Abstract
Pyruvate, a pivotal glucose metabolite, is an α-ketoacid that reacts with hydrogen peroxide (H2O2). Its pharmacological precursor, ethyl pyruvate, has shown anti-inflammatory/anti-tissue injury effects in various animal models of disease, but failed in a multicenter clinical trial. Since rodents, but not humans, can convert ethyl pyruvate to pyruvate in blood plasma, this additional source of extracellular pyruvate may have contributed to the discrepancy between the species. To examine this possibility, we investigated the kinetics of the reaction under biological conditions and determined the second order rate constant k as 2.360 ± 0.198 M−1 s−1. We then calculated the time required for H2O2 elimination by pyruvate. The results show that, with an average intracellular concentration of pyruvate (150 µM), elimination of 95% H2O2 at normal to pathological concentrations (0.01–50 µM) requires 141–185 min (2.4–3 hour). With 1,000 µM pyruvate, a concentration that can only exist extracellularly or in cell culture media, 95% elimination of H2O2 at 5–200 µM requires 21–25 min. We conclude that intracellular pyruvate, or other α-ketoacids, whose endogenous concentration is controlled by metabolism, have little role in H2O2 clearance. An increased extracellular concentration of pyruvate, however, does have remarkable peroxide scavenging effects, considering minimal peroxidase activity in this space.
Subject terms: Cardiovascular biology, Chemical biology, Pharmacology
Introduction
In recent years, ethyl pyruvate, an ethyl ester of pyruvate, has been shown to exert protective effects in various rodent models of acute/chronic inflammation and oxidant-induced tissue/organ injury1. These effects, however, were not observed in humans as examined in a phase II clinical trial2. This discrepancy between animals and humans might be related to the different in vivo activation site of ethyl pyruvate, and its interaction with hydrogen peroxide (H2O2) in each environment. This study investigated the reaction rate of pyruvate and H2O2 under these conditions to evaluate such differences.
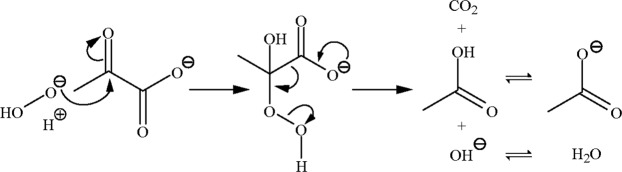
The reaction of pyruvate and H2O2 produces acetate, carbon dioxide (CO2), and water; its transition intermediate has been recently confirmed3,4. As shown in Fig. 1, the intermediate is an adduct of H2O2 and pyruvate, and its subsequent rearrangement is initiated by the release of CO2 from the carboxyl group. In ethyl pyruvate, ester formation blocks the pyruvate carboxyl and, therefore, the rearrangement cannot proceed to form these products. Ethyl pyruvate can be activated in vivo by carboxylesterase, an esterase with specificity towards carboxylate. Carboxylesterase is widely distributed within mammalian cells. In blood plasma, however, it is abundant in rodents but absent in humans5–7. Thus, ethyl pyruvate activation occurs in the plasma of rodents once absorbed or injected, but only in the intracellular environment of human cells.
Figure 1.
Mechanism of pyruvate and H2O2 reaction. Nucleophilic addition of H2O2 to the α-carbonyl group in pyruvate forms an unstable intermediate, 2-hydroperoxy-2-hydroxypropanoate, which subsequently undergoes rearrangement to produce CO2, acetate, and water at neutral pH.
Hydrogen peroxide is formed by superoxide dismutation, with superoxide arising from two primary sources: electron leak along the electron transport chain in mitochondria and the enzymatic action of NADPH oxidases (NOXs)8. NOXs are mainly localized to the plasma membrane and release superoxide/H2O2 at high concentrations to the extracellular space. Hydrogen peroxide enters cells through aquaporin transporters, and is removed by three peroxidase systems: glutathione peroxidases, catalase, and the peroxiredoxins. The average intracellular concentration of H2O2 is less than 10 nM, but that of blood is 1–5 µM9. Under inflammatory conditions, plasma H2O2 concentrations can reach 50 µM9.
The concentration of pyruvate is 77–201 µM intracellularly10–12, and 47–118 µM in plasma11,13,14. As a pivotal metabolite, pyruvate is continuously produced and removed by various metabolic pathways: after being produced by glycolysis, pyruvate is decarboxylated to acetyl CoA, which can either enter the tricarboxylic acid (TCA) cycle and generate ATP via oxidation, or participate in fatty acid synthesis. Pyruvate can also be carboxylated to oxaloacetate, which replenishes the TCA cycle or enters the gluconeogenesis pathway to generate glucose. Transamination converts pyruvate to alanine, which participates in protein synthesis. Under oxygen-deficient conditions, pyruvate is reduced to lactate. These pathways comprise regulated enzymatic reactions, which result in maintaining a relatively constant concentration of pyruvate intracellularly.
In the present study, we first examined the reaction order of pyruvate and H2O2 to confirm the concentration dependence of the reactants. The rate constant (k) was then determined, since there are no reports in the literature of the k obtained under physiological temperature, pH, and ionic strength. With this biomarker rate constant in hand and the biologically relevant concentrations of H2O2 and pyruvate described above, the rates of H2O2 elimination by pyruvate were calculated and confirmed by experimental measurements.
Results
Reaction order with respect to pyruvate and H2O2
The rate law of the pyruvate and H2O2 reaction is shown in Eq. (1), in which [Pyr] and [H2O2] represent the concentrations of pyruvate and H2O2, respectively; k is the rate constant; and a and b are the reaction orders with respect to pyruvate and H2O2, respectively.
| 1 |
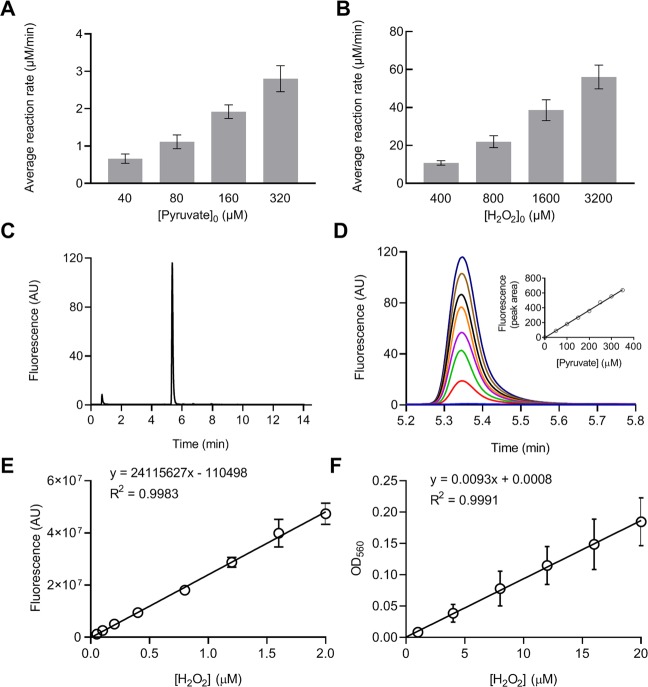
To evaluate the reaction under biological conditions, experiments in this report were all carried out at 37 °C in Dulbecco’s phosphate-buffered saline (DPBS), which has a pH of 7.3 ± 0.2 and ionic strength of 165 mM. The reaction order of pyruvate was estimated by reacting a fixed concentration of H2O2 (20 µM) with increasing concentrations of pyruvate (40, 80, 160, and 320 µM) for 5 min. Hydrogen peroxide concentration in the solutions was subsequently measured by a peroxidase-Amplex Red colorimetric assay. As shown in Fig. 2A, the increase in the average reaction rate was approximately proportional to the increase in pyruvate concentration, indicating that the reaction was first order with respect to pyruvate, or a =1. To determine the reaction order of H2O2, 200 µM pyruvate was reacted with increasing concentrations of H2O2 (400, 800, 1600, or 3200 µM) for 3 min. Pyruvate concentration was then measured using an HPLC method. The average reaction rate was found to increase proportionally to that of H2O2 concentration, indicating a first order reaction with respect to H2O2, or b = 1 (Fig. 2B). Taken together, the overall reaction order is two. Note that the reaction rate was measured as an average rate rather than an instantaneous rate and is less accurate than the latter; this approach should not, however, affect the estimation of the reaction order. As a demonstration of the method used for the above measurements, Fig. 2C,D show the HPLC chromatograms of the pyruvate and its standard curve. Pyruvate was derivatized to become fluorescent prior to HPLC analysis. Hydrogen peroxide was measured by a peroxidase-Amplex Red assay. The color product of the assay exhibits both fluorescence and visible color. Hydrogen peroxide samples at low concentration range (0–2 µM) were read with fluorescence and high concentrations (0–20 µM) with absorbance (OD560) in this assay. The standard curves in both ranges are linear (Fig. 2E,F).
Figure 2.
Reaction order with respect to pyruvate and H2O2. (A) The reaction order with respect to pyruvate was analyzed by reacting 20 µM H2O2 with increasing concentrations of pyruvate. The reaction was carried out for 5 min and the average reaction rate was calculated as the change of H2O2 concentration per min. (B) The reaction order of H2O2 was determined by reacting 200 µM pyruvate with increasing concentrations of H2O2. The reaction was carried out for 3 min and the rate was calculated as the change in pyruvate concentration per min. (C) and (D) HPLC chromatograms and the standard curve for pyruvate. (E) and (F) Standard curves of H2O2 measurement by a peroxidase-Amplex Red assay. The color product of the assay was read by absorption at OD560 or by fluorescence at λex 350 nm and λem 410 nm for the high or low concentration range of H2O2, respectively. Data were obtained from 3 separate experiments and presented as mean ± SD.
The rate constant of pyruvate and H2O2 reaction
To determine the rate constant k of the reaction, Eq. (1) was transformed to Eq. (2), in which [Pyr] represents the concentration of both reactants, and [Pyr] = [H2O2]. Integration of Eq. (2) from [Pyr]0 to [Pyr] and from 0 to t gives Eq. (3):
| 2 |
| 3 |
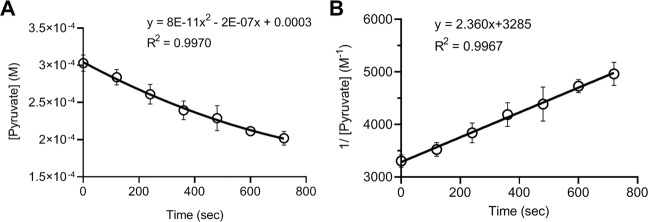
The rate constant k was determined by reacting pyruvate with H2O2, at the initial concentration of 300 µM, for 0, 2, 4, 6, 8, 10, and 12 min; pyruvate concentration was then measured by the HPLC analysis. As shown in Fig. 3, pyruvate concentration decreased over time, which fits a second-degree polynomial equation (Fig. 3A); plotting the inverse concentration of pyruvate vs. time gives a linear relationship, with a slope of 2.360 and an intercept of 3285 (Fig. 3B). Compared to Eq. (3), the plot indicates that the reaction rate constant k is 2.360 M−1 s−1, with [Pyr]0 is 0.000304 M or 304 µM by this measurement. Figure 3 was plotted with data sets from 6 replicate experiments. When graphed individually, the average and standard deviation of the k was 2.360 ± 0.198 M−1 s−1, or 0.000142 ± 0.000012 µM−1 m−1.
Figure 3.
Rate constant of pyruvate and H2O2 reaction. Reactions were carried out with 300 µM each of pyruvate and H2O2 in DPBS at 37 °C. At the indicated timepoint, pyruvate concentration in the reaction solution was measured using the HPLC method. (A) Data are graphed as pyruvate concentration vs. time and fit with a second-degree polynomial equation (B) Data are graphed as inverse pyruvate concentration vs. time and fit with a linear equation. The slope of the linear line is the rate constant k of the reaction according to Eq. (3). Data were obtained from 6 separate experiments and presented as mean ± SD.
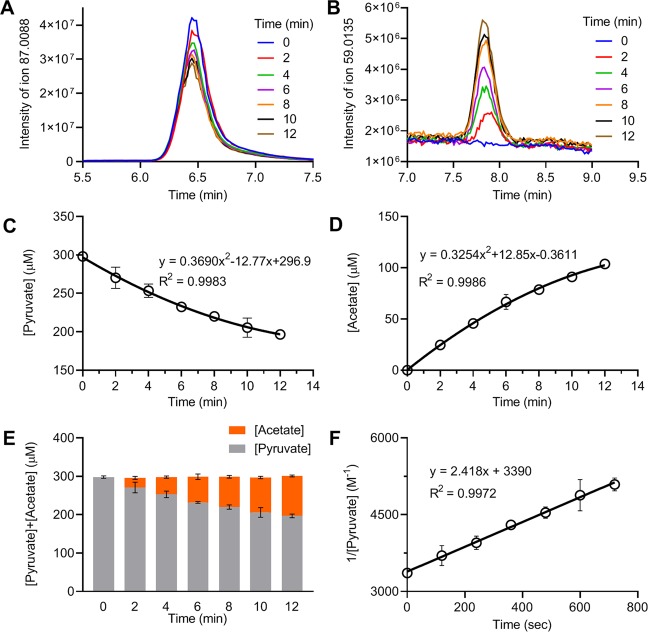
To confirm these HPLC measurements, a LC-MS method was used to measure the concentrations of pyruvate and acetate in the reaction solutions. As shown in Fig. 4A,B, the LC-MS chromatograms demonstrated distinct peaks for both pyruvate and acetate in the samples obtained at various timepoints. The decrease in pyruvate concentration over time was associated with an increase in the production of acetate (Fig. 4C,D). The total concentrations of pyruvate and acetate at each time point were constant (Fig. 4E), indicating an equimolar conversion of pyruvate to acetate. Plotting the inverse concentration of pyruvate over time showed a linear relationship with a slope of 2.418 and intercept of 3390 (Fig. 4F), consistent with the k as 2.418 M−1 s−1 and [Pyr]0 as 295 µM by the LC-MS measurement.
Figure 4.
LC-MS measurement of pyruvate and acetate concentrations. Reactions were carried out with 300 µM each of pyruvate and H2O2 in DPBS at 37 °C. At the indicated time, aliquots of the reaction mixture were removed for LC-MS analysis. (A,B) LC-MS chromatograms showing the detection of pyruvate and acetate, respectively, in the samples at the indicated times. (C,D) Concentrations of pyruvate and acetate, respectively, were calculated according to standard curves, graphed vs. time, and fit to second-degree polynomial equations. (E) Data are graphed as the sum of pyruvate and acetate concentrations vs. time, showing an unchanged total concentration over time. (F) Data are graphed as the inverse concentration of pyruvate vs. time and fit to a linear equation. The slope of the line indicates the k of the reaction. Data were obtained from 3 separate experiments and presented as mean ± SD.
Rate of H2O2 elimination by pyruvate under biological conditions
As mentioned above, the in vivo concentration of pyruvate is greater than that of H2O2. To calculate the rate of H2O2 elimination, Eq. (1) was transformed into Eq. (4), in which x represents the amount of the reactants that had undergone transformation.
| 4 |
The integration of Eq. (4) from 0 to t and from 0 to x gives Eq. (5), which allows for the calculation of the time required for x amount of H2O2 to be reacted.
| 5 |
Rearranging Eq. (5) gives Eq. (6), which allows for the calculation of the amount of H2O2 reacted at a specified time.
| 6 |
For calculations of the rate of H2O2 elimination with various amounts of [Pyr]0 and [H2O2]0, Excel spreadsheets were used with the above equations embedded. Examples are shown in Tables 1 and 2.
Table 1.
Calculation of time required for H2O2 elimination at indicated percentage*.
| k | [Pyr]0 | [H2O2]0 | R | x | [Pyr] | [H2O2] | P | Q | t |
|---|---|---|---|---|---|---|---|---|---|
| (µM−1 m−1) | (µM) | (µM) | (%) | (µM) | (µM) | (µM) | (min) | ||
| 0.000142 | 150 | 50 | 50 | 25 | 125 | 25 | −0.010 | −0.511 | 36.08 |
| 0.000142 | 150 | 50 | 95 | 47.5 | 103 | 2.5 | −0.010 | −2.615 | 184.67 |
| 0.000142 | 1,000 | 50 | 50 | 25 | 975 | 25 | −0.001 | −0.668 | 4.96 |
| 0.000142 | 1,000 | 50 | 95 | 47.5 | 953 | 2.5 | −0.001 | −2.947 | 21.91 |
*The values for k, [Pyr]0, [H2O2]0, and R were entered, R indicates the percentage of H2O2 elimination. The remaining terms are calculated using the following equations: x = R*[H2O2]0; [Pyr] = [Pyr]0 − x; [H2O2] = [H2O2]0 − x; P = ; Q = ; and t = P*Q/k.
Table 2.
Calculation of the amount of H2O2 eliminated (x) at indicated time t.
| k | t | [Pyr]0 | [H2O2]0 | D | E | x | [H2O2] |
|---|---|---|---|---|---|---|---|
| (µM−1 m−1) | (min) | (µM) | (µM) | (µM) | (µM) | (µM) | |
| 0.000142 | 33.0 | 150 | 50 | −100 | 0.6267 | 23.59 | 26.41 |
| 0.000142 | 22.0 | 1,000 | 50 | −950 | 0.0518 | 47.53 | 2.47 |
*The values for k, t, [Pyr]0, and [H2O2]0 were entered. The remaining terms are calculated using the following equations: D = [H2O2]0 − [Pyr]0; E = ; ; and [H2O2] = [H2O2]0 − x.
The time for elimination of 25, 50, 75, and 95% of H2O2 of its initial concentration, [H2O2]0, was calculated with Eq. (5). Two initial pyruvate concentrations were used: a normal intracellular concentration, 150 µM, and an increased extracellular concentration, 1,000 µM. Each concentration of pyruvate was reacted with serial concentrations of H2O2, ranging from normal to extreme pathological concentrations. The calculated results are shown in Fig. 5. The data were fitted to exponential decay equations, as [Pyr]0 was much greater than [H2O2]0 in each set of reactions. At the concentration differences of ≥10 fold (the lower four values of [H2O2]0 in each set), the reactions became pseudo-first order and the R2 of the data fitting ≥0.9998. As shown in Fig. 5A, at [Pyr]0 = 150 µM, the time required for eliminating 25, 50, 75, and 95% of H2O2 with initial concentrations of 0.01–50 µM ranged as 13.5–14.2, 33–36, 65–78, and 141–185 min, respectively. At [Pyr]0 = 1,000 µM (Fig. 5B), the time required for eliminating 25, 50, 75, and 95% H2O2 at initial concentrations of 5–200 µM ranged as 2.0–2.1, 4.9–5.2, 10–11, and 21–25 min, respectively.
Figure 5.
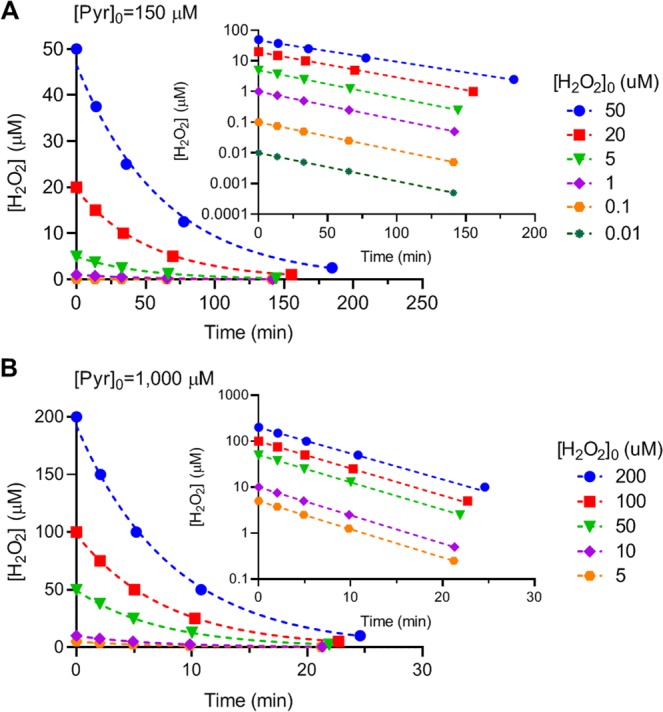
Calculated time course of H2O2 elimination by pyruvate. The time required for elimination of 25, 50, 75, or 95% of H2O2 of its initial concentration by pyruvate was calculated based on Eq. (5) and Table 1. (A) The initial concentration of pyruvate is 150 µM and of [H2O2]0 is 50, 20, 5, 1, 0.1, or 0.01 µM. (B) The initial concentration of pyruvate is 1,000 µM and of [H2O2]0 is 200, 100, 50, 10, or 5 µM. Data are graphed as [H2O2] vs. time, and fit to exponential decay equations. Inserts show the same graphs on a semilog scale.
To confirm these calculations, experiments were carried out by reacting 150 µM pyruvate with 0.1, 1, 5, or 50 µM H2O2. After a 33 min incubation, the H2O2 concentration in the reaction solution was measured. The H2O2 concentrations were also calculated under these conditions using the method shown in Table 2. The measured values were consistent with what was calculated (Table 3). Similarly, 1,000 µM pyruvate was reacted with 5, 10, 50, 100, and 200 µM H2O2 for 22 min, and the H2O2 concentrations were subsequently measured and compared with calculated values (Table 3).
Table 3.
Comparison of measured and calculated H2O2 concentrations at indicated time of reaction*.
| [Pyr]0 | [H2O2]0 | Reaction time | Measured [H2O2] | Calculated [H2O2] |
|---|---|---|---|---|
| (µM) | (µM) | (min) | (µM) | (µM) |
| 150 | 0.1 | 33 | 0.05 ± 0.02 | 0.05 |
| 150 | 1 | 33 | 0.50 ± 0.06 | 0.50 |
| 150 | 5 | 33 | 2.74 ± 0.24 | 2.50 |
| 150 | 50 | 33 | 27.2 ± 1.64 | 26.4 |
| 1,000 | 5 | 22 | 0.22 ± 0.07 | 0.22 |
| 1,000 | 10 | 22 | 0.39 ± 0.08 | 0.45 |
| 1,000 | 50 | 22 | 3.06 ± 0.33 | 2.47 |
| 1,000 | 100 | 22 | 5.95 ± 0.35 | 5.49 |
| 1,000 | 200 | 22 | 16.2 ± 1.11 | 13.5 |
*Measured data were obtained from 3 separate experiments and are presented as mean ± SD.
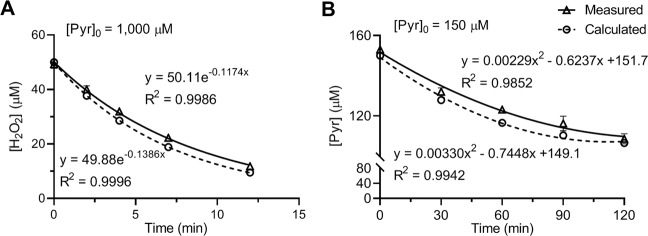
Confirmation was also carried out by reacting 50 µM H2O2 with 1,000 µM (Fig. 6A) or 150 µM pyruvate (Fig. 6B) and measuring the concentrations of H2O2 and pyruvate, respectively, after various incubation times. The corresponding time course was calculated according to Table 2. As shown in the figure, the measured and calculated time courses of the reaction aligned with each other. The data sets were fitted to exponential decay equations in Fig. 6A and second-degree polynomial equations in Fig. 6B.
Figure 6.
Comparison of measured and calculated time course of pyruvate and H2O2 reaction. Reactions were carried out by reacting 1,000 µM (A) or 150 µM (B) pyruvate with 50 µM H2O2 in DPBS at 37 °C. Concentrations of H2O2 (A) or pyruvate (B) in the reaction solutions were measured at indicated timepoints. Calculated [H2O2] or [Pyr] values were obtained based on Eq. 6 and Table 2. Data were fitted to exponential decay (A) and to second-degree polynomial (B) equations as shown within the plots. Measured data were obtained from 3 separate experiments and are presented as mean ± SD.
Discussion
This study examined the reaction rate of pyruvate and H2O2 under biological conditions. The reaction was determined to be first-order with respect to each reactant (Fig. 2), and the second order reaction rate constant k was 2.360 ± 0.198 M−1 s−1 or 0.000142 ± 0.000012 µM−1 m−1 (Figs. 3 and 4). Reaction rates were calculated with the determined k value using the concentrations of pyruvate and H2O2 observed under various biological conditions. The analysis showed that in the presence of an average intracellular concentration of pyruvate (150 µM), 50% elimination of H2O2 at normal to pathological concentrations (0.01–50 µM) requires 33–36 min, and 95% elimination takes 141–185 min (2.4–3 hr). In the presence of 1,000 µM pyruvate, 50% elimination of H2O2 at 5–200 µM requires only 5 min (4.9–5.2 min), and 95% elimination takes 21–25 min (Fig. 5). Experimental measurements confirmed these calculations (Table 3 and Fig. 6).
These data indicate that intracellular pyruvate has little role in H2O2 clearance. The rate of H2O2 elimination by 150 µM pyruvate (95%, 2.4–3 h) is too slow compared with intracellular peroxidase activities: The second order rate constants for reactions between H2O2 and glutathione peroxidases, catalase, or peroxiredoxins are in the 107–108 M−1 s−1 range15–17. Taking cytosolic glutathione peroxidase (GPx-1) alone, at a concentration of ~2 µM18 and rate constant of 4.1 ×107 M−1 s−1 17, the enzyme eliminates 95% of 1 µM H2O2 in 0.06 seconds, as calculated with Eq. 5 or Table 1. The same extent of elimination by 150 µM pyruvate requires 8,501 seconds (2.36 h). Low-molecular-weight thiols can also react with H2O2 directly, but the reaction rates are similar to that of pyruvate (within an order of magnitude). The second order rate constants for glutathione and cysteine, for example, are 0.89 and 2.9 M−1 s−1 (pH 7.4, 37 °C), respectively19. At average intracellular concentrations of 1–2 mM20 and 20–100 µM21, glutathione and cysteine eliminate 95% of 1 µM H2O2 in 1,684–3,368 seconds (0.5–0.9 h) and 10,401–53,486 seconds (2.9–14.9 h), respectively, as calculated with Eq. 5 or Table 1.
In the extracellular space, the rate of H2O2 elimination by 1,000 µM pyruvate (95%, 21–25 min) is rapid, considering minimal peroxidase activities present in this compartment and a 100- to 500-fold higher H2O2 concentration in plasma compared to the intracellular environment. Inflammation-activated NOXs can result in an extracellular accumulation of H2O2. Removing this pool of extracellular H2O2 by pyruvate would attenuate the oxidative stress/injury on the cells.
Ethyl pyruvate has been administered to mice and rats at doses of 40–100 mg/kg1, which would translate to blood concentrations of 4.4–12.3 mM using a blood-to-body weight ratio of 7%22. If ethyl pyruvate is activated by carboxylesterase in blood plasma, it will achieve an extracellular concentration close to 1,000 µM. At this concentration, pyruvate can markedly reduce the extracellular H2O2 level before entering cells to be metabolized. By contrast, when ethyl pyruvate can only be activated intracellularly (due to lack of plasma carboxylesterase), the generated pyruvate would be rapidly metabolized, having little impact on H2O2 levels. Thus, the conflicting results of the effects of ethyl pyruvate comparing rodent studies and the human trial may be related to the site of in vivo activation of the compound.
Based on the mechanism shown in Fig. 1, pyruvate could react with lipid peroxides and peroxynitrite in the same manner, forming an α-carbon adduct intermediate and releasing lipid hydroxide and nitrite as products, respectively. Thus, pharmacological concentrations of pyruvate could also scavenge these reactive oxygen species in the cell membrane, as well as other compartments. The same mechanism could also apply to other α-ketoacids, e.g., α-ketoglutarate and oxaloacetate. Nevertheless, the rate constant for these α-ketoacids would be lower than that of pyruvate owing to a bulkier alkyl chain23. The low intracellular concentrations of these metabolites, as with pyruvate, would, therefore, prevent them from having any practical “antioxidant” effects in that compartment.
It is also worthwhile noting that pyruvate is vital for primary cell culture and is added to various cell culture media at a concentration of 1,000 µM. Many oxidative stress studies add H2O2 to culture medium as an oxidant stress for cells, usually with an extended incubation period. As the added H2O2 is eliminated rapidly by pyruvate in these media, 50% in 5 min and 95% in 25 min, caution must be taken in the interpretation of the results, especially when comparing different studies.
Methods
Materials
Sodium pyruvate (≥99% purity, #P2256), 30% hydrogen peroxide (#H1009), acetic acid (#338826), sodium acetate trihydrate (#71188-250 G), O-phenylenediamine (#P23938), catalase (#C40), and Dulbecco’s phosphate buffered saline (DPBS, #D8537) were purchased from Sigma. Formic acid (#A11750), 3% hydrogen peroxide (#AC426001000), Amplex® Red hydrogen peroxide/peroxidase assay kit (#A22188), HPLC grade water (#W5-4), and HPLC grade acetonitrile (#A955-4) were obtained from Thermo Fisher Scientific.
Reaction of pyruvate and H2O2
Pyruvate and H2O2 reactions were carried out in DPBS at 37 °C in a total volume of 5 mL. Before the reaction, DPBS was incubated in a 37 °C water bath for at least 30 min to attain a stable temperature. Small aliquots of pyruvate and H2O2 stock solutions (<0.2 mL) were added to the DPBS solution to achieve the final concentration. Reactions were carried out at 37 °C for the designated times, and aliquots were removed for subsequent analysis. Owing to the time-sensitivity of the measurements, mixing and removing the reaction solution was staggered in order. For the measurements of pyruvate and acetate concentrations (by HPLC or LC-MS methods, see below), the reactions were stopped by adding 20 units catalase (10 unit/µL) to a 150 µl reaction mixture. For measurement of H2O2 concentration (by peroxidase-Amplex Red method, see below), aliquots of the reaction solution were directly loaded into a 96 well plate and immediately mixed with horseradish peroxidase and Amplex Red reagents to stop the reaction. Corresponding standards were included in each experiment and processed in the same way as samples.
HPLC measurement of pyruvate
The HPLC measurement of pyruvate followed a previously reported method with modifications14. Samples and pyruvate standards were derivatized by combining 1:1 by volume with O-phenylenediamine (OPD) dissolved at 25 mM in 2 M HCl, and incubated at 80 °C for 30 min. Derivatized pyruvate was quantified by HPLC with fluorescence detection using an Agilent 1260 Infinity II system with binary pump (G7112B) and fluorescence detector (G7121A). Samples were separated using an InfinityLab Poroshell 120 HPH C18 column (2.1 ×100 mm, 2.7 µm) with a guard column. Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. The injection volume was 5 µL. The mobile phase flow rate was 420 µL/min. The autosampler sample compartment was maintained at 4 °C, and the column oven was set at 40 °C. The mobile phase gradient (%B) was 0 min, 5%; 11 min, 60%; 11.1 min, 5%; 14.1 min, 5%. The fluorescence λex was 350 nm and λem 410 nm with 100 LU attenuation. Peak areas were integrated using OpenLab CDS ChemStation (Agilent) and sample pyruvate concentrations calculated from a standard curve.
LC-MS measurement of pyruvate and acetate concentrations
LC-MS analysis was performed on a Vanquish ultra-high performance liquid chromatography system coupled to a Q Exactive mass spectrometer (Thermo) that was equipped with an Ion Max source and HESI II probe adapting previously described methods24,25. External mass calibration was performed every seven days. Metabolites were separated using a ZIC-pHILIC stationary phase column (2.1 ×150 mm, 5 µm; Merck) with a guard column. Mobile phase A was 20 mM ammonium carbonate and 0.1% ammonium hydroxide. Mobile phase B was acetonitrile. The injection volume was 1 µL, the mobile phase flow rate was 100 µL/min, the column compartment temperature was set at 25 °C, and the autosampler compartment was set at 4 °C. The mobile phase gradient (%B) was 0 min, 80%; 10 min, 50%; 10.5 min, 8%; 14 min, 8%; 14.5 min, 80%; 25 min, 80%. The column effluent was introduced to the mass spectrometer with the following ionization source settings: sheath gas 40, auxiliary gas 15, sweep gas 1, spray voltage - 3.0 kV, capillary temperature 275 °C, S-lens RF level 40, probe temperature 350 °C. The mass spectrometer was operated in targeted selective ion monitoring mode for pyruvate (m/z 87.0088) and acetate (m/z 59.0135) with 2 m/z isolation window. The resolution was set to 140,000, and the AGC target was 3 × 106 ions. Data were acquired and analyzed using TraceFinder software (Thermo). Metabolite concentrations were calculated from the corresponding standard curve.
Amplex red assay
An Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit was used for the H2O2 concentration measurement, following the manufacturer’s instruction with modifications. Hydrogen peroxide standards and samples (10–50 µL) were loaded in triplicate into a 96 well black polystyrene microplate with a clear bottom (Corning). A working solution was immediately added for a final volume of 100 µL after each triplicate loading. The final concentration of horseradish peroxidase in the mixture was 0.1 U/mL, and Amplex Red was 0.05 mM. After 5–10 min incubation at room temperature in the dark, the plate was read with a microplate reader, SpectraMax MiniMax 300 Imaging Cytometer, equipped with SoftMax Pro 7 software (Molecular Devices). Fluorescence detection was set at λex 540 nm and λem 580 nm and optical absorbance detection at 560 nm. Sample concentrations were calculated according to standard curves included in each experiment.
Statistics
Data were obtained from 3–6 replicate experiments, each of which was performed in duplicate or triplicate. Data are presented as mean ± standard deviation. Curve fitting was performed with nonlinear least-squares regression, and R2 of the fitting is presented when necessary in figure plots.
Acknowledgements
The authors thank Stephanie Tribuna for expert secretarial assistance. This work was supported, in part, by NIH grants Hl128802 (WO); NIH grants HG007690, HL119145, and GM107618 (JL); and American Heart Association grant D700382 (JL).
Author contributions
V.A.G., W.M.O. and Y.Y.Z. designed and performed the experiments, J.L. supervised the project, and all the authors contributed to the writing and final approval of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang R, Zhu S, Tonnessen TI. Ethyl pyruvate is a novel anti-inflammatory agent to treat multiple inflammatory organ injuries. J Inflamm (Lond) 2016;13:37. doi: 10.1186/s12950-016-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett-Guerrero E, et al. A phase II multicenter double-blind placebo-controlled study of ethyl pyruvate in high-risk patients undergoing cardiac surgery with cardiopulmonary bypass. J. Cardiothorac. Vasc. Anesth. 2009;23:324–329. doi: 10.1053/j.jvca.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Asmus C, Mozziconacci O, Schoneich C, Low-temperature NMR. characterization of reaction of sodium pyruvate with hydrogen peroxide. J. Phys. Chem. A. 2015;119:966–977. doi: 10.1021/jp511831b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopalco A, et al. Mechanism of Decarboxylation of Pyruvic Acid in the Presence of Hydrogen Peroxide. J. Pharm. Sci. 2016;105:705–713. doi: 10.1002/jps.24653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry LM, Wollenberg L, Zhao Z. Esterase activities in the blood, liver and intestine of several preclinical species and humans. Drug Metab Lett. 2009;3:70–77. doi: 10.2174/187231209788654081. [DOI] [PubMed] [Google Scholar]
- 6.Bahar FG, Ohura K, Ogihara T, Imai T. Species difference of esterase expression and hydrolase activity in plasma. J. Pharm. Sci. 2012;101:3979–3988. doi: 10.1002/jps.23258. [DOI] [PubMed] [Google Scholar]
- 7.Li B, et al. Butyrylcholinesterase, paraoxonase, and albumin esterase, but not carboxylesterase, are present in human plasma. Biochem. Pharmacol. 2005;70:1673–1684. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 9.Forman HJ, Bernardo A, Davies KJ. What is the concentration of hydrogen peroxide in blood and plasma? Arch. Biochem. Biophys. 2016;603:48–53. doi: 10.1016/j.abb.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Joshi A, Palsson BO. Metabolic dynamics in the human red cell. Part IV–Data prediction and some model computations. J. Theor. Biol. 1990;142:69–85. doi: 10.1016/S0022-5193(05)80013-X. [DOI] [PubMed] [Google Scholar]
- 11.Mathioudakis D, et al. Pyruvate: immunonutritional effects on neutrophil intracellular amino or alpha-keto acid profiles and reactive oxygen species production. Amino Acids. 2011;40:1077–1090. doi: 10.1007/s00726-010-0731-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu A, Romero R, Petty HR. A sensitive fluorimetric assay for pyruvate. Anal. Biochem. 2010;396:146–151. doi: 10.1016/j.ab.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landon J, Fawcett JK, Wynn V. Blood pyruvate concentration measured by a specific method in control subjects. J. Clin. Pathol. 1962;15:579–584. doi: 10.1136/jcp.15.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pailla K, Blonde-Cynober F, Aussel C, De Bandt JP, Cynober L. Branched-chain keto-acids and pyruvate in blood: measurement by HPLC with fluorimetric detection and changes in older subjects. Clin. Chem. 2000;46:848–853. [PubMed] [Google Scholar]
- 15.Peskin AV, et al. The high reactivity of peroxiredoxin 2 with H(2)O(2) is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 16.Manta B, et al. The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch. Biochem. Biophys. 2009;484:146–154. doi: 10.1016/j.abb.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 17.Toppo S, Flohe L, Ursini F, Vanin S, Maiorino M. Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim. Biophys. Acta. 2009;1790:1486–1500. doi: 10.1016/j.bbagen.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Winterbourn CC, Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic. Biol. Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 20.Forman HJ, Zhang H, Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol. Aspects Med. 2009;30:1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stipanuk MH, Dominy JE, Jr., Lee JI, Coloso RM. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr. 2006;136:1652S–1659S. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- 22.Lee HB, Blaufox MD. Blood volume in the rat. J. Nucl. Med. 1985;26:72–76. [PubMed] [Google Scholar]
- 23.Lopalco A, Stella VJ. Effect of Molecular Structure on the Relative Hydrogen Peroxide Scavenging Ability of Some alpha-Keto Carboxylic Acids. J. Pharm. Sci. 2016;105:2879–2885. doi: 10.1016/j.xphs.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 24.Chen WW, Freinkman E, Wang T, Birsoy K, Sabatini DM. Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell. 2016;166:1324–1337 e1311. doi: 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Watson DG. Evaluation of the technical variations and the suitability of a hydrophilic interaction liquid chromatography-high resolution mass spectrometry (ZIC-pHILIC-Exactive orbitrap) for clinical urinary metabolomics study. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016;1022:199–205. doi: 10.1016/j.jchromb.2016.04.017. [DOI] [PubMed] [Google Scholar]