Abstract
A yeast expression plasmid was constructed containing a cardenolide biosynthetic module, referred to as CARD II, using the AssemblX toolkit, which enables the assembly of large DNA constructs. The genes cloned into the vector were (a) a Δ5‐3β‐hydroxysteroid dehydrogenase gene from Digitalis lanata, (b) a steroid Δ5‐isomerase gene from Comamonas testosteronii, (c) a mutated steroid‐5β‐reductase gene from Arabidopsis thaliana, and (d) a steroid 21‐hydroxylase gene from Mus musculus. A second plasmid bearing an ADR/ADX fusion gene from Bos taurus was also constructed. A Saccharomyces cerevisiae strain bearing these two plasmids was generated. This strain, termed “CARD II yeast”, was capable of producing 5β‐pregnane‐3β,21‐diol‐20‐one, a central intermediate in 5β‐cardenolide biosynthesis, starting from pregnenolone which was added to the culture medium. Using this approach, five consecutive steps in cardenolide biosynthesis were realized in baker's yeast.
Keywords: baker's yeast, biosynthesis, cardenolides, pathway engineering, steroids
We describe the construction of a novel yeast strain capable to reconstitute an important part of the cardenolide pathway in Digitalis plants. The intermediates are synthesized from a simple steroid in correct stereospecific manner with the help of five heterologously expressed genes from different bacteria, plants, and mammals.
1. INTRODUCTION
Cardenolides represent a group of natural products used in the treatment of congestive heart failure for a long time. Although the drugs are no longer relevant for the treatment of cardiac insufficiency due to their small therapeutic index, (Eichhorn & Gheorghiade, 2002) the focus of recent research shifted to their potential use in treating various other diseases such as cancer, diabetes, cystic fibrosis, herpes simplex, and HIV infections (Gurel, Karvar, Yucesan, Eker, & Sameeullah, 2017; Krishna, Manikyam, Sharma, & Sharma, 2015).
Cardenolides continue to be obtained by extraction from plants, with Digitalis lanata being the predominant source (Clemente et al., 2011). Attempts to produce cardiac glycosides in cell tissue cultures on a commercial scale have failed and their complex chemical structure hampers chemical synthesis. D. lanata cardiac glycosides are 5β‐cardenolides, which are characterized by a steroid scaffold with its rings connected cis‐trans‐cis and containing hydroxyl groups in C‐14β and C‐3β. The steroid H‐17β is substituted for an unsaturated five‐membered lactone ring. At position C‐3β a sugar side chain with up to five carbohydrate units is attached. Ten enzymes involved in the formation of 5β‐cardenolides have been identified, but only a few of the corresponding genes have been cloned and expressed as heterologuously. The enzymes in the early pathway, that is, the steps leading from a sterol precursor to the 5β‐cardenolide aglycone digitoxigenin, are progesterone 5β‐reductase (St5βR) and 3β‐hydroxysteroid dehydrogenase (3βHSD) and are characterized extensively. A putative biosynthetic pathway leading to digitoxigenin is outlined in Kreis and Müller‐Uri (Kreis & Müller‐Uri, 2012). Kreis and Müller‐Uri proposed a metabolic grid instead of a pathway which better reflects the situation in the cell and shows several compounds that may be intermediates or side products of cardenolide formation. Many of these compounds were tested as substrates for the known cardenolide biosynthetic enzymes, and the relaxed substrate specificity of St5βR and 3βHSD was demonstrated (Kreis et al., 2012).
In recent years, microorganisms including Saccharomyces cerevisiae were genetically engineered to potentially synthesize natural products usually derived from plants. Inside these engineered pathways a variety of target structures such as phenols, isoprenoids, alkaloids, and polyketides can be found (Siddiqui, Thodey, Trenchard, & Smolke, 2012). Pathway engineering also seems to be a promising approach for 5β‐cardenolide aglyca formation starting from pregnenolone or even a simple carbon source. Not all cardenolide biosynthetic enzymes are known but mammalian or bacterial genes can be transferred that code for enzymes that may masquerade as “genuine” proteins when attempting to realize a cardenolide pathway in a transgenic organism. The assembly of large DNA constructs coding for entire pathways poses a major challenge in the field of pathway engineering. AssemblX, a novel software‐assisted method, allows for the design and testing of multifunctional DNA constructs (Hochrein et al., 2017). It uses scar‐free, overlap‐based and sequence‐independent methods relying on rare cutting restriction enzymes and optimized adapter sequences.
Using the AssemblX approach, we here established a yeast plasmid bearing a cardenolide biosynthetic module termed CARD II. It comprises Δ5‐3β‐hydroxysteroid dehydrogenase (3βHSD) from D. lanata, steroid Δ5‐isomerase (3KSI) from Comamonas testosteronii, progesterone‐5β‐reductase (St5βR) from A. thaliana, and steroid 21‐hydroxylase (CYP21A1) from Mus musculus. CARD II yeast was capable of producing 5β‐pregnane‐3β,21‐diol‐20‐one, a central intermediate in 5β‐cardenolide biosynthesis, from pregnenolone in a 5‐step reaction (Figure 1).
Figure 1.
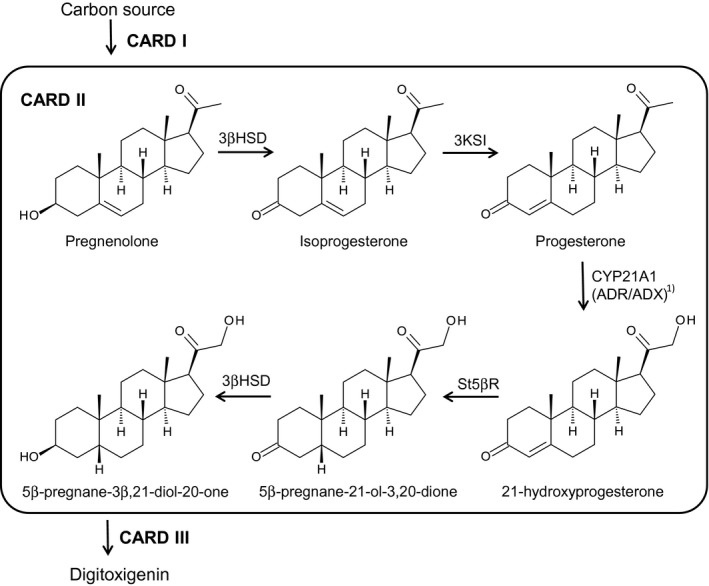
Overview of the CARD modules. CARD I: module connecting yeast sterol metabolism with the initial cardenolide pathway; CARD II (as described above): module comprising the five steps of the cardenolide pathway leading from pregnenolone to 5β‐pregnane‐3β,21‐diol‐20‐one; CARD III: module allowing for C‐14β‐hydroxylation and butenolide ring formation. 1) The fusion gene ADR/ADX was omitted at this stage to further investigate its benefit in the CYP21A1 step. 3βHSD, Δ5‐3β‐hydroxysteroid dehydrogenase; 3KSI, steroid Δ5‐isomerase; ADR, adrenodoxin reductase; ADX, adrenodoxin; CYP21A1, steroid 21‐hydroxylase; St5βR, steroid‐5β‐reductase
2. MATERIALS AND METHODS
2.1. Strains and plasmids
Escherichia coli strains BL21 (DE3) and M15 (pREP4; Qiagen GmbH) were used for heterologous protein expression. The strains NEB5α (New England Biolab) and JM109 (Promega) were utilized for cloning purposes. Bacteria were grown in LB media at 37°C containing ampicillin (100 µg/ml) and/ or ampicillin (100 µg/ml) and kanamycin (25 µg/ml) as selection markers.
S. cerevisiae strains YPH500 or BY4741 ΔAPAT (Euroscarf) were used for in vivo studies. Yeast cells were grown at 30°C, either in YPD or SC media.
The plasmids pQE30Dl3βHSD, pQE30AtSt5βR (containing the N‐truncated form PDB: 6EL3_A; Schmidt et al., 2018), pQE30AtSt5βR_full length (containing the full length form GenBank: ABU55811.1), pQE30AtSt5βR_R64D_F342A_F153A (AtSt5βR_DMC), and pQE30AtSt5βR_F342A_F153A (AtSt5βR_DM), pYesDest52MmCYP21A1, from our own collection were used. The plasmid pKK22‐3 containing the coding sequence for the 3KSI of C. testosteronii (Schwans, Sunden, Gonzalez, Tsai, & Herschlag, 2013) was a gift from Daniel Herschlag, Stanford University, USA. The plasmid pBAR_Twin containing the ADR/ADX fusion gene (Hannemann, Virus, & Bernhardt, 2006) was a gift from Rita Bernhardt, Saarland University, Germany.
2.2. RNA isolation from Saccharomyces cerevisiae
To isolate RNA from bakers' yeast, 2 ml of a logarithmic preculture was harvested by centrifugation (4,000× g, 3 min) and the obtained cell pellets were treated with the YeaStar™ RNA Kit (Zymo Research Europe GmbH) as described in the manufacturer's protocol.
2.3. Construction of the yeast expression vectors
The construction of the yeast expression vector pL was performed using the AssemblX toolkit (Hochrein et al., 2017). To construct the Level 0 modules, each of the Level 0 backbones was linearized by NotI‐HF digestion. The overlapping homology regions for each subunit were attached by PCR amplification with particularly designed primers. The generated transcription units were amplified via PCR using the extracted Level 0 plasmids as template. The products were verified by sequencing (Eurofins Genomics Germany GmbH) and subsequently used in a TAR reaction to assemble them in a Level 1 high‐copy plasmid and a Level 1 low‐copy plasmid. 100 ng of each transcription unit and 100 ng of a PacI linearized Level 1 backbone were applied in this reaction.
The plasmid pL1_hc bears a 2 µ origin of replication qualifying it as a high‐copy vector, whereas pL1_lc has an ARS/CEN origin of replication making it a low‐copy vector.
For assembly, the SLiCE (Zhang, Werling, & Edelmann, 2012). Gibson and NEBuilder HIFI DNA assembly (New England Biolab) methods were used. The molar ratio of linearized plasmids to amplified DNA fragment was either 1:3 (Gibson assembly, NEBuilder HIFI DNA assembly) or 1:10 (SLiCE assembly). As per the manufacturer's manual reaction, mixtures were incubated for 15 min at 50°C (Gibson, NEBuilder) or 1 hr at 37°C (SLiCE).
2 µl of each reaction was transformed into NEB5α cells (NEB) following the high efficiency transformation protocol provided by New England Biolabs (NEB). Putative positive clones were selected by a plasmid‐mediated selection marker and verified by colony PCR using the HotStart Green PCR‐Master Mix (Thermo Fisher Scientific). The positive clones were inoculated into 10 ml liquid medium and cultivated overnight at 37°C. Plasmids were extracted and prepared using the NucleoSpin® Plasmid kit (Macherey & Nagel).
For the construction of pCR601, a pESC‐Leu vector was used. The GAL10 promotor was substituted with the TEF2 promoter by restriction‐free cloning (http://www.rf-cloning.org). The ADR/ADX fusion gene was cloned between the TEF2 promotor and ADH1 terminator also by restriction‐free cloning (Figure 2).
Figure 2.
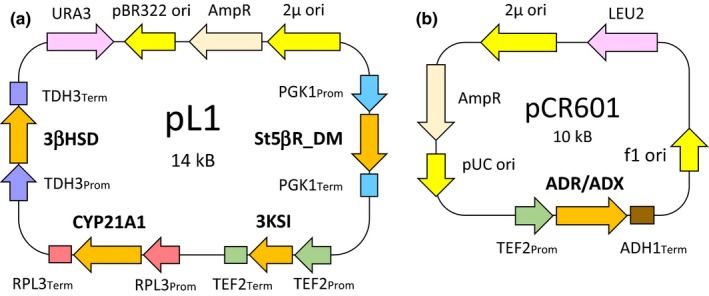
(a) Expression plasmid pL1. (b) Expression plasmid pCR601 containing the ADR/ADX fusion gene. TDH3 = triose‐phosphate dehydrogenase; TEF2 = translational elongation factor EF‐1 alpha; PGK1 = 3‐phosphoglycerate kinase; RPL3 = protein component of the large (60S) ribosomal subunit; ADH1 = alcohol dehydrogenase; Prom = Promotor; Term = Terminator. Other abbreviations as in Figure 1
The transformation of yeast cells with vectors was done using the LiAc/SS carrier DNA/PEG method (Gietz, St Jean, Woods, & Schiestl, 1992). Putative positive colonies were verified by colony PCR and cultivated agar plates containing selective media.
2.4. Extraction of plasmid DNA from yeast cells
The isolation of plasmid DNA from S. cerevisiae cells was performed using the Zymoprep™ Yeast Plasmid Miniprep II kit (Zymo Research Europe GmbH). 1.5 ml of a yeast cell suspension was harvested by centrifugation for 2 min at 800× g. The following steps were executed according to the user's manual. The obtained DNA was eluted in 10 µl Milli‐Q water (Millipore) and stored at 20°C.
2.5. In vivo biotransformation of pregnanes in Saccharomyces cerevisiae
Yeast cells containing Card II module plasmids were spread on synthetic drop‐out media agar plates (omitting uracil and/or leucine) and incubated at 30°C. In order to inoculate a preculture, a small batch of the cells was resuspended in 2 ml liquid selective media and incubated in an orbital shaker (220 rpm) at 30°C for 16–18 hr. For the main broth culture, 100 µl of the cell suspension was applied to 5 ml selective media together with 50 µl of pregnanes in ethanol to a concentration of 0.1–0.3 mM. Samples were taken at different times. Each time 5 ml of the cell suspension was taken and centrifuged at 10,000× g for 60 s. Cells and supernatant were subsequently processed for analytical purposes.
2.6. GC‐MS
Pregnanes were extracted from the medium with dichloromethane (5:2 v/v) applying a vortex for 30 s followed by centrifugation for 5 min at 13,000 rpm. The organic phase was separated and dried under N2. The dried pregnanes are resuspended in 100 µl dichloromethane and analyzed by GC‐MS.
Quantitative analysis was performed with a Shimadzu GCMS‐QP‐2010S equipped with a DB‐5ms column (30 m × 0.25 mm × 0.25 µm). Helium was used as the mobile phase with a flow rate of 48.1 cm/s. Injection temperature was 250°C, interface temperature 280°C, and ion source temperature 200°C. The oven was programmed as follows 100°C (hold for 0.5 min) and 320°C (increase 10°C/min, hold for 10 min). The injection volume was 1 µl. The scan range was set from 40 to 700 m/z. Since pregnenolone and 5β‐pregnane‐3,20‐dione could not be separated, samples were silylated with Silyl‐991 (BSTFA with 1% TCMS; Macherey & Nagel) and again analyzed by GC‐MS to discriminate between these two substances.
2.7. Quantitative real‐time PCR analysis of gene expression
To analyze the transcription rates of the cloned genes in yeast, 1 µg of total RNA was applied in a reverse transcriptase reaction using the peqGOLD cDNA Synthesis Kit H Plus (PeqLab). The qRT‐PCR samples were prepared using the SYBR Green Mastermix Kit (Applied Biosystems) according to the user's manual. Real‐time PCR was carried out with the StepOnePlus Real‐Time PCR System (Life Technologies), and the relative gene expression levels were calculated using the 2−ΔΔCt method adopting TFC1 as reference gene (Livak & Schmittgen, 2001).
2.8. Heterologous protein expression in Escherichia coli
Transformed E. coli cells were cultivated at 37°C and at 180 rpm to an oD600 of 0.5–0.7 in an orbital shaker (Multitron, Infors HT, Suisse) for the heterologous production of recombinant proteins. The expression of the recombinant proteins was induced by adding isopropyl‐β‐D‐thiogalactoside (IPTG). The final concentration of IPTG and the cultivation conditions needed depended on the expression vector used or the expression strain (Table 1). Cells were harvested by centrifugation (5,000× g, 20 min, and 4°C) and subsequently lysed by resuspending them in 20 ml lysis buffer (50 mM NaPi, 300 mM NaCl, 10 mM imidazole, pH 8.0) containing 1 mg/ml of lysozyme. After incubation for 1 hr at 4°C, while shaking as described above, cells were sonicated (6 x 30 s). After centrifugation (30 min, 13,000× g; 4°C), the recombinant proteins were separated by Ni‐NTA (nickel‐nitrilotriacetic acid) agarose purification using an ÄKTApure system (GE Healthcare). The clear supernatant was loaded on a 1 ml HisTrap FF Crude column (GE Healthcare) equilibrated with lysis buffer. After removing unbound compounds with a wash buffer (50 mM NaH2PO4, 300 mM NaCl, 40 mM Imidazole, pH 8.0), the affinity‐bound proteins were eluted using a gradient from 0% – 100% elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM Imidazole, pH 8.0) over 15 CV at a flow rate of 1 ml/min followed by 100% elution buffer for 5 CV at the same flow rate. The fraction volume during elution was set to 1 ml. Before using the recombinant proteins, the elution buffer was exchanged for incubation buffer (20 mM NaPi, pH 7.0) using PD‐10 columns (GE Healthcare).
Table 1.
Protein expression conditions
| Enzyme | Vector | Strain | IPTG (mM) | Temperature (°C) | Time (hr) |
|---|---|---|---|---|---|
| rDl3βHSD | pQE30 | M15[pREP4] | 0.1 | 4 | 96 |
| rCt3KSI | pDEST17 | BL21 (DE3) | 0.4 | 30 | 16 |
| rAtSt5βR | pQE30 | M15[pREP4] | 1.0 | 37 | 4 |
| rAtSt5βR_DMa | pQE30 | M15[pREP4] | 1.0 | 37 | 4 |
| rAtSt5βR_DMCb | pQE30 | M15[pREP4] | 1.0 | 37 | 4 |
| rAtSt5βR_R64D | pQE30 | M15[pREP4] | 1.0 | 37 | 4 |
AtSt5βR_DM: AtSt5βR_F342A_F153A.
AtSt5βR_DMC:AtSt5βR_R64D_F342A_F153A.
2.9. In vitro combinatorial enzyme assay
The combinatorial enzyme assay to reconstitute the Card II module of cardenolide biosynthesis in vitro was composed of rDl3βHSD, rCt3KSI, and rAtSt5βR_F342A_F153A or rAtSt5βR_R64D_F342A_F153A, respectively. The standard protein concentration in the assay was 0.05 mg/ml. Different ratios of the proteins were tested. As cosubstrates, NAD+ and NADH/H+ solutions were used in a standard concentration of 0.4 mM and tested in different ratios. Pregnenolone was used as substrate at 0.3 mM. Assays were incubated for 2 hr at 40°C and 550 rpm, and the reaction was stopped by adding 1 ml of dichloromethane. The mixture of protein solutions and cosubstrate(s) was preincubated at 40°C for 10 min before the reaction was started by the addition of substrate solution (pregnenolone in 96% ethanol) to a final concentration of 0.3 mM in a volume of 1 ml. The assay was incubated for 2 hr at 40°C and 550 rpm. The enzymatic reaction was terminated by the addition of 1 ml of dichloromethane, and the pregnanes were extracted as described above.
2.10. AtSt5βR muteins
AtSt5βR muteins were generated following the protocols described by Petersen et al (Zhang et al., 2012). Basically, a modified Stratagene QuickChange® SDM method was employed. Primers contained one to three mismatches flanked by 10–15 nucleotides specific to a well‐defined part of the respective St5βR ORF and a GC content of at least 40%. cDNA synthesis was completed after 30 PCR cycles. The respective mutant cDNAs were then restricted by DpnI and transformed into highly competent E. coli DH5α cells.
3. RESULTS
3.1. Identifying candidate genes
To achieve the overall project, we separated the cardenolide biosynthetic pathway into three modules (see Discussion). CARD II, our focus here, comprised the five steps of the cardenolide biosynthetic pathway leading from pregnenolone to 5β‐pregnane‐3β, 21‐diol‐20‐one (Figure 1). When using wild‐type yeast, esterification of pregnenolone yielded pregnenolone acetate as the main product which was not converted further. This reaction is catalyzed by acetyl‐CoA:pregnenolone acetyltransferase (APAT) coded by the ATF2 (YGR177c) gene (Cauet, Degryse, Ledoux, Spagnoli, & Achstetter, 1999). Because of that a ΔAPAT yeast strain with the ATF2 gene deleted was chosen. In order to evaluate the endogenous capacities of baker's yeast, we added several pregnanes supposed to be intermediates into the cardenolide pathway. Extracts were prepared after 48 hr of incubation and analyzed by GC‐MS. We demonstrated endogenous steroid dehydrogenase activity reducing the keto group of pregnanes but not pregn‐4‐enes (Figure 3). Oxidation of 3‐hydroxypregnanes or 3‐hydroxypregnenes to their respective 3‐keto derivatives was not seen. Steroid C‐14 and/or C‐21 hydroxylation was not observed, and also, 5β‐reduction of pregn‐4‐enes did not occur. Hence, the following 4 genes were identified as candidates suited to generate a CARD II yeast capable of carrying out the biosynthetic steps necessary to form 5β‐pregnane‐3β, 21‐diol‐20‐one from pregnenolone: (a) β‐hydroxysteroid dehydrogenase (Dl3βHSD) from D. lanata (GenBank: DQ466890.1), (b) 3‐ketosteroid isomerase (Ct3KSI) from C. testosteronii (UniProtKB ‐ P00947) steroid 5β‐reductase (AtStβR) from Arabidopsis thaliana (PDB: 6EL3_A), and steroid 21‐hydroxylase (CYP21A1) from M. musculus (MGI: 88591).
Figure 3.

Administering pregnanes supposed to be intermediates in the cardenolide pathway. Blue arrows indicate the few steps that can be carried out by yeast; open arrows indicate reactions not carried out by yeast
3.2. Combining cardenolide pathway enzymes in vitro
We used different combinations of the enzymes identified as candidates for pathway engineering to find suitable combinations to be introduced into yeast. For this purpose, rDl3βHSD, rCt3KSI, and various mutants of steroid 5β‐reductase of A. thaliana (rAtSt5βR_F342A_F153A and rAtSt5βR_R64D_F342A_F153A) were expressed in E. coli (Table 2). The AtSt5βR mutants were generated previously in an approach to modify substrate preferences of the enzyme (Petersen, Lanig, et al., 2016). The mutant rAtSt5βR_F342A_F153A used here was initially optimized for progesterone 5β‐reduction barely accepting small enone substrates (Petersen, Ernst, et al., 2016). It also converts 21‐hydroxprogesterone to 5β‐pregnane‐21‐ol‐3,20‐dione, a possible intermediate in the yeast cardenolide pathway to be constructed (Figure 4), about 40% better than wild‐type rAtSt5βR.
Table 2.
rDl3βHSD, rCt3KSI, and rAtSt5βR_F342A_F153A or rAtSt5βR_R64D_F342A_F153A were combined (1:1:1)
| Enzymes combined | Cosubstrate (mM) | After incubation | ||
|---|---|---|---|---|
| Progesterone |
5β‐Pregnane‐ 3,20‐dione |
5β‐Pregnane−3β‐ol−20‐one | ||
|
3βHSD 3KSI AtSt5βR_DMCb |
0.4 NAD+ | 25.8 ± 2.6 | n.d.a | n.d. |
|
3βHSD 3KSI AtSt5βR_DMC |
0.4 NAD+ 0.4 NADH/H+ | 17.2 ± 0.7 | n.d. | 0.1 ± 0.03 |
|
3βHSD 3KSI AtSt5βR_DMC |
0.8 NAD+ | 31.4 ± 0.3 | n.d. | 0.2 ± 0.07 |
|
3βHSD 3KSI AtSt5βR_DMC |
0.4 NAD+ | 31.4 ± 0.3 | n.d. | 0.2 ± 0.07 |
|
3βHSD 3KSI AtSt5βR_DMc |
0.4 NAD+ 0.4 NADPH/H+ |
5.0 ± 0.04 | 8.5 ± 0.3 | 67.5 ± 1.2 |
Pregnenolone (0.3 mM) was added, and the assays were incubated for 2 hr at 40°C and then analyzed by GC‐MS. Amounts of substrate and products were combined and set to equal 100%.
n.d.: not detectable.
AtSt5βR_DMC:AtSt5βR_R64D_F342A_F153A.
AtSt5βR_DM: AtSt5βR_F342A_F153A.
Figure 4.
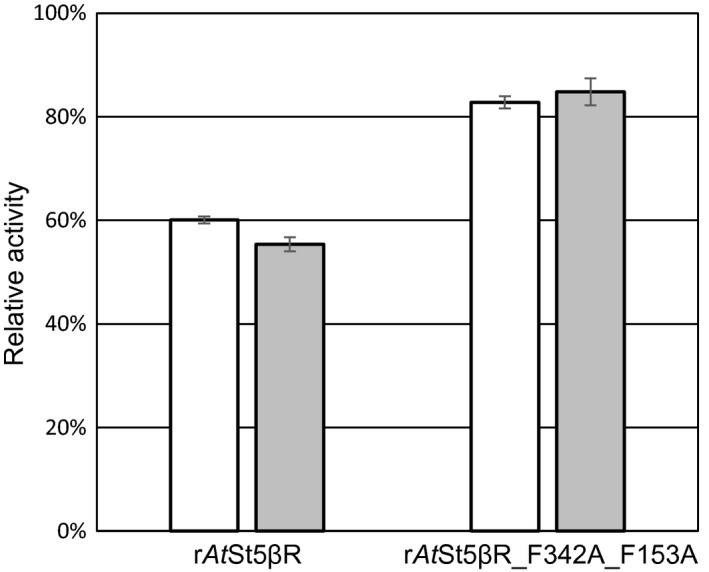
Enzymatic activity of rAtSt5βR and rAtSt5βR_F342A_F153A with two different substrates (open bars: progesterone, shaded bars: 21‐hydroxyprogesterone). The activities were measured in triplicate. Abbreviations as in Table 1
In the first reaction of the sequence, rDl3βHSD uses NAD+ as cosubstrate and releases NADH/H+ while isoprogesterone is formed. Progesterone is converted to 5β‐pregnane‐3,20‐dione by rAtSt5βR using NADPH/H+ as the cosubstrate (Figure 5). Actually, the sequence from pregnenolone to progesterone can be accomplished in the presence of rDl3βHSD and rAtSt5βR, NAD + and NADPH/H+. The intermediate isoprogesterone nonenzymatically isomerizes to progesterone. We added a bacterial 3KSI to allow for rapid enzymatic isomerization (Table 2). NADH/H+ can also be used for the reduction of the intermediate 5β‐pregnane‐3,20‐dione finally yielding 5β‐pregnane‐3β‐ol‐20‐dione.
Figure 5.
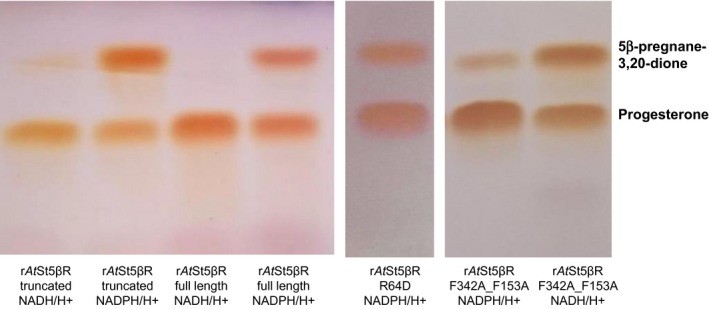
TLC analyses of various AtSt5βR muteins generated and tested during this study. The enzyme assays were carried out as described in Material and Methods
Special emphasis was put on the cosubstrate dependencies. As for rDl3βHSD, NAD + and NADH/H+ (for reduction) were the preferred cosubstrate(s) but NADP + and NADPH/H+ (for reduction) were also accepted (Herl, Frankenstein, Meitinger, Müller‐Uri, & Kreis, 2007). rAtSt5βR_full length strictly requires NADPH/H + as the hydride donor, whereas the truncated rAtSt5βR form used to generate all other muteins also showed weak activity with NADH/H+ (Figure 5). In several AtSt5βR muteins, the cosubstrate specificity was altered from NADPH/H+ to NADH/H+. One of the cosubstrate‐binding motifs (motif II) is GVARR (Pérez‐Bermúdez, Moya García, Tuñón, & Gavidia, 2010; Thorn et al., 2008) which was modified in the mutants rAtSt5βR_A62D_R63M_R64N and rAtSt5βR_R64D. In another mutein, rAtSt5βR_R64D_F342A_F153A, substrate‐ and cosubstrate‐tailoring was combined. Among the motif II cosubstrate mutants, rAtSt5βR_R64D was the most efficient accepting NADH/H+ (as estimated from TLC analyses; Figure 5). Quite unexpectedly, it turned out that rAtSt5βR_F342A_F153A (a mutein not modified in the cosubstrate binding site) also accepted NADH/H+ as a cosubstrate (Figure 5). Although it was possible to generate cosubstrate mutants in this way providing a cosubstrate‐regenerating system for the reaction sequence from pregnenolone to 5β‐pregnane‐3,20‐dione, the overall conversion rates were low when only NAD+ was added (Table 2). An efficient conversion to 5β‐pregnane‐3β‐ol‐20‐one was only seen when NADPH/H+ was provided. NAD+ could even be omitted. In the presence of rDl3βHSD1, rCt3KSI, rAtSt5βR_F342A_F153A, and NADPH/H+ about two‐third of the pregnenolone added was converted to 5β‐pregnane‐3,20‐dione in a four‐step enzymatic reaction using NADPH/H+ for reductive steps and NADP+ for the oxidative step at the beginning of the sequence. Finally, we decided to use AtSt5βR_F342A_F153A as the candidate gene to be introduced into yeast.
3.3. Combining cardenolide pathway enzymes in vivo
Dl3βHSD, Ct3KSI, AtSt5βR_F342A_F153A, and MmCYP21A1 genes were transferred into a ΔAPAT yeast strain using appropriate expression vectors. Each gene was set under the control of different constitutive promoters (Materials and Methods, Figure 2) and two different origins of replication (2 µ for high‐copy and ARS/CEN for low‐copy vectors) and termed pL1_hc or pL1_lc, respectively, using the AssemblX toolkit (Hochrein et al., 2017).
The four derived transcription units were initially assembled in S. cerevisiae YPH500 cells. Sequencing of the resulting plasmids verified the correct assembly. The individual enzymes were shown to be active in vivo by adding the respective pathway intermediates and incubating for several days. Product formation was verified by TLC. We then transformed the pL1_hc or pL1_lc plasmids into ΔAPAT yeast cells. Pregnenolone (0.2 mM) was administered, and product formation was verified after incubation for 8 days. The plasmid pL1_hc was used in the further experiments because it allowed for higher production rates than pL1_lc. The yeast strain ΔAPAT_pL1_hc was capable of producing 5β‐pregnane‐3β,21‐diol‐20‐one amounting to up to 12% of the steroidal products. Progesterone, 5β‐pregnane‐3,20‐dione, 5β‐pregnane‐3β‐ol‐20‐one, 21‐hydroxyprogesterone, 5β‐pregnane‐21‐ol‐3,20‐one accumulated as well. Progesterone and 21‐hydroxyprogesterone accumulated in large amounts and represented almost 65% of the pregnane pool.
Samples were analyzed by qPCR for the expression of the introduced genes immediately after the inoculation of pregnenolone, and again after 8 d and 16 d of cultivation. All genes introduced (AtSt5βR_F342A_F153A, Dl3βHSD, Ct3KSI, MmCYP21A1) were expressed. TFC1 (transcription factor class C) was used as the reference gene. The comparatively weak expression of Dl3βHSD was not investigated further since the reaction was not slowing substrate flow significantly. Gene expression was also detected after long periods of cultivation (Figure 6).
Figure 6.
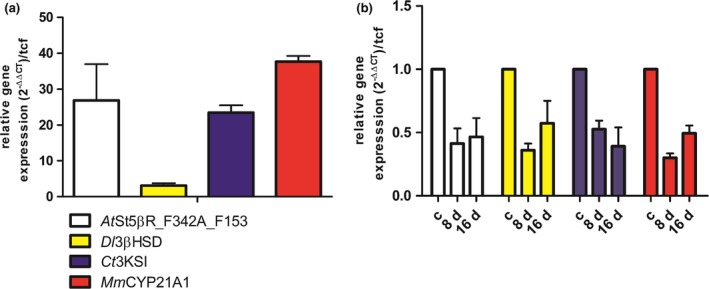
(a) Relative gene expression of AtSt5βR_F342A_F153A, Dl3βHSD, Ct3KSI and the MmCYP21A1 in ΔAPAT_pL1_hc. Immediately after inoculation and addition of the substrate (b) after 8 d and 16 d with the expression data in A set to equal 1. TFC1 was used as the reference gene
3.4. Improving pregnane flux in vivo
The conversion of progesterone to 21‐hydroxylated derivatives was quite low. It was demonstrated that progesterone but not pregnenolone and 5β‐pregnane‐3β‐ol‐20‐one were converted by a yeast strain bearing the MmCYP21A1 only (Figure 7). MmCYP21A1 seems to accept few steroid substrates only (see Discussion). In order to improve the electron flux in yeast mitochondria for the CYP21A1 gene (Szczebara et al., 2003), adrenodoxin reductase (ADR) and adrenodoxin (ADX) were subcloned as an ADR/ADX fusion gene construct into a high‐copy yeast expression vector resulting in pCR601. ΔAPAT yeast cells were then transformed with either a single plasmid carrying the CYP21A1 gene or with two plasmids, one carrying the CYP21A1 and the other one an ADR/ADX fusion gene. Progesterone was added to cells of both yeast lines in order to investigate whether the expressed ADR/ADX fusion gene would have a beneficial effect on the flux of progesterone into the cardenolide pathway. Actually, the activity of the steroid 21‐hydroxylase doubled in the presence of ADR/ADX. Therefore, we transformed ΔAPAT_pL1_hc yeast with a second plasmid (pCR601), yielding the yeast strain ΔAPAT_pL1601.
Figure 7.
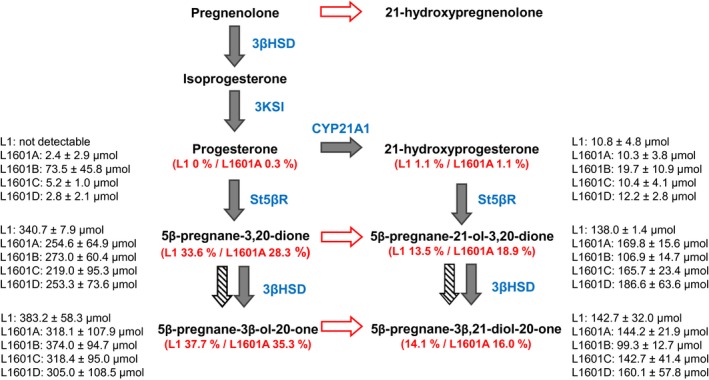
Summarizing various observations. 1) Black arrows show the flux of pregnane intermediates in ΔAPAT_pL1_hc yeast (L1); 2) Hashed arrows indicate reactions possibly sustained by yeast‐borne enzymes; 3) Pregnenolone metabolism in ΔAPAT_pL1_hc yeast and four lines derived thereof that were cotransformed with a plasmid containing an ADR/ADX gene (L1601). The pregnane patterns (%) detected in the medium after 5 d of incubation at 30 °C are shown (n= mean ± SD), for better comparison values of two strains (L1 and L1601A are shown in red as percentages); 4) Red arrows show conversions not observed in ΔAPAT_CYP21A1, only progesterone seems to be a substrate for MmCYP21A1
The yeast strains ΔAPAT_pL1_hc and ΔAPAT_pL1601 were grown in selective media, and pregnenolone (0.13 mM) was added. Complete depletion of pregnenolone was observed. A large amount of 5β‐pregnane‐3β,21‐diol‐20‐one was formed as also observed in the experiments without the plasmid pCR601. 5β‐pregnane‐3,20‐dione and 5β‐pregnane‐3β‐ol‐20‐one were abundant (Figure 8).
Figure 8.
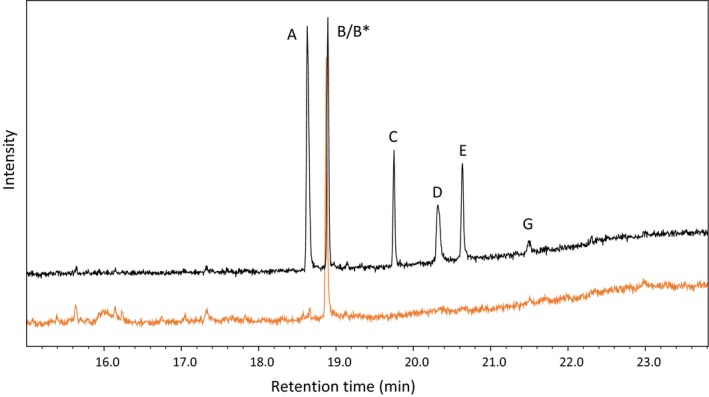
GC/MS Profile of a typical steroid extract from ΔAPAT_pL1601 (black) or ΔAPAT (orange) yeast cells fed with pregnenolone. A) 5β‐Pregnane‐β‐ol‐20‐one, B) 5β‐pregnane‐3,20‐dione, C) progesterone, D) 5β‐pregnane‐3β,21‐diol‐20‐one, E) 5β‐pregnane‐21‐ol‐3,20‐dione, G) 21‐hydroxyprogesterone
Four different colonies (named A, B, C, and D) of ΔAPAT_pL1601 yeast strain were analyzed. The clone ΔAPAT_pL1601_B showed only weak steroid 21‐hydroxylation activity and accumulated progesterone as the main product. The ΔAPAT_pL1601 clones A, C,and D behaved very similar, and the ADR/ADX gene had a slightly beneficial effect on C‐21‐hydroxylation. For example, ΔAPAT_pL1601_B contained 35% 21‐hydroxylated pregnanes, whereas ΔAPAT_pL1_hc cells contained only 28.7% (Figure 7).
4. DISCUSSION
Metabolic engineering may turn unicellular microorganisms into drug‐synthesizing “biofactories”. S. cerevisiae cells are well suited for building up heterologous biosynthetic pathways of complex natural compounds like hydrocortisone or opioids (Dumas, Brocard‐Masson, Assemat‐Lebrun, & Achstetter, 2006; Galanie, Thodey, Trenchard, Interrante, & Smolke, 2015). S. cerevisiae offers a unique mix of the simplicity of a recombinant organism combined with the complexity of a eukaryotic cell. Though many attempts have been tried in the past to produce cardenolides by plant cell tissue culture, farming of foxglove still remains the sole source of cardenolides (Clemente et al., 2011). The construction of a yeast‐based biomanufacturing platform for cardenolides has not yet been described. Inspired by the successful work of converting baker's yeast into a microorganism capable of biosynthesizing hydrocortisone (Dumas et al., 2006), we expect that this approach will also be applicable for cardenolides which share several biosynthetic steps with hydrocortisone biosynthesis.
First of all, we defined three biosynthetic modules to be realized in separate approaches. CARD I was defined as the module connecting yeast sterol metabolism with the initial cardenolide pathway by using the sterol‐cleaving system designed by Duport (Duport, Spagnoli, Degryse, & Pompon, 1998). Starting from a simple carbon source, CARD I would provide pregnenolone to be channeled into the cardenolide pathway. CARD II, our focus here, comprised the five steps of the cardenolide biosynthetic pathway leading from pregnenolone to 5β‐pregnane‐3β, 21‐diol‐20‐one (Figure 1). CARD III will include C‐14β‐hydroxylation and butenolide ring formation but cannot be designed yet, since (a) genes encoding steroid C‐14β‐hydroxylases have not been described and (b) butenolide ring formation has not been elucidated in detail (Pádua et al., 2016).
Then, we checked for cardenolide biosynthetic steps that can be carried out by yeast itself. Consistent with other findings hydroxylation at C‐14 and C‐21 was not observed (Charney & Herzog, 1967; Pajič et al., 1999). We demonstrated endogenous steroid dehydrogenase activity reducing the keto group of pregn‐5‐anes but not pregn‐4‐enes. Oxidation of 3‐hydroxypregnanes or 3‐hydroxypregnanes to their respective 3‐keto derivatives was not seen in vivo. This corroborates the findings of Camerino et al (Camerino, Alberti, & Vercellone, 1953) who reported that only nonconjugated 3‐keto groups can be reduced by S. cerevisiae. A cytosolic hydroxysteroid dehydrogenase was also identified in Schizosaccharomyces pombe which, in addition to nonconjugated 3‐keto groups, also converted progesterone, testosterone, and 4‐androstene‐3,17‐dione (possessing conjugated 3‐keto groups) to the respective 3‐hydroxy derivatives.
We identified 3β‐hydroxysteroid dehydrogenase from D. lanata (Dl3βHSD), 3‐ketosteroid isomerase from C. testosteronii (Ct3KSI) and a steroid 5β‐reductase (AtStβ5R) of A. thaliana as candidates to bridge the biosynthetic gap from pregnenolone to 5β‐pregane‐3β,21‐diol‐20‐one. In previous approaches, mammalian 3βHSD was used to introduce a pregnane pathway into a yeast host (e.g., Szczebara et al., 2003). We here used the Dl3βHSD from D. lanata. 3βHSD activity is not associated with 3KSI activity in Dl3βHSD (Meitinger et al., 2016), whereas the mammalian gene is a bifunctional 3βHSD/3KSI (Labrie et al., 1996). Isomerization of isoprogesterone to progesterone is achieved in aqueous solution without the addition of an isomerase, although much less pronounced than in the presence of 3KSI (Meitinger et al., 2016). It is assumed that rapid isomerization of isoprogesterone to progesterone enhances the flux through the cardenolide pathway. Progesterone is only a weak substrate for the back reaction (reduction of the 3‐keto group) catalyzed by NADH/H+, whereas 5β‐pregnane‐3,20‐dione, one of the downstream intermediates, is reduced to the respective alcohol about 10 times more efficiently than progesterone. This has also been demonstrated for the 3βHSD of Erysimum crepidifolium (Munkert, Ernst, Müller‐Uri, & Kreis, 2014). Bacterial 3KSI has been the subject of intensive studies as a prototype for understanding the catalytic mechanism of the allylic rearrangement. Ct3KSI is a very proficient enzyme with extremely high turn‐over rates (Feierberg & Åqvist, 2002). Introduction of Ct3KSI and AtSt5βR genes into baker's yeast had not been not reported so far.
The ∆4,5 double bond of progesterone stereoselectively reduced by AtSt5βR which belongs to the PRISE subfamily of SDR enzymes (Petersen, Ernst, et al., 2016). These 1,4‐enone reductases use NADPH/H+ as cosubstrate and accept a wide range of steroid and nonsteroid substrates (Burda et al., 2009; Durchschein et al., 2012). We here generated muteins with altered cosubstrate specificity to achieve the enzymatic conversion of pregnenolone to 5β‐pregnane‐3‐ol‐20 one in vitro by just adding pregnenolone and NAD+ to the enzyme mixture. However, the AtSt5βR_F342A_F153A mutant generated by Petersen et al (Petersen, Lanig, et al., 2016) showed a 10‐fold higher progesterone conversion efficiency than AtSt5βR_R64D_F342A_F153A, which explains the low overall conversion rates when using rAtSt5βR_R64D_F342A_F153A (Table 2; Petersen, Ernst, et al., 2016). Progesterone‐5β‐reduction seems to be the rate limiting step of this reaction sequence when only NAD+ is used as the cosubstrate. We here demonstrated for the first time that four consecutive steps of the cardenolide pathway can be realized in vitro when combining Dl3βHSD, rCt3KSI, rAtSt5βR_F342A_F153A, and NADPH/H+. NADPH/H+ plays a crucial role in the metabolism of S. cerevisiae. It is also essential for cardenolide biosynthesis. The availability of NADPH/H+ can affect artificial pathways. We here tried to find AtSt5βR mutants that can use NADH/H+ as cosubstrate. However, as far as progesterone reduction was concerned the mutants generated were too weak to be considered as components of the CARD II module. Recently, Kim, Jang, Sung, Kim, and Lee (2018) enhanced the availability of NADPH in vivo by replacing a NADH‐generating enzyme (ALD2) with (ALD6) generating NADPH instead. This may be considered when improving our CARD II yeast in further studies (Kim et al., 2018).
We then transferred the respective Dl3βHSD, Ct3KSI, AtSt5βR_F342A_F153A, and MmCYP21A1 genes into a ΔAPAT yeast using the AssemblX toolkit (Hochrein et al., 2017). Only CYP21A1, known to convert progesterone into 21‐hydroxyprogesterone, had previously been expressed in baker's yeast (Wu, Hu, & Chung, 1991) as well as in fission yeast (Drăgan, Hartmann, & Bureik, 2006). It was demonstrated that all foreign genes were expressed in ΔAPAT_pL1601 yeast (Figure 5) and that expression is still high after prolonged cultivation. TFC1, which was used as the reference gene, was noted to display a very stable expression under various growth conditions (Teste, Duquenne, François, & Parrou, 2009).
One big advantage of multienzymatic synthesis in vivo is that the cosubstrates necessary are provided by the microbial production host. Most biosynthetic pathways reconstructed to date are in vivo approaches, and only in a few cases complex syntheses were carried out in vitro.
Besides the target product, 5β‐pregnane‐3,21‐diol‐20‐one several side products accumulate (Figure 8), and the product pattern was analyzed and discussed to identify bottle‐necks and find explanations for the substrate flow seen. Since 21‐hydroxyprogesterone is a substrate for rAtSt5βR (Herl et al., 2009) we propose that 21‐hydroxypregnane formation is achieved using 21‐hydroxyprogesterone and 5β‐pregnane‐21‐ol‐3,20‐dione as substrates in vivo. Hence, the other 5β‐pregnanes have to be regarded as side products that are probably not accepted by MmCYP21A1. In other words, C‐21 steroid hydroxylation in ΔAPAT_pL1_hc or ΔAPAT_pL1601 yeasts seem to take place directly after progesterone formation. In contrast, it is assumed that progesterone 5β‐reduction precedes pregnane 21‐hydroxylation in Digitalis (Clemente et al., 2011; Kreis et al., 2012). The activity of the 21‐hydroxylase doubled in the presence of ADR/ADX (known to improve the electron flux in yeast mitochondria (Szczebara et al., 2003)) in a yeast strain only transformed with MmCYP21A1. It only had a weak yet significant effect in ΔAPAT_pL1_hc (Figure 7). CYP21A1 and its human functional homologue CYP21A2 were characterized as steroid 21‐hydroxylases, converting progesterone to 21‐hydroxyprogesterone and 17α‐hydroxyprogesterone to 11‐deoxycortisol. Although more than hundred CYP21A2 mutations have been reported, the substrate preferences of these proteins have not been investigated in detail (Haider et al., 2013). Twelve CYP21A2 muteins were analyzed, and in each catalytic activity was found to be much lower than that of the wild‐type enzyme. None of the muteins discriminated for either of the two substrates (progesterone, 17α‐hydroxyprogesterone) tested (Wang et al., 2017). This issue should be addressed in future studies by the generation and characterization of CYP21A2/A1 mutants with more relaxed substrate specificity, also hydroxylating the side products. Studies in vivo have demonstrated that 5β‐pregnane‐3,20‐dione and 5β‐pregnane‐3β‐ol‐20‐one are incorporated into cardenolides in D. lanata shoot cultures indicating a more relaxed substrate specificity of endogenous steroid hydroxylases (Haussmann, Kreis, Stuhlemmer, & Reinhard, 1997). Unfortunately, pregnane 21‐hydroxylating enzymes have not yet been isolated from plants but they should anyway be considered as candidates in further attempts to improve substrate flux.
5. CONCLUSION
In summary, we were able to provide proof of concept that the reconstitution of CARD II, a module for cardenolide biosynthesis, can successfully work in vivo. The focus of future experiments concerning the CARD II module is the elimination of side reactions and/or the channeling of side products into 5β‐pregnane‐3β,21‐diol‐20‐one by modifying the MmCYP21A1 by site‐directed mutagenesis or the identification of a more appropriate (plant) CYP21A1 gene.
CONFLICT OF INTEREST
The authors have no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceptualization: Wolfgang Kreis. Investigation: Daniel Geiger, Jennifer Munkert, Jan Petersen, Christoph Rieck, Juliane Strasser. Resources: Nadine Meitinger, Katrin Messerschmidt. Writing—Original Draft Preparation: Daniel Geiger, Wolfgang Kreis, Christoph Rieck. Writing—Review & Editing: Wolfgang Kreis, Jennifer Munkert, Christoph Rieck.
ETHICS STATEMENT
None required.
ACKNOWLEDGMENTS
We thank Daniel Herschlag for providing plasmid pKK22‐3::3KSI and Rita Bernhard for providing plasmid pBAR_Twin::ADR/ADX. We thank Barbara White for proofreading this manuscript.
Rieck C, Geiger D, Munkert J, et al. Biosynthetic approach to combine the first steps of cardenolide formation in Saccharomyces cerevisiae . MicrobiologyOpen. 2019;8:e925 10.1002/mbo3.925
Christoph Rieck and Daniel Geiger authors contributed equally to this manuscript.
Data Availability Statement: Not applicable.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- Burda, E. , Kraußer, M. , Fischer, G. , Hummel, W. , Müller‐Uri, F. , Kreis, W. , & Gröger, H. (2009). Recombinant Δ4, 5‐steroid 5 β‐reductases as biocatalysts for the reduction of activated C=C‐double bonds in monocyclic and acyclic molecules. Advanced Synthesis & Catalysis, 351(17), 2787–2790. [Google Scholar]
- Camerino, B. , Alberti, C. G. , & Vercellone, A. (1953). Selektive Reduktion der Carbonylgruppe in Stellung 3 von Steroiden durch Hefe. Helvetica Chimica Acta, 36(7), 1945–1948. 10.1002/hlca.19530360733 [DOI] [Google Scholar]
- Cauet, G. , Degryse, E. , Ledoux, C. , Spagnoli, R. , & Achstetter, T. (1999). Pregnenolone esterification in Saccharomyces cerevisiae: A potential detoxification mechanism. European Journal of Biochemistry, 261(1), 317–324. 10.1046/j.1432-1327.1999.00282.x [DOI] [PubMed] [Google Scholar]
- Charney, W. , & Herzog, H. L. (1967). Microbiological transformations of steroids; a handbook. New York, NY: Academic Press. [Google Scholar]
- Clemente, E. S. , Müller‐Uri, F. , Nebauer, S. G. , Segura, J. , Kreis, W. , & Arrillaga, I. (2011). Digitalis In Chittaranjan K. (Ed.), Wild crop relatives: Genomic and breeding resources, plantation and ornamental crops (pp. 73–112). Berlin/Heidelberg, Germany: Springer. [Google Scholar]
- Drăgan, C. A. , Hartmann, R. W. , & Bureik, M. (2006). A fission yeast‐based test system for the determination of IC50 values of anti‐prostate tumor drugs acting on CYP21. Journal of Enzyme Inhibition and Medicinal Chemistry, 21(5), 547–556. [DOI] [PubMed] [Google Scholar]
- Dumas, B. , Brocard‐Masson, C. , Assemat‐Lebrun, K. , & Achstetter, T. (2006). Hydrocortisone made in yeast: Metabolic engineering turns a unicellular microorganism into a drug‐synthesizing factory. Biotechnology Journal: Healthcare Nutrition Technology, 1(3), 299–307. 10.1002/biot.200500046 [DOI] [PubMed] [Google Scholar]
- Duport, C. , Spagnoli, R. , Degryse, E. , & Pompon, D. (1998). Self‐sufficient biosynthesis of pregnenolone and progesterone in engineered yeast. Nature Biotechnology, 16(2), 186 10.1038/nbt0298-186 [DOI] [PubMed] [Google Scholar]
- Durchschein, K. , Wallner, S. , Macheroux, P. , Schwab, W. , Winkler, T. , Kreis, W. , & Faber, K. (2012). Nicotinamide‐dependent Ene reductases as alternative biocatalysts for the reduction of activated alkenes. European Journal of Organic Chemistry, 2012(26), 4963–4968. 10.1002/ejoc.201200776 [DOI] [Google Scholar]
- Eichhorn, E. J. , & Gheorghiade, M. (2002). Digoxin. Progress in Cardiovascular Diseases, 44(4), 251–266. 10.1053/pcad.2002.31591 [DOI] [PubMed] [Google Scholar]
- Feierberg, I. , & Åqvist, J. (2002). Computational modeling of enzymatic keto‐enol isomerization reactions. Theoretical Chemistry Accounts, 108(2), 71–84. 10.1007/s00214-002-0365-7 [DOI] [Google Scholar]
- Galanie, S. , Thodey, K. , Trenchard, I. J. , Interrante, M. F. , & Smolke, C. D. (2015). Complete biosynthesis of opioids in yeast. Science, 349(6252), 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D. , St Jean, A. , Woods, R. A. , & Schiestl, R. H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Research, 20(6), 1425 10.1093/nar/20.6.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel, E. , Karvar, S. , Yucesan, B. , Eker, I. , & Sameeullah, M. (2017). An overview of cardenolides in digitalis‐more than a cardiotonic compound. Current Pharmaceutical Design, 23(34), 5104–5114. [DOI] [PubMed] [Google Scholar]
- Haider, S. , Islam, B. , D'Atri, V. , Sgobba, M. , Poojari, C. , Sun, L. , … New, M. I. (2013). Structure–phenotype correlations of human CYP21A2 mutations in congenital adrenal hyperplasia. Proceedings of the National Academy of Sciences, 110(7), 2605–2610. 10.1073/pnas.1221133110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannemann, F. , Virus, C. , & Bernhardt, R. (2006). Design of an Escherichia coli system for whole cell mediated steroid synthesis and molecular evolution of steroid hydroxylases. Journal of Biotechnology, 124(1), 172–181. 10.1016/j.jbiotec.2006.01.009 [DOI] [PubMed] [Google Scholar]
- Haussmann, W. , Kreis, W. , Stuhlemmer, U. , & Reinhard, E. (1997). Effects of various pregnanes and two 23‐nor‐5‐cholenic acids on cardenolide accumulation in cell and organ cultures of Digitalis lanata . Planta Medica, 63(05), 446–453. [DOI] [PubMed] [Google Scholar]
- Herl, V. , Fischer, G. , Reva, V. A. , Stiebritz, M. , Muller, Y. A. , Müller‐Uri, F. , & Kreis, W. (2009). The VEP1 gene (At4g24220) encodes a short‐chain dehydrogenase/reductase with 3‐oxo‐Δ4, 5‐steroid 5β‐reductase activity in Arabidopsis thaliana L. Biochimie, 91(4), 517–525. 10.1016/j.biochi.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Herl, V. , Frankenstein, J. , Meitinger, N. , Müller‐Uri, F. , & Kreis, W. (2007). Δ5‐3β‐Hydroxysteroid dehydrogenase (3βHSD) from Digitalis lanata. Heterologous expression and characterisation of the recombinant enzyme. Planta Medica, 73(07), 704–710. [DOI] [PubMed] [Google Scholar]
- Hochrein, L. , Machens, F. , Gremmels, J. , Schulz, K. , Messerschmidt, K. , & Mueller‐Roeber, B. (2017). AssemblX: A user‐friendly toolkit for rapid and reliable multi‐gene assemblies. Nucleic Acids Research, 45(10), e80 10.1093/nar/gkx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. E. , Jang, I. S. , Sung, B. H. , Kim, S. C. , & Lee, J. Y. (2018). Rerouting of NADPH synthetic pathways for increased protopanaxadiol production in Saccharomyces cerevisiae . Scientific Reports, 8(1), 15820 10.1038/s41598-018-34210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis, W. , & Müller‐Uri, F. (2012). Cardenolide aglycone formation in Digitalis In Bach T. J., & Rohmer M. (Eds.), Isoprenoid synthesis in plants and microorganisms (pp. 425–438). New York, NY: Springer. [Google Scholar]
- Krishna, A. B. , Manikyam, H. K. , Sharma, V. K. , & Sharma, N. (2015). Plant cardenolides in therapeutics. International Journal of Indigenous Medicinal Plants, 48, 1871–1896. [Google Scholar]
- Labrie, F. , Simard, J. , Bélanger, A. , Pelletier, G. , Morel, Y. , Mebarki, F. , … Rhéaume, E. (1996). The 3β‐hydroxysteroid dehydrogenase/isomerase gene family: Lessons from type II 3β‐HSD congenital deficiency. In signal transduction in testicular cells (pp. 185–218). Berlin, Heidelberg: Springer. [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25(4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Meitinger, N. , Munkert, J. , Maia de Pádua, R. , de Souza Filho, J. D. , Maid, H. , Bauer, W. , … Kreis, W. (2016). The catalytic mechanism of the 3‐ketosteroid isomerase of Digitalis lanata involves an intramolecular proton transfer and the activity is not associated with the 3β‐hydroxysteroid dehydrogenase activity. Tetrahedron Letters, 57(14), 1567–1571. 10.1016/j.tetlet.2016.02.099 [DOI] [Google Scholar]
- Munkert, J. , Ernst, M. , Müller‐Uri, F. , & Kreis, W. (2014). Identification and stress‐induced expression of three 3β‐hydroxysteroid dehydrogenases from Erysimum crepidifolium Rchb. and their putative role in cardenolide biosynthesis. Phytochemistry, 100, 26–33. 10.1016/j.phytochem.2014.01.006 [DOI] [PubMed] [Google Scholar]
- Pádua, R. M. , Meitinger, N. , Hennemann, M. , Schebitz, P. , Waibel, R. , Löber, S. , … Kreis, W. (2016). Spontaneous butenolide ring formation of pregnane‐21‐O‐malonyl hemiesters under mild reaction conditions is facilitated by the 14β‐hydroxy group present in all natural cardenolides. Tetrahedron, 72(30), 4556–4563. 10.1016/j.tet.2016.06.024 [DOI] [Google Scholar]
- Pajič, T. , Vitas, M. , Žigon, D. , Pavko, A. , Kelly, S. L. , & Komel, R. (1999). Biotransformation of steroids by the fission yeast Schizosaccharomyces pombe . Yeast, 15(8), 639–645. [DOI] [PubMed] [Google Scholar]
- Pérez‐Bermúdez, P. , Moya García, A. A. , Tuñón, I. , & Gavidia, I. (2010). Digitalis purpurea P5βR2, encoding steroid 5β‐reductase, is a novel defense‐related gene involved in cardenolide biosynthesis. New Phytologist, 185(3), 687–700. 10.1111/j.1469-8137.2009.03080.x [DOI] [PubMed] [Google Scholar]
- Petersen, J. , Ernst, M. , Gärtner, E. , Klein, J. , Müller‐Uri, F. , Munkert, J. , & Kreis, W. (2016). PRISE (progesterone 5β‐reductases/iridoid synthases): Their roles in specialized plant metabolism. Planta Medica, 82(Suppl. 01), P781 10.1055/s-0036-1596807 [DOI] [Google Scholar]
- Petersen, J. , Lanig, H. , Munkert, J. , Bauer, P. , Müller‐Uri, F. , & Kreis, W. (2016). Progesterone 5β‐reductases/iridoid synthases (PRISE): Gatekeeper role of highly conserved phenylalanines in substrate preference and trapping is supported by molecular dynamics simulations. Journal of Biomolecular Structure and Dynamics, 34(8), 1667–1680. 10.1080/07391102.2015.1088797 [DOI] [PubMed] [Google Scholar]
- Schmidt, K. , Petersen, J. , Munkert, J. , Egerer‐Sieber, C. , Hornig, M. , Muller, Y. A. , & Kreis, W. (2018). PRISEs (progesterone 5β‐reductase and/or iridoid synthase‐like 1, 4‐enone reductases): Catalytic and substrate promiscuity allows for realization of multiple pathways in plant metabolism. Phytochemistry, 156, 9–19. 10.1016/j.phytochem.2018.08.012 [DOI] [PubMed] [Google Scholar]
- Schwans, J. P. , Sunden, F. , Gonzalez, A. , Tsai, Y. , & Herschlag, D. (2013). Uncovering the determinants of a highly perturbed tyrosine pKa in the active site of ketosteroid isomerase. Biochemistry, 52(44), 7840–7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, M. S. , Thodey, K. , Trenchard, I. , & Smolke, C. D. (2012). Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Research, 12(2), 144–170. 10.1111/j.1567-1364.2011.00774.x [DOI] [PubMed] [Google Scholar]
- Szczebara, F. M. , Chandelier, C. , Villeret, C. , Masurel, A. , Bourot, S. , Duport, C. , … Dumas, B. (2003). Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nature Biotechnology, 21(2), 143 10.1038/nbt775 [DOI] [PubMed] [Google Scholar]
- Teste, M. A. , Duquenne, M. , François, J. M. , & Parrou, J. L. (2009). Validation of reference genes for quantitative expression analysis by real‐time RT‐PCR in Saccharomyces cerevisiae . BMC Molecular Biology, 10(1), 99 10.1186/1471-2199-10-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn, A. , Egerer‐Sieber, C. , Jäger, C. M. , Herl, V. , Müller‐Uri, F. , Kreis, W. , & Muller, Y. A. (2008). The crystal structure of progesterone 5β‐reductase from Digitalis lanata defines a novel class of short chain dehydrogenases/reductases. Journal of Biological Chemistry, 283(25), 17260–17269. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Pallan, P. S. , Zhang, W. , Lei, L. I. , Yoshimoto, F. K. , Waterman, M. R. , … Guengerich, F. P. (2017). Functional analysis of human cytochrome P450 21A2 variants involved in congenital adrenal hyperplasia. Journal of Biological Chemistry, 292(26), 10767–10778. 10.1074/jbc.M117.792465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D. A. , Hu, M. C. , & Chung, B. C. (1991). Expression and functional study of wild‐type and mutant human cytochrome P450c21 in Saccharomyces cerevisiae . DNA and Cell Biology, 10(3), 201–209. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Werling, U. , & Edelmann, W. (2012). SLiCE: A novel bacterial cell extract‐based DNA cloning method. Nucleic Acids Research, 40(8), e55 10.1093/nar/gkr1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


