Abstract
Streptococcus mutans and Candida albicans are often isolated from plaques associated with early childhood caries. However, there are limited studies examining how these microorganisms interact with one another and how best to manage them. Recent studies have shown that curcumin (CUR), a natural compound, has the potential to independently control both of these microorganisms. The purpose of this study was to investigate how S. mutans and C. albicans respond in mono‐ and dual‐species biofilms challenged with CUR. Quantitative biofilm biomass and viability were first evaluated and supported by live–dead PCR to assess biofilm composition. Confocal laser scanning microscopy (CLSM) was used to evaluate the exopolysaccharide (EPS) content and thickness of the biofilms, and the structure of the biofilms and morphology of the cells were observed by scanning electron microscopy (SEM). Quantitative real‐time PCR (qRT‐PCR) was applied to assess relative gene expression. The 50% minimum biofilm eradication concentration (MBEC50) of CUR against S. mutans and C. albicans was 0.5 mM. The biomass and viability decreased after treatment with CUR both in dual‐species biofilms and in mono‐species biofilm. CUR inhibited S. mutans and C. albicans in both mono‐ and dual‐species biofilms. Streptococcus mutans was more sensitive to CUR in dual‐species biofilm than in mono‐species biofilms, whereas C. albicans was less sensitive in dual‐species biofilms. EPS production was decreased by CUR in both mono‐ and dual‐species biofilms, which coincided with the downregulation of glucosyltransferase and quorum sensing‐related gene expression of S. mutans. In C. albicans, the agglutinin‐like sequence family of C. albicans was also downregulated in dual‐species biofilms. Collectively, these data show the potential benefit of using a natural antimicrobial, CUR, to control caries‐related dual‐species plaque biofilms.
Keywords: biofilm, Candida albicans, curcumin, Streptococcus mutans
Curcumin, a natural compound, can inhibit oral dual‐species biofilms. The results here show that curcumin has not only inhibited the mono‐species biofilms from Streptococcus mutans and Candida albicans, but also the dual‐species biofilm formed by those two species. Exopolysaccharide production was decreased by curcumin treatment in both mono‐ and dual‐species biofilms.
1. INTRODUCTION
Polymicrobial infection occurs during the process of dental caries development. Streptococcus mutans interacts with Candida albicans to form cross‐kingdom biofilms associated with early childhood caries (ECC) (Hajishengallis, Parsaei, Klein, & Koo, 2017). The amount of C. albicans is dramatically higher in children with ECC than in those without tooth decay (Ghasempour, Sefidgar, Eyzadian, & Gharakhani, 2011). Studies have shown that the presence of C. albicans can augment exopolysaccharides (EPS) in biofilms, resulting in an increase in biomass related to mono‐species biofilms of S. mutans alone (Falsetta et al., 2014; Hwang et al., 2017). S. mutans–C. albicans associations enhance S. mutans infection, modulate the structure of biofilms, and thus influence the onset and severity of dental caries (Falsetta et al., 2014).
Eradicating these bacterial–fungal biofilms in vitro is challenging (Heitman, Metwalli, Khan, Krom, & Jabra‐Rizk, 2013). Most of the clinically used therapeutic approaches are monotherapies, based on either antibacterial or antifungal agents despite the polymicrobial nature of disease‐causing biofilms (Xiao et al., 2018). Fluoride is widely used in the prevention of dental caries. However, fluoride does not offer protection against the infectious aspects and instead is involved in the remineralization process (Ten Cate, 2012). The long‐term use of chlorhexidine will lead to staining of the tongue and taste disorders (Neturi, 2014; Rahmani‐Badi, Sepehr, & Babaie‐Naiej, 2015). Antibiotic resistance also limits the application of therapeutic approaches to control biofilm infection (Marc et al., 2018). Finding new therapeutic methods for disrupting bacterial–fungal interkingdom biofilms could help improve anticaries strategies and oral health. Naturally derived chemotherapeutic agents are an attractive option. They have advantages over synthetic derivatives due to their natural evolution and diminished likelihood of resistance. Curcumin (CUR), a food‐grade natural product extracted from the root of turmeric, is widely used as a flavoring and coloring agent in food. It is also used in clinical practice due to its anti‐inflammatory and antitumor activities (Esatbeyoglu et al., 2012). Our group has shown that CUR exerts promising anticaries effects on S. mutans by altering the EPS synthesis mechanism (Li, Li, Lin, & Zhou, 2018).
Since dual‐species biofilms of S. mutans and C. albicans have the advantage of better simulating the pathogenic biofilms in ECC relative to mono‐species biofilms, exploration of the effect of CUR on dual‐species biofilms should be prioritized. In this study, we establish an in vitro biofilm model by using S. mutans and C. albicans, and investigate the reaction of dual‐species biofilms to CUR.
2. MATERIALS AND METHODS
2.1. Growth conditions and MBEC assay to determine the effects of CUR on S. mutans and C. albicans
Streptococcus mutans UA159 (ATCC 700610) and C. albicans SC5314 (ATCC MYA‐2876) were selected for this study. Candida albicans was grown in Sabouraud's dextrose broth (SDB, HKM) at 37°C with shaking at 200 rpm for 18 hr. Streptococcus mutans was cultured in brain heart infusion broth (BHI, Difco) for 18 hr at 37°C. CUR was dissolved in dimethyl sulfoxide (DMSO) for liquid storage.
Streptococcus mutans and C. albicans were cultured in BHI and SDB separately for 18 hr. Cellular microorganisms were collected by centrifugation (13,201 g, 5 min, 4°C). Phosphate‐buffered saline (PBS) was applied to wash the cultures three times. Suspensions of microorganisms were standardized to OD values equal to 1 × 107 CFUs/ml using a microplate spectrophotometer (Infinite 200, TECAN). Biofilms were grown in 96‐well flat‐bottom plates. The minimum biofilm eradication concentration (MBEC) for S. mutans and C. albicans biofilms was determined by the colony counting method. The biofilms were cultured following the methodology described above for 24 hr in 96‐well flat‐bottom plates. The medium was refreshed, and serial dilutions of CUR were added to the biofilms. Medium with the corresponding concentrations of DMSO without CUR was used as a control. After 24 hr of cultivation, the supernatants were removed, and sterile PBS was gently applied three times to wash the biofilms that formed on the plates. The biofilms were scraped and then resuspended in PBS, and the CFU counts were used to confirm the MBEC of the microorganisms.
2.2. The effects of CUR on mono‐ or dual‐species biofilms in vitro
The biofilms were grown as described in previous studies with some modification (Willems, Kos, Jabra‐Rizk, Krom, & Bjarnsholt, 2016; Zhou et al., 2018). The cells were cultured as previously described and standardized by artificial saliva solution (ASS, NOVON) supplemented with 1% sucrose and 10% fetal bovine serum to OD600 = 0.1. Three test groups with CUR (S. mutans biofilm, C. albicans biofilm, and S. mutans‐C. albicans biofilm) and corresponding control groups without CUR were established.
The biofilms formed on 6‐well flat‐bottom microtiter plates (Coster Corning). For the dual‐species biofilms, 1.5 ml of S. mutans and 1.5 ml of C. albicans were added to the microtiter plate. To form mono‐species biofilms, 3 ml of S. mutans and 3 ml of C. albicans were added separately. The plates were cultured for 48 hr at 37°C and 5% CO2. The medium was refreshed every 24 hr. Biofilms that formed on the plates were washed gently three times with sterile PBS. Then, CUR was added to the experimental groups and the corresponding concentrations of DMSO were added to the control groups, followed by incubation for 24 hr.
Crystal violet (CV) and 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide (MTT) assays were employed to verify the effects of CUR on the biomass and viability of biofilms. For CV assays, the biofilms that formed on the plates were fixed with methanol after being gently washed three times with sterile PBS. Then, the samples were washed again and stained with CV solution for 15 min. The supernatants were thoroughly discarded, and the microplates were allowed to air‐dry overnight. Finally, 200 μl of 95% ethanol was used to dissolve the CV in the microplates for 20 min. For each sample, 100 μl of the solution was analyzed on a spectrophotometer, and the absorbance at 600 nm was recorded. For MTT assays, 200 μl of 0.5 mg/ml MTT was added to the wells and incubated at 37°C for 3 hr in the dark (Li et al., 2018). Then, the supernatants were displaced by 100 μl of 100% DMSO for 10 min while protected from light. For each sample, 75 μl of the solution was analyzed on a spectrophotometer and the absorbance at 570 nm was recorded using a microplate reader.
2.3. Analysis of the composition of bacteria in biofilms by live–dead PCR
Live–dead PCR was used to analyze the composition of microorganisms in biofilms. Propidium monoazide (PMA) was applied to examine the quantity of live and dead microorganisms in biofilms as previously described in the literature (Sherry et al., 2016; Yasunaga et al., 2013). Because PMA is unable to permeate intact membranes of viable cells, it can only penetrate dead cells following membrane damage and bind to the DNA of dead cells following exposure to a halogen light source. This covalent bonding to DNA prevents amplification during quantitative PCR (qPCR); therefore, only live cells can be detected. No PMA‐added groups were included for each sample to determine total cells.
The biofilms were collected with a cell scraper and added to 2 ml microcentrifuge tubes with 1 ml of PBS. After resuspension, 5 μl aliquots of 10 μM PMA were added to the samples. Next, the samples were collected by centrifugation (13,201 g, 5 min, 4°C). Genomic DNA was extracted by a DNeasy Blood & Tissue Kit (QIAGEN) and analyzed to confirm the density and purity. Samples were analyzed using quantitative PCR (qPCR), and the Ct values were input into the formula for a standard curve to determine the quantity of microorganisms in each of the biofilms. Details were described in a previous study (Zhou et al., 2018). The primer sequences used were from previous studies in Table 1.
Table 1.
Nucleotide sequence of primers used for PCR
| Gene | Primer sequence (5′−3′) | |
|---|---|---|
| Forward | Reverse | |
| Streptococcus mutans | GATACATAGCCGACCTGAG | CCATTGCCGAAGATTCC (Sherry et al., 2016)a |
| Candida albicans | GGGTTTGCTTGAAAGACGGTA | TTGAAGATATACGTGGTGGACGTTA (Sherry et al., 2016)a |
| gtfB | ACACTTTCGGGTGGCTTG | GCTTAGATGTCACTTCGGTTG (Li et al., 2018)a |
| gtfC | CCAAAATGGTATTATGGCTGTCG | GAGTCTCTATCAAAGTAACGCAGT (Li et al., 2018)a |
| gbpB | AGCAACAGAAGCACAACCATCAG | CCACCATTACCCCAGTAGTTTCC (Li et al., 2018)a |
| comC | GACTTTAAAGAAATTAAGACTG | AAGCTTGTGTAAAACTTCTGT (Li et al., 2018)a |
| comD | CTCTGATTGACCATTCTTCTGG | CATTCTGAGTTTATGCCCCTC (Li et al., 2018)a |
| comE | CCTGAAAAGGGCAATCACCAG | GGGGCATAAACTCAGAATGTGTCG (Li et al., 2018)a |
| 16S rRNA | CTTACCAGGTCTTGACATCCCG | ACCCAACATCTCACGACACGAG (Li et al., 2018)a |
| als1 | TTCTCATGAATCAGCATCCACAA | CAGAATTTTCACCCATACTTGGTTTC (Alalwan et al., 2017)a |
| als3 | CAACTTGGGTTATTGAAACAAAAACA | AGAAACAGAAACCCAAGAACAACCT (Alalwan et al., 2017)a |
| 18S rRNA | AAACGGCTACCACATCCAAG | CCAAGCCCAAGGTTCAACTA (Barker et al., 2004)a |
The numbers in the brackets after the primer sequences were references.
2.4. The effect of CUR on the EPS of biofilms by CLSM
Confocal laser scanning microscopy was used to examine the amount of EPS in biofilms. The EPS matrix was labeled with Alexa Fluor® 647‐labeled dextran conjugate, a kind of red fluorescent dye (Invitrogen Corp), at the beginning of biofilm formation and was washed gently with sterile normal saline (NS) after 72 hr (Klein et al., 2009; Yang, Liu, He, Chen, & Li, 2016). Then, SYTO9 green fluorescent dye (Molecular Probes) was applied to stain the microorganism cells for 15 min at room temperature. For image collection, the gates were set to 655–668 nm for Alexa Fluor® 647‐labeled dextran conjugate and 480–500 nm for SYTO9 (Li et al., 2018). Images were collected by CLSM and analyzed by COMSTAT (Liu et al., 2017).
2.5. Observation of the morphology of the microorganisms in biofilms
Scanning electron microscopy (SEM) was applied to observe the structure of biofilms. The treated biofilms were collected as previously mentioned. Then, the biofilms were fixed with 2.5% glutaraldehyde overnight. The biofilms were washed with sterile PBS and dehydrated by an alcohol gradient (30%, 50%, 70%, 90%, and 100%). After that, the biofilms were treated with tert‐butanol three times. Finally, samples were dried by lyophilization and metal spraying. The samples were observed at 2,000× and 10,000× magnification by SEM.
2.6. Detection of gene expression in the biofilms
The biofilms were collected by centrifugation (13,201 g, 5 min, 4°C). Next, the total RNA was extracted by an miRNeasy Mini Kit (QIAGEN) and analyzed by a Nanodrop 2000 spectrophotometer to confirm the density and purity. Reverse transcription of the total RNA was performed. A cDNA library was constructed, and PCR was performed with reference to previous studies (Zhou et al., 2018). Table 1 shows the primer sequences (Alalwan et al., 2017; Barker et al., 2004; Li et al., 2018; Sherry et al., 2016). The results were calculated by the 2−ΔΔCt method.
2.7. Statistical analysis
All experiments were independently performed in triplicate. GraphPad Prism version 7.0 (GraphPad Software) and SPSS 17.0 were employed to analyze the data. An unpaired t test was applied to analyze the data. A p value < .05 was set as the significance level.
3. RESULTS
3.1. Inhibition of biofilm formation by CUR
The MBEC was shown to be 0.5 mM CUR for S. mutans as well as C. albicans by assay. According to the CV and MTT assays, the biomass and viability of biofilms were greater in dual‐species biofilms despite the addition of CUR (Figure 1). After 24 hr of incubation with CUR, the biomasses of both mono‐ and dual‐species biofilms were significantly reduced compared with that of the control groups, specifically by 23% for S. mutans, 36% for C. albicans, and 24% for dual‐species biofilms (p < .05, Figure 1a). The viability of the biofilms decreased significantly in all treated groups (p < .05, Figure 1b). The viability of the S. mutans biofilms decreased by 54%, while those of the C. albicans and dual‐species biofilms decreased by 29% and 30%, respectively, relative to the control group (Figure 1b).
Figure 1.
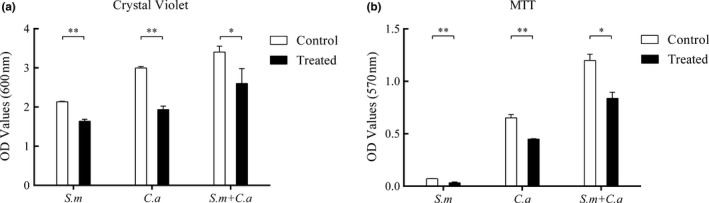
Effect of 0.5 mM CUR on mono‐ and dual‐species biofilms. Different types of mature biofilms were incubated with CUR for 24 hr. The biofilm biomass was evaluated by CV assay (a), while the viability of biofilms was evaluated by MTT assay (b). The asterisks (*) indicate significant differences (*p < .05; **p < .01). CUR decreased the biomass and viability of all treated groups
3.2. Inhibition of live/dead cells in biofilms by CUR
Curcumin decreased total/live cells in both mono‐species and dual‐species biofilms (Figure A1), but the percent reductions with CUR in S. mutans and C. albicans biofilms were different.
For the total S. mutans cells, CUR inhibited 79% of S. mutans in dual‐species biofilms while inhibiting 47% in mono‐species biofilms (p < .001). A similar trend was observed for live S. mutans cells: CUR inhibited 69% of S. mutans in dual‐species biofilms while inhibiting 58% in mono‐species biofilm (p < .001) (Figure 2a). For the total C. albicans cells, CUR inhibited 38% of C. albicans in dual‐speices biofilms while inhibiting 60% in mono‐species biofilms (p < .001). The same trend was observed for live C. albicans cells, with 52% reduction of C. albicans in dual‐speices biofilms and 86% inhibition in mono‐species biofilm (p < .001) (Figure 2b).
Figure 2.
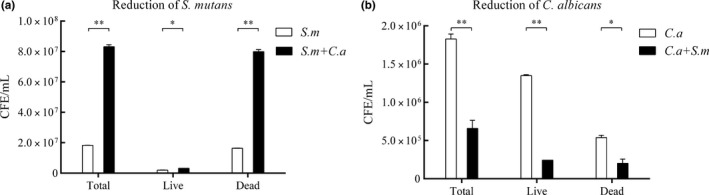
Effect of CUR on live/dead microorganisms in different types of biofilms. Dual‐species biofilm models were grown for 48 hr, and spent supernatants were replaced with fresh ASS every 24 hr. After mature biofilms formed, the biofilms were treated with 0.5 mM CUR for 24 hr. The net reduction (net reduction equals the treated group minus the control group) of Streptococcus mutans (a) and Candida albicans (b) in mono‐ and dual‐species biofilms was evaluated by species‐specific qPCR. The asterisks (*) indicate significant differences (*p < .05; **p < .01)
3.3. CUR‐induced EPS reduction of mono‐ and dual‐species biofilms by CLSM
The amount of EPS in mono‐ and dual‐species biofilms was quantified by CLSM. The production of EPS was decreased both in mono‐ and dual‐species biofilms by CUR, as determined by CLSM images (Figure 3). The EPS in the treated group was sparser than that in the control group (Figure 3a). This result was consistent with Figure 3b, which showed a reduction in EPS in both mono‐ and dual‐species biofilms. The dual‐species group was reduced by 24% relative to the control group. Streptococcus mutans biofilms had a 22% reduction in EPS, and C. albicans biofilms had an 80% reduction in EPS. Likewise, the thickness of the biofilms in the treated groups was overtly attenuated compared with that in the control groups (Figure 3c).
Figure 3.
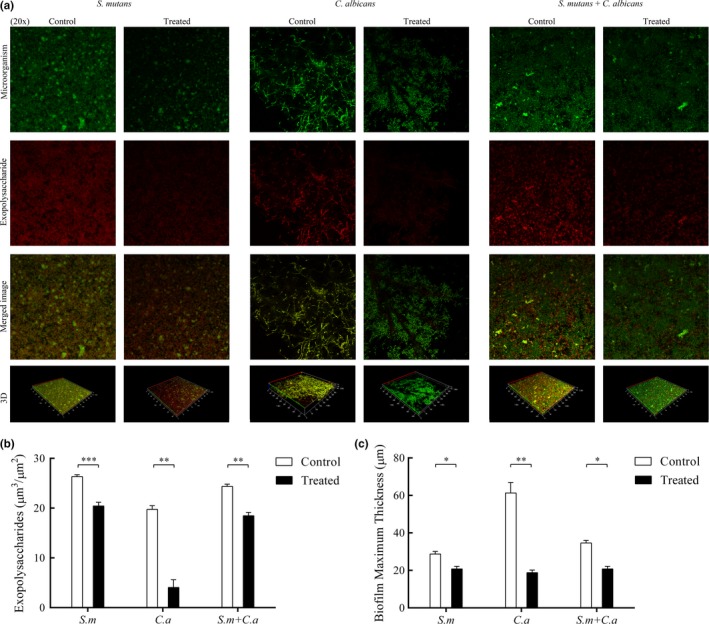
Effect of CUR on the EPS of mono‐ and dual‐species biofilms by CLSM. After incubation and staining, images of the biofilms were collected. The green channel was used for microorganism (a). The red channel was used for EPS (a). The EPS and thickness of the biofilms were quantified and compared (b, c). The asterisks (*) indicate significant differences (*p < .05; **p < .01; ***p < .001)
3.4. Morphology of microorganisms in biofilms by SEM
Scanning electron microscopy images revealed biofilm morphologies and supported the CLSM results. In S. mutans mono‐species biofilms, the biofilm structures in the treated group were lost (Figure 4a). The data also showed that CUR treatment not only led to changes in biofilm structure but also caused slackening of the matrix in C. albicans mono‐species biofilms (Figure 4b). The structure of dual‐species biofilm seemed more disordered in the treated group than in the control group. Notably, the arrangement of hypha was altered, and less matrix was observed in the treated group of dual‐species biofilms (Figure 4c).
Figure 4.
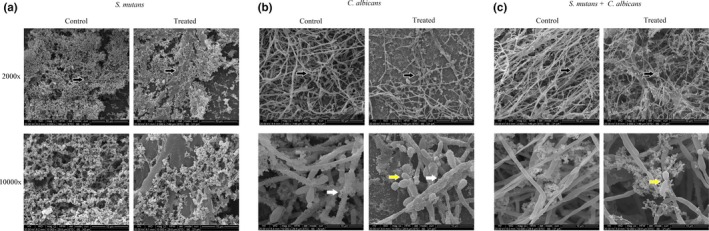
Morphological characteristics of mono‐ and dual‐species biofilms under CUR treatment. Representative SEM images of Streptococcus mutans (a), Candida albicans (b), and dual‐species biofilms (c). Each field of vision was magnified 2,000× and 10,000×. The black arrows indicate the magnified viewing area. The yellow arrows indicate the yeast. The white arrows indicate the EPS in the biofilms
3.5. Gene expression changes in biofilms
The mRNA levels of all genes tested were decreased in the CUR‐treated group relative to the control group. CUR decreased the expression of virulence‐related genes in both mono‐ and dual‐species biofilms (Figure 5).
Figure 5.
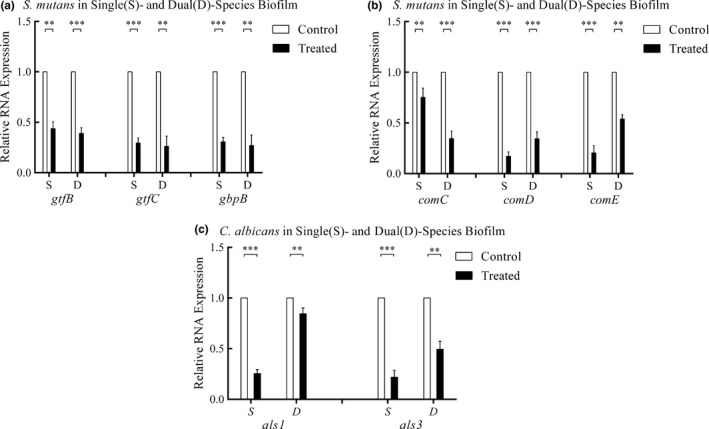
Change in gene expression in biofilms under the effects of CUR. The mRNA levels of genes in different virulence systems of Streptococcus mutans and Candida albicans are shown in Figure 5. Different levels of gene expression were standardized to 16 sRNA levels. (a) Relative RNA expression of gtfs in S. mutans, (b) relative RNA expression of two‐component signal transduction system in S. mutans, and (c) relative RNA expression of als in C. albicans. The asterisks (*) indicated significant differences (*p < .05; **p < .01; ***p < .001)
For S. mutans, the relative expression levels of the virulence genes gtfB, gtfC, and gbpB in the mono‐species biofilms were 44%, 29%, and 31%, respectively, compared with the levels in the control groups, while in the dual‐species biofilm they were 39%, 26%, and 27%, respectively (Figure 5a). The genes of the two‐component signal transduction system, including comC, comD, and comE, were evaluated. The relative gene expression levels of comC, comD, and comE were downregulated by 25%, 83%, and 80%, respectively, in mono‐species biofilms and by 65%, 66%, and 46%, respectively, in dual‐species biofilms (Figure 5b).
In C. albicans, the expression levels of genes related to biofilm formation, als1 and als3, were downregulated by 75% and 78%, respectively, in mono‐species biofilms and by 56% and 69%, respectively, in dual‐species biofilms (Figure 5c).
4. DISCUSSION
Early childhood caries has been treated using different modalities but with little success. Bacterial–fungal interactions commonly occur in the process of ECC formation. Combating this cross‐kingdom infection is a challenge in the control of ECC (Klinke et al., 2014; Yang et al., 2012). Natural products are an alternative option that results in less antibiotic resistance, and they are widely used in clinical settings (Jeon, Rosalen, Falsetta, & Koo, 2011; Nijampatnam et al., 2018; Townsley & Shank, 2017). The present study showed the value of applying CUR to address oral dual‐species biofilms.
Curcumin inhibited the biomass and viability of both mono‐ and dual‐species biofilms. A previous study reported that CUR exerted both short‐ and long‐term effects on the viability of S. mutans biofilms at 0.5 mM (Li et al., 2018). Alalwan et al. found that 200 μg/ml CUR (approximately equal to 0.543 mM) caused an 80% reduction in the metabolic activity of C. albicans biofilms (Alalwan et al., 2017). This finding is consistent with the present study. To determine the biological basis of the effect of CUR on dual‐species biofilms, live–dead PCR was conducted to assay the composition of cells in the biofilms (Sherry et al., 2016). CUR affected S. mutans and C. albicans in both dual‐ and mono‐species biofilms. However, the two microorganisms showed different responses to CUR. Many more cells of S. mutans were eradicated in dual‐species biofilms. The interaction between S. mutans and C. albicans enhanced the effect of CUR on S. mutans in dual‐species biofilms. Bacteria–fungi interactions and their relevance in health are well established (Arvanitis & Mylonakis, 2015). Dual‐species biofilms achieve higher biomass and cell numbers than mono‐species biofilms (Sztajer et al., 2014). Streptococcus mutans‐derived glucosyltransferase B (GtfB) itself can promote C. albicans biofilm development (Ellepola, Liu, Cao, Koo, & Seneviratne, 2017). The enhanced inhibition of S. mutans by CUR in dual‐species biofilms shown here is interesting.
Exopolysaccharides increases in dual‐species biofilms and enhances the cariogenic potential of biofilms (Mitchell et al., 2017). CUR influences the amount of EPS in all biofilms, which was confirmed here by CLSM. A previous study indicated that the presence of C. albicans augmented the production of EPS, and dual‐species biofilms accrued more biomass and harbored more viable S. mutans cells than mono‐species biofilms (Falsetta et al., 2014). Similarly, the presence of Pseudomonas aeruginosa protected Salmonella cells in biofilms from disinfection treatment by generating greater EPS production in dual‐species biofilms than mono‐species biofilms (Pang, Yang, & Yuk, 2017). Kim et al. (2018) found that bacteria‐derived EPS enhanced antifungal drug tolerance in a cross‐kingdom oral biofilm. In mono‐species biofilms, we found that CUR disrupted EPS secretion not only from S. mutans but also from C. albicans biofilms. It is likely that CUR affects EPS production, either by fungi or bacteria, to disrupt dual‐species biofilms.
Streptococcus mutans extracellular glucosyltransferases (Gtfs), particularly GtfB and GtfC, synthesize predominantly water‐insoluble glucans, which contribute to the structural scaffold of biofilms (Bowen & Koo, 2011). A previous study showed that deletion of gtfB and gtfC significantly disrupted biofilm formation, which corresponded with the results in this study (Ooshima et al., 2001). In our study, gtfB and gtfC were inhibited by CUR relative to the control group, and a reduction in EPS and structural looseness were observed in biofilms treated with CUR. Quorum sensing (QS) systems have a key role in coordinating biofilm formation and activating virulence factors in many bacteria and are considered an enticing target for fighting biofilm infection (Suntharalingam & Cvitkovitch, 2005; Worthington, Richards, & Melander, 2012). Among these, the competence‐stimulating peptide‐QS (CSP‐QS) system is the most common intraspecific example, including a competence‐stimulating peptide (encoded by comC), a histidine kinase sensor protein (encoded by comD), and a cognate response regulator (encoded by comE) (Suntharalingam & Cvitkovitch, 2005; Yue et al., 2018). A previous study found that in the presence of chlorhexidine, the upregulation of immB in a comC mutant was largely inhibited compared with that in the wild‐type strain, implying that the function of the QS system is one of the mechanisms regulating S. mutans antimicrobial sensitivity (Wang, Liu, Huo, & Ling, 2013). In the present study, the expression of comC, comD, and comE was suppressed by CUR in all biofilms. The results indicated that CUR inhibition occurs through the QS system.
The agglutinin‐like sequence (Als) family plays an important role in adhesion and aggregation with other substances, and it is a key element for C. albicans in biofilm formation (Hoyer & Cota, 2016). The gene expression of als1 and als3 decreased sharply after CUR treatment. This may have affected the function of the Als family and alleviated the interaction of C. albicans with bacteria. Lee et al. suggested that CUR exerted antifungal activity by inducing disruption of the fungal plasma membrane (Lee & Lee, 2014). CUR most likely hampers the Als family of C. albicans to exert antibacterial effects on S. mutans in dual‐species biofilms.
5. CONCLUSIONS
Curcumin induces effects on S. mutans and C. albicans, such as their EPS formation, through different biological systems. Candida albicans is less sensitive to CUR in fungi–bacteria interkingdom biofilms.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTION
Xinlong Li and Luoping Yin participated in the study design, carried out the experimental studies on biofilms, and performed statistical analysis. Gordon Ramage helped to draft the manuscript. Binchun Li, Ye Tao, and Qinghui Zhi participated in the study design and assisted with statistical support. Huancai Lin and Yan Zhou conceived the study, participated in the study design and data analysis, and were responsible for writing and submitting the final manuscript. All authors read and approved the manuscript.
ETHICAL APPROVAL
None required.
ACKNOWLEDGMENTS
This work was supported by Science and Technology Planning Project of Guangdong Province, China (2017A020215064).
APPENDIX 1.
1.1.
Figure A1.
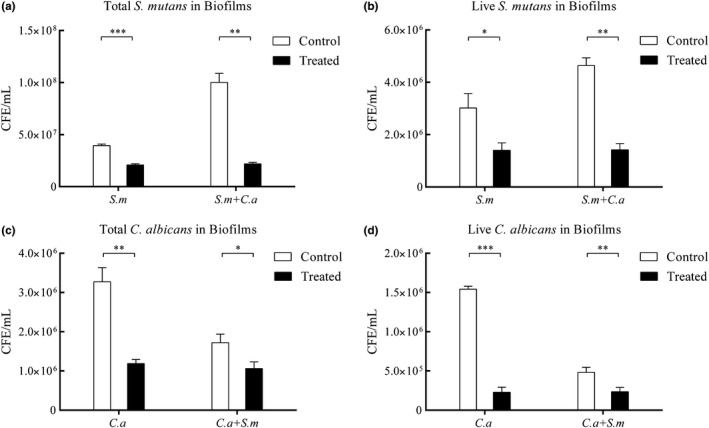
Effect of CUR on total/live microorganisms in different biofilms. Dual‐species biofilm models were grown for 48 h, and spent supernatants were replaced with fresh AS every 24 h. After mature biofilm formed, the biofilms were treated with 0.5 mM CUR for 24 h. Biofilms were assessed for total and viable composition of C. albicans and S. mutans using viability qRT‐PCR
[Correction added on 21 October 2019 after first online publication: Figure S1 from Supporting Information has been moved to Appendix section as Figure A1]
Li X, Yin L, Ramage G, et al. Assessing the impact of curcumin on dual‐species biofilms formed by Streptococcus mutans and Candida albicans . MicrobiologyOpen. 2019;8:e937 10.1002/mbo3.937
Xinlong Li and Luoping Yin contributed equally to this study.
Contributor Information
Huancai Lin, Email: linhc@mail.sysu.edu.cn.
Yan Zhou, Email: zhouy75@mail.sysu.edu.cn, Email: zhouy10.3@163.com.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article.
REFERENCES
- Alalwan, H. , Rajendran, R. , Lappin, D. F. , Combet, E. , Shahzad, M. , Robertson, D. , … Ramage, G. (2017). The anti‐adhesive effect of curcumin on Candida albicans biofilms on denture materials. Frontiers in Microbiology, 8, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis, M. , & Mylonakis, E. (2015). Fungal‐bacterial interactions and their relevance in health. Cellular Microbiology, 17, 1442–1446. [DOI] [PubMed] [Google Scholar]
- Barker, K. S. , Crisp, S. , Wiederhold, N. , Lewis, R. E. , Bareither, B. , Eckstein, J. , … Rogers, P. D. (2004). Genome‐wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans . Journal of Antimicrobial Chemotherapy, 54, 376–385. 10.1093/jac/dkh336 [DOI] [PubMed] [Google Scholar]
- Bowen, W. H. , & Koo, H. (2011). Biology of Streptococcus mutans‐derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Research, 45, 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellepola, K. , Liu, Y. , Cao, T. , Koo, H. , & Seneviratne, C. J. (2017). Bacterial GtfB augments Candida albicans accumulation in cross‐kingdom biofilms. Journal of Dental Research, 96, 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esatbeyoglu, T. , Huebbe, P. , Ernst, I. M. A. , Chin, D. , Wagner, A. E. , & Rimbach, G. (2012). Curcumin‐from molecule to biological function. Angewandte Chemie International Edition, 51, 5308–5332. [DOI] [PubMed] [Google Scholar]
- Falsetta, M. L. , Klein, M. I. , Colonne, P. M. , Scott‐Anne, K. , Gregoire, S. , Pai, C. H. , … Koo, H. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infection and Immunity, 82, 1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasempour, M. , Sefidgar, S. A. , Eyzadian, H. , & Gharakhani, S. (2011). Prevalence of Candida albicans in dental plaque and caries lesion of early childhood caries (ECC) according to sampling site. Caspian Journal of Internal Medicine, 2, 304–308. [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, E. , Parsaei, Y. , Klein, M. I. , & Koo, H. (2017). Advances in the microbial etiology and pathogenesis of early childhood caries. Molecular Oral Microbiology, 32, 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman, J. , Metwalli, K. H. , Khan, S. A. , Krom, B. P. , & Jabra‐Rizk, M. A. (2013). Streptococcus mutans, Candida albicans, and the human mouth: A sticky situation. PLoS Path, 9, e1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer, L. L. , & Cota, E. (2016). Candida albicans agglutinin‐like sequence (Als) family vignettes: A review of Als protein structure and function. Frontiers in Microbiology, 7, 280 10.3389/fmicb.2016.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, G. , Liu, Y. , Kim, D. , Li, Y. , Krysan, D. J. , & Koo, H. (2017). Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross‐kingdom biofilm development in vivo. PLoS Path, 13, e1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J. G. , Rosalen, P. L. , Falsetta, M. L. , & Koo, H. (2011). Natural products in caries research: Current (limited) knowledge, challenges and future perspective. Caries Research, 45, 243–263. 10.1159/000327250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Liu, Y. , Benhamou, R. I. , Sanchez, H. , Simon‐Soro, A. , Li, Y. , … Koo, H. (2018). Bacterial‐derived exopolysaccharides enhance antifungal drug tolerance in a cross‐kingdom oral biofilm. ISME Journal, 12, 1427–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, M. I. , Duarte, S. , Xiao, J. , Mitra, S. , Foster, T. H. , & Koo, H. (2009). Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development. Applied and Environment Microbiology, 75, 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke, T. , Urban, M. , Luck, C. , Hannig, C. , Kuhn, M. , & Kramer, N. (2014). Changes in Candida spp., mutans streptococci and lactobacilli following treatment of early childhood caries: A 1‐year follow‐up. Caries Research, 48, 24–31. [DOI] [PubMed] [Google Scholar]
- Lee, W. , & Lee, D. G. (2014). An antifungal mechanism of curcumin lies in membrane‐targeted action within Candida albicans . IUBMB Life, 66, 780–785. [DOI] [PubMed] [Google Scholar]
- Li, B. , Li, X. , Lin, H. , & Zhou, Y. (2018). Curcumin as a promising antibacterial agent: Effects on metabolism and biofilm formation in S. mutans . BioMed Research International, 2018, 4508709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Qiu, W. , Zhang, K. , Zhou, X. , Ren, B. , He, J. , … Li, M. (2017). Nicotine enhances interspecies relationship between Streptococcus mutans and Candida albicans . BioMed Research International, 2017, 7953920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc, G. , Araniciu, C. , Oniga, S. D. , Vlase, L. , Pirnau, A. , Duma, M. , … Oniga, O. (2018). New N‐(oxazolylmethyl)‐thiazolidinedione active against Candida albicans biofilm: Potential Als proteins inhibitors. Molecules, 23(10), 2522 10.3390/molecules23102522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, T. J. , Hwang, G. , Liu, Y. , Kim, D. , Li, Y. , Krysan, D. J. , & Koo, H. (2017). Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross‐kingdom biofilm development in vivo. PLoS Path, 13, e1006407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neturi, R. S. (2014). Effects of green tea on Streptococcus mutans counts – A randomised control trail. Journal of Clinical and Diagnostic Research, 8(11), ZC128–ZC130. 10.7860/JCDR/2014/10963.5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijampatnam, B. , Zhang, H. , Cai, X. , Michalek, S. M. , Wu, H. , & Velu, S. E. (2018). Inhibition of Streptococcus mutans Biofilms by the Natural Stilbene Piceatannol Through the Inhibition of Glucosyltransferases. ACS Omega, 3, 8378–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooshima, T. , Matsumura, M. , Hoshino, T. , Kawabata, S. , Sobue, S. , & Fujiwara, T. (2001). Contributions of three glycosyltransferases to sucrose‐dependent adherence of Streptococcus mutans . Journal of Dental Research, 80, 1672–1677. [DOI] [PubMed] [Google Scholar]
- Pang, X. Y. , Yang, Y. S. , & Yuk, H. G. (2017). Biofilm formation and disinfectant resistance of Salmonella sp. in mono‐ and dual‐species with Pseudomonas aeruginosa . Journal of Applied Microbiology, 123, 651–660. [DOI] [PubMed] [Google Scholar]
- Rahmani‐Badi, A. , Sepehr, S. , & Babaie‐Naiej, H. (2015). A combination of cis‐2‐decenoic acid and chlorhexidine removes dental plaque. Archives of Oral Biology, 60, 1655–1661. [DOI] [PubMed] [Google Scholar]
- Sherry, L. , Lappin, G. , O'Donnell, L. E. , Millhouse, E. , Millington, O. R. , Bradshaw, D. J. , … Ramage, G. (2016). Viable compositional analysis of an eleven species oral polymicrobial biofilm. Frontiers in Microbiology, 7, 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam, P. , & Cvitkovitch, D. G. (2005). Quorum sensing in streptococcal biofilm formation. Trends in Microbiology, 13, 3–6. [DOI] [PubMed] [Google Scholar]
- Sztajer, H. , Szafranski, S. P. , Tomasch, J. , Reck, M. , Nimtz, M. , Rohde, M. , & Wagner‐Dobler, I. (2014). Cross‐feeding and interkingdom communication in dual‐species biofilms of Streptococcus mutans and Candida albicans . ISME Journal, 8, 2256–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Cate, J. M. (2012). Novel anticaries and remineralizing agents: Prospects for the future. Journal of Dental Research, 91, 813–815. [DOI] [PubMed] [Google Scholar]
- Townsley, L. , & Shank, E. A. (2017). Natural‐product antibiotics: Cues for modulating bacterial biofilm formation. Trends in Microbiology, 25, 1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. L. , Liu, J. , Huo, Y. B. , & Ling, J. Q. (2013). Bacteriocin immunity proteins play a role in quorum‐sensing system regulated antimicrobial sensitivity of Streptococcus mutans UA159. Archives of Oral Biology, 58, 384–390. [DOI] [PubMed] [Google Scholar]
- Willems, H. M. , Kos, K. , Jabra‐Rizk, M. A. , Krom, B. P. , & Bjarnsholt, T. (2016). Candida albicans in oral biofilms could prevent caries. Pathogens and Disease, 74, ftw039. [DOI] [PubMed] [Google Scholar]
- Worthington, R. J. , Richards, J. J. , & Melander, C. (2012). Small molecule control of bacterial biofilms. Organic & Biomolecular Chemistry, 10, 7457–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J. , Grier, A. , Faustoferri, R. C. , Alzoubi, S. , Gill, A. L. , Feng, C. , … Gill, S. R. (2018). Association between oral candida and bacteriome in children with severe ECC. Journal of Dental Research, 97, 1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. Q. , Zhang, Q. , Lu, L. Y. , Yang, R. , Liu, Y. , & Zou, J. (2012). Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Archives of Oral Biology, 57, 1048–1053. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Liu, S. , He, Y. , Chen, Z. , & Li, M. (2016). Effect of LongZhang Gargle on biofilm formation and acidogenicity of Streptococcus mutans in vitro. BioMed Research International, 2016, 5829823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga, A. , Yoshida, A. , Morikawa, K. , Maki, K. , Nakamura, S. , Soh, I. , … Ansai, T. (2013). Monitoring the prevalence of viable and dead cariogenic bacteria in oral specimens and in vitro biofilms by qPCR combined with propidium monoazide. BMC Microbiology, 13, 157 10.1186/1471-2180-13-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, J. , Yang, H. , Liu, S. , Song, F. , Guo, J. , & Huang, C. (2018). Influence of naringenin on the biofilm formation of Streptococcus mutans . Journal of Dentistry, 76, 24–31. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Millhouse, E. , Shaw, T. , Lappin, D. F. , Rajendran, R. , Bagg, J. , … Ramage, G. (2018). Evaluating Streptococcus mutans strain dependent characteristics in a polymicrobial biofilm community. Frontiers in Microbiology, 9, 1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


