Abstract
The possibility of introducing a reliable assay for a quick identification and differentiation of the main species of Mycobacterium tuberculosis complex (MTBC) supports the improvement of efficient tuberculosis combating strategies worldwide. Commercially available assays are often based on cultured samples; however, due to the long cultivation time of mycobacteria, results are delayed. Developed PCR approaches have been published previously, though, when testing intricate veterinary samples, the complex composition of multiplex qPCRs frequently leads to assay failure. In order to overcome those limits, a paradigm of a three‐reaction high‐resolution melting (HRM) assay for the simultaneous identification and differentiation of the main members of MTBC was established. The assay is based on single nucleotide polymorphisms within gyrB and gyrA, which have been used as target for the establishment of two highly specific HRM assays (HRM assays 1 and 2) discriminating M. tuberculosis/ Mycobacterium canetti, Mycobacterium bovis/M. bovis BCG, Mycobacterium caprae/rare M. caprae/M. bovis ecotypes, Mycobacterium africanum/Mycobacterium orygis/ Mycobacterium pinnipedii/Clade A1, Mycobacterium microti, and a rare subtype of M. canettii followed by a third HRM assay (HRM assay 3) allowing a further differentiation of M. bovis, M. bovis BCG, and a rare subtype of M. caprae/M. bovis, which is considered to be a novel ecotype. High‐resolution melting assay 1 is described in a previously published report. High‐resolution melting assay 2 showed 100% correlation of all 39 examined isolates with the results of a commercial identification kit. 96% of the clinical samples tested demonstrated concordant results. High‐resolution melting assay 3 showed an accordance of 100% with the results of the commercially available identification kit of all 22 samples analyzed. The proposed strategy of the three‐reaction HRM assay can be used for an accurate differentiation of up to seven groups of MTBC and potentially to identify a rare subtype of M. canettii either on isolates or on clinical samples.
The HRM assay enables rapid identification and differentiation of the clinically most relevant species of MTBC namely M. tuberculosis, M. africanum, M. microti, M. bovis, M. caprae, and M. bovis BCG from directly extracted samples and cultured material. The use of this powerful assay may save several months of cultivation time and supports improvement of efficient tuberculosis combating strategies worldwide.
![]()
1. INTRODUCTION
Tuberculosis is a major cause of human death induced by only one infectious agent resulting in approximately 10 million new infections per year along with about 1.6 million deaths in 2017 (WHO, 2018). Tuberculosis persists as a major health concern not only in humans but also in veterinary medicine.
Mycobacterium tuberculosis complex (MTBC) comprises the closely related species Mycobacterium (M.) tuberculosis, M. bovis, M. bovis Bacillus Calmette and Guérin (BCG), M. caprae, M. africanum, M. microti, M. pinnipedii, M. orygis, and four further species known as animal‐adapted Clade A1 (Dassie bacillus, M. mungi, Chimpanzee bacillus, and M. suricattae) (Brites et al., 2018). Furthermore, Mycobacterium canettii is a genetically more diverse and recombinogenic organism as observed earlier (Fabre et al., 2010), only leading to opportunistic human infections from time to time (Boritsch et al., 2016; Supply et al., 2013). Although its similarity of the nucleotide codes to the species of MBTC, it is not considered to be part of MTBC (Brites et al., 2018). Mycobacterium canettii is mainly limited to the horn of Africa and most of the known strains were isolated in the Republic of Djibouti (Blouin et al., 2014). Mycobacterium tuberculosis is known to be the major source of human tuberculosis; however, numerous cases of infection with other members of the complex are known. Mycobacterium bovis and more rarely M. caprae are the causative agents for bovine tuberculosis, which is recognized to be an important zoonosis responsible for significant economic loss (Rodriguez‐Campos, Smith, Boniotti, & Aranaz, 2014). A recent study (Loiseau et al., 2019) revealed two rare M. caprae/M. bovis ecotypes with no intrinsic pyrazinamide (PZA) resistance, in contrast to the common M. bovis strains, detected in samples from Malawi (Guerra‐Assunção et al., 2015), Germany (Friedrich Loeffler Institute) (defined in this study as ecotype I), and China (Orloski, Robbe‐Austerman, Stuber, Hench, & Schoenbaum, 2018) (defined in this study as ecotype II). These findings resulted in a proposed revision of interpretation of the GenoType MTBC test (Hain Lifescience). In regions where tuberculosis is endemic, neonates were vaccinated with the attenuated M. bovis strain BCG. In immunocompromised children, this procedure can cause a disease pattern similar to the one of tuberculosis (Hesseling et al., 2006). In order to evaluate a zoonotic risk of MTBC, it is important to rely on a fast and accurate method capable of identification and differentiation of the species of MTBC leading to improved programs in public health surveillance and enhanced food safety.
Various molecular assays exist to differentiate species within MTBC. Nevertheless, many approaches have several limits. Most of the methods as, for example, the commercial GenoType MTBC test are not validated for use on clinical samples (Costa, Amaro, et al., 2014; Kasai, Ezaki, & Harayama, 2000; Niemann, Harmsen, Rüsch‐Gerdes, & Richter, 2000; Pinsky & Banaei, 2008; Pounder et al., 2010; Reddington et al., 2012). Since Mycobacteria require many months to obtain considerable growth in culture, methods based on cultivated isolates require much time to determine the correct MTBC species. Other methods are very laborious (Kamerbeek et al., 1997), require expensive equipment (Jagielski et al., 2014), or rely on multiplex qPCR assays (Costa, Amaro, et al., 2014; Halse, Escuyer, & Musser, 2011; Pinsky & Banaei, 2008; Pounder et al., 2010; Reddington et al., 2012). However, the complex composition of a multiplex qPCR can often lead to PCR inhibition due to intricate organ samples of animal tissues.
High‐resolution melting (HRM) approaches are cheap and rapid assays, which are able to detect single nucleotide polymorphism (SNP) according to altered melting temperatures (Tm) of dissociating PCR amplicons (Vossen, Aten, Roos, & Dunnen, 2009). A fluorescent nucleic acid dye is intercalating with the resulting PCR amplicons, which are dissociating upon increase in temperature and thus resulting in a decrease in fluorescence intensity. The determinate of Tm is based on its nucleotide sequence, length, and level of GC. The user‐friendly single‐plex HRM assay can be completed within roughly 2 hr. Moreover, as a major advantage, HRM assays can be performed using samples directly extracted from clinical tissue. High‐resolution melting assays have been used to discriminate various bacteria species (Esteves et al., 2018; Jeffery, Gasser, Steer, & Noormohammadi, 2007; Robertson et al., 2009; Stephens, Inman‐Bamber, Giffard, & Huygens, 2008; Winchell, Wolff, Tiller, Bowen, & Hoffmaster, 2010) or to analyze antibiotic resistance in M. tuberculosis (Anthwal et al., 2017; Chen et al., 2011; Yadav et al., 2012). Moreover, HRM assays have been established for differentiation of nontuberculous mycobacteria (NTM) and confining them from MTBC (Issa et al., 2014; Khosravi, Hashemzadeh, Hashemi Shahraki, & Teimoori, 2017; Perng et al., 2012). Some studies combined HRM with multiplex qPCR assays targeting at the region of difference (RD) (Pinsky & Banaei, 2008; Pounder et al., 2010).
We have previously reported the design and evaluation of a HRM assay (HRM assay 1) for the identification and differentiation of MTBC into three groups most relevant for veterinarians (Landolt, Stephan, & Scherrer, 2019). In the present study, two additional HRM assays (HRM assays 2 and 3) were developed with the aim to discern the main members of MTBC. By combining HRM assays 1 and 2 targeting six SNPs on the gyrB gene, a two‐step paradigm was obtained, distinguishing M. tuberculosis/M. canettii, M. canettii (rare subtype), M. africanum/M. orygis/M. pinnipedii/Clade A1, M. bovis/M. bovis BCG, M. caprae/rare M. caprae/M. bovis ecotypes, and M. microti in cultured isolates and clinical samples. If requested, it is possible to further differentiate M. bovis, M. bovis BCG, and one of the two rare M. caprae/M. bovis ecotypes (ecotype I) by conducting another HRM assay (HRM assay 3) based on two SNPs within gyrA and thus completing the three‐step paradigm. For most diagnostic applications, however, a combination of HRM assays 1 and 2 will be sufficient.
2. MATERIALS AND METHODS
2.1. Samples and reference strains
Sixty‐one samples positive for MTBC were collected from 39 different animals (Table 1, Table A1). One wild boar isolate was put at our disposal by courtesy of Lucía de Juan Ferré and Beatriz Romero Martínez. In total, 62 samples including 39 cultured isolates and 23 directly extracted clinical samples were tested with HRM assay 2. High‐resolution melting assay 3 was validated to distinguish between M. bovis and M. bovis BCG using 15 M. bovis isolates and 7 M. bovis clinical samples. Positive control samples (M. microti ATCC 19422, M. bovis BCG Pasteur ATCC 35734, M. bovis BCG Tice ATCC 27289, and M. tuberculosis H37Rv) were used in each qPCR run. Clinical samples were received in the Laboratory of Veterinary Bacteriology, University of Zurich, between 2013 and 2016.
Table 1.
MTBC‐positive samples used for the development of the HRM assays 2 and 3
| Species | Host | HRM assay 2 | HRM assay 3 |
|---|---|---|---|
| No. of isolates | No. of isolates | ||
| Cultured material (n = 39) | |||
| M. tuberculosis | Elephant | 3 | |
| M. caprae | Cow | 7 | |
| M. bovis | Cow | 15 | 15 |
| M. microti | Cat | 8 | |
| M. microti | Alpaca | 3 | |
| M. microti | Llama | 2 | |
| M. microti | Wild boar (Spain) | 1 | |
| Clinical samples (n = 23) | |||
| M. tuberculosis | Elephant | 2 | |
| M. caprae | Cow | 4 | |
| M. bovis | Cow | 7 | 7 |
| M. microti | Cat | 5 | |
| M. microti | Alpaca | 3 | |
| M. microti | Llama | 2 | |
| Total | 62 | 22 | |
Thirty‐nine isolates obtained from cultured material, whereas 23 samples were clinical samples directly extracted from tissue samples. Sixty‐one samples were derived from Switzerland, whereas one isolate originated from Spain.
2.2. Culture and DNA extraction
Sample preparation, culture, and DNA extraction were conducted as described in a former study (Ghielmetti et al., 2017). GenoType MTBC test (Hain Lifescience), spoligotyping (Ruettger et al., 2012), and multilocus variable number tandem repeat analysis using an internationally established 24‐loci panel (Supply et al., 2006) were used for species identification of cultured isolates. Standard biosecurity procedures were followed for handling of samples.
2.3. HRM development
High‐resolution melting development of assay 1 was described in a former study (Landolt et al., 2019). The differentiation of the main members of MTBC is performed applying a paradigm of a three‐reaction HRM assay based on four SNPs located on gyrB (base pair positions 675, 756, 1,410, and 1,450 [Niemann et al., 2000] and one SNP on gyrA [base pair position 1,323]). In order to extend the obtained SNP differentiation scheme with the recently described two rare ecotypes of M. caprae/M. bovis (ecotypes I and II), a rare M. canettii subtype (Loiseau et al., 2019) and the animal‐adapted MTBC clades (Brites et al., 2018), two additional SNPs on gyrB (base pair positions 1,437 and 1,439), and one SNP on gyrA (base pair position 1,359) were included in the assay design (Figure 1).
Figure 1.
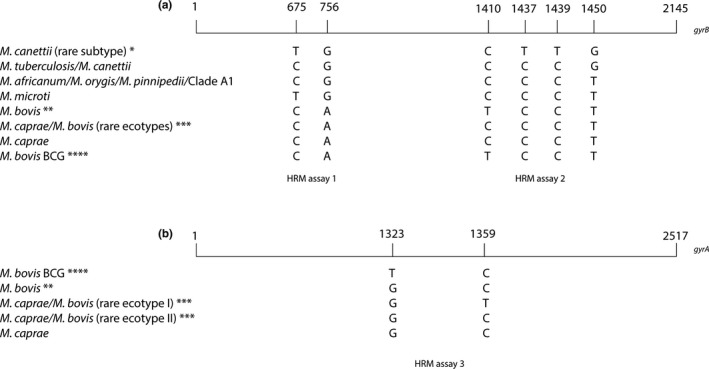
Differentiation of MTBC based on eight different single nucleotide polymorphisms (SNPs). Numbers represent the position of the SNP in relation to the start codon of gyrB and gyrA, respectively. (a) Six SNPs on gyrB are represented. HRM assay 1 (Landolt et al., 2019) allows the distinction between M. tuberculosis/M. canettii/M. africanum/M. orygis/M. pinnipedii/Clade A1, M. microti/M. canettii (rare subtype) and M. bovis/M. bovis BCG/M. caprae/rare M. caprae/M. bovis ecotypes. HRM assay 2 can differentiate between M. tuberculosis/M. canettii, M. canettii (rare subtype), M. africanum/M. orygis/M. pinnipedii/Clade A1/M. microti/M. caprae/rare M. caprae/M. bovis ecotypes, and M. bovis/M. bovis BCG. Combining the results of HRM assays 1 and 2, six groups can be distinguished: M. canettii (rare subtype), M. tuberculosis/M. canettii, M. africanum/M. orygis/M. pinnipedii/Clade A1, M. microti, M. caprae/rare M. caprae/M. bovis ecotypes, and M. bovis/M. bovis BCG. (b) Two SNPs on positions 1,323 and 1,359 of gyrA are illustrated indicating the differentiation between M. bovis, M. bovis BCG, and rare M. caprae/M. bovis ecotype I. *Rare subtype, highly recombinogenic strain, intrinsic pyrazinamide (PZA) resistance. **Intrinsic PZA resistance. ***No intrinsic PZA resistance. ****Intrinsic PZA and cycloserine resistance
2.4. Primer of HRM assays
Primers were designed based on alignments of the available sequences of gyrB and gyrA from MTBC (GenBank accession numbers: gyrB M. bovis AB014184.1, gyrB M. microti AB014205.1, gyrB M. tuberculosis X78888.1, gyrB M. africanum FR878060.1, gyrB M. caprae CP016401.1, gyrA M. bovis LT708304.1, and gyrA M. bovis BCG Pasteur AM408590.1). The designed primers amplify a conserved 107‐base pair (bp) fragment within gyrB comprising two SNPs and gyrA comprising one SNP, respectively (Figure 1). Gene specificity of all primers was confirmed by NCBI BLAST searches. Primers were HPLC‐purified and synthesized by Microsynth. Three primer pairs were designed for the three‐reaction HRM paradigm (Table 2).
Table 2.
Primers used for the three‐reaction HRM paradigm
| Assay | Primer | Target | Amplicon | Primer sequence (5′ → 3′) | Reference |
|---|---|---|---|---|---|
| HRM assay 1 |
HRM_gyrB_for HRM_gyrB_rev |
gyrB | 144 bp |
CGGCTCGAAGTCGAGATCAAG TTCGAAAACAGCGGGGTCG |
Landolt et al. (2019) |
| HRM assay 2 |
HRM_gyrB2_for HRM_gyrB2_rev |
gyrB | 107 bp |
CAAATCGTTTGTGCAGAAGGTCTG CTTGCGCCGAGGACACAG |
This study |
| HRM assay 3 |
HRM_gyrA_for HRM_gyrA_rev |
gyrA | 107 bp |
AGGCAATCCTGGACATGCAG GATGTCTTCCAGATCGGCGATC |
This study |
2.5. qPCR and melting conditions
Each HRM assay was processed separately on a Rotor‐Gene Q system (Qiagen) with the Type‐it HRM PCR Kit (Qiagen). The qPCRs were performed as described previously (Landolt et al., 2019). High‐resolution melting ramping from 76°C to 93°C was applied. Fluorescence data were measured every 2 s at 0.1°C increments generating specific melting curves. Reference strains M. microtii ATCC 19422, M. bovis BCG Pasteur ATCC 35734, and M. tuberculosis H37Rv were used as melting curve standards and positive controls. Additionally, for HRM assay 3, M. bovis BCG Tice ATCC 27289 was used. As a negative control, ultrapure water was included in each experiment. Rotor‐Gene Q Software 2.3.1 (Qiagen) was used for data analysis to generate normalized and difference plots as described in a previous study (Landolt et al., 2019). To prevent false‐negative results possibly deriving from inhibition, clinical samples were analyzed in duplicate undiluted and in the form of a 1:5 dilution. The cultured isolates were analyzed at concentrations between 100 pg and 10 ng.
To investigate the intra‐ and interassay variability of the Tm, illustrating the repeatability of the developed HRM assays, a randomly chosen subset of 22 cultured isolates and 18 clinical specimens for HRM assays 2 and 9 cultured isolates and 7 clinical specimens for HRM assay 3 were tested, respectively. The variability assays were conducted in triplicates in three single runs at three different days.
2.6. Specificity
To check for possible nonspecific signals of HRM assays 2 and 3, 41 different NTM, Nocardia paucivorans, Escherichia coli, and Streptococcus suis were tested (Table A2).
2.7. Sensitivity
The analytical sensitivity of HRM assay 2 was measured by triplicate testing of a 10‐fold dilution series of DNA isolated from strains M. microti ATCC 19422, M. bovis BCG Pasteur ATCC 35734, M. caprae clinical isolate ZH22914, and M. tuberculosis H37Rv with known concentrations in genome equivalents (GE). The analytical sensitivity of HRM assay 3 was tested for M. bovis BCG Pasteur ATCC 35734 and the clinical isolate M. bovis ZH20665 in an analogous manner by analyzing 10‐fold DNA dilution series. Based on an estimated genome size of 4.4 Mb for members of MTBC, 1 GE correlates with a DNA quantity of 4.8 fg. The slope of the resulting standard curve corresponded to the amplification efficiency of each tested strain. The limit of detection (LOD) was defined as lowest dilution for samples with successful PCR amplification of all triplicates having a Ct < 38 and a standard deviation of ≤ 0.5.
3. RESULTS
3.1. HRM of cultured isolates
All cultured isolates tested were amplified successfully yielding a melting curve. The resulting species‐specific Tm (Tables 3 and 4, Tables A3 and A4) of corresponding melting curves (Figures 2 and 3) from sample subsets used for the determination of the intra‐ and interassay variability clearly represent three independent groups in the case of HRM assay 2 and two distinct groups with HRM assay 3, respectively. Obtained Tm ranges between the different groups are very close to each other impeding a clear differentiation between members of MTBC solely based on Tm. In contrast, the illustration of dissociating curves in the form of difference plots (Figures 2c and 3c) as well as normalized plots (Figures 2b and 3b) allowed a clear differentiation between MTBC species.
Table 3.
Intra‐ and interassay variability of HRM assay 2 of cultured samples
| Run 1 | Run 2 | Run 3 | Interassay | |||||
|---|---|---|---|---|---|---|---|---|
| Tm | CV% | Tm | CV% | Tm | CV% | Tm | CV% | |
| M. tuberculosis H37Rv | 83.55 | 83.75 | 83.6 | 83.63 | 0.12 | |||
| M. bovis BCG Pasteur ATCC 35734 | 82.62 | 82.77 | 82.70 | 82.7 | 0.09 | |||
| M. microti ATCC 19422 | 83.13 | 83.25 | 83.28 | 83.22 | 0.10 | |||
| M. tuberculosis (n = 3) | 83.56 ± 0.04 | 0.01 | 83.84 ± 0.04 | 0.02 | 83.66 ± 0.06 | 0.04 | 83.70 ± 0.18 | 0.17 |
| M. bovis (n = 6) | 82.70 ± 0.10 | 0.04 | 82.93 ± 0.08 | 0.03 | 82.74 ± 0.06 | 0.02 | 82.80 ± 0.20 | 0.14 |
| M. caprae (n = 6) | 83.12 ± 0.09 | 0.03 | 83.27 ± 0.09 | 0.02 | 83.23 ± 0.06 | 0.02 | 83.19 ± 0.16 | 0.11 |
| M. microti (n = 7) | 83.10 ± 0.08 | 0.02 | 83.34 ± 0.09 | 0.04 | 83.29 ± 0.04 | 0.01 | 83.23 ± 0.21 | 0.14 |
Mean values and standard deviation of melting temperatures (Tm) of a randomly chosen subset of cultured samples are listed. Corresponding coefficients of variation (CV) in % are indicated for each MTBC species tested.
Table 4.
Intra‐ and interassay variability of HRM assay 3 of cultured samples
| Run 1 | Run 2 | Run 3 | Interassay | |||||
|---|---|---|---|---|---|---|---|---|
| Tm | CV% | Tm | CV% | Tm | CV% | Tm | CV% | |
| M. tuberculosis H37Rv | 86.82 | 86.82 | 86.85 | 86.83 | 0.02 | |||
| M. microti ATCC 19422 | 86.83 | 86.83 | 86.83 | 86.83 | 0 | |||
| M. bovis BCG Pasteur ATCC 35734 | 86.42 | 86.38 | 86.42 | 86.41 | 0.03 | |||
| M. bovis (n = 7) | 86.83 ± 0.50 | 0.01 | 86.83 ± 0.08 | 0.02 | 86.83 ± 0.10 | 0.03 | 86.83 ± 0.10 | 0.03 |
| M. bovis BCG (n = 2) | 86.39 ± 0.06 | 0.03 | 86.41 ± 0.03 | 0.03 | 86.45 ± 0.03 | 0.03 | 86.41 ± 0.08 | 0.04 |
Mean values and standard deviation of melting temperatures (Tm) of a randomly chosen subset of cultured samples are listed. Corresponding coefficients of variation (CV) in % are indicated for each MTBC species tested.
Figure 2.
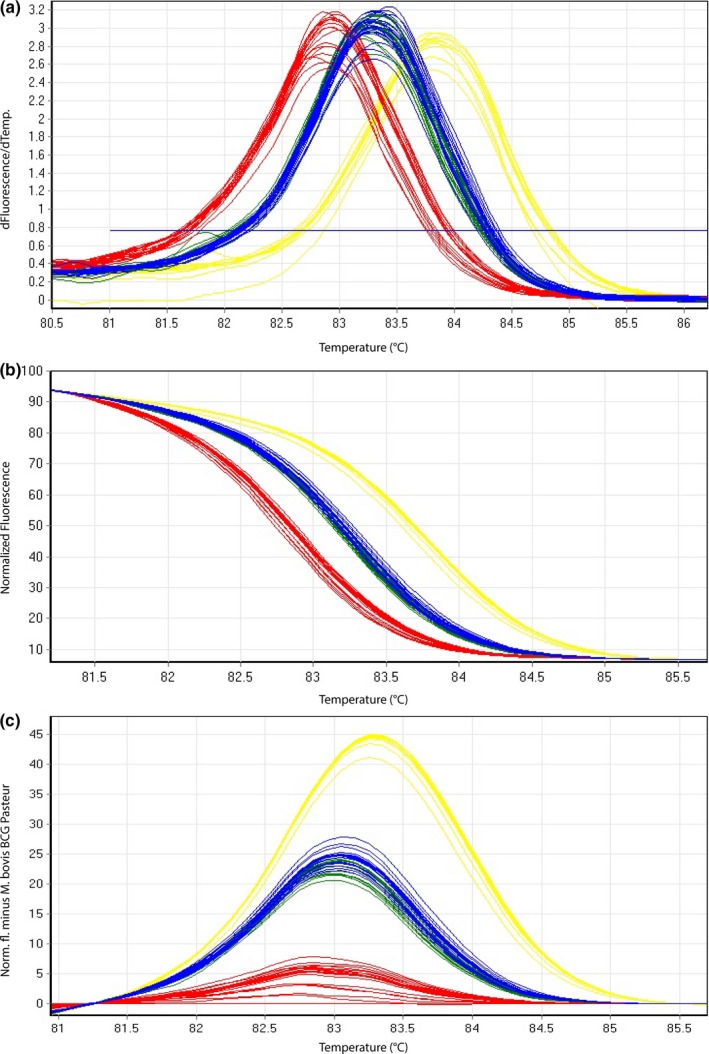
Representative high‐resolution melting graphs corresponding to one high‐resolution melting analysis of a subset of cultured samples (n = 22) of HRM assay 2. Curves of tested samples previously identified as M. tuberculosis are shown in yellow, M. microti in blue, M. bovis/M. bovis BCG in red, and M. caprae in green. (a) Melting curves; (b) normalized plot; and (c) difference plot in relation to M. bovis BCG Pasteur ATCC 35734
Figure 3.
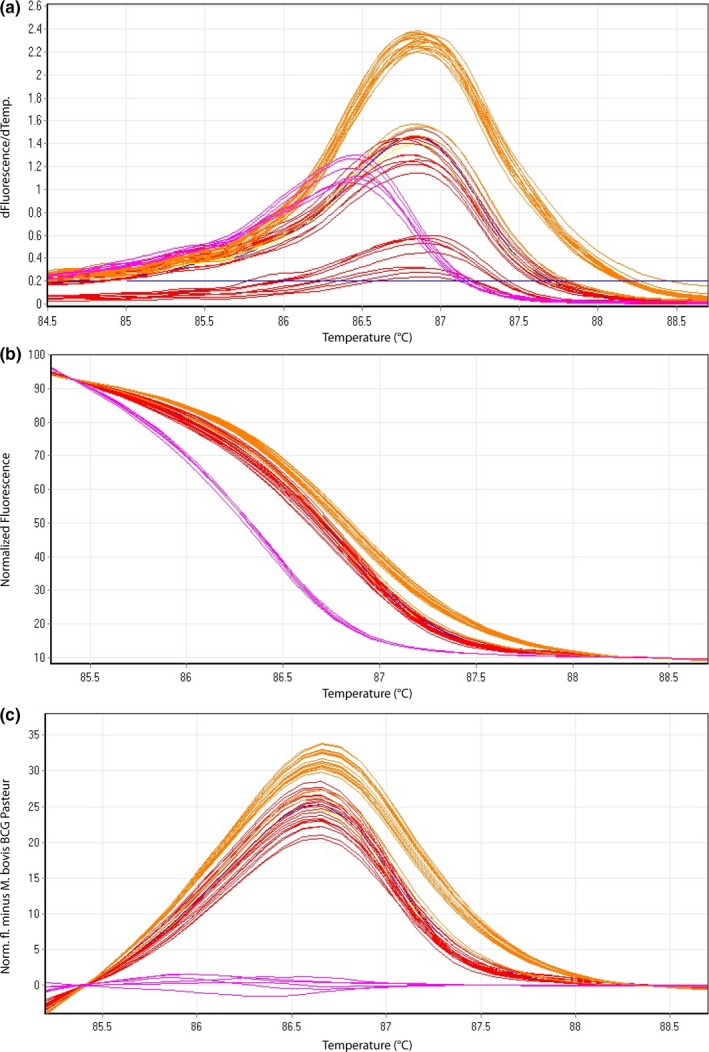
Representative high‐resolution melting graphs corresponding to one high‐resolution melting analysis of a subset of cultured samples (n = 9) and clinical specimens (n = 7) of HRM assay 3. Curves of tested samples previously identified as M. bovis BCG are shown in pink, cultured samples of M. bovis in red, and clinical specimen of M. bovis in orange. (a) Melting curves; (b) normalized plot; and (c) difference plot in relation to M. bovis BCG Pasteur ATCC 35734
High‐resolution melting assay 2 differentiates three groups, namely M. tuberculosis, M. microti/M. caprae, and M. bovis/M. bovis BCG. The obtained intra‐assay coefficients of variation (CVs) and the interassay CVs were ranging between 0.01%–0.04% and 0.11%–0.17%, respectively (Table 3, Table A3), demonstrating highly reproducible and robust assays. Species identification results of all 39 (100%) tested cultured isolates were in accordance with the species classification of GenoType MTBC test (Hain Lifescience) results.
High‐resolution melting assay 3 differentiates M. bovis from M. bovis BCG. The intra‐assay CVs and the interassay CVs yielded Tm values ranging between 0.01%–0.03% and 0.03%–0.04%, respectively (Table 4, Table A4). Species identification results of all 15 (100%) tested cultured isolates were in agreement with the GenoType MTBC test (Hain Lifescience) results.
3.2. HRM of clinical samples
For 22/23 (96%) clinical samples tested in HRM assay 2, obtained normalized and difference plots (Figure 4b,c) showed the appearance of three distinct groups, namely M. tuberculosis, M. microti/M. caprae, and M. bovis/M. bovis BCG in accordance with the results of GenoType MTBC test (Hain Lifescience). One sample revealed significantly lower Tm values compared with the other results. This clinical sample (samples 17–1,063), however, has not yet been successfully cultured and is in the progress of further investigations. High‐resolution melting assay 3 showed a clear distinction of all cultured isolates as well as directly isolated clinical samples of M. bovis from M. bovis BCG in 100% concordance with the GenoType MTBC test (Hain Lifescience) results.
Figure 4.
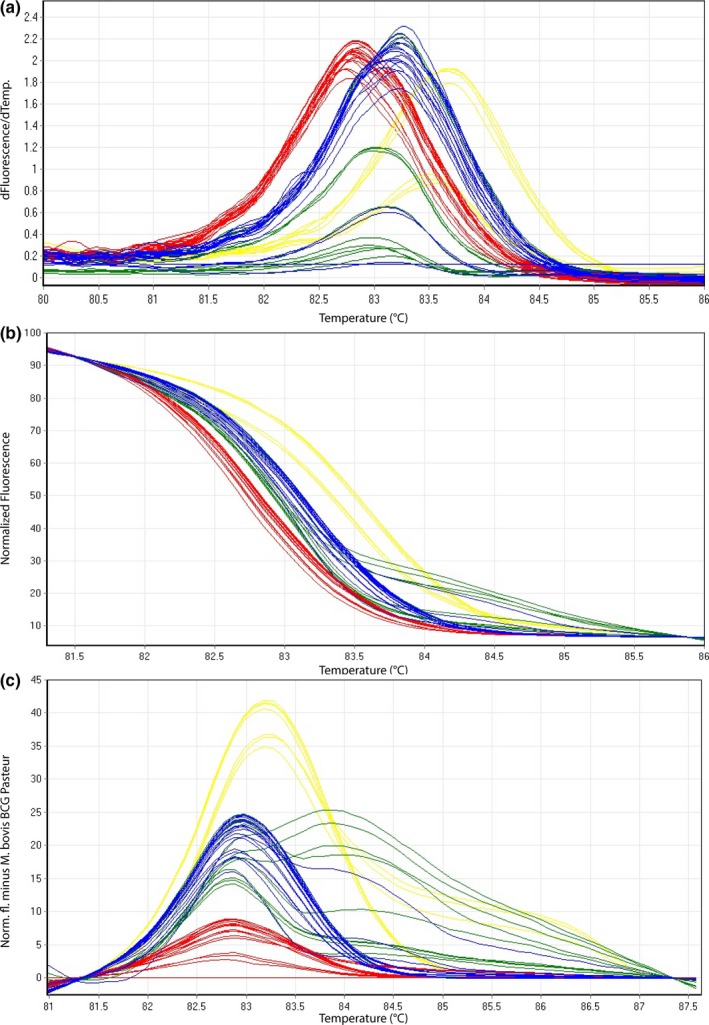
Representative high‐resolution melting graphs corresponding to one high‐resolution melting analysis of a subset of clinical specimens (n = 18) for HRM assay 2. Curves of tested samples previously identified as M. tuberculosis are shown in yellow, M. microti in blue, M. bovis/M. bovis BCG in red, and M. caprae in green. (a) Melting curves; (b) normalized plot; and (c) difference plot in relation to M. bovis BCG Pasteur ATCC 35734
In HRM assay 2, intra‐ and interassay CVs were between 0.02%–0.04% and between 0.12%–0.19%, respectively (Table 5, Table A5).
Table 5.
Intra‐ and interassay variability of HRM assay 2 of clinical specimens
| Run 1 | Run 2 | Run 3 | Interassay | |||||
|---|---|---|---|---|---|---|---|---|
| Tm | CV% | Tm | CV% | Tm | CV% | Tm | CV% | |
| M. tuberculosis H37Rv | 83.85 | 83.67 | 83.58 | 83.70 | 0.16 | |||
| M. bovis BCG Pasteur ATCC 35734 | 82.82 | 82.73 | 82.62 | 82.72 | 0.12 | |||
| M. microti ATCC 19422 | 83.30 | 83.25 | 83.08 | 83.21 | 0.14 | |||
| M. tuberculosis (n = 2) | 83.71 ± 0.13 | 0.03 | 83.60 ± 0.10 | 0.03 | 83.52 ± 0.09 | 0.03 | 83.63 ± 0.20 | 0.12 |
| M. bovis (n = 6) | 82.97 ± 0.10 | 0.04 | 82.82 ± 0.07 | 0.02 | 82.70 ± 0.10 | 0.03 | 82.84 ± 0.24 | 0.19 |
| M. caprae (n = 4) | 83.24 ± 0.14 | 0.02 | 83.11 ± 0.16 | 0.02 | 82.97 ± 0.12 | 0.03 | 83.11 ± 0.26 | 0.15 |
| M. microti (n = 6) | 83.29 ± 0.09 | 0.02 | 83.19 ± 0.09 | 0.03 | 83.00 ± 0.10 | 0.04 | 83.14 ± 0.24 | 0.16 |
Mean values and standard deviation of melting temperatures (Tm) of a randomly chosen subset of clinical specimen are listed. Corresponding coefficients of variation (CV) in % are indicated for each MTBC species tested.
High‐resolution melting assay 3 showed intra‐assay CVs between 0.01% and 0.02% and an interassay CV of 0.04% (Table 6, Table A4).
Table 6.
Intra‐ and interassay variability of HRM assay 3 of clinical specimens
| Run 1 | Run 2 | Run 3 | Interassay | |||||
|---|---|---|---|---|---|---|---|---|
| Tm | CV% | Tm | CV% | Tm | CV% | Tm | CV% | |
| M. tuberculosis H37Rv | 86.82 | 86.82 | 86.85 | 86.83 | 0.02 | |||
| M. microti ATCC 19422 | 86.83 | 86.83 | 86.83 | 86.83 | 0 | |||
| M. bovis BCG Pasteur ATCC 35734 | 86.45 | 86.42 | 86.47 | 86.41 | 0.03 | |||
| M. bovis (n = 7) | 86.86 ± 0.04 | 0.01 | 86.84 ± 0.07 | 0.02 | 86.88 ± 0.08 | 0.02 | 86.86 ± 0.09 | 0.04 |
Mean values and standard deviation of melting temperatures (Tm) of a randomly chosen subset of clinical specimen are listed. Corresponding coefficients of variation (CV) in % are indicated for each MTBC species tested.
3.3. Specificity
Forty‐one NTM and N. paucivorans, E. coli, and S. suis were tested for specificity of HRM assay 2 resulting in no melting curves or melting curves with entirely different Tm in respect to Tm deriving from samples of MTBC. In HRM assay 3, some NTM showed similar curves and Tm values. Therefore, HRM assay 3 is recommended to be applied only after identification of M. bovis or M. bovis BCG.
3.4. Sensitivity
For HRM assay 2, efficiencies of the qPCR were 94% for M. microti, 97% for M. bovis, 112% for M. caprae, and 91% for H37Rv (Figure A1). In HRM assay 3, efficiencies were determined to be 89% for M. bovis BCG and 103% for M. bovis (Figure A2).
High‐resolution melting assay 2 showed a LOD for the lowest dilution of which the acceptance criteria (standard deviation < 0.5 and Ct value < 38) were complied with 10 GE, corresponding to 50 fg of template DNA in the qPCR, for M. tuberculosis, M. caprae, M. microti, and M. bovis (Table 7). The LOD of HRM assay 3 was 100 GE for both M. bovis and M. bovis BCG (Table 8).
Table 7.
Limit of detection (LOD) of the qPCR of HRM assay 2
| MTBC member | Genome equivalents | Ct | SD |
|---|---|---|---|
| M. tuberculosis H37Rv | 1,000,000 | 13.37 | 0.01 |
| 100,000 | 16.49 | 0.28 | |
| 10,000 | 19.99 | 0.08 | |
| 1,000 | 23.96 | 0.17 | |
| 100 | 27.46 | 0.22 | |
| 10 | 30.92 | 0.40 | |
| 1 | 33.06 | 1.15 | |
| M. bovis BCG pasteur ATCC 35734 | 1,000,000 | 12.62 | 0.02 |
| 100,000 | 16.02 | 0.03 | |
| 10,000 | 19.69 | 0.24 | |
| 1,000 | 23.82 | 0.19 | |
| 100 | 26.83 | 0.24 | |
| 10 | 30.11 | 0.43 | |
| 1 | 32.6 | 0.61 | |
| M. microti ATCC 19422 | 1,000,000 | 13.68 | 0.05 |
| 100,000 | 17.20 | 0.13 | |
| 10,000 | 20.80 | 0.33 | |
| 1,000 | 24.34 | 0.25 | |
| 100 | 27.77 | 0.04 | |
| 10 | 31.01 | 0.49 | |
| 1 | 33.19 | 0.52 | |
| M. caprae ZH 22914 | 1,000,000 | 17.96 | 0.01 |
| 100,000 | 21.31 | 0.07 | |
| 10,000 | 25.02 | 0.06 | |
| 1,000 | 28.28 | 0.07 | |
| 100 | 31.79 | 0.29 | |
| 10 | 32.42 | 0.38 | |
| 1 | 34.09 | 0.98 |
Determination of Ct values and its standard deviation (SD) of 3 replicates for a dilution series ranging from 1 to 1,000,000 genome equivalents using reference strains M. tuberculosis H37Rv, M. bovis Pasteur ATCC 35734, M. microti ATCC 19422, and the clinical specimen M. caprae ZH 22914. Bold represents the determined LOD for the lowest dilution of which the acceptance criteria (standard deviation < 0.5 and Ct value < 38) were fulfilled.
Table 8.
Limit of detection (LOD) of the qPCR of HRM assay 3
| MTBC member | Genome equivalents | Ct | SD |
|---|---|---|---|
| M. bovis ZH 20655 | 1,000,000 | 19.19 | 0.07 |
| 100,000 | 23.57 | 0.04 | |
| 10,000 | 28.41 | 0.31 | |
| 1,000 | 32.45 | 0.39 | |
| 100 | 36.37 | 0.10 | |
| 10 | 37.46 | 0.80 | |
| 1 | 37.67 | 0.28 | |
| M. bovis BCG pasteur ATCC 35734 | 1,000,000 | 16.41 | 0.14 |
| 100,000 | 20.72 | 0.18 | |
| 10,000 | 25.06 | 0.31 | |
| 1,000 | 29.34 | 0.20 | |
| 100 | 32.83 | 0.24 | |
| 10 | 35.57 | 1.84 | |
| 1 | 37.00 | 2.10 |
Determination of Ct values and its standard deviation (SD) of 3 replicates for a dilution series ranging from 1 to 1,000,000 genome equivalents using reference strains M. bovis BCG Pasteur ATCC 35734 and the clinical specimen M. bovis ZH 20665. Bold represents the determined LOD for the lowest dilution of which the acceptance criteria (standard deviation < 0.5 and Ct value < 38) were fulfilled.
4. DISCUSSION
In the present study, the establishment of a three‐reaction HRM paradigm in the form of three HRM assays is described, which can rapidly differentiate the main species of MTBC. M. microti, M. tuberculosis, M. caprae, M. bovis, and M. bovis BCG were demonstrated to undoubtedly and consistently be differentiated from each other by distinctive difference plots (Figures 2c, 3c, and 4c). Based on the recently published suggestion of revising the Hain GenoType MTBC test interpretation, proposing eight possible binding patterns including novel ecotypes/subtypes (Loiseau et al., 2019), the three‐reaction HRM paradigm can potentially reveal the same eight different groups. However, the Hain test is only validated for cultured samples and remains a very costly and time‐consuming approach comprising 13 different probes.
The clear advantage of the developed three‐reaction HRM paradigm approach compared with previous studies (Costa, Amaro, et al., 2014; Costa, Botelho, Couto, Viveiros, & Inácio, 2014; Halse et al., 2011; Pinsky & Banaei, 2008; Pounder et al., 2010; Reddington et al., 2012) is the achievement of an inexpensive, rapid as well as easy to use single‐plex method, which can be used for cultured samples as well as for clinical samples. In approximately 2 hr, a sample can be identified as member of MTBC and assigned to the correct species, which is a benefit comparing to methods, which are based on time‐consuming procedures (Kamerbeek et al., 1997) or require cultured samples (Kasai et al., 2000; Niemann et al., 2000). An additional advantage of this three‐reaction HRM paradigm aiming at different loci lays in the fact that it is an adaptive approach, which opens the possibility for individual combinations of primer pairs depending on the question raised. In case of a probable detection of M. orygis strains and therefore a desired differentiation from M. africanum, there is potential to design a further primer pair, which would allow detecting a mutation at codon 329 of gyrB (Huard et al., 2006) representing a unique SNP in M. orygis.
The only drawback of the described HRM approach compared to the Hain test is the inability to differentiate three strains originating from China (Orloski et al., 2018) (M. caprae/M. bovis ecotype II) from M. caprae. To resolve this problem, it would be possible to expand the assay by designing a novel primer pair covering a SNP at base pair position 1,310 of gyrB and thus reliably identifying M. caprae/M. bovis ecotype II. Since M. caprae/M. bovis ecotype II is a rare type of strains, it can be neglected in routine diagnostic laboratories, where it is essential to rely on simply performable assays. Moreover, an additional differentiation of M. caprae/M. bovis ecotype II from M. caprae has no advantage in respect to the choice of antibiotic treatment, because both strain types are not intrinsically resistant to PZA.
By a stepwise combination of three independent HRM assays, it is possible to differentiate the species of MTBC firstly into three groups (M. microti/M. canettii (rare subtype), M. tuberculosis/M. africanum/M. orygis/M. pinnipedii/Clade A1, and M. caprae/M. bovis/M. bovis BCG/rare M. caprae/M. bovis ecotypes), secondly into six groups (M. microti, M. tuberculosis/M. canettii, M. canettii (rare subtype), M. africanum/M. orygis/M. pinnipedi/Clade A1, M. caprae/rare M. caprae/M. bovis ecotypes, and M. bovis/M. bovis BCG), and finally on gyrA, M. bovis BCG, M. bovis, and rare M. caprae/M. bovis ecotype I can further be separated leading to a clear differentiation into the main human‐ and veterinary‐associated MTBC species (Figure 5). The three HRM assays can be performed either consecutively or in parallel since the qPCR conditions are equal. The interpretation of the three‐reaction paradigm is straightforward and simple to achieve. For routine laboratories, a simple combination of HRM assays 1 and 2 will lead to a rapid detection and differentiation of the most significant agents of tuberculosis appearing worldwide.
Figure 5.
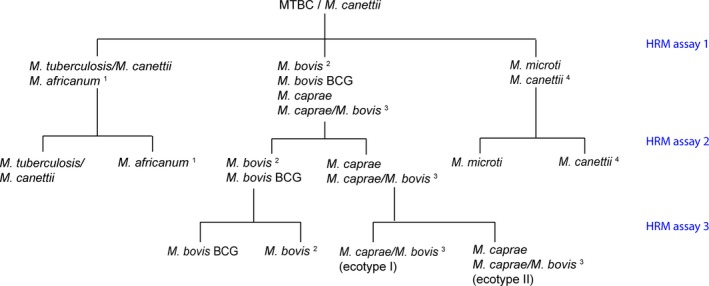
HRM assay 1 (Landolt et al., 2019) allows the distinction between M. tuberculosis/M. canettii/M. africanum/M. orygis/M. pinnipedii/Clade A1, M. microti/M. canettii (rare subtype), and M. bovis/M. bovis BCG/M. caprae/rare M. caprae/M. bovis ecotypes. A combination of HRM assays 1 and 2 is leading to six groups (M. tuberculosis/M. canettii, M. africanum/M. orygis/M. pinnipedii/Clade A1, M. microti, M. canettii (rare subtype), M. caprae/rare M. caprae/M. bovis ecotypes I and II, and M. bovis/M. bovis BCG). By performing HRM assay 3, M. bovis, M. bovis BCG, and rare M. caprae/M. bovis ecotype I can further be distinguished. 1. M. africanum not distinguishable in gyrA and gyrB from M. orygis, M. pinnipedii, Clade A1 (Dassie bacillus, M. mungi, Chimpanzee bacillus, M. suricattae) (Brites et al., 2018). 2. Frequent subtype, intrinsic pyrazinamide (PZA) resistance. 3. Rare ecotypes, no intrinsic PZA resistance. 4. Rare subtype, highly recombinogenic
Tm ranges deriving from HRM assays are partially overlapping (Tables 3, 4, 5, 6). Therefore, based solely on Tm it is not possible to clearly differentiate the members of MTBC. However, by transforming melting curves into normalized and difference plots using algorithms of the Rotor‐Gene Q Software 2.3.1 (Qiagen), the members of MTBC can be separated into distinct groups (Figures 2, 3, 4). Thereby, the species‐specific melting profiles showed an explicit behavior. The presented HRM assays identified MTBC‐positive cultured isolates in accordance with the results of the GenoType MTBC test.
The clinical samples were in agreement of 96% with respect to the GenoType MTBC test. One sample (4%) deriving from an alpaca, however, showed a nonspecific melting curve preventing a correct assessment of the sample to MTBC applying the developed HRM assays. A possible explanation of this interference of the melting curve could be correlated to the inhibitory substances of the complex sample mixture originating from different tissues including lymph node, lung, heart, liver, and cervical vertebra. Investigations to unravel this finding were not successful yet. Although all remaining 22 clinical samples deriving from lymph nodes, lung, or liver tissues showed an unambiguous and correct result (Table A1), it is important to mention that samples containing very little amount of target DNA material (as obtained in two tested M. caprae samples and one M. microti clinical sample) display a weak fluorescence signal (Figure 4a) and therefore may lead to irregular shapes of the normalized—(Figure 4b) and difference—(Figure 4c) plots in comparison with all other samples enclosing high amounts of MTBC DNA. Such irregular patterns of normalized and difference plots visualize the detection limit of a HRM assay when testing directly extracted clinical samples.
Summarizing the obtained data, both HRM assays 2 and 3 showed a very good reproducibility with small variations of Tm, which is demonstrated in very low values of intra‐ and interassay CVs. Since HRM assays 1 and 2 are proven to be 100% specific, they can be used unambiguously for the identification and differentiation of MTBC. Moreover, the assays yielded PCR efficiencies of more than 91% and 89%, respectively. The sensitivity of HRM assay 2 showing a LOD of 10 GE is adequate. The LOD of HRM assay 3 is slightly higher with a measured LOD of 100 GE. However, since the tested collective of samples did not cover species of all eight genotype groups, it is suggested to further evaluate the assay by testing a more extensive collection of isolates.
5. CONCLUSION
The developed three‐reaction HRM paradigm is a quick, sensitive, and specific assay for differentiation of MTBC between the main species highly relevant in human and veterinary diagnostics namely M. tuberculosis/M. canettii, M. canettii (rare subtype), M. africanum/M. orygis/M. pinnipedii/Clade A1, M. microti, M. caprae/rare M. caprae/M. bovis ecotype II, M. bovis, M. bovis BCG, and rare M. caprae/M. bovis ecotype I extracted from clinical samples and from isolates. Several months of cultivation time may be saved by using these potent HRM assays. Since most species within MTBC are implicated in human infections (Huard et al., 2006), it is of advantage to have early knowledge of transmission of tuberculosis for consequently choosing an appropriate drug therapy for humans or a proper eradication strategy when dealing with veterinary samples. Tuberculosis surveillance policies and public health management depend on powerful and affordable diagnostic tools such as this paradigm of a three‐reaction HRM assay, which could be easily implemented in laboratories worldwide.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
PL and SS conceptualized, drafted, and investigated the data; involved in formal analysis; and wrote the original manuscript. MJAS edited and provided sequencing data. RS and SS wrote, reviewed, and edited the manuscript.
ETHICS STATEMENT
The recommendations of Swiss federal regulations (TSV 916.401 and VSFK 817.190) were followed. The animal samples were analyzed in the context of a monitoring program of lymph nodes aiming at an early recognition of bovine tuberculosis and NTM infections. No animals were killed for the purposes of this research project, and no ethical approval was required.
ACKNOWLEDGMENTS
We are grateful to the kind provision of a M. microti wild boar isolate from Lucía de Juan Ferré and Beatriz Romero Martínez, European Union Reference Laboratory for Bovine Tuberculosis, Spain.
APPENDIX A.
Figure A1.
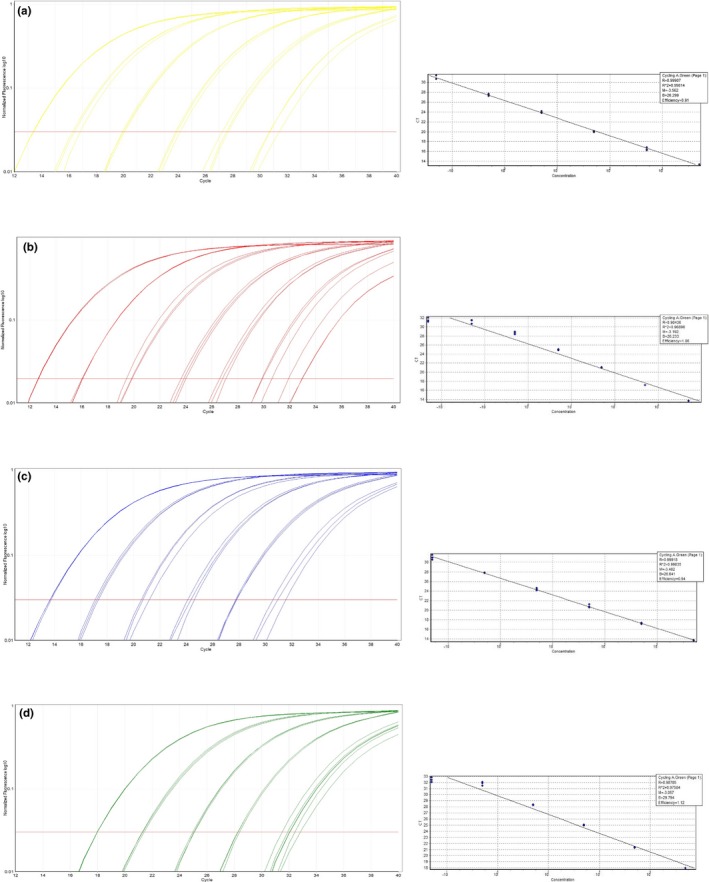
qPCR amplification curves and standard curves of the serial dilution using (a) M. tuberculosis H37Rv; (b) M. bovis BCG Pasteur ATCC 35734; (c) M. microti ATCC 19422; and (d) M. caprae ZH 22914 for high‐resolution melting (HRM) assay 2
Figure A2.
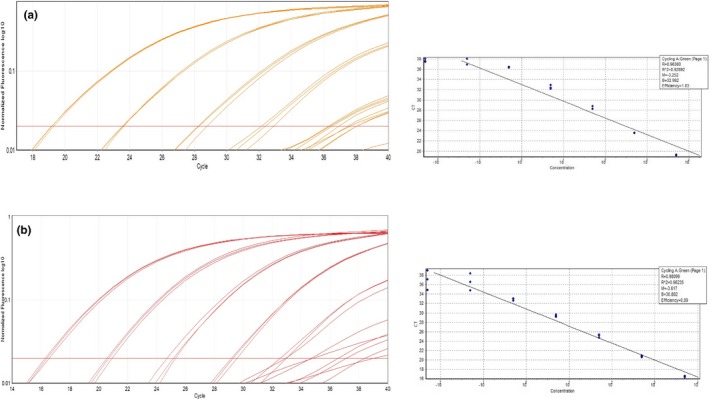
qPCR amplification curves and standard curves of the serial dilution using (a) M. bovis BCG Pasteur ATCC 35734 and (b) M. bovis for high‐resolution melting (HRM) assay 3
Table A1.
MTBC‐positive isolates used for the development of the high‐resolution melting (HRM) assays 2 and 3
| Sample | Species | Origin | Specimen | Culture? | Clinical specimen? |
|---|---|---|---|---|---|
| M. bovis (15) | |||||
| 20175 | M. bovis | Cow | Lung | Cultured | Not available |
| 20482 | M. bovis | Cow | Lung, lymph node pool | Cultured | Not available |
| 20531 | M. bovis | Cow | Lymph node pool | Cultured | Not available |
| 20593 | M. bovis | Cow | Lymph node pool | Cultured | Clinical specimen |
| 20594 | M. bovis | Cow | Lymph node pool | Cultured | Clinical specimen |
| 20596 | M. bovis | Cow | Lymph node pool | Cultured | Clinical specimen |
| 20597 | M. bovis | Cow | Lymph node pool | Cultured | Not available |
| 20599 | M. bovis | Cow | Lymph node pool | Cultured | Not available |
| 20600 | M. bovis | Cow | Lymph node pool | Cultured | Clinical specimen |
| 20606 | M. bovis | Cow | Lymph node pool | Cultured | Clinical specimen |
| 20608 | M. bovis | Cow | Lymph node pool | Cultured | Clinical specimen |
| 20609 | M. bovis | Cow | Lymph node pool | Cultured | Clinical specimen |
| 20665 | M. bovis | Cow | Lymph node pool | Cultured | Not available |
| 22539 | M. bovis | Cow | Lymph node | Cultured | Not available |
| 22667 | M. bovis | Cow | Lymph node | Cultured | Not available |
| M. caprae (7) | |||||
| 14‐13 | M. caprae | Cow | Lymph node | Cultured | Not available |
| 22914 | M. caprae | Cow | Lymph node pool | Cultured | Clinical specimen |
| 22971 | M. caprae | Cow | Lymph node pool | Cultured | Clinical specimen |
| 22848 | M. caprae | Cow | Liver, lung, lymph node pool | Cultured | Clinical specimen |
| 22966 | M. caprae | Cow | Lymph node pool | Cultured | Not available |
| 13‐450 | M. caprae | Cow | Lymph node | Cultured | Not available |
| 13‐162 | M. caprae | Cow | Lymph node | Cultured | Clinical specimen |
| M. microti (15) | |||||
| 22928 | M. microti | Cat | Lung, lymph node pool | Cultured | Clinical specimen |
| 15‐1765 | M. microti | Cat | Lung | Cultured | Clinical specimen |
| 15‐342 | M. microti | Alpaca | Lymph node | Cultured | Clinical specimen |
| 14‐58 | M. microti | Cat | Lung | Cultured | Clinical specimen |
| 14‐690 | M. microti | Alpaca | Liver | Cultured | Clinical specimen |
| 17‐2287 | M. microti | Alpaca | Spleen | Cultured | Not available |
| 15817 | M. microti | Lama | Lymph node | Cultured | Clinical specimen |
| 15‐1955 | M. microti | Cat | Lymph node | Cultured | Clinical specimen |
| 16‐2156 | M. microti | Cat | Bronchoalveolar lavage | Cultured | Not available |
| 1522744 | M. microti | Cat | Lung, lymph node pool | Cultured | Not available |
| 16‐1347 | M. microti | Cat | Lung | Cultured | Not available |
| 17‐1084 | M. microti | Cat | Lymph node, skin | Cultured | Clinical specimen |
| 17‐1063 | M. microti | Alpaca | Lymph node, lung, heart, liver, cervical vertebra | Culture ongoing | Clinical specimen |
| 17‐549 | M. microti | Lama | Liver | Cultured | Clinical specimen |
| MI16 | M. microti | Wildboar Spain | Unknown | Cultured | Not available |
| M. tuberculosis (4) | |||||
| 15‐961‐2 | M. tuberculosis | Elephant 1 | Lung | Cultured | Clinical specimen |
| 15‐961‐1 | M. tuberculosis | Elephant 1 | Pharyngeal swab | Not cultivated | Clinical specimen |
| 15‐1115‐2 | M. tuberculosis | Elephant 2 | Lung | Cultured | Not available |
| 15‐1221‐1 | M. tuberculosis | Elephant 3 | Lung | Cultured | Not available |
Table A2.
Exclusivity panel consisting of 41 nontuberculous mycobacteria and 3 nonmycobacterial species for specificity testing of the high‐resolution melting (HRM) assays 2 and 3
| Species | No. of isolates | Species | No. of isolates |
|---|---|---|---|
| M. abscessus sp. | 2 | M. malmoense | 1 |
| M. avium subsp. avium | 2 | M. marinum | 1 |
| M. avium subsp. hominissuis | 32 | M. monacense | 3 |
| M. avium subsp. paratuberculosis | 1 | M. nebraskense | 1 |
| M. avium subsp. silvaticum | 1 | M. neoaurum | 5 |
| M. bourgelatii | 1 | M. nonchromogenicum | 7 |
| M. celatum | 1 | M. palustre | 1 |
| M. chelonae subsp. chelonae | 1 | M. parafortuitum | 2 |
| M. chimaera/intracellulare/youngonense | 1 | M. paragordonae | 6 |
| M. chitae | 1 | M. peregrinum | 2 |
| M. elephantis | 1 | M. persicum | 2 |
| M. engbaekii | 1 | M. phlei | 3 |
| M. europaeum | 1 | M. scrofulaceum | 1 |
| M. fortuitum/porcinum | 1 | M. simiae | 1 |
| M. goodii | 1 | M. smegmatis | 1 |
| M. gordonae | 2 | M. szulgai | 1 |
| M. hassiacum | 1 | M. terrae | 1 |
| M. interjectum/paraense | 1 | M. vaccae | 4 |
| M. intermedium | 1 | M. xenopi | 5 |
| M. intracellulare | 3 | Nocardia paucivorans | 1 |
| M. kansasii | 10 | Escherichia coli | 1 |
| M. lymphaticum | 1 | Streptococcus suis | 1 |
Table A3.
Raw data set and statistical parameters generated from the intra‐ and interassay variability of high‐resolution melting (HRM) assay 2 using a randomly chosen subset of 22 cultured samples
| Isolate | Run 1 | Run 2 | Run 3 | Inter‐Assay | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MTBC member | Sample no | Origin | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Mean Tm | SD | CV% | Inter‐assay CV% |
| M. microti | ATCC 19422 | 20.65 | 83.13 | 20.87 | 83.25 | 19.74 | 83.28 | 83.22 | 0.08 | 0.10 | ||||||||||||||
| M. bovis BCG Pasteur | ATCC 35734 | 24.67 | 82.62 | 24.82 | 82.77 | 23.57 | 82.70 | 82.70 | 0.08 | 0.09 | ||||||||||||||
| M. tuberculosis | H37RV | 22.41 | 83.55 | 22.44 | 83.75 | 20.97 | 83.60 | 83.63 | 0.10 | 0.12 | ||||||||||||||
| M. tuberculosis | 15‐961 2B | Elephant | 14.87 | 83.60 | 83.58 | 0.02 | 0.02 | 15.11 | 83.80 | 83.80 | 0.00 | 0.00 | 14.01 | 83.70 | 83.69 | 0.02 | 0.02 | 83.69 | 0.11 | 0.13 | ||||
| 14.95 | 83.58 | 15.13 | 83.80 | 14.05 | 83.70 | |||||||||||||||||||
| 14.96 | 83.57 | 15.14 | 83.80 | 13.94 | 83.67 | |||||||||||||||||||
| M. tuberculosis | 15‐1115‐2 | Elephant | 16.34 | 83.55 | 83.54 | 0.01 | 0.01 | 16.69 | 83.87 | 83.86 | 0.03 | 0.03 | 15.32 | 83.63 | 83.65 | 0.03 | 0.03 | 83.69 | 0.16 | 0.19 | ||||
| 16.71 | 83.55 | 16.65 | 83.83 | 15.47 | 83.65 | |||||||||||||||||||
| 16.73 | 83.53 | 16.75 | 83.88 | 15.46 | 83.68 | |||||||||||||||||||
| M. tuberculosis | 15‐1221‐1 | Elephant | 19.59 | 83.53 | 83.53 | 0.01 | 0.01 | 0.01 | 19.98 | 83.87 | 83.83 | 0.04 | 0.04 | 0.02 | 18.45 | 83.60 | 83.67 | 0.06 | 0.07 | 0.04 | 83.68 | 0.15 | 0.18 | 0.17 |
| 19.80 | 83.52 | 20.02 | 83.83 | 18.45 | 83.68 | |||||||||||||||||||
| 19.77 | 83.53 | 19.75 | 83.80 | 18.47 | 83.72 | |||||||||||||||||||
| M. caprae | 229171 | Cow | 20.74 | 83.05 | 83.05 | 0.00 | 0.00 | 21.20 | 83.23 | 83.24 | 0.01 | 0.01 | 19.79 | 83.23 | 83.23 | 0.02 | 0.02 | 83.17 | 0.11 | 0.13 | ||||
| 37.30 | 21.09 | 83.25 | 19.76 | 83.22 | ||||||||||||||||||||
| 21.07 | 83.05 | 21.28 | 83.23 | 19.55 | 83.25 | |||||||||||||||||||
| M. caprae | 22914 | Cow | 20.60 | 83.10 | 83.12 | 0.03 | 0.03 | 20.97 | 83.30 | 83.26 | 0.04 | 0.04 | 19.52 | 83.27 | 83.25 | 0.02 | 0.02 | 83.21 | 0.08 | 0.10 | ||||
| 20.68 | 83.10 | 21.02 | 83.25 | 19.59 | 83.25 | |||||||||||||||||||
| 20.71 | 83.15 | 20.79 | 83.23 | 19.48 | 83.23 | |||||||||||||||||||
| M. caprae | 22848 | Cow | 22.93 | 83.07 | 83.04 | 0.02 | 0.03 | 23.24 | 83.20 | 83.19 | 0.01 | 0.01 | 21.68 | 83.23 | 83.20 | 0.03 | 0.04 | 83.14 | 0.09 | 0.10 | ||||
| 22.94 | 83.03 | 23.20 | 83.18 | 21.77 | 83.20 | |||||||||||||||||||
| 22.90 | 83.03 | 23.34 | 83.18 | 21.53 | 83.17 | |||||||||||||||||||
| M. caprae | 22966 | Cow | 19.89 | 83.13 | 83.15 | 0.02 | 0.02 | 20.04 | 83.35 | 83.33 | 0.04 | 0.05 | 18.60 | 83.25 | 83.26 | 0.02 | 0.02 | 83.25 | 0.09 | 0.11 | ||||
| 19.84 | 83.15 | 19.97 | 83.35 | 18.46 | 83.25 | |||||||||||||||||||
| 19.81 | 83.17 | 20.04 | 83.28 | 18.84 | 83.28 | |||||||||||||||||||
| M. caprae | 13‐450 | Cow | 18.04 | 83.20 | 83.16 | 0.04 | 0.04 | 18.23 | 83.30 | 83.31 | 0.02 | 0.02 | 17.00 | 83.28 | 83.28 | 0.01 | 0.01 | 83.25 | 0.08 | 0.09 | ||||
| 18.04 | 83.13 | 18.10 | 83.33 | 16.99 | 83.28 | |||||||||||||||||||
| 18.13 | 83.15 | 18.21 | 83.30 | 16.93 | 83.27 | |||||||||||||||||||
| M. caprae | 13‐162 | Cow | 22.08 | 83.13 | 83.10 | 0.03 | 0.03 | 0.03 | 22.43 | 83.23 | 83.24 | 0.01 | 0.01 | 0.03 | 20.91 | 83.25 | 83.26 | 0.01 | 0.01 | 0.02 | 83.20 | 0.08 | 0.10 | 0.11 |
| 21.93 | 83.10 | 22.23 | 83.25 | 20.75 | 83.25 | |||||||||||||||||||
| 22.01 | 83.08 | 22.33 | 83.23 | 20.87 | 83.27 | |||||||||||||||||||
| M. bovis | 20593 | Cow | 26.50 | 82.65 | 82.64 | 0.01 | 0.01 | 27.13 | 82.88 | 82.93 | 0.06 | 0.08 | 25.69 | 82.68 | 82.69 | 0.01 | 0.01 | 82.75 | 0.15 | 0.18 | ||||
| 24.89 | 82.65 | 23.85 | 83.00 | 24.93 | 82.70 | |||||||||||||||||||
| 25.63 | 82.63 | 24.38 | 82.90 | 25.46 | 82.70 | |||||||||||||||||||
| M. bovis | 20594 | Cow | 25.56 | 82.60 | 82.61 | 0.02 | 0.02 | 26.03 | 82.88 | 82.88 | 0.03 | 0.03 | 24.80 | 82.72 | 82.71 | 0.02 | 0.03 | 82.73 | 0.14 | 0.16 | ||||
| 24.81 | 82.60 | 25.75 | 82.85 | 24.77 | 82.72 | |||||||||||||||||||
| 25.43 | 82.63 | 25.79 | 82.90 | 23.95 | 82.68 | |||||||||||||||||||
| M. bovis | 20175 | Cow | 18.77 | 82.80 | 82.74 | 0.05 | 0.06 | 18.90 | 82.98 | 82.93 | 0.07 | 0.09 | 17.68 | 82.80 | 82.78 | 0.03 | 0.03 | 82.82 | 0.10 | 0.12 | ||||
| 18.80 | 82.70 | 18.92 | 82.97 | 17.53 | 82.80 | |||||||||||||||||||
| 18.81 | 82.73 | 18.94 | 82.85 | 17.66 | 82.75 | |||||||||||||||||||
| M. bovis | 20596 | Cow | 21.91 | 82.67 | 82.67 | 0.08 | 0.09 | 22.37 | 82.95 | 82.94 | 0.01 | 0.01 | 21.14 | 82.80 | 82.76 | 0.04 | 0.04 | 82.79 | 0.14 | 0.17 | ||||
| 22.42 | 82.75 | 22.47 | 82.93 | 20.81 | 82.75 | |||||||||||||||||||
| 22.56 | 82.60 | 22.43 | 82.95 | 21.01 | 82.73 | |||||||||||||||||||
| M. bovis | 22667 | Cow | 19.91 | 82.80 | 82.80 | 0.00 | 0.00 | 20.04 | 82.93 | 82.92 | 0.01 | 0.01 | 18.95 | 82.78 | 82.78 | 0.01 | 0.01 | 82.83 | 0.08 | 0.10 | ||||
| 19.84 | 82.80 | 20.05 | 82.92 | 18.79 | 82.77 | |||||||||||||||||||
| 19.81 | 82.80 | 20.05 | 82.92 | 19.09 | 82.78 | |||||||||||||||||||
| M. bovis | 22539 | Cow | 21.42 | 82.73 | 82.76 | 0.04 | 0.04 | 0.04 | 21.80 | 82.92 | 82.92 | 0.00 | 0.00 | 0.04 | 20.49 | 82.77 | 82.76 | 0.01 | 0.01 | 0.02 | 82.81 | 0.09 | 0.11 | 0.14 |
| 21.55 | 82.75 | 21.44 | 82.92 | 20.44 | 82.75 | |||||||||||||||||||
| 21.38 | 82.80 | 21.60 | 82.92 | 20.52 | 82.75 | |||||||||||||||||||
| M. microti | 22928 | Cat | 16.90 | 83.17 | 83.17 | 0.02 | 0.02 | 17.13 | 83.40 | 83.42 | 0.02 | 0.02 | 16.01 | 83.30 | 83.31 | 0.01 | 0.01 | 83.30 | 0.13 | 0.15 | ||||
| 16.93 | 83.18 | 17.13 | 83.43 | 15.89 | 83.30 | |||||||||||||||||||
| 17.01 | 83.15 | 17.08 | 83.43 | 16.13 | 83.32 | |||||||||||||||||||
| M. microti | 15‐1765 | Cat | 21.77 | 83.18 | 83.16 | 0.03 | 0.03 | 22.21 | 83.32 | 83.30 | 0.02 | 0.02 | 20.79 | 83.28 | 83.28 | 0.02 | 0.02 | 83.25 | 0.08 | 0.09 | ||||
| 21.44 | 83.17 | 21.93 | 83.30 | 19.99 | 83.27 | |||||||||||||||||||
| 21.78 | 83.13 | 22.05 | 83.28 | 20.59 | 83.30 | |||||||||||||||||||
| M. microti | 14‐58 | Cat | 24.40 | 83.02 | 83.02 | 0.01 | 0.01 | 24.39 | 83.28 | 83.27 | 0.02 | 0.02 | 23.44 | 83.28 | 83.28 | 0.02 | 0.02 | 83.19 | 0.15 | 0.18 | ||||
| 24.39 | 83.02 | 24.64 | 83.28 | 23.18 | 83.30 | |||||||||||||||||||
| 24.31 | 83.03 | 24.43 | 83.25 | 23.45 | 83.27 | |||||||||||||||||||
| M. microti | 14‐690 | Alpaca | 15.10 | 83.05 | 83.05 | 0.03 | 0.03 | 15.33 | 83.30 | 83.29 | 0.01 | 0.01 | 14.28 | 83.30 | 83.29 | 0.01 | 0.01 | 83.21 | 0.14 | 0.17 | ||||
| 15.13 | 83.07 | 15.27 | 83.28 | 14.20 | 83.30 | |||||||||||||||||||
| 15.21 | 83.02 | 15.29 | 83.30 | 14.24 | 83.28 | |||||||||||||||||||
| M. microti | 16‐2156 | Cat | 23.07 | 83.07 | 83.08 | 0.02 | 0.02 | 23.48 | 83.30 | 83.31 | 0.01 | 0.01 | 22.32 | 83.27 | 83.27 | 0.00 | 0.00 | 83.22 | 0.12 | 0.14 | ||||
| 23.49 | 83.08 | 23.60 | 83.30 | 22.17 | 83.27 | |||||||||||||||||||
| 23.49 | 83.10 | 23.57 | 83.32 | 22.01 | 83.27 | |||||||||||||||||||
| M. microti | 1522744 | Cat | 22.79 | 83.12 | 83.09 | 0.03 | 0.03 | 22.89 | 83.33 | 83.29 | 0.03 | 0.04 | 21.82 | 83.27 | 83.26 | 0.01 | 0.01 | 83.21 | 0.11 | 0.13 | ||||
| 22.67 | 83.07 | 22.97 | 83.27 | 21.74 | 83.27 | |||||||||||||||||||
| 22.68 | 83.08 | 22.94 | 83.27 | 21.49 | 83.25 | |||||||||||||||||||
| M. microti | 16‐1347 | Cat | 18.92 | 83.13 | 83.15 | 0.03 | 0.03 | 0.02 | 19.20 | 83.30 | 83.27 | 0.03 | 0.03 | 0.02 | 17.52 | 83.30 | 83.30 | 0.00 | 0.00 | 0.01 | 83.24 | 0.08 | 0.10 | 0.14 |
| 18.94 | 83.13 | 18.99 | 83.25 | 17.21 | 83.30 | |||||||||||||||||||
| 18.87 | 83.18 | 18.86 | 83.25 | 17.32 | 83.30 | |||||||||||||||||||
Table A4.
Raw data set and statistical parameters generated from the intra‐ and interassay variability of high‐resolution melting (HRM) assay 3 using a randomly chosen subset of 9 cultured samples and 7 clinical specimens
| Isolate | Run 1 | Run 2 | Run 3 | Inter‐Assay | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MTBC member | Sample no | Origin | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Mean Tm | SD | CV% | Inter‐assay CV% |
| M. microti | ATCC 19422 | 26.37 | 86.83 | 28.03 | 86.83 | 28.56 | 86.83 | 86.83 | 0.00 | 0.00 | ||||||||||||||
| M. tuberculosis | H37RV | 29.35 | 86.82 | 29.36 | 86.82 | 29.03 | 86.85 | 86.83 | 0.02 | 0.02 | ||||||||||||||
| M. bovis isolate | 20593 | Cow | 35.04 | 86.88 | 86.86 | 0.02 | 0.02 | 33.95 | 86.85 | 86.85 | 0.00 | 0.00 | 33.73 | 86.87 | 86.90 | 0.03 | 0.03 | 86.87 | 0.02 | 0.03 | ||||
| 35.10 | 86.85 | 33.59 | 86.85 | 33.52 | 86.92 | |||||||||||||||||||
| 35.36 | 86.85 | 33.70 | 86.85 | 32.61 | 86.90 | |||||||||||||||||||
| M. bovis isolate | 20594 | Cow | 32.92 | 86.85 | 86.85 | 0.00 | 0.00 | 32.73 | 86.85 | 86.86 | 0.02 | 0.02 | 32.36 | 86.83 | 86.88 | 0.05 | 0.05 | 86.86 | 0.02 | 0.02 | ||||
| 33.48 | 86.85 | 32.56 | 86.85 | 32.62 | 86.90 | |||||||||||||||||||
| 33.28 | 86.85 | 32.83 | 86.88 | 32.60 | 86.92 | |||||||||||||||||||
| M. bovis isolate | 20609 | Cow | 35.18 | 86.83 | 86.83 | 0.02 | 0.02 | 34.71 | 86.87 | 86.87 | 0.03 | 0.03 | 33.73 | 86.90 | 86.87 | 0.06 | 0.07 | 86.86 | 0.02 | 0.02 | ||||
| 34.74 | 86.85 | 33.89 | 86.85 | 33.87 | 86.80 | |||||||||||||||||||
| 34.54 | 86.82 | 34.39 | 86.90 | 33.60 | 86.90 | |||||||||||||||||||
| M. bovis isolate | 20608 | Cow | 28.92 | 86.82 | 86.82 | 0.01 | 0.01 | 28.69 | 86.83 | 86.84 | 0.01 | 0.01 | 27.96 | 86.78 | 86.77 | 0.04 | 0.04 | 86.81 | 0.04 | 0.04 | ||||
| 28.55 | 86.83 | 28.50 | 86.85 | 28.36 | 86.73 | |||||||||||||||||||
| 29.49 | 86.82 | 28.75 | 86.85 | 28.46 | 86.80 | |||||||||||||||||||
| M. bovis isolate | 20596 | Cow | 29.93 | 86.78 | 86.79 | 0.01 | 0.01 | 29.35 | 86.83 | 86.84 | 0.01 | 0.01 | 29.37 | 86.80 | 86.80 | 0.00 | 0.00 | 86.81 | 0.02 | 0.03 | ||||
| 29.69 | 86.80 | 29.25 | 86.85 | 28.64 | 86.80 | |||||||||||||||||||
| 29.87 | 86.80 | 29.30 | 86.83 | 28.92 | 86.80 | |||||||||||||||||||
| M. bovis isolate | 20600 | Cow | 27.83 | 86.83 | 86.83 | 0.00 | 0.00 | 29.51 | 86.85 | 86.83 | 0.02 | 0.02 | 29.82 | 86.85 | 86.84 | 0.01 | 0.01 | 86.84 | 0.01 | 0.01 | ||||
| 29.66 | 86.83 | 30.07 | 86.83 | 29.91 | 86.83 | |||||||||||||||||||
| 30.09 | 86.83 | 30.49 | 86.82 | 29.91 | 86.85 | |||||||||||||||||||
| M. bovis isolate | 20606 | Cow | 28.36 | 86.80 | 86.81 | 0.02 | 0.02 | 0.01 | 27.94 | 86.80 | 86.78 | 0.03 | 0.03 | 0.02 | 27.42 | 86.83 | 86.84 | 0.01 | 0.01 | 0.03 | 86.81 | 0.03 | 0.03 | 0.03 |
| 27.86 | 86.80 | 28.01 | 86.78 | 27.34 | 86.85 | |||||||||||||||||||
| 28.12 | 86.83 | 27.88 | 86.75 | 27.47 | 86.83 | |||||||||||||||||||
| M. bovis specimen | 20593 | Cow | 19.04 | 86.83 | 86.84 | 0.01 | 0.01 | 18.82 | 86.82 | 86.80 | 0.02 | 0.02 | 18.64 | 86.88 | 86.92 | 0.04 | 0.04 | 86.85 | 0.06 | 0.07 | ||||
| 18.84 | 86.85 | 19.19 | 86.80 | 18.80 | 86.92 | |||||||||||||||||||
| 18.65 | 86.85 | 18.95 | 86.78 | 18.68 | 86.95 | |||||||||||||||||||
| M. bovis specimen | 20594 | Cow | 36.86 | 86.88 | 0.01 | 0.01 | 16.22 | 86.78 | 86.78 | 0.02 | 0.02 | 16.16 | 86.90 | 86.91 | 0.01 | 0.01 | 86.86 | 0.06 | 0.07 | |||||
| 16.18 | 86.88 | 16.51 | 86.80 | 16.18 | 86.92 | |||||||||||||||||||
| 16.75 | 86.87 | 16.55 | 86.77 | 16.07 | 86.90 | |||||||||||||||||||
| M. bovis specimen | 20609 | Cow | 29.46 | 86.85 | 86.85 | 0.00 | 0.00 | 28.99 | 86.80 | 86.82 | 0.03 | 0.03 | 28.66 | 86.88 | 86.85 | 0.03 | 0.03 | 86.84 | 0.02 | 0.02 | ||||
| 29.75 | 86.85 | 29.35 | 86.80 | 28.51 | 86.85 | |||||||||||||||||||
| 29.04 | 86.85 | 29.45 | 86.85 | 28.57 | 86.83 | |||||||||||||||||||
| M. bovis specimen | 20608 | Cow | 17.18 | 86.85 | 86.88 | 0.03 | 0.03 | 17.34 | 86.85 | 86.86 | 0.01 | 0.01 | 17.03 | 86.90 | 86.88 | 0.03 | 0.03 | 86.87 | 0.01 | 0.02 | ||||
| 17.43 | 86.90 | 17.60 | 86.85 | 17.13 | 86.90 | |||||||||||||||||||
| 17.14 | 86.88 | 17.64 | 86.87 | 17.22 | 86.85 | |||||||||||||||||||
| M. bovis specimen | 20596 | Cow | 16.60 | 86.85 | 86.86 | 0.04 | 0.05 | 16.99 | 86.90 | 86.90 | 0.00 | 0.00 | 16.66 | 86.87 | 86.86 | 0.01 | 0.01 | 86.87 | 0.03 | 0.03 | ||||
| 17.02 | 86.90 | 17.01 | 86.90 | 16.54 | 86.85 | |||||||||||||||||||
| 16.84 | 86.82 | 17.14 | 86.90 | 16.42 | 86.85 | |||||||||||||||||||
| M. bovis specimen | 20600 | Cow | 16.88 | 86.85 | 86.85 | 0.00 | 0.00 | 17.15 | 86.88 | 86.88 | 0.03 | 0.03 | 16.97 | 86.85 | 86.84 | 0.01 | 0.01 | 86.86 | 0.02 | 0.02 | ||||
| 16.83 | 86.85 | 17.47 | 86.90 | 16.96 | 86.85 | |||||||||||||||||||
| 16.74 | 86.85 | 17.24 | 86.85 | 16.64 | 86.83 | |||||||||||||||||||
| M. bovis specimen | 20606 | Cow | 17.22 | 86.85 | 86.85 | 0.00 | 0.00 | 0.01 | 17.18 | 86.85 | 86.85 | 0.02 | 0.02 | 0.02 | 16.72 | 86.85 | 86.83 | 0.03 | 0.03 | 0.02 | 86.84 | 0.01 | 0.02 | 0.04 |
| 17.34 | 86.85 | 17.31 | 86.83 | 16.77 | 86.80 | |||||||||||||||||||
| 16.90 | 86.85 | 17.23 | 86.87 | 16.87 | 86.83 | |||||||||||||||||||
| M. bovis BCG Tice isolate | ATCC 27289 | 30.43 | 86.40 | 86.37 | 0.04 | 0.04 | 30.70 | 86.40 | 86.40 | 0.02 | 0.02 | 30.26 | 86.43 | 86.44 | 0.01 | 0.01 | 86.40 | 0.04 | 0.04 | |||||
| 30.84 | 86.38 | 30.51 | 86.38 | 30.14 | 86.45 | |||||||||||||||||||
| 30.73 | 86.33 | 30.22 | 86.42 | 29.81 | 86.45 | |||||||||||||||||||
| M. bovis BCG Pasteur isolate | ATCC 35734 | 30.62 | 86.45 | 86.44 | 0.02 | 0.02 | 0.03 | 31.32 | 86.42 | 86.41 | 0.03 | 0.03 | 0.03 | 30.56 | 86.47 | 86.46 | 0.03 | 0.04 | 0.03 | 86.44 | 0.02 | 0.03 | 0.04 | |
| 30.72 | 86.42 | 31.15 | 86.38 | 30.23 | 86.42 | |||||||||||||||||||
| 30.78 | 86.45 | 31.14 | 86.43 | 30.27 | 86.48 | |||||||||||||||||||
Table A5.
Raw data set and statistical parameters generated from the intra‐ and interassay variability of high‐resolution melting (HRM) assay 2 using a randomly chosen subset of 18 clinical specimens
| Isolate | Run 1 | Run 2 | Run 3 | Inter‐Assay | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MTBC member | Sample no | Origin | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Ct | Tm | Mean Tm | SD | CV% | Intra‐assay CV% | Mean Tm | SD | CV% | Inter‐assay CV% |
| M. microti | ATCC 19422 | 21.28 | 83.30 | 21.82 | 83.25 | 21.25 | 83.08 | 83.21 | 0.12 | 0.14 | ||||||||||||||
| M. bovis BCG Pasteur | ATCC 35734 | 25.78 | 82.82 | 25.33 | 82.73 | 25.23 | 82.62 | 82.72 | 0.10 | 0.12 | ||||||||||||||
| M. tuberculosis | H37RV | 22.87 | 83.85 | 22.65 | 83.67 | 22.62 | 83.58 | 83.70 | 0.14 | 0.16 | ||||||||||||||
| M. tuberculosis | 15‐961 2A | Elephant | 29.98 | 83.58 | 83.59 | 0.01 | 0.01 | 29.48 | 83.50 | 83.53 | 0.03 | 0.03 | 29.16 | 83.47 | 83.44 | 0.02 | 0.03 | 83.52 | 0.08 | 0.09 | ||||
| 29.90 | 83.60 | 29.72 | 83.55 | 29.33 | 83.43 | |||||||||||||||||||
| 30.23 | 83.60 | 29.32 | 83.53 | 29.19 | 83.43 | |||||||||||||||||||
| M. tuberculosis | 19‐691 1A + B | Elephant | 24.51 | 83.77 | 83.81 | 0.03 | 0.04 | 0.03 | 24.78 | 83.68 | 83.68 | 0.03 | 0.03 | 0.03 | 24.44 | 83.60 | 83.57 | 0.03 | 0.03 | 0.03 | 83.68 | 0.12 | 0.14 | 0.12 |
| 24.84 | 83.82 | 24.64 | 83.70 | 24.49 | 83.55 | |||||||||||||||||||
| 24.63 | 83.83 | 24.78 | 83.65 | 24.78 | 83.55 | |||||||||||||||||||
| Microsoft. caprae | 22971 | Cow | 31.58 | 83.20 | 83.22 | 0.02 | 0.02 | 31.45 | 83.13 | 83.13 | 0.02 | 0.02 | 31.21 | 83.08 | 82.99 | 0.08 | 0.09 | 83.11 | 0.11 | 0.14 | ||||
| 31.37 | 83.23 | 30.75 | 83.12 | 31.27 | 82.95 | |||||||||||||||||||
| 31.39 | 83.22 | 30.96 | 83.15 | 31.04 | 82.95 | |||||||||||||||||||
| M. caprae | 22914 | Cow | 29.37 | 83.17 | 83.16 | 0.01 | 0.01 | 28.86 | 83.00 | 83.02 | 0.03 | 0.03 | 28.57 | 82.88 | 82.88 | 0.02 | 0.02 | 83.02 | 0.14 | 0.17 | ||||
| 29.04 | 83.15 | 28.59 | 83.05 | 28.93 | 82.87 | |||||||||||||||||||
| 28.91 | 83.17 | 28.54 | 83.00 | 28.40 | 82.90 | |||||||||||||||||||
| M. caprae | 22848 | Cow | 19.00 | 83.35 | 83.34 | 0.04 | 0.04 | 19.60 | 83.25 | 83.26 | 0.01 | 0.01 | 19.69 | 83.08 | 83.07 | 0.01 | 0.01 | 83.22 | 0.14 | 0.16 | ||||
| 19.33 | 83.37 | 19.27 | 83.27 | 19.58 | 83.07 | |||||||||||||||||||
| 19.85 | 83.30 | 19.16 | 83.25 | 19.57 | 83.07 | |||||||||||||||||||
| M. caprae | 13‐162 | Cow | 32.47 | 83.10 | 83.11 | 0.02 | 0.02 | 0.02 | 31.56 | 82.98 | 82.98 | 0.03 | 0.03 | 0.02 | 31.10 | 82.87 | 82.87 | 0.02 | 0.02 | 0.03 | 82.98 | 0.12 | 0.15 | 0.15 |
| 33.03 | 83.10 | 31.89 | 82.95 | 32.23 | 82.85 | |||||||||||||||||||
| 31.85 | 83.13 | 31.93 | 83.00 | 31.75 | 82.88 | |||||||||||||||||||
| M. bovis | 20593 | Cow | 14.57 | 82.90 | 82.98 | 0.07 | 0.08 | 14.31 | 82.80 | 82.79 | 0.01 | 0.01 | 14.18 | 82.68 | 82.69 | 0.01 | 0.01 | 82.82 | 0.15 | 0.18 | ||||
| 14.67 | 83.02 | 14.32 | 82.80 | 14.27 | 82.70 | |||||||||||||||||||
| 14.58 | 83.02 | 14.23 | 82.78 | 14.81 | 82.70 | |||||||||||||||||||
| M. bovis | 20594 | Cow | 12.65 | 83.07 | 83.02 | 0.04 | 0.05 | 12.54 | 82.85 | 82.85 | 0.03 | 0.03 | 11.91 | 82.80 | 82.78 | 0.02 | 0.02 | 82.89 | 0.12 | 0.15 | ||||
| 12.60 | 83.00 | 12.86 | 82.83 | 11.48 | 82.77 | |||||||||||||||||||
| 12.49 | 83.00 | 12.58 | 82.88 | 11.69 | 82.77 | |||||||||||||||||||
| M. bovis | 20609 | Cow | 22.46 | 82.87 | 82.89 | 0.03 | 0.04 | 22.84 | 82.78 | 82.76 | 0.02 | 0.02 | 22.50 | 82.65 | 82.63 | 0.03 | 0.03 | 82.76 | 0.13 | 0.16 | ||||
| 22.49 | 82.93 | 22.65 | 82.75 | 22.62 | 82.65 | |||||||||||||||||||
| 22.29 | 82.88 | 22.45 | 82.75 | 22.55 | 82.60 | |||||||||||||||||||
| M. bovis | 20608 | Cow | 13.54 | 83.03 | 83.03 | 0.02 | 0.02 | 13.30 | 82.85 | 82.84 | 0.01 | 0.01 | 13.11 | 82.68 | 82.70 | 0.02 | 0.02 | 82.86 | 0.17 | 0.20 | ||||
| 13.19 | 83.02 | 13.46 | 82.85 | 13.29 | 82.72 | |||||||||||||||||||
| 13.31 | 83.05 | 13.16 | 82.83 | 13.13 | 82.70 | |||||||||||||||||||
| M. bovis | 20596 | Cow | 12.86 | 83.02 | 83.05 | 0.03 | 0.03 | 12.81 | 82.83 | 82.83 | 0.00 | 0.00 | 13.02 | 82.65 | 82.68 | 0.04 | 0.04 | 82.85 | 0.18 | 0.22 | ||||
| 12.93 | 83.05 | 12.82 | 82.83 | 12.97 | 82.68 | |||||||||||||||||||
| 13.10 | 83.07 | 12.84 | 82.83 | 12.99 | 82.72 | |||||||||||||||||||
| M. bovis | 20600 | Cow | 13.19 | 83.00 | 83.00 | 0.00 | 0.00 | 0.04 | 12.90 | 82.82 | 82.83 | 0.02 | 0.02 | 0.02 | 13.18 | 82.65 | 82.66 | 0.04 | 0.05 | 0.03 | 82.83 | 0.17 | 0.21 | 0.19 |
| 13.17 | 83.00 | 13.08 | 82.82 | 12.77 | 82.62 | |||||||||||||||||||
| 13.07 | 83.00 | 13.06 | 82.85 | 12.95 | 82.70 | |||||||||||||||||||
| M. microti | 22928 | Cat | 21.04 | 83.28 | 83.32 | 0.04 | 0.04 | 21.00 | 83.22 | 83.22 | 0.01 | 0.01 | 20.82 | 83.03 | 83.04 | 0.01 | 0.01 | 83.20 | 0.14 | 0.17 | ||||
| 21.25 | 83.33 | 21.16 | 83.22 | 20.88 | 83.05 | |||||||||||||||||||
| 21.00 | 83.35 | 20.93 | 83.23 | 20.82 | 83.05 | |||||||||||||||||||
| M. microti | 15‐342 | Alpaca | 15.26 | 83.35 | 83.36 | 0.01 | 0.01 | 14.98 | 83.27 | 83.24 | 0.02 | 0.03 | 15.10 | 83.03 | 83.06 | 0.02 | 0.03 | 83.22 | 0.15 | 0.18 | ||||
| 15.15 | 83.35 | 14.60 | 83.23 | 14.91 | 83.07 | |||||||||||||||||||
| 15.19 | 83.37 | 15.04 | 83.23 | 15.15 | 83.07 | |||||||||||||||||||
| M. microti | 17‐1084 | Cat | 22.93 | 83.27 | 83.29 | 0.02 | 0.02 | 22.99 | 83.20 | 83.19 | 0.02 | 0.02 | 23.08 | 83.05 | 83.07 | 0.03 | 0.03 | 83.18 | 0.11 | 0.13 | ||||
| 22.82 | 83.30 | 22.95 | 83.20 | 22.87 | 83.05 | |||||||||||||||||||
| 22.90 | 83.30 | 23.20 | 83.17 | 23.15 | 83.10 | |||||||||||||||||||
| M. microti | 15‐1955 | Cat | 25.48 | 83.23 | 83.23 | 0.02 | 0.02 | 25.79 | 83.10 | 83.12 | 0.04 | 0.05 | 26.19 | 83.00 | 82.99 | 0.04 | 0.04 | 83.12 | 0.12 | 0.15 | ||||
| 25.21 | 83.22 | 25.61 | 83.10 | 25.74 | 82.95 | |||||||||||||||||||
| 25.37 | 83.25 | 25.65 | 83.17 | 26.35 | 83.02 | |||||||||||||||||||
| M. microti | 14‐690 | Alpaca | 20.03 | 83.32 | 83.32 | 0.00 | 0.00 | 20.00 | 83.20 | 83.22 | 0.02 | 0.02 | 20.13 | 83.05 | 83.03 | 0.03 | 0.03 | 83.19 | 0.15 | 0.17 | ||||
| 19.84 | 83.32 | 19.90 | 83.23 | 19.94 | 83.05 | |||||||||||||||||||
| 19.90 | 83.32 | 20.01 | 83.23 | 19.98 | 83.00 | |||||||||||||||||||
| M. microti | 17‐549 | Llama | 30.79 | 83.22 | 83.23 | 0.04 | 0.05 | 0.02 | 30.07 | 83.10 | 83.13 | 0.03 | 0.03 | 0.03 | 30.24 | 82.90 | 82.96 | 0.05 | 0.06 | 0.04 | 83.11 | 0.14 | 0.17 | 0.16 |
| 30.23 | 83.20 | 30.62 | 83.15 | 30.03 | 83.00 | |||||||||||||||||||
| 30.78 | 83.28 | 30.20 | 83.13 | 29.89 | 82.98 | |||||||||||||||||||
Landolt P, Stephan R, Stevens MJA, Scherrer S. Three‐reaction high‐resolution melting assay for rapid differentiation of Mycobacterium tuberculosis complex members. MicrobiologyOpen. 2019;8:e919 10.1002/mbo3.919
Data Availability Statement: Raw data sets from intra‐ and interassay variability runs are comprehended in the appendix. On request, additional raw data can be obtained from the corresponding author.
DATA AVAILABILITY STATEMENT
Raw data sets from intra‐ and interassay variability runs are comprehended in the appendix. On request, additional raw data can be obtained from the corresponding author.
REFERENCES
- Anthwal, D. , Gupta, R. K. , Bhalla, M. , Bhatnagar, S. , Tyagi, J. S. , & Haldar, S. (2017). Direct detection of rifampin and isoniazid resistance in sputum samples from tuberculosis patients by high‐resolution melt curve analysis. Journal of Clinical Microbiology, 55, 1755–1766. 10.1128/JCM.02104-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin, Y. , Cazajous, G. , Dehan, C. , Soler, C. , Vong, R. , Hassan, M. O. , … Vergnaud, G. (2014). Progenitor “Mycobacterium canettii” clone responsible for lymph node tuberculosis epidemic, Djibouti. Emerging Infectious Diseases, 20, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boritsch, E. C. , Khanna, V. , Pawlik, A. , Honoré, N. , Navas, V. H. , Ma, L. , … Brosch, R. (2016). Key experimental evidence of chromosomal DNA transfer among selected tuberculosis‐causing mycobacteria. Proceedings of the National Academy of Sciences of the United States of America, 113, 9876–9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites, D. , Loiseau, C. , Menardo, F. , Borrell, S. , Boniotti, M. B. , Warren, R. , … Gagneux, S. (2018). A new phylogenetic framework for the animal‐adapted Mycobacterium tuberculosis complex. Frontiers in Microbiology, 9, 2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Kong, F. , Wang, Q. , Li, C. , Zhang, J. , & Gilbert, G. L. (2011). Rapid detection of isoniazid, rifampin, and ofloxacin resistance in Mycobacterium tuberculosis clinical isolates using high‐resolution melting analysis. Journal of Clinical Microbiology, 49, 3450–3457. 10.1128/JCM.01068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, P. , Amaro, A. , Ferreira, A. S. , Machado, D. , Albuquerque, T. , Couto, I. , … Inácio, J. (2014). Rapid identification of veterinary‐relevant Mycobacterium tuberculosis complex species using 16S rDNA, IS6110 and regions of difference‐targeted dual‐labelled hydrolysis probes. Journal of Microbiol Methods, 107, 13–22. 10.1016/j.mimet.2014.08.017 [DOI] [PubMed] [Google Scholar]
- Costa, P. , Botelho, A. , Couto, I. , Viveiros, M. , & Inácio, J. (2014). Standing of nucleic acid testing strategies in veterinary diagnosis laboratories to uncover Mycobacterium tuberculosis complex members. Frontiers in Molecular Biosciences, 1, 16 10.3389/fmolb.2014.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves, L. M. , Bulhões, S. M. , Branco, C. C. , Carreira, T. , Vieira, M. L. , Gomes‐Solecki, M. , & Mota‐Vieira, L. (2018). Diagnosis of human leptospirosis in a clinical setting: Real‐time PCR high resolution melting analysis for detection of Leptospira at the onset of disease. Scientific Reports, 8, 9213 10.1038/s41598-018-27555-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, M. , Hauck, Y. , Soler, C. , Koeck, J.‐L. , van Ingen, J. , van Soolingen, D. , … Pourcel, C. (2010). Molecular characteristics of “Mycobacterium canettii” the smooth Mycobacterium tuberculosis bacilli. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 10, 1165–1173. [DOI] [PubMed] [Google Scholar]
- Ghielmetti, G. , Scherrer, S. , Friedel, U. , Frei, D. , Suter, D. , Perler, L. , & Wittenbrink, M. M. (2017). Epidemiological tracing of bovine tuberculosis in Switzerland, multilocus variable number of tandem repeat analysis of Mycobacterium bovis and Mycobacterium caprae . PLoS ONE, 12, e0172474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra‐Assunção, J. A. , Houben, R. M. G. J. , Crampin, A. C. , Mzembe, T. , Mallard, K. , Coll, F. , … Glynn, J. R. (2015). Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: A whole‐genome sequencing approach in a large, population‐based cohort with a high HIV infection prevalence and active follow‐up. Journal of Infectious Diseases, 211, 1154–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halse, T. A. , Escuyer, V. E. , & Musser, K. A. (2011). Evaluation of a single‐tube multiplex real‐time PCR for differentiation of members of the Mycobacterium tuberculosis complex in clinical specimens. Journal of Clinical Microbiology, 49, 2562–2567. 10.1128/JCM.00467-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesseling, A. C. , Rabie, H. , Marais, B. J. , Manders, M. , Lips, M. , Schaaf, H. S. , … Beyers, N. (2006). Bacille Calmette‐Guérin vaccine‐induced disease in HIV‐infected and HIV‐uninfected children. Clinical Infectious Diseases, 42, 548–558. [DOI] [PubMed] [Google Scholar]
- Huard, R. C. , Fabre, M. , de Haas, P. , Lazzarini, L. C. , van Soolingen, D. , Cousins, D. , & Ho, J. L. (2006). Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. Journal of Bacteriology, 188, 4271–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa, R. , Abdul, H. , Hashim, S. H. , Seradja, V. H. , Shaili, N. A. , & Hassan, N. A. M.(2014). High resolution melting analysis for the differentiation of Mycobacterium species. Journal of Medical Microbiology, 63, 1284–1287. 10.1099/jmm.0.072611-0 [DOI] [PubMed] [Google Scholar]
- Jagielski, T. , van Ingen, J. , Rastogi, N. , Dziadek, J. , Mazur, P. K. , & Bielecki, J. (2014). Current methods in the molecular typing of Mycobacterium tuberculosis and other mycobacteria. BioMed Research International, 2014, 645802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery, N. , Gasser, R. B. , Steer, P. A. , & Noormohammadi, A. H. (2007). Classification of Mycoplasma synoviae strains using single‐strand conformation polymorphism and high‐resolution melting‐curve analysis of the vlhA gene single‐copy region. Microbiology, 153, 2679–2688. 10.1099/mic.0.2006/005140-0 [DOI] [PubMed] [Google Scholar]
- Kamerbeek, J. , Schouls, L. , Kolk, A. , van Agterveld, M. , van Soolingen, D. , Kuijper, S. , … van Embden, J. (1997). Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. Journal of Clinical Microbiology, 35, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai, H. , Ezaki, T. , & Harayama, S. (2000). Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. Journal of Clinical Microbiology, 38, 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi, A. D. , Hashemzadeh, M. , Hashemi Shahraki, A. , & Teimoori, A. (2017). Differential identification of mycobacterial species using high‐resolution melting analysis. Frontiers in Microbiology, 8, 2045 10.3389/fmicb.2017.02045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt, P. , Stephan, R. , & Scherrer, S. (2019). Development of a new High Resolution Melting (HRM) assay for identification and differentiation of Mycobacterium tuberculosis complex samples. Scientific Reports, 9, 1850 10.1038/s41598-018-38243-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau, C. , Brites, D. , Moser, I. , Coll, F. , Pourcel, C. , Robbe‐Austerman, S. , … Köser, C. U. (2019). Revised Interpretation of the Hain Lifescience GenoType MTBC to differentiate Mycobacterium canettii and members of the Mycobacterium tuberculosis complex . Antimicrobial Agents and Chemotherapy, 63, e00159‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann, S. , Harmsen, D. , Rüsch‐Gerdes, S. , & Richter, E. (2000). Differentiation of clinical Mycobacterium tuberculosis complex Isolates by gyrB DNA sequence polymorphism analysis. Journal of Clinical Microbiology, 38, 3231–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orloski, K. , Robbe‐Austerman, S. , Stuber, T. , Hench, B. , & Schoenbaum, M. (2018). Whole genome sequencing of Mycobacterium bovis isolated from livestock in the United States, 1989–2018. Frontiers in Veterinary Science, 5, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng, C.‐L. , Chen, H.‐Y. , Chiueh, T.‐S. , Wang, W.‐Y. , Huang, C.‐T. , & Sun, J.‐R. (2012). Identification of non‐tuberculous mycobacteria by real‐time PCR coupled with a high‐resolution melting system. Journal of Medical Microbiology, 61, 944–951. 10.1099/jmm.0.042424-0 [DOI] [PubMed] [Google Scholar]
- Pinsky, B. A. , & Banaei, N. (2008). Multiplex real‐time PCR assay for rapid identification of Mycobacterium tuberculosis complex members to the species level. Journal of Clinical Microbiology, 46, 2241–2246. 10.1128/JCM.00347-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounder, J. I. , Anderson, C. M. , Voelkerding, K. V. , Salfinger, M. , Dormandy, J. , Somoskovi, A. , … Petti, C. A. (2010). Mycobacterium tuberculosis complex differentiation by genomic deletion patterns with multiplex polymerase chain reaction and melting analysis. Diagnostic Microbiology and Infectious Disease, 67, 101–105. 10.1016/j.diagmicrobio.2009.12.014 [DOI] [PubMed] [Google Scholar]
- Reddington, K. , Zumla, A. , Bates, M. , van Soolingen, D. , Niemann, S. , Barry, T. , & O'Grady, J. (2012). SeekTB, a two‐stage multiplex real‐time‐PCR‐based method for differentiation of the Mycobacterium tuberculosis complex. Journal of Clinical Microbiology, 50, 2203–2206. 10.1128/JCM.00718-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, T. , Bibby, S. , O'Rourke, D. , Belfiore, T. , Lambie, H. , & Noormohammadi, A. H. (2009). Characterization of Chlamydiaceae species using PCR and high resolution melt curve analysis of the 16S rRNA gene. Journal of Applied Microbiology, 107, 2017–2028. [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Campos, S. , Smith, N. H. , Boniotti, M. B. , & Aranaz, A. (2014). Overview and phylogeny of Mycobacterium tuberculosis complex organisms: Implications for diagnostics and legislation of bovine tuberculosis. Research in Veterinary Science, 97, S5–S19. [DOI] [PubMed] [Google Scholar]
- Ruettger, A. , Nieter, J. , Skrypnyk, A. , Engelmann, I. , Ziegler, A. , Moser, I. , … Sachse, K. (2012). Rapid spoligotyping of Mycobacterium tuberculosis complex bacteria by use of a microarray system with automatic data processing and assignment. Journal of Clinical Microbiology, 50, 2492–2495. 10.1128/JCM.00442-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A. J. , Inman‐Bamber, J. , Giffard, P. M. , & Huygens, F. (2008). High‐resolution melting analysis of the spa repeat region of Staphylococcus aureus . Clinical Chemistry, 54, 432–436. 10.1373/clinchem.2007.093658 [DOI] [PubMed] [Google Scholar]
- Supply, P. , Allix, C. , Lesjean, S. , Cardoso‐Oelemann, M. , Rüsch‐Gerdes, S. , Willery, E. , … van Soolingen, D. (2006). Proposal for standardization of optimized mycobacterial interspersed repetitive unit‐variable‐number tandem repeat typing of Mycobacterium tuberculosis . Journal of Clinical Microbiology, 44, 4498–4510. 10.1128/JCM.01392-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply, P. , Marceau, M. , Mangenot, S. , Roche, D. , Rouanet, C. , Khanna, V. , … Brosch, R. (2013). Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis . Nature Genetics, 45, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen, R. H. A. M. , Aten, E. , Roos, A. , & den Dunnen, J. T. (2009). High‐resolution melting analysis (HRMA): More than just sequence variant screening. Human Mutation, 30, 860–866. [DOI] [PubMed] [Google Scholar]
- WHO (2018). Global tuberculosis report 2018 8. Retrieved from http://www.who.int/tb/publications/global_report/en/ [Google Scholar]
- Winchell, J. M. , Wolff, B. J. , Tiller, R. , Bowen, M. D. , & Hoffmaster, A. R. (2010). Rapid identification and discrimination of Brucella isolates by use of real‐time PCR and high‐resolution melt analysis. Journal of Clinical Microbiology, 48, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, R. , Sethi, S. , Mewara, A. , Dhatwalia, S. K. , Gupta, D. , & Sharma, M. (2012). Rapid detection of rifampicin, isoniazid and streptomycin resistance in Mycobacterium tuberculosis clinical isolates by high‐resolution melting curve analysis. Journal of Applied Microbiology, 113, 856–862. 10.1111/j.1365-2672.2012.05379.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data sets from intra‐ and interassay variability runs are comprehended in the appendix. On request, additional raw data can be obtained from the corresponding author.


