lundgren SN, Morrison HG, Sogin ML, Williams SM, Moore JH, Karagas MR, Hoen AG. The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth. Pediatr Res. 2018;84:71–79. doi: 10.1038/s41390-018-0022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G, Vlachou A, Verbrugghe K, De Vuyst L. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol. 2006;72:7835–7841. doi: 10.1128/Aem.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss-Coray T, Sierra F. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlova EV, Smeianov VV, Efimov BA, Kafarskaia LI, Pavlova SI, Shkoporov AN. Anti-inflammatory properties of intestinal Bifidobacterium strains isolated from healthy infants. Microbiol Immunol. 2012;56:27–39. doi: 10.1111/j.1348-0421.2011.00398.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Nguyen SG, Guevarra RB, Lee I, Unno T. Analysis of swine fecal microbiota at various growth stages. Arch Microbiol. 2015;197:753–759. doi: 10.1007/s00203-015-1108-1. [DOI] [PubMed] [Google Scholar]
- Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Supplement 1):4578–4785. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MY, Rho M, Song YM, Lee K, Sung J, Ko G. Stability of gut enterotypes in Korean monozygotic twins and their association with biomarkers and diet. Sci Rep. 2014;4:7348. doi: 10.1038/srep07348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Tiezzi F, Schillebeeckx C, McNulty NP, Schwab C, Shull C, Maltecca C. Host contributes to longitudinal diversity of fecal microbiota in swine selected for lean growth. Microbiome. 2018;6:4. doi: 10.1186/s40168-017-0384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney JK. Advances in swine biomedical model genomics. Int J Biol Sci. 2007;3:179–184. doi: 10.7150/ijbs.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N, Berri M, Estellé J, Levenez F, Lemonnier G, Denis C, Leplat JJ, Chevaleyre C, Billon Y, Doré J, Rogel-Gaillard C, Lepage P. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ Microbiol Rep. 2015;7:554–569. doi: 10.1111/1758-2229.12285. [DOI] [PubMed] [Google Scholar]
- Metzler-Zebeli BU, Schmitz-Esser S, Mann E, Grull D, Molnar T, Zebeli Q. Adaptation of the cecal bacterial microbiome of growing pigs in response to resistant starch type 4. Appl Environ Microbiol. 2015;81:8489–8499. doi: 10.1128/AEM.02756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council . Nutrient requirements of swine. 11. Washington, DC: The National Academies Press; 2012. [Google Scholar]
- Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajarillo EAB, Chae JP, Balolong MP, Kim HB, Seo KS, Kang DK. Characterization of the fecal microbial communities of Duroc pigs using 16S rRNA gene pyrosequencing. Asian Austral J Anim. 2015;28:584–591. doi: 10.5713/ajas.14.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada F, Taniyama Y, Muratsu J, Otsu R, Shimizu H, Rakugi H, Morishita R. Source of chronic inflammation in aging. Front Cardiovasc Med. 2018;5:12. doi: 10.3389/fcvm.2018.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Backhed F. The gut microbiota - masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- Thevaranjan N, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21(455-466):e454. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Estellé J, Kiilerich P, Ramayo-Caldas Y, Xia Z, Feng Q, Liang S, Pedersen AØ, Kjeldsen NJ, Liu C, Maguin E, Doré J, Pons N, le Chatelier E, Prifti E, Li J, Jia H, Liu X, Xu X, Ehrlich SD, Madsen L, Kristiansen K, Rogel-Gaillard C, Wang J. A reference gene catalogue of the pig gut microbiome. Nat Microbiol. 2016;1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One. 2015;10:e0117441. doi: 10.1371/journal.pone.0117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 710 kb)
(XLSX 41 kb)