Abstract
The coral holobiont is a complex ecosystem consisting of coral animals and a highly diverse consortium of associated microorganisms including algae, fungi, and bacteria. Several studies have highlighted the importance of coral‐associated bacteria and their potential roles in promoting the host fitness and survival. Recently, dynamics of coral‐associated microbiomes have been demonstrated to be linked to patterns of coral heat tolerance. Here, we examined the effect of elevated seawater temperature on the structure and diversity of bacterial populations associated with Porites lutea, using full‐length 16S rRNA sequences obtained from Pacific Biosciences circular consensus sequencing. We observed a significant increase in alpha diversity indices and a distinct shift in microbiome composition during thermal stress. There was a marked decline in the apparent relative abundance of Gammaproteobacteria family Endozoicomonadaceae after P. lutea had been exposed to elevated seawater temperature. Concomitantly, the bacterial community structure shifted toward the predominance of Alphaproteobacteria family Rhodobacteraceae. Interestingly, we did not observe an increase in relative abundance of Vibrio‐related sequences in our heat‐stressed samples even though the appearance of Vibrio spp. has often been detected in parallel with the increase in the relative abundance of Rhodobacteraceae during thermal bleaching in other coral species. The ability of full‐length 16S rRNA sequences in resolving taxonomic uncertainty of associated bacteria at a species level enabled us to identify 24 robust indicator bacterial species for thermally stressed corals. It is worth noting that the majority of those indicator species were members of the family Rhodobacteraceae. The comparison of bacterial community structure and diversity between corals in ambient water temperature and thermally stressed corals may provide a better understanding on how bacteria symbionts contribute to the resilience of their coral hosts to ocean warming.
Keywords: coral bleaching, coral microbiome, coral‐associated bacteria, Endozoicomonas, heat stress, PacBio sequencing, Porites lutea, Rhodobacteraceae, thermal bleaching, thermal stress
We examined the effect of elevated seawater temperature on the structure and diversity of bacterial populations associated with Porites lutea, using full‐length 16S rRNA sequences obtained from Pacific Biosciences circular consensus sequencing. The bacterial community structure shifted toward the predominance of Alphaproteobacteria family Rhodobacteraceae. On the other hand, there was a marked decline in the apparent relative abundance of Gammaproteobacteria family Endozoicomonadaceae after P. lutea had been exposed to elevated seawater temperature.

1. INTRODUCTION
Coral reefs are among the most productive and biologically diverse ecosystems on Earth (Connell, 1978). They are estimated to cover only 0.1%–0.5% of the ocean floor (Copper, 1994; Spalding & Grenfell, 1997); nevertheless, coral reefs harbor almost a third of all marine fish species and support the productivity of ~25% of marine fisheries (Moberg & Folke, 1999). The coral “holobiont” is a complex ecological unit consisting of coral animals and a suite of associated symbionts including the dinoflagellate Symbiodiniaceae, fungi and bacteria (Bourne & Munn, 2005; Koren & Rosenberg, 2006; Pantos et al., 2003; Rohwer, Breitbart, Jara, Azam, & Knowlton, 2001; Rohwer, Seguritan, Azam, & Knowlton, 2002; Rosenberg, Koren, Reshef, Efrony, & Zilber‐Rosenberg, 2007). A number of studies have shown that corals harbor a dynamic and highly diverse consortium of microbes that have coevolved with their hosts and contributed important functions to the holobionts (Ainsworth et al., 2015; Bourne, Morrow, & Webster, 2016; Bourne & Munn, 2005; Krediet, Ritchie, Paul, & Teplitski, 2013; Rohwer et al., 2001, 2002; Rosenberg & Loya, 2013; Thompson, Rivera, Closek, & Medina, 2014). Coral‐associated bacteria appear to be involved in the provisioning and cycling of carbon, nitrogen and sulfur in coral reefs (Cardini et al., 2015; Ceh et al., 2013; Chimetto et al., 2008; Lema, Willis, & Bourne, 2012; Lesser et al., 2007; Rädecker, Pogoreutz, Voolstra, Wiedenmann, & Wild, 2015; Raina, Tapiolas, Willis, & Bourne, 2009; Siboni, Ben‐Dov, Sivan, & Kushmaro, 2008). In addition, members of the coral microbiome may have beneficial roles in protecting their hosts against pathogenic and opportunistic bacteria by preventing their colonization through the production of antibacterial compounds or through physical occupation of otherwise available niches (Kelman, Kashman, Rosenberg, Kushmaro, & Loya, 2006; Koh, 1997; Krediet et al., 2013; Reshef, Koren, Loya, Zilber‐Rosenberg, & Rosenberg, 2006; Ritchie & Smith, 2004).
Thermally induced coral bleaching has resulted in significant decline in live coral cover at regional scales (Brown, 1997; Bruno & Selig, 2007; Fitt, Brown, Warner, & Dunne, 2001; Gardner et al., 2019; Gardner, Côté, Gill, Grant, & Watkinson, 2003; Hughes et al., 2018; Jones, 2008). Earlier attempts to understand the coral host responses to ocean warming have mainly focused on the partnership between corals and their photosynthetic symbionts (Baker, 2003; Boulotte et al., 2016; Fautin & Buddemeier, 2004; Howells et al., 2012; Hume et al., 2016; Keshavmurthy et al., 2014; Rowan, 2004). Adaptive mechanisms to elevated water temperature such as Symbiodiniaceae “shuffling” (an adjustment in relative abundance of Symbiodiniaceae type in‐hospite) and Symbiodiniaceae “switching” (an acquisition of new Symbiodiniaceae types from the surrounding environment) have been proposed (Baker, 2003; Baker, Starger, Mcclanahan, & Glynn, 2004; Berkelmans & van Oppen, 2006; Boulotte et al., 2016; Fautin & Buddemeier, 2004; Jones, Berkelmans, Oppen, Mieog, & Sinclair, 2008; Tamar, 2006). The roles of bacterial consortium in the coral holobiont symbioses during thermal stress have largely been overlooked until recently. A study by Gilbert, Hill, Doblin, and Ralph (2012) examined the response of a coral holobiont to thermal stress when the bacterial community was treated with antibiotics and demonstrated that a disruption to the microbial consortium diminished the resilience of the holobiont. Intact microbial communities offered coral holobionts resilience to thermal stress and increased the rate of recovery after bleaching events (Gilbert et al., 2012). More recently, dynamics of coral‐associated microbiomes have been demonstrated to be linked to patterns of coral heat tolerance (Ziegler, Seneca, Yum, Palumbi, & Voolstra, 2017). Ziegler et al. (2017) showed that the composition of Acropora hyacinthus‐associated bacterial community was different across thermally variable habitats and adapted to the new environment when corals were reciprocally transplanted. Subsequent exposure of those transplanted corals to thermal stress conditions changed the bacterial community of heat‐sensitive corals from a more stable, cooler environment, whereas heat‐tolerant corals from a highly variable, warmer environment harbored a stable bacterial community (Ziegler et al., 2017).
Branching coral species, especially acroporids, are generally fast‐growing and known to be more susceptible to thermal bleaching compared to massive Porites corals, which are slow‐growing and more “stress‐tolerant” (Darling, Alvarez‐Filip, Oliver, Mcclanahan, & Côté, 2012; Loya et al., 2001; Pisapia et al., 2016). Shifts in coral‐associated microbiomes during bleaching events have been documented mostly in Acropora species: Acropora millepora and Acropora tenuis from the Great Barrier Reef, Acropora millepora from Fiji and Acropora muricata in Nan‐wan, Taiwan (Bourne, Iida, Uthicke, & Smith‐Keune, 2008; Grottoli et al., 2018; Lee, Davy, Tang, & Kench, 2016, 2017; Littman, Willis, Pfeffer, & Bourne, 2009). Here, we investigated the dynamics of bacterial communities associated with a massive stony coral Porites lutea, a dominant reef‐builder widely distributed in the Indo‐Pacific, under short‐term thermal stress. Our survey of P. lutea microbiomes utilized a long‐read PacBio SMRT sequencing technology to capture full‐length 16S rRNA sequences in order to examine the shifts in coral‐associated bacterial community structure and diversity. Full‐length 16S rRNA sequences have a benefit of spanning all hypervariable regions, enabling high‐resolution taxonomic classification of bacterial species (Pootakham et al., 2017). The investigation of coral‐microbial interactions and their dynamics during environmental stress will help further our understanding of the involvement of bacterial symbionts in the acclimatization and resilience of coral holobiont to climate change.
2. MATERIALS AND METHODS
2.1. Sample collection and DNA extraction
Five P. lutea colonies were collected from the Maiton Island in the Andaman Sea (7°45′42.5″N 98°28′51.3″E) at the depth of 7–10 m and immediately placed in containers with aerated seawater. We chose colonies that were at least 10 m apart from each other to reduce the possibility of collecting clonal colonies. P. lutea colonies were transported back to shore (Phuket Marine Biological Center) within an hour of collection and transferred to flow‐through aquaria (transparent glass tanks with the dimension of 120 × 25 × 25 cm, holding approximately 75 L), which circulated seawater pumped from the reef to the Phuket Marine Biological Center Research Station. Each colony was fragmented into four nubbins using a hammer and chisel and acclimated for 14 days in flow‐through aquaria at ambient temperature (29°C) with a 12‐hr light–dark cycle and irradiance of 150 μmol m−2 s−1. Two nubbins from each colony were placed in the “control” aquarium, and the other two were placed in the “heated” aquarium. The position of ten nubbins (two from each of the five colonies) in the aquarium was randomized. During the experiment, one aquarium was maintained at the ambient water temperature while the other aquarium was exposed to a heat treatment, which involved an incremental ramping of seawater temperature at a rate of 0.5°C/day for six consecutive days using one 100‐W submersible aquarium heater (Eheim). From day 7 to day 13, the temperature was maintained at 34°C in the “heat treatment” aquarium (Figure 1a). Coral nubbins were removed from the aquaria for sampling 6 (D6C and D6H) and 13 (D13C and D13H) days after the onset of the thermal stress treatment (Figure 1b). Since the coral nubbins in the control and heated aquaria were from the same parent colonies (one fragment from each colony in each tank), changes in the microbiome were due to treatment effects and not genetic differences between colonies. It is worth noting that we used one aquarium for each condition (control and heated); hence, there was no technical replicate in this study. However, we closely monitored the temperature in both aquaria to ensure that it remained stable in the control aquarium (29°C) and was steadily increasing until it reached 34°C in the heated aquarium (Figure 1a).
Figure 1.
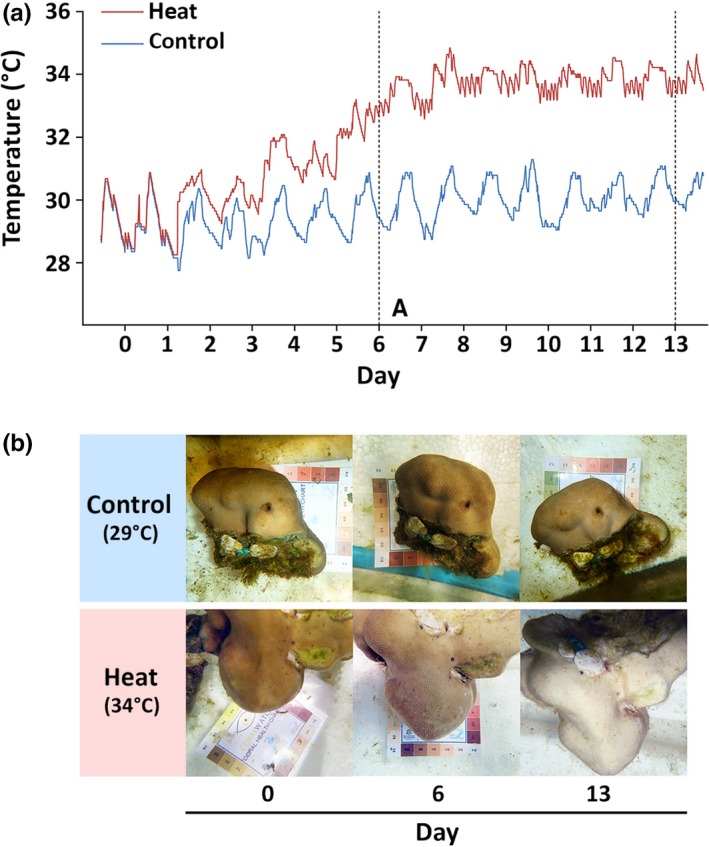
Temperature logs and physiological response of Porites lutea subjected to a heat treatment. (a) Records of daily water temperatures in control (blue line) and heated (red line) aquaria during the 13‐day experiment. Vertical dotted lines indicate the days on which the samples were collected (day 6 and day 13). (b) Pictures of representative coral fragments from the control (~29°C) and heated (~34°C) aquaria taken prior to (day 0) and following the imposition of short‐term thermal stress (day 6 and day 13)
Coral tissues were collected using scalpel blades, placed in sterile screw‐capped tubes, and immediately frozen in liquid nitrogen. DNA extraction was performed using the CTAB protocol reported in Pootakham et al. (2017). DNA samples were quantified using the NanoDrop ND‐1000 Spectrophotometer (Thermo Fisher Scientific) and diluted to 50 ng/μl for PCR amplification.
2.2. 16S rRNA amplification and PacBio sequencing
To obtain full‐length 16S rRNA amplicons, we carried out the PCR using 50 ng of genomic DNA template and two bacterial‐specific primers 27F (5′‐AGAGTTTGATCMTGGCTCAG) and 1492R (5′‐TACGGYTACCTTGTTACGACTT). To multiplex amplicons from nine samples into one SMRTbell library, we used 27F and 1492R primers tailed with 16‐base PacBio barcodes at their 5’ ends (://github.com/PacificBiosciences/Bioinformatics-Training/blob/master/barcoding/pacbio_384_barcodes.fasta). Amplification reactions were carried out using barcoded forward (27F) and reverse (1492R) primers in a final volume of 20 μl consisting of 0.4 U of Phusion High‐Fidelity DNA Polymerase (Thermo Fisher Scientific), 0.2 mM dNTPs, 1× Phusion HF Buffer, 0.1 μM of each primer and 1.5 mM MgCl2. The PCR conditions used were the same as those reported in Pootakham et al. (2017). Since we were unable to amplify 16S rRNA fragments from samples D13C‐5 and D13H‐5 due to their low DNA quality, the number of independent data points for each condition is as follows: D6C n = 5; D6H n = 5; D13C n = 4; and D13H n = 4. PCR products were subsequently purified with Agencourt AMPure XP magnetic beads (Beckman Coulter) and eluted in 20 μl elution buffer (EB). The Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific) was used to quantify the purified PCR products. Barcoded 16S amplicons were multiplexed in equimolar concentrations, and a total of 500 ng of pooled DNA was used for library preparation. SMRTbell libraries were constructed (each containing amplicons from nine samples) and sequenced on a PacBio RSII system using the P6‐C4 chemistry with a 6‐hr movie time (Pacific Biosciences).
2.3. 16S rRNA sequence data analysis
To obtain the circular consensus sequences (CCS) with a minimum of five full passes, PacBio raw reads were demultiplexed and processed using the RS‐ReadsOfInsert protocol (SMRT Analysis software version 2.3). Consensus 16S rRNA sequences that were shorter than 1,000 nt (likely generated as a result of partial amplification) were removed from further analyses. Based on the CCS read length distribution (Figure A1), we arbitrarily chose 1,000 nt as a cutoff to avoid using sequences that contained nonoverlapping hypervariable regions for downstream analyses since they might affect the taxonomic classification as well as the diversity index calculation. Both strands of the consensus sequences were screened for the presence of chimeric sequences using the abundance‐based algorithm in UCHIME (Edgar, 2010) and a reference dataset from Ribosomal Database Project (RDP) (Cole et al., 2014). Filtered sequences were analyzed as previously described in Pootakham et al. (2017) using QIIME software version 1.9.1 (Caporaso et al., 2010). Sequences classified as unknown or chloroplasts were removed prior to subsequent analyses. Alpha and beta diversity analyses were performed using the QIIME pipeline based on sequence data that had been clustered into OTUs at a 97% similarity level. The OTU table was rarefied to an even depth of 5,652 sequences per sample corresponding to the lowest number of sequences present in a sample. Taxonomic classification was assigned to the representative sequence of each OTU using the RDP classifier retrained toward the Greengenes database V13.8 (DeSantis et al., 2006). Since we had full‐length 16S rRNA sequence information, “unassigned” OTUs were subsequently classified based on their representative sequences using local sequence alignment (BLAST search) against the nt database. If the representative sequences of the OTUs returned the best hits with known taxonomy, those OTUs were classified accordingly. However, if the representative sequences of the OTUs returned “uncultured environmental sample” as their best hits, those OTUs remained categorized as “unassigned.” We further investigated the nature of “unassigned” sequences by constructing the maximum‐likelihood phylogenetic tree using MEGA7 software (Kumar, Stecher, & Tamura, 2016). The clustering of the sequences was tested by a bootstrap approach using 1,000 replications. Comparison between bacterial communities associated with P. lutea samples in this study (maintained in the aquarium) and those associated with P. lutea samples collected in situ from the same site (MT1‐MT3 samples from Pootakham et al. (2017) and MT‐Mar samples from Pootakham et al. (2018)) were analyzed as mentioned above using QIIME software version 1.9.1 (Caporaso et al., 2010).
White's nonparametric t test was used to test for differences in relative abundance of associated bacteria between samples collected in control and heated aquaria (White, Nagarajan, & Pop, 2009). To compare bacterial community compositions, we applied a nonparametric permutational multivariate analysis of variance (PERMANOVA) using the weighted UniFrac distance matrix as implemented in the vegan function Adonis (permutation = 1,000) (Lozupone & Knight, 2005; Oksanen, Blanchet, Kindt, Legendre, & Minchin, 2013). The UniFrac method measures the distance between communities based on the lineages they contain (Lozupone & Knight, 2005). The representative OTU sequences were aligned to generate a phylogenetic tree using the command pick_open_reference_otus.py (://qiime.org/scripts/pick_open_reference_otus.html). We used the weighted version of the UniFrac metric, which accounts for the relative abundance of each of the taxa within the communities (Lozupone & Knight, 2005).
2.4. Core microbiome and indicator species identification
For the identification of P. lutea core microbiome members and the identification of indicator species, we employed the information from the full‐length 16S rRNA sequences and followed the analysis pipeline reported in Pootakham et al. (2017). Members of the P. lutea core microbiome were identified at 100% sample coverage (the minimal percentage of all samples in which an OTU must be present to be considered part of the core microbiome) based on previous studies (Lawler et al., 2016; van de Water et al., 2016; Hadaidi et al., 2017; Pootakham et al., 2017).
2.5. Performance comparison between full‐length 16S rRNA and short hypervariable fragments
To compare the performance of short hypervariable regions (V3‐V4 and V5‐V6) and full‐length 16S rRNA sequences in resolving taxonomic classification, we obtained the V3‐V4 and V5‐V6 sequences in silico from their respective full‐length reads using 5′‐ACTCCTACGGGAGGCAGCAG and 5′‐CTACCAGGGTATCTAATC for the V3‐V4 region and 5′‐GGATTAGATACCCTGGTAGTCC and 5′‐CTCACGRCACGAGCTGACG for the V5‐V6 region. The extracted in silico V3‐V4 and V5‐V6 fragments and their counterpart full‐length sequences were aligned to the RDP type strain reference sequences using BLAST (e‐value cutoff of 10–10). Any sequences that yielded a minimum of two best hits from different species with identical e‐values, aligned sequences, and bit‐scores were categorized as “ineffective” for the species‐level classification. For classification at the genus level, any query sequence that had two or more best hits belonging to different genera was considered “unassigned.”
3. RESULTS
3.1. Evaluation of coral‐associated bacterial diversity during thermal stress using full‐length 16S rRNA gene sequences
The degree of bleaching was monitored using the Coral Health Chart from Coral Watch (https://coralwatch-old.org/web/guest/coral-health-chart). Coral samples that had experienced thermal stress were partially bleached (color scale: E3) after six days, and pigments were completely lost 13 days after the onset of heat treatment (color scale: E1; Figure 1b). Coral samples that were maintained in the control aquarium at 29°C remained pigmented throughout the experiment (color scale: E4‐E5) and did not show any sign of bleaching. Full‐length 16S rRNA genes were amplified from coral‐associated bacteria and sequenced using the PacBio platform. A total of 1,416,896 PacBio reads totaling 29.5 Gb were obtained from 16 SMRT cells. Polymerase reads were assembled and demultiplexed into 357,254 circular consensus sequencing (CCS) reads with a minimum of five full passes (Table 1). After removing 16S rRNA CCS reads shorter than 1,000 nucleotides and chimeric sequences, we obtained a total of 248,950 reads. The number of processed reads ranged from 5,652 to 26,091 per sample, with an average of 13,831 reads/sample (Table 1).
Table 1.
Number of circular consensus sequence (CCS) reads, OTUs, and alpha diversity indices of Porites lutea microbiome under thermal stress. D6C and D13C indicate that samples were taken from the control aquarium on day 6 and day 13, respectively. D6H and D13H indicate that samples were taken from the heated aquarium on day 6 and day 13, respectively
| Condition | Sample | Number of CCS reads | Number of processed readsa | Number of OTUs | Chao1 | Simpson | Shannon | Good's coverage |
|---|---|---|---|---|---|---|---|---|
| D6C | D6C‐1 | 18,213 | 13,317 | 768 | 989 | 0.9053 | 5.6155 | 0.9805 |
| D6C‐2 | 21,716 | 15,085 | 211 | 335 | 0.6844 | 2.4380 | 0.9928 | |
| D6C‐3 | 11,873 | 8,582 | 172 | 430 | 0.7662 | 2.9071 | 0.9878 | |
| D6C‐4 | 35,302 | 26,091 | 391 | 552 | 0.8109 | 3.7021 | 0.9937 | |
| D6C‐5 | 29,280 | 21,091 | 996 | 1,249 | 0.9261 | 6.0606 | 0.9845 | |
| D6H | D6H‐1 | 22,349 | 15,984 | 948 | 1,331 | 0.9397 | 6.4742 | 0.9769 |
| D6H‐2 | 16,886 | 11,476 | 746 | 1,108 | 0.9293 | 5.9003 | 0.9695 | |
| D6H‐3 | 11,952 | 8,463 | 1,072 | 1,751 | 0.9783 | 7.4745 | 0.9330 | |
| D6H‐4 | 13,447 | 9,438 | 573 | 772 | 0.9274 | 5.9100 | 0.9782 | |
| D6H‐5 | 23,381 | 15,547 | 914 | 1,334 | 0.8657 | 5.5290 | 0.9742 | |
| D13C | D13C‐1 | 21,138 | 15,480 | 234 | 417 | 0.7780 | 3.5362 | 0.9935 |
| D13C‐2 | 16,216 | 12,188 | 408 | 676 | 0.7915 | 3.6422 | 0.9833 | |
| D13C‐3 | 26,510 | 16,924 | 1,046 | 1,322 | 0.8877 | 5.2677 | 0.9750 | |
| D13C‐4 | 18,187 | 11,459 | 816 | 1,328 | 0.8103 | 4.5242 | 0.9588 | |
| D13H | D13H‐1 | 27,513 | 19,834 | 1,602 | 2,144 | 0.9661 | 7.7104 | 0.9688 |
| D13H‐2 | 21,953 | 14,759 | 1,187 | 1,725 | 0.9712 | 7.2585 | 0.9648 | |
| D13H‐3 | 10,342 | 5,652 | 829 | 1,507 | 0.9108 | 6.8358 | 0.8563 | |
| D13H‐4 | 10,996 | 7,580 | 772 | 1,104 | 0.9713 | 7.0518 | 0.9557 | |
| Total: | 357,254 | 248,950 |
After removal of chimeras and reads shorter than 1,000 bases.
A series of alpha diversity indices were calculated for each sample in order to evaluate the diversity of coral‐associated bacterial communities present within the samples. A wide range in numbers of operational taxonomic units (OTUs) at a 3% dissimilarity level was observed, with the lowest number of OTUs (172) associated with D6C‐3 sample and the highest number of OTUs (1,602) associated with D13H‐1 sample (Table 1). Good's coverage indices along with the rarefaction curves that have plateaued off for most samples indicated that sufficient sampling has been achieved to capture the total diversity of the bacterial communities under investigation (Table 1, Figure A2). The Shannon indices ranged from 2.43 (D6C‐2) to 7.71 (D13H‐1), similar to those reported for P. lutea from the South China Sea, Indo‐Pacific, and West Indian Ocean (Li et al., 2013; McKew et al., 2012; Sere et al., 2016; Shannon, 1948). A significant difference between the Shannon indices associated with samples from control and heated aquaria was observed on day 13 (D13C vs. D13H samples; Mann–Whitney U test, p‐value = .02857). The Simpson diversity indices ranged from 0.6844 (D6C2) to 0.9783 (D6H‐3), and significant differences in the values of Simpson diversity indices were observed between samples in the control and heated treatment groups on both day 6 and day 13 (Mann–Whitney U test; D6C vs. D6H, p‐value = .03175; D13C vs. D13H, p‐value = .02857). The values of both Shannon and Simpson diversity indices were significantly higher when coral samples were subjected to thermal stress.
To evaluate the diversity of bacterial community present among samples, the beta diversity indices were calculated (Figure 2). Principal coordinates analysis (PCoA) of weighted UniFrac distances based on RDP results demonstrated that control and heat‐treated coral samples harbored distinct bacterial communities, and this distinction became more evident after an extended period of thermal stress (PERMANOVA; D6C vs. D6H, pseudo‐F = 8.372, p = .007; D13C vs. D13H, pseudo‐F = 17.492, p = .032; D6H vs. D13H pseudo‐F = 2.872, p = .037; Figure 2).
Figure 2.
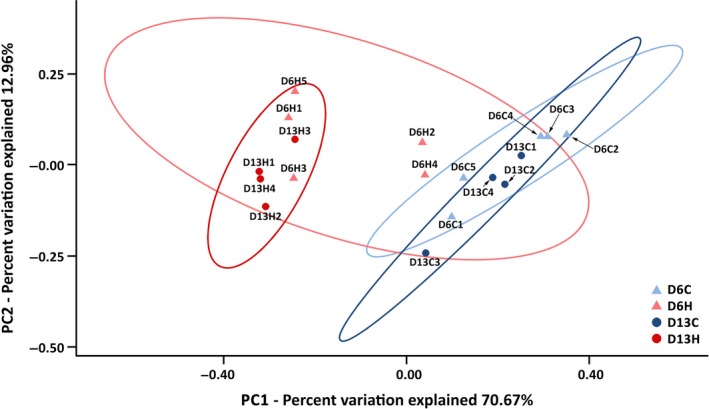
Beta diversity estimates of Porites lutea‐associated microbiomes during a thermal stress. A principal coordinates analysis (PCoA) plot of coral‐associated bacterial communities during a thermal stress based on weighted UniFrac distances. Blue symbols indicate samples from the control aquarium (~29°C), and red symbols indicate samples from the heated aquarium (~34°C)
3.2. Structure and composition of coral‐associated microbiomes under thermal stress
Using a confidence threshold of 80%, 220,698 out of 284,950 filtered reads from all 18 samples were assigned based on the RDP classifier (Wang, Garrity, Tiedje, & Cole, 2007). CCS reads were classified into 22 phyla (15 known and 7 candidate phyla), 60 classes, and 122 orders (Table S1). Proteobacteria represented a ubiquitous and dominant taxon in all P. lutea samples irrespective of the heat treatment (Figure 3). Microbial communities associated with P. lutea also consisted of members of Cyanobacteria and Bacteroidetes. We compared bacterial communities associated with P. lutea samples in the control aquarium with previously reported communities associated with P. lutea samples collected in situ from the same sampling site (Pootakham et al., 2017, 2018) and noticed that bacterial communities associated with coral samples that had been brought into aquaria harbored a significantly higher proportion of Cyanobacteria (Figure A3; White's nonparametric t test, p‐value = .013).
Figure 3.
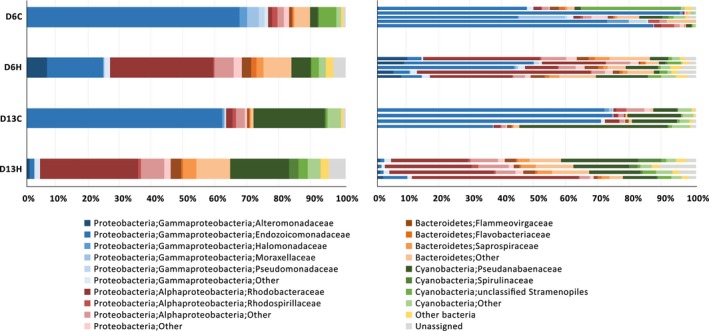
Dynamics of coral‐associated bacteria during a thermal stress. Bar charts displaying the composition of bacterial communities associated with Porites lutea in control and heated aquaria at a group level (left) and an individual colony level (right). Taxonomic classification of OTUs present in each sample at the phylum/class/family levels was based on the Greengenes database. Eleven most abundant families were depicted, and the remaining taxa were grouped together under “others.”
We observed a significant alteration in the composition of coral‐associated microbiomes 6 days (D6C vs. D6H; PERMANOVA, pseudo‐F = 1.906, p = .018) and 13 days (D13C vs. D13H; PERMANOVA, pseudo‐F = 2.389, p = .024) after the samples had been exposed to elevated water temperature (Figure 3). Interestingly, a substantial proportion (~12%) of the OTUs present in the microbiomes associated with heat‐stressed corals could not be assigned based on the RDP classifier. The full‐length nature of the 16S rRNA sequence data allowed us to further assign taxonomy by aligning the representative sequence of each OTU to the Genbank nt database using BLAST. Approximately 50% of the unassigned sequences matched unclassified bacterial sequences obtained from uncultured environmental samples (labeled as “unassigned” in Figure 3), while the other 40% and 10% of the previously unassigned sequences (based on RDP classifier) matched sequences belonging to Bacteroidetes and Proteobacteria phyla, respectively. In addition, we constructed the phylogenetic tree to investigate the relationship of the top four most abundant unassigned OTUs (OTU8, OTU67, OTU9974, and OTU12304) with known sequences. While OTU8 and OTU9974 appeared to be related to a member of Flammeovirgaceae and Planctomycetales (Pla4 lineage), respectively, OTU67 and OTU12304 did not seem to be related to any bacterial sequences with known classification (Figure A4).
Compared to samples maintained at ambient temperature, Gammaproteobacteria family Endozoicomonadaceae became significantly less abundant when corals experienced a thermal stress (White's nonparametric t test, p < .01; Figure 4). On the contrary, Alphaproteobacteria appeared to be dominant in the bacterial communities associated with corals in the heated aquarium. More specifically, Alphaproteobacteria family Rhodobacteraceae increased in their relative abundances significantly after P. lutea experienced a thermal stress (White's nonparametric t test, p < .01; Figure 4). Additionally, the proportions of Alteromonadaceae and Saprospiraceae present in the heat‐treated samples increased significantly compared to those present in the control samples (White's nonparametric t test, p < .05; Figure 4).
Figure 4.
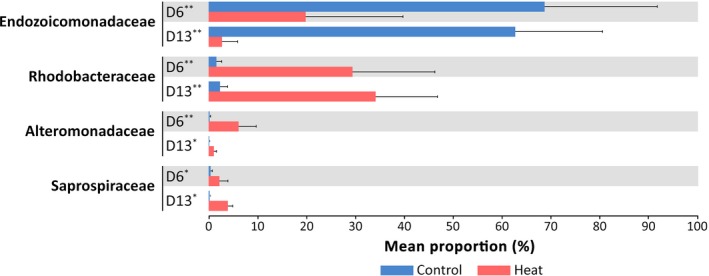
Changes in relative abundance of coral‐associated bacteria under thermal stress. A bar chart displaying average proportions of four bacterial families, the relative abundance of which was significantly different between control and heat treatment (White's nonparametric t test). A single asterisk (*) indicates a significance level of p < .05, and a double asterisk (**) indicates a significance level of p < .01
3.3. Identification of the P. lutea core microbiome
Previous studies have reported the association of similar bacterial populations with the same coral species inhabiting different geographic locations (Bourne & Munn, 2005; Frias‐Lopez, Zerkle, Bonheyo, & Fouke, 2002; Hong, Yu, Chen, Chiang, & Tang, 2009; Ritchie & Smith, 2004; Rohwer et al., 2001, 2002; Rohwer & Kelley, 2004), suggesting that corals hosted species‐specific microbiome regardless of the environmental factors. Even though the composition of bacterial communities associated with untreated and heat‐stressed corals appeared to be different, we identified a number of conserved bacterial species consistently present in 100% of the samples (regardless of their relative abundances in the communities). According to our stringent criterion, a total of ten species representing four orders and three phyla were identified as putative members of the core microbiome (Figure 5). The majority of core microbiome candidates were Proteobacteria, with Gammaproteobacteria (five species) and Alphaproteobacteria (three species) constituting 47.4% and 8.4% of the core microbiome population, respectively. Endozoicomonas elysicola (25.32%), Endozoicomonas euniceicola (11.23%), and Prochlorothrix hollandica (10.03%) were the top three most abundant species, while Pseudoruegeria aquimaris (0.78%), Kistimonas asteriae (1.71%), and Tropicibacter phthalicicus (2.51%) were among the least abundant species in the core microbiome (Figure 5). We also compared the data from this study to those from previous reports (Pootakham et al., 2017, 2018) and found one bacterial species, Endozoicomonas elysicola, to be exceptionally persistent in all 30 P. lutea colonies investigated across different geographical locations and seawater temperatures. Most of the species identified as putative members of P. lutea core microbiome were not present in high abundance. In fact, only three members had a relative abundance higher than 10% within the whole community while five had a relative abundance between 1% and 6%, and the remaining was found to have lower than 1% relative abundance within the community.
Figure 5.
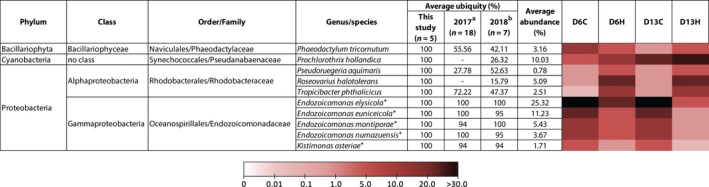
Porites lutea core microbiome. A list of bacterial species present in P. lutea core microbiome, their average ubiquity (the percentage of all P. lutea samples in which the species was detected) in this and previous studies [2017a refers to a study by Pootakham et al. (2017), and 2018b refers to a study by Pootakham et al. (2018)], and their average relative abundance across all conditions. The heat map shows the average abundance of the species under each condition. The asterisk (*) indicates species that have previously been identified as members of P. lutea core microbiome. The dash (‐) indicates that the sequences affiliated with that particular species were not detected
3.4. Indicator species associated with heat‐stressed corals
We examined the microbial communities from both untreated and heat‐stressed samples to see whether any specific bacterial species were significantly associated with corals that had been exposed to elevated water temperature. We applied the indicator value (INDVAL) method to analyze our data for the presence of indicator species for control and heated conditions (De Caceres & Legendre, 2009). A complete list of bacteria that were identified as indicator species for the control or heat‐stressed corals on either day 6 or day 13 is provided in Table S2. Bacterial species that were identified as indicator species (for either control or heat‐stressed condition) on both day 6 and day 13 are considered strong indicator species and listed in Table 2.
Table 2.
A list of indicator species significantly (p < .05) associated with corals under normal (D6C, D13C) or elevated temperature (D6H, D13H) as revealed by the indicator value (INDVAL) analysis
| Phylum | Class | Family | Indicator species | INDVAL (p‐value) | |||
|---|---|---|---|---|---|---|---|
| D6C | D13C | D6H | D13H | ||||
| Cryptophyta | Cryptophyceae | Pyrenomonadaceae | Rhodomonas salina | – | – | 0.800 (.048) | 0.991 (.026) |
| Proteobacteria | Alphaproteobacteria | Rhodobacteraceae | Actibacterium mucosum | – | – | 0.973 (.008) | 0.987 (.031) |
| Aestuariivita boseongensis | – | – | 0.984 (.013) | 0.811 (.033) | |||
| Aliiroseovarius crassostreae | – | – | 0.980 (.008) | 0.964 (.022) | |||
| Celeribacter neptunius | – | – | 0.958 (.010) | 0.927 (.029) | |||
| Confluentimicrobium lipolyticum | – | – | 0.990 (.009) | 0.955 (.035) | |||
| Donghicola eburneus | – | – | 0.994 (.011) | 0.997 (.036) | |||
| Leisingera aquimarina | – | – | 1.000 (.008) | 0.989 (.032) | |||
| Lutimaribacter pacificus | – | – | 0.800 (.038) | 1.000 (.029) | |||
| Marivita litorea | – | – | 0.947 (.011) | 1.000 (.030) | |||
| Planktomarina temperata | – | – | 0.911 (.019) | 0.957 (.031) | |||
| Ponticoccus lacteus | – | – | 0.986 (.031) | 0.976 (.034) | |||
| Poseidonocella sedimentorum | – | – | 0.769 (.046) | 0.893 (.048) | |||
| Pseudoruegeria aquimaris | – | – | 0.868 (.017) | 0.917 (.035) | |||
| Pseudoruegeria lutimaris | – | – | 1.000 (.005) | 0.937 (.026) | |||
| Roseovarius halotolerans | – | – | 0.982 (.005) | 0.973 (.028) | |||
| Roseovarius pacificus | – | – | 0.986 (.009) | 0.977 (.040) | |||
| Ruegeria atlantica | – | – | 0.913 (.026) | 0.978 (.028) | |||
| Ruegeria pomeroyi | – | – | 0.978 (.013) | 0.989 (.031) | |||
| Sulfitobacter geojensis | – | – | 0.800 (.044) | 1.000 (.024) | |||
| Thalassobius mediterraneus | – | – | 0.800 (.046) | 0.984 (.028) | |||
| Thalassococcus lentus | – | – | 0.968 (.014) | 0.865 (.029) | |||
| Tropicibacter phthalicicus | – | – | 0.988 (.011) | 0.958 (.038) | |||
| Gammaproteobacteria | Cellvibrionaceae | Eionea nigra | – | – | 0.983 (.042) | 0.941 (.035) | |
| Endozoicomonadaceae | Endozoicomonas elysicola | 0.828 (.014) | 0.981 (.030) | – | – | ||
| Endozoicomonas euniceicola | 0.880 (.011) | 0.970 (.023) | – | – | |||
| Endozoicomonas gorgoniicola | 0.866 (.007) | 0.982 (.020) | – | – | |||
| Endozoicomonas montiporae | 0.846 (.008) | 0.972 (.031) | – | – | |||
| Endozoicomonas numazuensis | 0.786 (.047) | 0.986 (.031) | – | – | |||
| Kistimonas asteriae | 0.858 (.026) | 0.980 (.048) | – | – | |||
All six indicator species for corals maintained at ambient temperature (D6C and D13C) belonged to Gammaproteobacteria family Endozoicomonadaceae, consistent with the information that members of this family were significantly more abundant in the microbiomes associated with corals from control aquarium (Figure 4 and Table 2). Interestingly, with the exception of Rhodomonas salina and Eionea nigra, all indicator species for coral microbiomes that had experienced a thermal stress (D6H and D13H) were members of Alphaproteobacteria family Rhodobacteraceae, with several species detected exclusively in either D6H or D13H samples (species with an index value of 1; Table 2).
3.5. Taxonomic classification at species resolution using full‐length 16S rRNA sequence information
Classifying the taxonomy of 16S rRNA sequences based on short hypervariable fragments of the 16S rRNA gene can be difficult. In this study, we adopted the PacBio CCS technology to obtain full‐length 16S rRNA sequences covering all hypervariable regions of the 16S rRNA genes. In order to demonstrate the benefit of utilizing full‐length sequences in 16S rRNA gene‐based community surveys, we extracted the commonly used V3‐V4 and V5‐V6 regions from the full‐length reads and aligned both of the in silico simulated 420‐nucleotide V3‐V4 and 255‐nucleotide V5‐V6 amplicons and their respective full‐length counterparts against reference sequences in the RDP (Cole et al., 2014). A much higher proportion of full‐length 16 rRNA sequences could be classified at the species level compared to those of V3‐V4 and V5‐V6 fragments (Figure 6). We were able to assign taxonomic classification at the species level for 99.8%, 86.2%, and 82.0% for the full‐length 16S rRNA sequences, V3‐V4 hypervariable fragments, and V5‐V6 hypervariable fragments, respectively. Furthermore, we assigned taxonomy (at the genus level) to each in silico (V3‐V4 and V5‐V6) amplicon extracted from the full‐length sequence and compared taxonomic profiles obtained from short amplicons to those obtained from the full‐length 16S sequences (Table S3). Taxonomic classification based on hypervariable V3‐V4 or V5‐V6 regions yielded different microbiome profiles from the classification based on full‐length 16S rRNA sequences. For example, Roseovarius was one of the two most prevalent genera associated with heat‐stressed corals (D6H and D13H) when using full‐length 16S rRNA sequences for classification (Table S3). However, when V5‐V6 fragments were used instead of their full‐length counterparts, Pseudoruegeria was erroneously represented as the most abundant genus affiliated with heat‐stressed microbiomes (when the taxonomic classification was performed with full‐length 16S data, the relative abundance of Pseudoruegeria in the heat‐stressed microbiomes was <2%; Table S3). The proportion of unassigned sequences was also considerably higher when partial 16S fragments were used instead of their full‐length counterparts, suggesting that the commonly used hypervariable V3‐V4 and V5‐V6 regions were identical among several bacterial 16S rRNA sequences in the reference database (Table S3).
Figure 6.
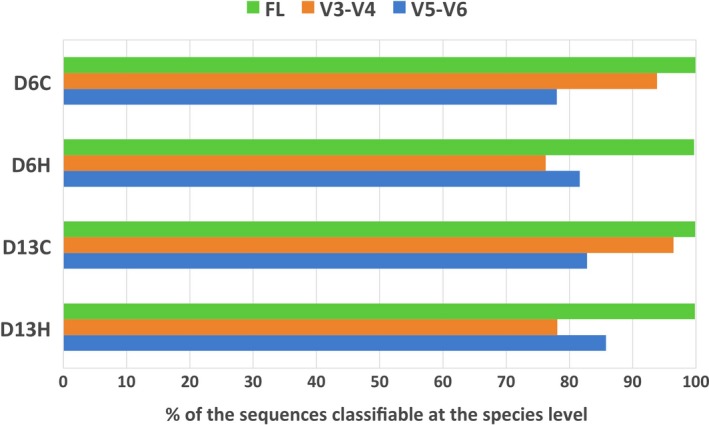
Performance comparison between full‐length 16S rRNA and hypervariable fragments (V3‐V4 or V5‐V6). A bar graph illustrating proportions of sequence reads from each treatment that are classifiable at the species level using full‐length (FL; green) or partial 16S rRNA gene fragments (V3‐V4, orange; V5‐V6, blue).
4. DISCUSSION
4.1. Thermal stress affects the structure and diversity of coral‐associated microbiomes
A diverse group of bacteria are known to associate with corals, and dynamics of bacterial community have recently been shown to be linked to patterns of heat tolerance in their hosts (Ziegler et al., 2017). Here, we investigated the effects of elevated water temperature on the structure and diversity of microbiomes associated with P. lutea using full‐length 16 rRNA gene sequences. We employed the aquarium system since it allowed us to control other environmental factors (i.e., the only difference between controlled and heated aquaria was the water temperature). The microbiomes of corals have been known to shift their compositions when brought into aquaria. Kooperman, Ben‐Dov, Kramarsky‐Winter, Barak, and Kushmaro (2007) previously reported a decrease in relative abundance of Cyanobacteria in Fungia granulosa that had been maintained in the aquarium (Kooperman et al., 2007). Our observation with P. lutea revealed the opposite trend (Figure A3), with Cyanobacteria being more abundant in aquarium samples. The relative abundance of Cyanobacteria appeared to increase with the amount of time that P. lutea samples had been maintained in the aquarium (White's nonparametric t test; D6 vs. D13, p‐value = .023; Figure 3). However, the shift in the relative abundance of Cyanobacteria was independent of an increase in water temperature since it was observed in both control and heat‐treated samples (White's nonparametric t test; D6C vs. D6H and D13C vs. D13H, p‐value > .05).
At an average sequencing depth of 13,831 reads per sample, the rarefaction curves have plateaued off for most samples, suggesting that sufficient sampling has been achieved to capture the total diversity of bacterial communities under investigation. Simpson and Shannon diversity estimates revealed that thermal stress treatments had significantly higher bacterial alpha diversity compared to the controls (Mann–Whitney U test; D6C vs. D6H, p‐value = .03175; D13C vs. D13H, p‐value = .02857), similar to previous observations in Acropora millepora (Bourne et al., 2008), Acropora muricata (Lee, Davy, Tang, & Kench, 2016; Shiu et al., 2017), and Pocillopora damicornis (Tout et al., 2015). A number of studies have provided emerging evidence that environmental stressors including elevated water temperature led to an increase in alpha diversity within coral‐associated microbiomes (Bourne et al., 2008; Lee et al., 2016; McDevitt‐Irwin, Baum, Garren, & Vega Thurber, 2017; Meron et al., 2011; Shiu et al., 2017; Tout et al., 2015). Meron et al. (2011) demonstrated that corals exposed to environment with lower pH exhibited higher microbial diversity (Meron et al., 2011). Corals experiencing a high level of anthropogenic disturbance have also been shown to harbor microbiomes with higher diversity than those farther away from the disturbance (Morrow, Moss, Chadwick, & Liles, 2012). Furthermore, a major coral microbiome restructuring and a marked increase in microbial diversity were evident when F. granulosa corals were exposed to long‐term high salinity (Röthig, Ochsenkühn, Roik, Merwe, & Voolstra, 2016).
Besides an increase in the alpha diversity observed, the community composition also changed dramatically especially after 13 days of heat treatment (Figure 2). There was a notable increase in the relative abundance of Alphaproteobacteria and a simultaneous decline in the relative abundance of Gammaproteobacteria during thermal stress (Figure 3). A clear shift in Gammaproteobacteria community was observed in conjunction with the reduction in Endozoicomonadaceae abundance, which became more pronounced after 13 days of heat treatment (Figure 4). Similar to our results, a marked decline in the proportion of Endozoicomonadaceae in the coral‐associated microbiome during thermal stress has been reported in P. damicornis (Tout et al., 2015) as well as in P. lutea (Pootakham et al., 2018). Endozoicomonadaceae has been reported to inhabit both coral mucus and coral tissue of several scleractinian species including Acropora cervicornis, P. astreoides, and P. furcate (Ainsworth et al., 2015; Bayer et al., 2013; Chu & Vollmer, 2016). Fluorescent in situ hybridization (FISH) experiments conducted by Bayer et al. (2013) showed that members of the Endozoicomonadaceae family (genus Endozoicomonas) resided in close proximity to Symbiodiniaceae cells within the coral endoderm, suggesting an intimate association with their coral hosts. Researchers have proposed Endozoicomonas to be beneficial symbionts (McDevitt‐Irwin et al., 2017), playing an important role in sulfur recycling (Raina et al., 2009) and protein/carbohydrate transport for the host (Neave, Michell, Apprill, & Voolstra, 2017). The decline in relative abundance of these potentially symbiotic taxa was also induced by other environmental stressors besides elevated water temperature. Previous studies have found that the relative occurrence of sequences associated with Endozoicomonadaceae decreased in Acropora and Porites corals in response to ocean acidification (Morrow et al., 2015) and in Porites and Paramuricea corals in response to diseases (Meyer, Paul, & Teplitski, 2014; Vezzulli, Pezzati, Huete‐Stauffer, Pruzzo, & Cerrano, 2013).
The structural abundance of dominant microbiome members under normal condition did not seem to be maintained under thermal stress. While there was a significant reduction in the relative abundance of Endozoicomonadaceae following the exposure to thermal stress, we observed a concomitant increase in the relative abundance of Rhodobacteraceae, Alteromonadaceae, and Saprospiraceae (Figures 3 and 4). Ziegler et al. (2017) also noticed a higher proportion of Rhodobacteraceae in the bacterial communities associated with the highly variable pool (with a temperature range of 25–35°C) compared to the moderately variable pool (with temperature range of 26–32°C). A dramatic community shift during thermal stress, which primarily involved an increase in the relative abundance of Rhodobacterales, was also observed in P. damicornis (Tout et al., 2015). Similarly, a study by van de Water et al. (2018) showed that members of Rhodobacteraceae (genera Rhodovulum and Dinoroseobacter) became more abundant in Montipora aequituberculata‐associated microbiome after an extended period of elevated water temperature treatment (van de Water et al., 2018). Patterns of increased Rhodobacterales‐affiliated sequences in the tissue layer have also been reported when A. muricata corals were exposed to high temperature for three days (Lee, Davy, Tang, & Kench, 2017). The order Rhodobacterales appears to be opportunistic bacteria, flourishing and increasing their relative abundance when coral hosts experience biotic (e.g., white band and white plaque diseases) and abiotic (e.g., thermal stress and ocean acidification) stress during which there may be open niche space (Cardenas, Rodriguez, Pizarro, Cadavid, & Arevalo‐Ferro, 2012; Meron et al., 2011; Roder et al., 2014; Welsh et al., 2017). Interestingly, even though Rhodobacteraceae became more abundant in coral‐associated communities under thermal stress (Tout et al., 2015; Lee et al., 2017; van de Water et al., 2018) and low pH (Meron et al., 2011), Grottoli et al. (2018) observed a decrease in sequences affiliated with this bacterial family when coral hosts were simultaneously exposed to both elevated seawater temperature and ocean acidification (Grottoli et al., 2018).
Several studies have reported an increase in relative abundance of Rhodobacteraceae in the microbiomes of diseased corals (Kellogg et al., 2013; Pollock, Wada, Torda, Willis, & Bourne, 2016; Sunagawa et al., 2009). Using PhyloChip™ G3, Kellogg et al. (2013) showed that a number of Rhodobacteraceae OTUs were consistently and significantly enriched in white plaque samples of Orbicella faveolata, similar to the work reported by Sunagawa et al. (2009) using an earlier generation PhyloChip™ (Kellogg et al., 2013; Sunagawa et al., 2009). Rhodobacteraceae OTUs were also represented in greater proportions in diseased samples of Diploria strigosa and Siderastrea siderea compared to the samples from healthy colonies (Cardenas et al., 2012). Rhodobacterales are fast‐growing taxa, exhibiting a quick response to increasing availability of amino acids and other resources (Mayali, Weber, Mabery, & Pett‐Ridge, 2014) that could be made available from diseased cells damaged by pathogens such as Vibrio spp (Welsh et al., 2017). The appearance of Vibrio‐related sequences has been observed in parallel with the increase in the relative abundance of Rhodobacteraceae during thermal bleaching in A. millepora (Bourne et al., 2008), A. muricata (Lee et al., 2016), A. gemmifera (Gardner et al., 2019), and P. damicornis (Tout et al., 2015). Members of the genus Vibrio are known for their roles as opportunistic or pathogenic bacteria associated with white band, yellow blotch/band disease, and white‐syndrome‐like diseases (Barneah, Ben‐Dov, Kramarsky‐Winter, & Kushmaro, 2007; Cervino et al., 2004; Gil‐Agudelo, Fonseca, Weil, Garzon‐Ferreira, & Smith, 2007; Gil‐Agudelo, Smith, & Weil, 2006; Luna, Bongiorni, Gili, Biavasco, & Danovaro, 2010; Richie & Smith, 1998; Ritchie, 1994; Rosenberg & Falkovitz, 2004; Sussman, Willis, Victor, & Bourne, 2008). Interestingly, despite the similarity in the bacterial profile shifts associated with diseases and thermal stress, we did not observe an increase in relative abundance of Vibrio‐related sequences in our heat‐treated samples. Indeed, we hardly detected any Vibrio OTUs in our samples across treatments and time periods. The absence of Vibrio species in both healthy and thermally bleached P. lutea colonies was consistent with previous study (Pootakham et al., 2017, 2018).
4.2. Porites lutea core microbiome
The concept of core microbiome has recently been introduced in coral‐microbial studies (Ainsworth et al., 2015; Chu & Vollmer, 2016; Hernandez‐Agreda, Leggat, Bongaerts, & Ainsworth, 2016; Lawler et al., 2016; Robertson, Haltli, Mccauley, Overy, & Kerr, 2016; van de Water et al., 2016; Hernandez‐Agreda, Gates, & Ainsworth, 2017; Sweet & Bulling, 2017), and it is characterized as the stable populations consistently present across complex microbial assemblages (Shade & Handelsman, 2012). Members of a core microbiome are likely to be important for the development, health, and functioning of their hosts (Ley, Turnbaugh, Klein, & Gordon, 2006; Round & Mazmanian, 2009). We stringently defined the members of the P. lutea core microbiome as bacterial species that were present in 100% of all samples based on the percentages of sample coverage previously used in coral microbiome studies (Lawler et al., 2016; van de Water et al., 2016). With full‐length 16S rRNA sequence data, we were able to achieve the taxonomic classification of the core microbiome members at the species resolution. Most of P. lutea core microbiome members belonged to Alphaproteobacteria family Rhodobacteraceae and Gammaproteobacteria family Endozoicomonadaceae (Figure 5). Rhodobacterales are thought to be among the most abundant and diverse bacteria in marine environments, accounting for a quarter of the total bacteria present in coastal and polar regions (Wagner‐Dobler & Biebl, 2006). They are functionally diverse and involved in a range of biological activities from the production of antibiotic compounds to the reduction of trace metals (Brinkhoff et al., 2004). They also contribute to ocean carbon and sulfur cycling through the oxidation of carbon monoxide and the production of dimethylsulfide (Wagner‐Dobler & Biebl, 2006).
Another prominent taxon that constituted the majority of P. lutea core microbiome was the family Endozoicomonadaceae, which has been reported as dominant members of the core microbiomes in P. furcate, A. cervicornis, P. verrucosa, P. astreoides, and Stylophora pistillata (Chu & Vollmer, 2016; Glasl, Webster, & Bourne, 2017). Notably, all five Endozoicomonadaceae species designated as core microbiome members in this study have previously been identified in the P. lutea core microbiome (Pootakham et al., 2017, 2018). Moreover, E. elysicola was identified in all 30 P. lutea samples collected from different sampling sites, during different seasons and under varying temperature conditions (Figure 5). Members of the family Endozoicomonadaceae, especially genus Endozoicomonas, are ubiquitous and highly abundant in the marine environment. While they do not appear to be common in deep‐sea corals, members of this family are frequently found associated with tropical and temperate coral species (Cardenas et al., 2012; La Rivière, Garrabou, & Bally, 2015; Lee et al., 2012; Morrow et al., 2012; Sunagawa et al., 2009). Based on their widespread abundance in marine invertebrates and their dominance in the microbiomes of several coral species (Chu & Vollmer, 2016; Glasl et al., 2017), Endozoicomonas species are likely to play important biological and ecological roles in coral holobionts. Endozoicomonas has been detected on the surface mucus layer and in coral skeleton of Acropora and Porites hosts, and it has been suggested that members of this taxon may be involved in biofilm production that promotes surface colonization of other bacteria (Jessen et al., 2013; Speck & Donachie, 2012). The Endozoicomonas genomes encoded a complete set of putative proteins in the synthetic pathways for various amino acids including alanine, cysteine, glycine, lysine, methionine, serine, and threonine, raising the possibility that the microbial symbionts, especially members of the core microbiome, may provide essential amino acids that cannot be synthesized by the host (Neave et al., 2017; Price et al., 2014).
4.3. Members of the Rhodobacteraceae family are indicator species for thermal stress in Porites lutea
To determine whether specific microbial taxa were significantly associated with corals under thermal stress, we applied an indicator species analysis (INDVAL) to identify representative or indicative bacterial species for elevated water temperature (De Caceres & Legendre, 2009). Interestingly, the microbial communities of heat‐stressed corals were characterized primarily (59 out of 81 species) by a group of bacterial indicator species belonging to 19 different Rhodobacteraceae genera (Table 2; Table S2). On the contrary, nearly all indicator species for normal water temperature were Endozoicomonadaceae (genera Endozoicomonas and Kistimonas). Members of Rhodobacteraceae have previously been reported as indicator taxa for polluted environment and sites impacted by sedimentation and local sewage or municipal wastewater (Ziegler et al., 2016). Several OTUs with sequences related to Rhodobacteraceae have also been identified as indicative microbial taxa for diseased coral tissues (Roder et al., 2014; Sere et al., 2013; Sunagawa et al., 2009). The results presented in this work suggested that members of Rhodobacteraceae family were robust indicator species for thermally stressed P. lutea colonies.
Scleractinian corals are among the largest producers of DMSP (Broadbent & Jones, 2004; Broadbent, Jones, & Jones, 2002), a compound that plays a role in alleviating cellular oxidative stress as DMSP and its breakdown products (dimethylsulfide [DMS] and dimethylsulfoxide [DMSO]) can readily scavenge hydroxyl radicals and other reactive oxygen species (Sunda, Kieber, Kiene, & Huntsman, 2002; Yoch, 2002). Interestingly, Garren et al. (2014) found that coral hosts under thermal stress exude mucus that was richer in DMSP, which appeared to trigger heightened pathogen response, suggesting that pathogenic (e.g., Vibrio coralliilyticus) or opportunistic (e.g., Rhodobacteraceae) bacteria may use this chemical cue to target stressed hosts (Garren et al., 2014). Ruegeria pomeroyi, one of the indicator species for heat‐stressed P. lutea, was capable of metabolizing DMSP utilizing both the demethylation pathway (releasing methanethiol) and the cleavage pathway (producing DMS) (Reisch et al., 2013). The ability to utilize DMSP exuded by stressed corals may allow R. pomeroyi to thrive and expand its niches within the coral holobiont especially during thermal stress. Another indicator species for thermal stressed microbiome was Aliiroseovarius crassostreae, a marine Rhodobacter that has been shown to be a causative agent of Roseovarius oyster disease, which resulted in high mortality rates in juvenile eastern oysters in the United States (Boettcher, Geaghan, Maloy, & Barber, 2005). To this point, there has been no report linking R. pomeroyi or A. crassastreae to disease conditions in P. lutea. Future studies are required to determine whether these two indicator species are capable of inducing signs of coral diseases.
4.4. Full‐length 16S rRNA sequences enable taxonomical classification at the species resolution
Classifying the taxonomy of 16S rRNA sequences based on short hypervariable fragments of the 16S rRNA gene can be challenging. Our results showed that the number of “unassigned” sequences was remarkably higher when partial 16S fragments were used instead of the full‐length sequences (Table S3). Taxonomical classification at the genus level yielded noticeably different microbiome profiles when short fragments encompassing hypervariable regions, especially the V5‐V6 regions, were used instead of the full‐length 16S rRNA sequences (Table S3). Misclassification of partial 16S rRNA sequences (V3‐V4 or V5‐V6 regions) can affect the accuracy of taxonomic profiles as well as bacterial community's estimates of species richness (Youssef et al., 2009). Youssef et al. (2009) compared the OTU estimates for each simulated hypervariable region to those for full‐length or nearly full‐length 16S rRNA sequences at different taxonomic cutoffs and demonstrated that the number of OTUs and species richness could be either underestimated or overestimated, depending on the hypervariable sequences used (Youssef et al., 2009). The bias in species richness estimates stemming from the use of partial 16S sequences can be explained by the percentages of conserved, variable, and hypervariable nucleotide positions in the sequences used to perform the analyses (Youssef et al., 2009).
5. CONCLUSIONS
Emerging evidence suggests that environmental stressors, especially the climate change‐associated pulse warming events, can induce changes in the coral microbiomes (Ainsworth & Gates, 2016; Bourne et al., 2008; Gardner et al., 2019; Grottoli et al., 2018; Hoegh‐Guldberg et al., 2007; Lee et al., 2016, 2017; Littman et al., 2009; Wang et al., 2018; Zhou et al., 2017; Ziegler et al., 2017). Several studies have highlighted the significance of coral‐associated microorganisms and their roles in fitness and survival of the host animals (Ainsworth et al., 2015; Bourne et al., 2008; Gilbert et al., 2012; Lesser, Mazel, Gorbunov, & Falkowski, 2004; Shieh & Lin, 1992; Tout et al., 2015). Our work examined the effect of thermal stress on the composition and structure of bacterial communities associated with P. lutea, one of the dominant reef‐builders widely distributed across the Indo‐West Pacific, including the Gulf of Thailand and Andaman Sea (Yeemin et al., 2009). We observed a significant increase alpha diversity indices and a prominent shift in microbiome composition during thermal stress. This is one of the first coral‐associated microbiome studies that utilized the long‐read PacBio SMRT sequencing technology to capture full‐length 16S rRNA gene, which allowed over 99.8% of the sequences to be classified at the species level. The ability to achieve species‐resolution classification enabled us to identify members of P. lutea core microbiome and a group of indicator species associated with thermal stress. The knowledge on how thermal stress impacts the diversity and composition of coral microbiomes may help us gain a better understanding of how these associated bacteria contribute to the acclimatization and resilience of the coral holobiont to thermal bleaching.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
WP and ST conceptualized the study and acquired funding. WP, TY, LP, NJ, and ST carried out investigation. WP, WM, CS, CN, and WK carried out formal analysis. WM and WK carried out visualization experiments. WP wrote the original draft of the manuscript. WP, WM, and WK reviewed and edited the manuscript.
ETHICAL APPROVAL
None required.
Supporting information
ACKNOWLEDGMENTS
We thank the following funding bodies: National Science and Technology Development Agency (NSTDA), Thailand (Grant Number P‐16‐50720), and UNESCO‐L'Oreal For Women in Science Fellowship (awarded to Pootakham). We would also like to thank the reviewers and editors whose comments and suggestions improved the manuscript.
APPENDIX 1.
Figure A1.
A histogram showing CCS read length distribution of 16S rRNA amplicons
Figure A2.
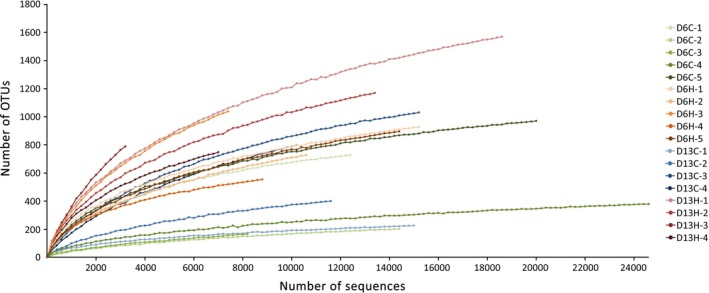
Rarefaction curves of OTUs for the coral‐associated bacterial community samples from control and heated aquaria
Figure A3.
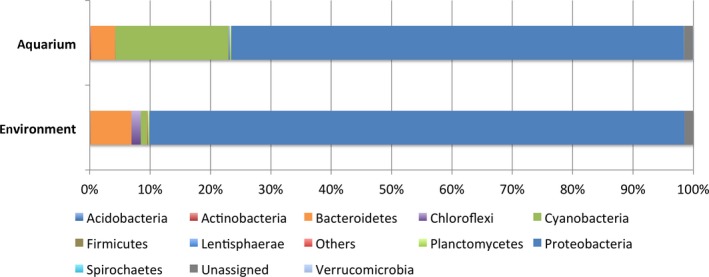
A bar chart showing the composition of bacterial communities associated with P. lutea in this study (aquarium) and those associated with P. lutea collected directly from the field (environment). Taxonomic classification of OTUs present in each sample at the phylum level was based on the Greengenes database
Figure A4.
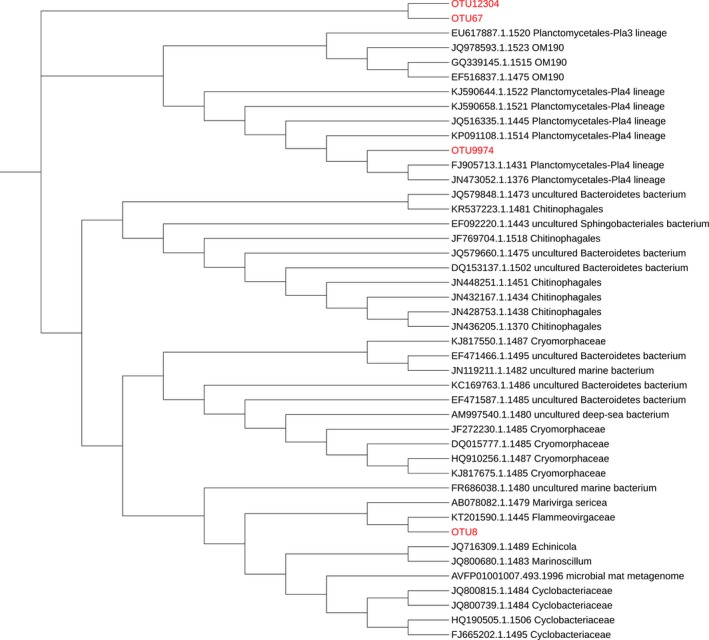
A maximum‐likelihood phylogenetic tree generated using MEGA7, showing the relationship of four “unassigned” OTUs with known bacterial sequences. The clustering of the sequences was tested by a bootstrap approach using 1,000 replications
Pootakham W, Mhuantong W, Yoocha T, et al. Heat‐induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea . MicrobiologyOpen. 2019;8:e935 10.1002/mbo3.935
DATA AVAILABILITY STATEMENT
The PacBio 16S rRNA sequence data were deposited in the GenBank NCBI Sequence Read Archive (SRA) database under the accession number https://www.ncbi.nlm.nih.gov/bioproject/PRJNA429083
REFERENCES
- Ainsworth, T. D. , & Gates, R. D. (2016). OCEAN BIOLOGY. Corals' Microbial Sentinels. Science, 352, 1518–1519. 10.1126/science.aad9957 [DOI] [PubMed] [Google Scholar]
- Ainsworth, T. D. , Krause, L. , Bridge, T. , Torda, G. , Raina, J. B. , Zakrzewski, M. , … Leggat, W. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME Journal, 9, 2261–2274. 10.1038/ismej.2015.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, A. C. (2003). Flexibility and specificity in coral‐algal symbiosis: diversity, ecology, and biogeography of symbiodinium. Annual Review of Ecology, Evolution, and Systematics, 34, 661–689. 10.1146/annurev.ecolsys.34.011802.132417 [DOI] [Google Scholar]
- Baker, A. C. , Starger, C. J. , Mcclanahan, T. R. , & Glynn, P. W. (2004). Coral reefs: Corals' adaptive response to climate change. Nature, 430, 741 10.1038/430741a [DOI] [PubMed] [Google Scholar]
- Barneah, O. , Ben‐Dov, E. , Kramarsky‐Winter, E. , & Kushmaro, A. (2007). Characterization of black band disease in Red Sea stony corals. Environmental Microbiology, 9, 1995–2006. 10.1111/j.1462-2920.2007.01315.x [DOI] [PubMed] [Google Scholar]
- Bayer, T. , Neave, M. J. , Alsheikh‐Hussain, A. , Aranda, M. , Yum, L. K. , Mincer, T. , … Voolstra, C. R. (2013). The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue‐associated Endozoicomonas bacteria. Applied and Environmental Microbiology, 79, 4759–4762. 10.1128/AEM.00695-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelmans, R. , & Van Oppen, M. J. (2006). The role of zooxanthellae in the thermal tolerance of corals: A 'nugget of hope' for coral reefs in an era of climate change. Proceedings of the Royal Society B: Biological Sciences, 273, 2305–2312. 10.1098/rspb.2006.3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher, K. J. , Geaghan, K. K. , Maloy, A. P. , & Barber, B. J. (2005). Roseovarius crassostreae sp. nov., a member of the Roseobacter clade and the apparent cause of juvenile oyster disease (JOD) in cultured Eastern oysters. International Journal of Systematic and Evolutionary Microbiology, 55, 1531–1537. 10.1099/ijs.0.63620-0 [DOI] [PubMed] [Google Scholar]
- Boulotte, N. M. , Dalton, S. J. , Carroll, A. G. , Harrison, P. L. , Putnam, H. M. , Peplow, L. M. , & Van Oppen, M. J. (2016). Exploring the symbiodinium rare biosphere provides evidence for symbiont switching in reef‐building corals. ISME Journal, 10, 2693–2701. 10.1038/ismej.2016.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, D. , Iida, Y. , Uthicke, S. , & Smith‐Keune, C. (2008). Changes in coral‐associated microbial communities during a bleaching event. ISME Journal, 2, 350–363. 10.1038/ismej.2007.112 [DOI] [PubMed] [Google Scholar]
- Bourne, D. G. , Morrow, K. M. , & Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annual Review of Microbiology, 70, 317–340. 10.1146/annurev-micro-102215-095440 [DOI] [PubMed] [Google Scholar]
- Bourne, D. G. , & Munn, C. B. (2005). Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environmental Microbiology, 7, 1162–1174. 10.1111/j.1462-2920.2005.00793.x [DOI] [PubMed] [Google Scholar]
- Brinkhoff, T. , Bach, G. , Heidorn, T. , Liang, L. , Schlingloff, A. , & Simon, M. (2004). Antibiotic production by a Roseobacter clade‐affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Applied and Environment Microbiology, 70, 2560–2565. 10.1128/AEM.70.4.2560-2565.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent, A. D. , & Jones, G. B. (2004). DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the Great Barrier Reef. Marine and Freshwater Research, 55, 849–855. 10.1071/MF04114 [DOI] [Google Scholar]
- Broadbent, A. D. , Jones, G. B. , & Jones, R. J. (2002). DMSP in corals and benthic algae from the great barrier reef. Estuarine, Coastal and Shelf Science, 55, 547–555. 10.1006/ecss.2002.1021 [DOI] [Google Scholar]
- Brown, B. E. (1997). Coral bleaching: Causes and consequences. Coral Reefs, 16, S129–S138. 10.1007/s003380050249 [DOI] [Google Scholar]
- Bruno, J. F. , & Selig, E. R. (2007). Regional decline of coral cover in the Indo‐Pacific: Timing, extent, and subregional comparisons. PLoS ONE, 2, e711 10.1371/journal.pone.0000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas, A. , Rodriguez, R. L. , Pizarro, V. , Cadavid, L. F. , & Arevalo‐Ferro, C. (2012). Shifts in bacterial communities of two Caribbean reef‐building coral species affected by white plague disease. ISME Journal, 6, 502–512. 10.1038/ismej.2011.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardini, U. , Bednarz, V. N. , Naumann, M. S. , Van Hoytema, N. , Rix, L. , Foster, R. A. , … Wild, C. (2015). Functional significance of dinitrogen fixation in sustaining coral productivity under oligotrophic conditions. Proceedings of the Royal Society B: Biological Sciences, 282, 20152257 10.1098/rspb.2015.2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceh, J. , Kilburn, M. R. , Cliff, J. B. , Raina, J.‐B. , Van Keulen, M. , & Bourne, D. G. (2013). Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecology and Evolution, 3, 2393–2400. 10.1002/ece3.642 [DOI] [Google Scholar]
- Cervino, J. M. , Hayes, R. L. , Polson, S. W. , Polson, S. C. , Goreau, T. J. , Martinez, R. J. , & Smith, G. W. (2004). Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in caribbean corals. Applied and Environmental Microbiology, 70, 6855–6864. 10.1128/aem.70.11.6855-6864.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimetto, L. A. , Brocchi, M. , Thompson, C. C. , Martins, R. C. R. , Ramos, H. R. , & Thompson, F. L. (2008). Vibrios dominate as culturable nitrogen‐fixing bacteria of the Brazilian coral Mussismilia hispida . Systematic and Applied Microbiology, 31, 312–319. 10.1016/j.syapm.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Chu, N. D. , & Vollmer, S. V. (2016). Caribbean corals house shared and host‐specific microbial symbionts over time and space. Environmental Microbiology Reports, 8, 493–500. 10.1111/1758-2229.12412 [DOI] [PubMed] [Google Scholar]
- Cole, J. R. , Wang, Q. , Fish, J. A. , Chai, B. , Mcgarrell, D. M. , Sun, Y. , … Tiedje, J. M. (2014). Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Research, 42, D633–642. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell, J. H. (1978). Diversity in tropical rain forests and coral reefs. Science, 199, 1302–1310. 10.1126/science.199.4335.1302 [DOI] [PubMed] [Google Scholar]
- Copper, P. (1994). Ancient reef ecosystem expansion and collapse. Coral Reefs, 13, 3–11. 10.1007/bf00426428 [DOI] [Google Scholar]
- Darling, E. S. , Alvarez‐Filip, L. , Oliver, T. A. , Mcclanahan, T. R. , & Côté, I. M. (2012). Evaluating life‐history strategies of reef corals from species traits. Ecology Letters, 15, 1378–1386. 10.1111/j.1461-0248.2012.01861.x [DOI] [PubMed] [Google Scholar]
- De Caceres, M. , & Legendre, P. (2009). Associations between species and groups of sites: Indices and statistical inference. Ecology, 90, 3566–3574. 10.1890/08-1823.1 [DOI] [PubMed] [Google Scholar]
- Desantis, T. Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E. L. , Keller, K. , … Andersen, G. L. (2006). Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Fautin, D. G. , & Buddemeier, R. W. (2004). Adaptive bleaching: A general phenomenon In Fautin D. G., Westfall J. A., Cartwrigh P., Daly M., & Wyttenbach C. R., (Eds.), Coelenterate biology 2003: Trends in research on cnidaria and ctenophora (pp. 459–467). Dordrecht, The Netherlands: Springer. [Google Scholar]
- Fitt, W. K. , Brown, B. E. , Warner, M. E. , & Dunne, R. P. (2001). Coral bleaching: Interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs, 20, 51–65. 10.1007/s003380100146 [DOI] [Google Scholar]
- Frias‐Lopez, J. , Zerkle, A. L. , Bonheyo, G. T. , & Fouke, B. W. (2002). Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Applied and Environment Microbiology, 68, 2214–2228. 10.1128/AEM.68.5.2214-2228.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, S. G. , Camp, E. F. , Smith, D. J. , Kahlke, T. , Osman, E. O. , Gendron, G. , … Suggett, D. J. (2019). Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecology and Evolution, 9, 938–956. 10.1002/ece3.4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, T. A. , Côté, I. M. , Gill, J. A. , Grant, A. , & Watkinson, A. R. (2003). Long‐term region‐wide declines in caribbean corals. Science, 301, 958–960. 10.1126/science.1086050 [DOI] [PubMed] [Google Scholar]
- Garren, M. , Son, K. , Raina, J. B. , Rusconi, R. , Menolascina, F. , Shapiro, O. H. , … Stocker, R. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat‐stressed corals. ISME Journal, 8, 999–1007. 10.1038/ismej.2013.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil‐Agudelo, D. L. , Fonseca, D. P. , Weil, E. , Garzon‐Ferreira, J. , & Smith, G. W. (2007). Bacterial communities associated with the mucopolysaccharide layers of three coral species affected and unaffected with dark spots disease. Canadian Journal of Microbiology, 53, 465–471. 10.1139/W07-002 [DOI] [PubMed] [Google Scholar]
- Gil‐Agudelo, D. L. , Smith, G. W. , & Weil, E. (2006). The white band disease type II pathogen in Puerto Rico. Revista De Biología Tropical, 54, 59–67.18457175 [Google Scholar]
- Gilbert, J. A. , Hill, R. , Doblin, M. A. , & Ralph, P. J. (2012). Microbial consortia increase thermal tolerance of corals. Marine Biology, 159, 1763–1771. 10.1007/s00227-012-1967-9 [DOI] [Google Scholar]
- Glasl, B. , Webster, N. S. , & Bourne, D. G. (2017). Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Marine Biology, 164, 91 10.1007/s00227-017-3097-x [DOI] [Google Scholar]
- Grottoli, A. G. , Dalcin Martins, P. , Wilkins, M. J. , Johnston, M. D. , Warner, M. E. , Cai, W. J. , … Schoepf, V. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS ONE, 13, e0191156 10.1371/journal.pone.0191156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadaidi, G. , Rothig, T. , Yum, L. K. , Ziegler, M. , Arif, C. , Roder, C. , … Voolstra, C. R. (2017). Stable mucus‐associated bacterial communities in bleached and healthy corals of Porites lobata from the Arabian Seas. Scientific Reports, 7, 45362 10.1038/srep45362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Agreda, A. , Gates, R. D. , & Ainsworth, T. D. (2017). Defining the Core Microbiome in Corals' Microbial Soup. Trends in Microbiology, 25, 125–140. 10.1016/j.tim.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Hernandez‐Agreda, A. , Leggat, W. , Bongaerts, P. , & Ainsworth, T. D. (2016). The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. MBio, 7(4), e00560-16 10.1128/mBio.00560-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh‐Guldberg, O. , Mumby, P. J. , Hooten, A. J. , Steneck, R. S. , Greenfield, P. , Gomez, E. , … Hatziolos, M. E. (2007). Coral reefs under rapid climate change and ocean acidification. Science, 318, 1737–1742. 10.1126/science.1152509 [DOI] [PubMed] [Google Scholar]
- Hong, M.‐J. , Yu, Y.‐T. , Chen, C. A. , Chiang, P.‐W. , & Tang, S.‐L. (2009). Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Applied and Environmental Microbiology, 75, 7797–7806. 10.1128/AEM.01418-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howells, E. J. , Beltran, V. H. , Larsen, N. W. , Bay, L. K. , Willis, B. L. , & Van Oppen, M. J. H. (2012). Coral thermal tolerance shaped by local adaptation of photosymbionts. Nature Climate Change, 2, 116–120. 10.1038/nclimate1330 [DOI] [Google Scholar]
- Hughes, T. P. , Anderson, K. D. , Connolly, S. R. , Heron, S. F. , Kerry, J. T. , Lough, J. M. , … Wilson, S. K. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science, 359, 80–83. 10.1126/science.aan8048 [DOI] [PubMed] [Google Scholar]
- Hume, B. C. C. , Voolstra, C. R. , Arif, C. , D’Angelo, C. , Burt, J. A. , Eyal, G. , … Wiedenmann, J. (2016). Ancestral genetic diversity associated with the rapid spread of stress‐tolerant coral symbionts in response to Holocene climate change. Proceedings of the National Academy of Sciences of the United States of America, 113, 4416–4421. 10.1073/pnas.1601910113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen, C. , Villa Lizcano, J. F. , Bayer, T. , Roder, C. , Aranda, M. , Wild, C. , & Voolstra, C. R. (2013). In‐situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea coral Acropora hemprichii . PLoS ONE, 8, e62091 10.1371/journal.pone.0062091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. M. , Berkelmans, R. , Van Oppen, M. J. , Mieog, J. C. , & Sinclair, W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: Field evidence of acclimatization. Proceedings of the Royal Society B: Biological Sciences, 275, 1359–1365. 10.1098/rspb.2008.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R. J. (2008). Coral bleaching, bleaching‐induced mortality, and the adaptive significance of the bleaching response. Marine Biology, 154, 65–80. 10.1007/s00227-007-0900-0 [DOI] [Google Scholar]
- Kellogg, C. A. , Piceno, Y. M. , Tom, L. M. , Desantis, T. Z. , Gray, M. A. , Zawada, D. G. , & Andersen, G. L. (2013). Comparing bacterial community composition between healthy and white plague‐like disease states in Orbicella annularis using PhyloChip G3 microarrays. PLoS ONE, 8, e79801 10.1371/journal.pone.0079801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman, D. , Kashman, Y. , Rosenberg, E. , Kushmaro, A. , & Loya, Y. (2006). Antimicrobial activity of Red Sea corals. Marine Biology, 149, 357–363. 10.1007/s00227-005-0218-8 [DOI] [Google Scholar]
- Keshavmurthy, S. , Meng, P. J. , Wang, J. T. , Kuo, C. Y. , Yang, S. Y. , Hsu, C. M. , … Chen, C. A. (2014). Can resistant coral‐Symbiodinium associations enable coral communities to survive climate change? A study of a site exposed to long‐term hot water input. PeerJ, 2, e327 10.7717/peerj.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, E. G. L. (1997). Do scleractinian corals engage in chemical warfare against microbes? Journal of Chemical Ecology, 23, 379–398. 10.1023/B:JOEC.0000006366.58633.f4 [DOI] [Google Scholar]
- Kooperman, N. , Ben‐Dov, E. , Kramarsky‐Winter, E. , Barak, Z. , & Kushmaro, A. (2007). Coral mucus‐associated bacterial communities from natural and aquarium environments. FEMS Microbiology Letters, 276, 106–113. 10.1111/j.1574-6968.2007.00921.x [DOI] [PubMed] [Google Scholar]
- Koren, O. , & Rosenberg, E. (2006). Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Applied and Environment Microbiology, 72, 5254–5259. 10.1128/AEM.00554-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krediet, C. J. , Ritchie, K. B. , Paul, V. J. , & Teplitski, M. (2013). Coral‐associated micro‐organisms and their roles in promoting coral health and thwarting diseases. Proceedings of the Royal Society B: Biological Sciences, 280, 20122328 10.1098/rspb.2012.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rivière, M. , Garrabou, J. , & Bally, M. (2015). Evidence for host specificity among dominant bacterial symbionts in temperate gorgonian corals. Coral Reefs, 34, 1087–1098. 10.1007/s00338-015-1334-7 [DOI] [Google Scholar]
- Lawler, S. N. , Kellogg, C. A. , France, S. C. , Clostio, R. W. , Brooke, S. D. , & Ross, S. W. (2016). Coral‐associated bacterial diversity is conserved across two deep‐sea anthothela species. Frontiers in Microbiology, 7, 458 10.3389/fmicb.2016.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, O. O. , Yang, J. , Bougouffa, S. , Wang, Y. , Batang, Z. , Tian, R. , … Qian, P. Y. (2012). Spatial and species variations in bacterial communities associated with corals from the Red Sea as revealed by pyrosequencing. Applied and Environment Microbiology, 78, 7173–7184. 10.1128/AEM.01111-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. T. M. , Davy, S. K. , Tang, S.‐L. , & Kench, P. S. (2016). Mucus sugar content shapes the bacterial community structure in thermally stressed acropora muricata. Frontiers in Microbiology, 7, 371 10.3389/fmicb.2016.00371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. T. , Davy, S. K. , Tang, S. L. , & Kench, P. S. (2017). Water flow buffers shifts in bacterial community structure in heat‐stressed Acropora muricata. Scientific Reports, 7, 43600 10.1038/srep43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema, K. A. , Willis, B. L. , & Bourne, D. G. (2012). Corals form characteristic associations with symbiotic nitrogen‐fixing bacteria. Applied and Environment Microbiology, 78, 3136–3144. 10.1128/AEM.07800-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser, M. P. , Falcón, L. I. , Rodríguez‐Román, A. , Enríquez, S. , Hoegh‐Guldberg, O. , & Iglesias‐Prieto, R. (2007). Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa . Marine Ecology Progress Series, 346, 143–152. 10.3354/meps07008 [DOI] [Google Scholar]
- Lesser, M. P. , Mazel, C. H. , Gorbunov, M. Y. , & Falkowski, P. G. (2004). Discovery of symbiotic nitrogen‐fixing cyanobacteria in corals. Science, 305, 997–1000. 10.1126/science.1099128 [DOI] [PubMed] [Google Scholar]
- Ley, R. E. , Turnbaugh, P. J. , Klein, S. , & Gordon, J. I. (2006). Microbial ecology: Human gut microbes associated with obesity. Nature, 444, 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Li, J. , Chen, Q. , Zhang, S. , Huang, H. , Yang, J. , Tian, X.‐P. , & Long, L.‐J. (2013). Highly heterogeneous bacterial communities associated with the South China Sea Reef corals Porites lutea, Galaxea fascicularis and Acropora millepora . PLoS ONE, 8, e71301 10.1371/journal.pone.0071301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman, R. A. , Willis, B. L. , Pfeffer, C. , & Bourne, D. G. (2009). Diversities of coral‐associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiology Ecology, 68, 152–163. 10.1111/j.1574-6941.2009.00666.x [DOI] [PubMed] [Google Scholar]
- Loya, Y. , Sakai, K. , Yamazato, K. , Nakano, Y. , Sambali, H. , & Van Woesik, R. (2001). Coral bleaching: The winners and the losers. Ecology Letters, 4, 122–131. 10.1046/j.1461-0248.2001.00203.x [DOI] [Google Scholar]
- Lozupone, C. , & Knight, R. (2005). UniFrac: A new phylogenetic method for comparing microbial communities. Applied and Environment Microbiology, 71, 8228–8235. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, G. M. , Bongiorni, L. , Gili, C. , Biavasco, F. , & Danovaro, R. (2010). Vibrio harveyi as a causative agent of the White Syndrome in tropical stony corals. Environmental Microbiology Reports, 2, 120–127. 10.1111/j.1758-2229.2009.00114.x [DOI] [PubMed] [Google Scholar]
- Mayali, X. , Weber, P. K. , Mabery, S. , & Pett‐Ridge, J. (2014). Phylogenetic patterns in the microbial response to resource availability: Amino acid incorporation in San Francisco Bay. PLoS ONE, 9, e95842 10.1371/journal.pone.0095842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdevitt‐Irwin, J. M. , Baum, J. K. , Garren, M. , & Vega Thurber, R. L. (2017). Responses of Coral‐Associated Bacterial Communities to Local and Global Stressors. Frontiers in Marine Science, 4, 262 10.3389/fmars.2017.00262 [DOI] [Google Scholar]
- Mckew, B. A. , Dumbrell, A. J. , Daud, S. D. , Hepburn, L. , Thorpe, E. , Mogensen, L. , & Whitby, C. (2012). Characterization of geographically distinct bacterial communities associated with coral mucus produced by Acropora spp. and Porites spp. Applied and Environment Microbiology, 78, 5229–5237. 10.1128/AEM.07764-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron, D. , Atias, E. , Iasur Kruh, L. , Elifantz, H. , Minz, D. , Fine, M. , & Banin, E. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME Journal, 5, 51–60. 10.1038/ismej.2010.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, J. L. , Paul, V. J. , & Teplitski, M. (2014). Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS ONE, 9, e100316 10.1371/journal.pone.0100316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg, F. , & Folke, C. (1999). Ecological goods and services of coral reef ecosystems. Ecological Economics, 29, 215–233. 10.1016/S0921-8009(99)00009-9 [DOI] [Google Scholar]
- Morrow, K. M. , Bourne, D. G. , Humphrey, C. , Botte, E. S. , Laffy, P. , Zaneveld, J. , … Webster, N. S. (2015). Natural volcanic CO2 seeps reveal future trajectories for host‐microbial associations in corals and sponges. ISME Journal, 9, 894–908. 10.1038/ismej.2014.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, K. M. , Moss, A. G. , Chadwick, N. E. , & Liles, M. R. (2012). Bacterial associates of two Caribbean coral species reveal species‐specific distribution and geographic variability. Applied and Environment Microbiology, 78, 6438–6449. 10.1128/AEM.01162-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave, M. J. , Michell, C. T. , Apprill, A. , & Voolstra, C. R. (2017). Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Scientific Reports, 7, 40579 10.1038/srep40579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Kindt, R. , Legendre, P. , & Minchin, P. R. (2013). vegan: Community Ecology Package. http://cran.r-project.org/web/packages/vegan/index.html [Google Scholar]
- Pantos, O. , Cooney, R. P. , Le Tissier, M. D. A. , Barer, M. R. , O'Donnell, A. G. , & Bythell, J. C. (2003). The bacterial ecology of a plague‐like disease affecting the Caribbean coral Montastrea annularis. Environmental Microbiology, 5, 370–382. 10.1046/j.1462-2920.2003.00427.x [DOI] [PubMed] [Google Scholar]
- Pisapia, C. , Burn, D. , Yoosuf, R. , Najeeb, A. , Anderson, K. D. , & Pratchett, M. S. (2016). Coral recovery in the central Maldives archipelago since the last major mass‐bleaching, in 1998. Scientific Reports, 6, 34720 10.1038/srep34720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock, F. J. , Wada, N. , Torda, G. , Willis, B. L. , & Bourne, D. G. (2016). White syndrome‐affected corals have a distinct microbiome at disease lesion fronts. Applied and Environment Microbiology, 83(2), e02799-16 10.1128/AEM.02799-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham, W. , Mhuantong, W. , Putchim, L. , Yoocha, T. , Sonthirod, C. , Kongkachana, W. , … Tangphatsornruang, S. (2018). Dynamics of coral‐associated microbiomes during a thermal bleaching event. MicrobiologyOpen, 7(5), e00604 10.1002/mbo3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham, W. , Mhuantong, W. , Yoocha, T. , Putchim, L. , Sonthirod, C. , Naktang, C. , … Tangphatsornruang, S. (2017). High resolution profiling of coral‐associated bacterial communities using full‐length 16S rRNA sequence data from PacBio SMRT sequencing system. Scientific Reports, 7, 2774 10.1038/s41598-017-03139-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, D. R. , Feng, H. , Baker, J. D. , Bavan, S. , Luetje, C. W. , & Wilson, A. C. (2014). Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proceedings of the National Academy of Sciences of the United States of America, 111, 320–325. 10.1073/pnas.1306068111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rädecker, N. , Pogoreutz, C. , Voolstra, C. R. , Wiedenmann, J. , & Wild, C. (2015). Nitrogen cycling in corals: The key to understanding holobiont functioning? Trends in Microbiology, 23, 490–497. 10.1016/j.tim.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Raina, J. B. , Tapiolas, D. , Willis, B. L. , & Bourne, D. G. (2009). Coral‐associated bacteria and their role in the biogeochemical cycling of sulfur. Applied and Environment Microbiology, 75, 3492–3501. 10.1128/AEM.02567-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisch, C. R. , Crabb, W. M. , Gifford, S. M. , Teng, Q. , Stoudemayer, M. J. , Moran, M. A. , & Whitman, W. B. (2013). Metabolism of dimethylsulphoniopropionate by Ruegeria pomeroyi DSS‐3. Molecular Microbiology, 89, 774–791. 10.1111/mmi.12314 [DOI] [PubMed] [Google Scholar]
- Reshef, L. , Koren, O. , Loya, Y. , Zilber‐Rosenberg, I. , & Rosenberg, E. (2006). The coral probiotic hypothesis. Environmental Microbiology, 8, 2068–2073. 10.1111/j.1462-2920.2006.01148.x [DOI] [PubMed] [Google Scholar]
- Richie, K. , & Smith, W. (1998). Description of type II white band disease in acroporid corals. Revista De Biologia Tropical, 46, 199–203. [Google Scholar]
- Ritchie, K. (1994). Bacteria associated with bleached and nonbleached areas of Montastrea annularis, in: Proceedings, 5th symposium on the Natural History of the Bahamas: Bahamian Field Station, San Salvador, 75–79. [Google Scholar]
- Ritchie, K. B. , & Smith, G. W. (2004). Microbial communities of coral surface mucopolysaccharide layers In Rosenberg E., & Loya Y. (Eds.), Coral health and disease (pp. 259‐264). Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- Robertson, V. , Haltli, B. , Mccauley, E. P. , Overy, D. P. , & Kerr, R. G. (2016). Highly variable bacterial communities associated with the octocoral Antillogorgia elisabethae. Microorganisms, 4(3), 23 10.3390/microorganisms4030023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roder, C. , Arif, C. , Bayer, T. , Aranda, M. , Daniels, C. , Shibl, A. , … Voolstra, C. R. (2014). Bacterial profiling of white plague disease in a comparative coral species framework. ISME Journal, 8, 31–39. 10.1038/ismej.2013.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer, F. , Breitbart, M. , Jara, J. , Azam, F. , & Knowlton, N. (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi . Coral Reefs, 20, 85–91. 10.1007/s003380100138 [DOI] [Google Scholar]
- Rohwer, F. , & Kelley, S. (2004). Culture‐independent analyses of coral‐associated microbes In Rosenberg E., & Loya Y. (Eds.), Coral health and disease (pp. 265–277). Berlin, Heidelberg: Springer Berlin Heidelberg. [Google Scholar]
- Rohwer, F. , Seguritan, V. , Azam, F. , & Knowlton, N. (2002). Diversity and distribution of coral‐associated bacteria. Marine Ecology Progress Series, 243, 1–10. 10.3354/meps243001 [DOI] [Google Scholar]
- Rosenberg, E. , & Falkovitz, L. (2004). The Vibrio shiloi/Oculina patagonica model system of coral bleaching. Annual Review of Microbiology, 58, 143–159. 10.1146/annurev.micro.58.030603.123610 [DOI] [PubMed] [Google Scholar]
- Rosenberg, E. , Koren, O. , Reshef, L. , Efrony, R. , & Zilber‐Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nature Reviews Microbiology, 5, 355–362. 10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Rosenberg, E. , & Loya, Y. (2013). Coral health and disease. Berlin, Heidelberg: Springer. [Google Scholar]
- Röthig, T. , Ochsenkühn, M. A. , Roik, A. , Merwe, R. , & Voolstra, C. R. (2016). Long‐term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Molecular Ecology, 25, 1308–1323. 10.1111/mec.13567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, J. L. , & Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology, 9, 313 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan, R. (2004). Coral bleaching: Thermal adaptation in reef coral symbionts. Nature, 430, 742 10.1038/430742a [DOI] [PubMed] [Google Scholar]
- Sere, M. G. , Tortosa, P. , Chabanet, P. , Turquet, J. , Quod, J. P. , & Schleyer, M. H. (2013). Bacterial communities associated with Porites white patch syndrome (PWPS) on three western Indian Ocean (WIO) coral reefs. PLoS ONE, 8, e83746 10.1371/journal.pone.0083746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sere, M. , Wilkinson, D. A. , Schleyer, M. H. , Chabanet, P. , Quod, J. P. , & Tortosa, P. (2016). Characterisation of an atypical manifestation of black band disease on Porites lutea in the Western Indian Ocean. PeerJ, 4, e2073 10.7717/peerj.2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade, A. , & Handelsman, J. (2012). Beyond the Venn diagram: The hunt for a core microbiome. Environmental Microbiology, 14, 4–12. 10.1111/j.1462-2920.2011.02585.x [DOI] [PubMed] [Google Scholar]
- Shannon, C. E. (1948). A mathematical theory of communication. Bell System Technical Journal, 27, 379–423. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- Shieh, W. Y. , & Lin, Y. M. (1992). Nitrogen fixation (acetylene reduction) associated with the zoanthid Palythoa tuberculosa Esper. Journal of Experimental Marine Biology and Ecology, 163, 31–41. 10.1016/0022-0981(92)90145-Z [DOI] [Google Scholar]
- Shiu, J. H. , Keshavmurthy, S. , Chiang, P. W. , Chen, H. J. , Lou, S. P. , Tseng, C. H. , … Tang, S. L. (2017). Dynamics of coral‐associated bacterial communities acclimated to temperature stress based on recent thermal history. Scientific Reports, 7, 14933 10.1038/s41598-017-14927-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboni, N. , Ben‐Dov, E. , Sivan, A. , & Kushmaro, A. (2008). Global distribution and diversity of coral‐associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environmental Microbiology, 10, 2979–2990. 10.1111/j.1462-2920.2008.01718.x [DOI] [PubMed] [Google Scholar]
- Spalding, M. D. , & Grenfell, A. M. (1997). New estimates of global and regional coral reef areas. Coral Reefs, 16, 225–230. 10.1007/s003380050078 [DOI] [Google Scholar]
- Speck, M. D. , & Donachie, S. P. (2012). Widespread Oceanospirillaceae Bacteria in Porites spp. Journal of Marine Biology, 2012, 7 10.1155/2012/746720 [DOI] [Google Scholar]
- Sunagawa, S. , Desantis, T. Z. , Piceno, Y. M. , Brodie, E. L. , Desalvo, M. K. , Voolstra, C. R. , … Medina, M. (2009). Bacterial diversity and White Plague Disease‐associated community changes in the Caribbean coral Montastraea faveolata. ISME Journal, 3, 512–521. 10.1038/ismej.2008.131 [DOI] [PubMed] [Google Scholar]
- Sunda, W. , Kieber, D. J. , Kiene, R. P. , & Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature, 418, 317–320. 10.1038/nature00851 [DOI] [PubMed] [Google Scholar]
- Sussman, M. , Willis, B. L. , Victor, S. , & Bourne, D. G. (2008). Coral pathogens identified for White Syndrome (WS) epizootics in the Indo‐Pacific. PLoS ONE, 3, e2393 10.1371/journal.pone.0002393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet, M. J. , & Bulling, M. T. (2017). On the importance of the microbiome and pathobiome in coral health and disease. Frontiers in Marine Science, 4, 9 10.3389/fmars.2017.00009 [DOI] [Google Scholar]
- Tamar, L. G. (2006). Most corals may not change their symbionts. Marine Ecology Progress Series, 321, 1–7. 10.3354/meps321001 [DOI] [Google Scholar]
- Thompson, J. R. , Rivera, H. E. , Closek, C. J. , & Medina, M. (2014). Microbes in the coral holobiont: Partners through evolution, development, and ecological interactions. Frontiers in Cellular and Infection Microbiology, 4, 176 10.3389/fcimb.2014.00176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tout, J. , Siboni, N. , Messer, L. F. , Garren, M. , Stocker, R. , Webster, N. S. , … Seymour, J. R. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Frontiers in Microbiology, 6, 432 10.3389/fmicb.2015.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Water, J. , Chaib De Mares, M. , Dixon, G. B. , Raina, J. B. , Willis, B. L. , Bourne, D. G. , & Van Oppen, M. J. H. (2018). Antimicrobial and stress responses to increased temperature and bacterial pathogen challenge in the holobiont of a reef‐building coral. Molecular Ecology, 27, 1065–1080. 10.1111/mec.14489 [DOI] [PubMed] [Google Scholar]
- van de Water, J. A. J. M. , Melkonian, R. , Junca, H. , Voolstra, C. R. , Reynaud, S. , Allemand, D. , & Ferrier‐Pagès, C. (2016). Spirochaetes dominate the microbial community associated with the red coral Corallium rubrum on a broad geographic scale. Scientific Reports, 6, 27277 10.1038/srep27277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli, L. , Pezzati, E. , Huete‐Stauffer, C. , Pruzzo, C. , & Cerrano, C. (2013). 16SrDNA pyrosequencing of the mediterranean gorgonian paramuricea clavata reveals a link among alterations in bacterial holobiont members, anthropogenic influence and disease outbreaks. PLoS ONE, 8, e67745 10.1371/journal.pone.0067745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner‐Dobler, I. , & Biebl, H. (2006). Environmental biology of the marine Roseobacter lineage. Annual Review of Microbiology, 60, 255–280. 10.1146/annurev.micro.60.080805.142115 [DOI] [PubMed] [Google Scholar]
- Wang, L. , Shantz, A. A. , Payet, J. P. , Sharpton, T. J. , Foster, A. , Burkepile, D. E. , & Vega Thurber, R. (2018). Corals and their microbiomes are differentially affected by exposure to elevated nutrients and a natural thermal anomaly. Frontiers in Marine Science, 5, 101 10.3389/fmars.2018.00101 [DOI] [Google Scholar]
- Wang, Q. , Garrity, G. M. , Tiedje, J. M. , & Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261–5267. 10.1128/aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh, R. M. , Rosales, S. M. , Zaneveld, J. R. , Payet, J. P. , Mcminds, R. , Hubbs, S. L. , & Vega Thurber, R. L. (2017). Alien vs. predator: Bacterial challenge alters coral microbiomes unless controlled by Halobacteriovorax predators. PeerJ, 5, e3315 10.7717/peerj.3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. R. , Nagarajan, N. , & Pop, M. (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Computational Biology, 5, e1000352 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeemin, T. , Saenghaisuk, C. , Sutthacheep, S. , Pengsakun, S. , Klinthong, W. , & Saengmanee, K. (2009). Conditions of coral communities in the Gulf of Thailand: A decade after the 1998 severe bleaching event. Journal of Coral Reef Studies, 11, 207–217. 10.3755/galaxea.11.207 [DOI] [Google Scholar]
- Yoch, D. C. (2002). Dimethylsulfoniopropionate: Its sources, role in the marine food web, and biological degradation to dimethylsulfide. Applied and Environment Microbiology, 68, 5804–5815. 10.1128/AEM.68.12.5804-5815.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef, N. , Sheik, C. S. , Krumholz, L. R. , Najar, F. Z. , Roe, B. A. , & Elshahed, M. S. (2009). Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing‐generated fragments in 16S rRNA gene‐based environmental surveys. Applied and Environment Microbiology, 75, 5227–5236. 10.1128/AEM.00592-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G. , Cai, L. , Yuan, T. , Tian, R. , Tong, H. , Zhang, W. , … Huang, H. (2017). Microbiome dynamics in early life stages of the scleractinian coral Acropora gemmifera in response to elevated pCO2. Environmental Microbiology, 19, 3342–3352. 10.1111/1462-2920.13840 [DOI] [PubMed] [Google Scholar]
- Ziegler, M. , Roik, A. , Porter, A. , Zubier, K. , Mudarris, M. S. , Ormond, R. , & Voolstra, C. R. (2016). Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Marine Pollution Bulletin, 105, 629–640. 10.1016/j.marpolbul.2015.12.045 [DOI] [PubMed] [Google Scholar]
- Ziegler, M. , Seneca, F. O. , Yum, L. K. , Palumbi, S. R. , & Voolstra, C. R. (2017). Bacterial community dynamics are linked to patterns of coral heat tolerance. Nature Communications, 8, 14213 10.1038/ncomms14213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PacBio 16S rRNA sequence data were deposited in the GenBank NCBI Sequence Read Archive (SRA) database under the accession number https://www.ncbi.nlm.nih.gov/bioproject/PRJNA429083


