Abstract
Site-specific introduction of bioorthogonal handles into biomolecules provides powerful tools for studying and manipulating the structures and functions of proteins. Recent advances in bioorthogonal chemistry demonstrate that tetrazine-based bioorthogonal cycloaddition is a particularly useful methodology due to its high reactivity, biological selectivity, and turn-on property for fluorescence imaging. Despite its broad applications in protein labeling and imaging, utilization of tetrazine-based bioorthogonal cycloaddition has been limited to date by the requirement of a hydrophobic strained alkene reactive moiety. Circumventing this structural requirement, we report the site-specific incorporation of noncanonical amino acids (ncAAs) with a small isocyanide (or isonitrile) group into proteins in both bacterial and mammalian cells. We showed that under physiological conditions and in the absence of a catalyst these isocyanide-containing ncAAs could react selectively with tetrazine molecules via [4 + 1]-cycloaddition, thus providing a versatile bioorthogonal handle for site-specific protein labeling and protein decaging. Significantly, these bioorthogonal reactions between isocyanides and tetrazines also provide a unique mechanism for the activation of tetrazine-quenched fluorophores. The addition of these isocyanide-containing ncAAs to the list of 20 commonly used, naturally occurring amino acids expands our repertoire of reagents for bioorthogonal chemistry, therefore enabling new biological applications ranging from protein labeling and imaging studies to the chemical activation of proteins.
Graphical Abstract
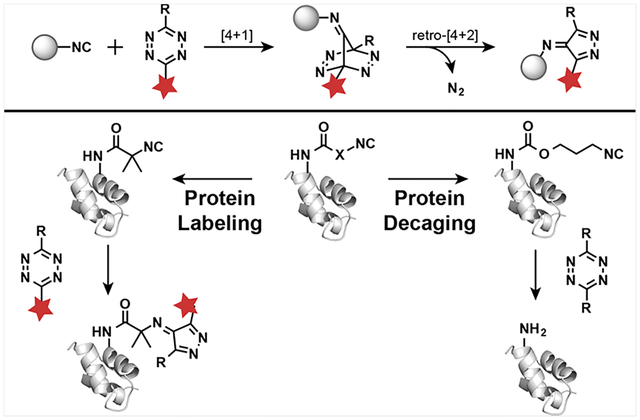
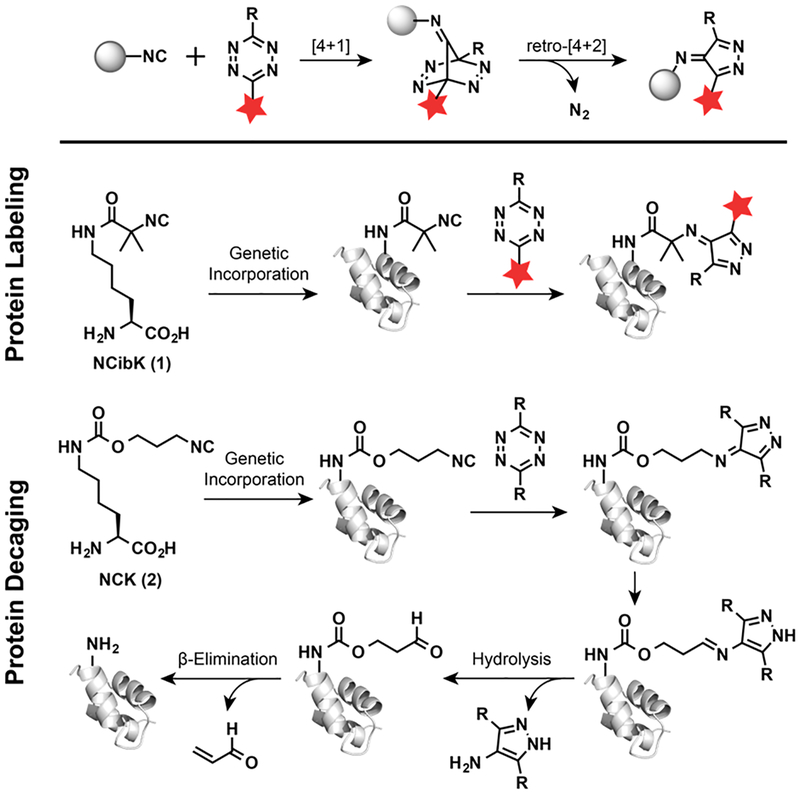
The development of bioorthogonal reactions and strategies for introducing “handles” into polypeptides has transformed our ability to study and manipulate proteins.1–8 There have been many improvements in the speed and selectivity of bioorthogonal reactions in the past several decades.9–12 Recent advances in bioorthogonal chemistry have focused on tetrazine-based inverse electron demand Diels-Alder (IEDDA) cycloaddition reactions due to their rapid reaction rates, high selectivity, and product stability.9,13–17 These reactions are particularly useful for precision protein labeling, decaging, and cellular protein imaging.13,14,17–24 For example, tetrazine-containing amino acids have been genetically incorporated into proteins, allowing for a rapid bioorthogonal conjugation with trans-cyclooctene (TCO)-labeled biomolecules.13,17 Taking advantage of the high reaction rate and good biocompatibility of IEDDA reactions, Chen and others have redirected this reaction for site-specific protein decaging.18–22 Proteins, including kinases and luciferase, have been caged by TCO-labeled lysine, followed by activation upon reaction with tetrazines.18,20 Besides acting as an IEDDA reaction partner, tetrazine can absorb visible light at around 500–525 nm, which makes it an ideal quencher toward a series of fluorophores. Based on this dual functionality of tetrazine, IEDDA reaction was also employed to design a variety of fluorogenic fluorophores.25–28 Despite the broad application of the IEDDA reactions, most dienophiles used in tetrazine-based cycloaddition reactions are strained hydrophobic alkenes or alkynes, such as TCO, cyclooctyne, norbornene, cyclobutene, cyclopropene, or spirohexene.13,15,16,29,30 In this report, we incorporated noncanonical amino acids (ncAAs) containing small and hydrophilic isocyano (or isonitrile) groups into proteins in both bacterial and mammalian cells using genetic code expansion technology. Site-specific protein labeling was then achieved via a [4 + 1]-cycloaddition reaction with various types of functional tetrazine moieties.
Isocyanide is a popular reactive group due to the presence of a resonance structure containing a negative carbanion and a positive nitrogen ion. This enables isocyanide to serve as either a nucleophile or an electrophile in chemical reactions. Due to their unique reactivity, isocyanides have been used as important building blocks in organic syntheses, especially in multicomponent reactions, such as Ugi reaction and Passerini reaction.31–33 Because of their small size, stability, and lack of toxicity, isocyanides also have attracted interest in several biological applications.34,35 In an early effort, Ziegler and co-workers demonstrated that isocyanide compounds can undergo Ugi reactions to functionalize the carboxyl and amino groups of native proteins under physiological conditions.36 For improved labeling selectivity of isocyano moieties, bioorthogonal [4 + 1]-cycloaddition reactions between isocyanides and tetrazines are of increasing interest (Figure 1).37 Leeper and co-workers have developed isocyano derivatives of galactosamine and mannosamine for metabolic glycan labeling.38,39 The incorporated isocyano glycans can subsequently be derivatized with tetrazine-labeled fluorophores to enable visualization of cancer cell glycoproteins. Most recently, Franzini and co-workers used 3-isocyanopropyl (ICPr) moieties as hydroxy masking groups for anticancer drugs and bioimaging reagents with no reported toxicity in vitro and in vivo.40 Upon addition of decaging tetrazines, ICPr undergoes a [4 + 1]-cycloaddition reaction, releasing N2 to yield a pyrazoleimine intermediate, followed by hydrolysis to yield a 3-oxopropyl moiety.41 β-Elimination of the resulting 3-oxopropyl-caged compound generates active biomolecules with hydroxyl groups.42 During the preparation of this manuscript, a tert-butyl isocyano group was also reported to react with bulky tetrazine for protein labeling, which has been used to achieve bioorthogonal labeling of three different proteins at same time, together with azide-dibenzocyclooctyne (DBCO) and less hindered tetrazine-TCO pairs.43 Despite the potential importance of isocyanides for modulating and studying biological functions, methods for site-specific incorporation of isocyano groups into proteins have not yet become available.
Figure 1.
Genetic incorporation of ε-N-2-isocyanoisobutyryl-lysine (NCibK, 1) and ε-N-isocyano-lysine (NCK, 2) for protein labeling and decaging, respectively.
RESULTS
Design and Synthesis of Isocyanide-Containing Amino Acids.
To genetically introduce isocyanide into protein, we designed a lysine amino acid analog containing isocyanide, capable of generating stable conjugation products following reaction with tetrazines. To prevent tautomerization and hydrolysis after the [4 + 1]-cycloaddition reaction with tetrazines, a tert-butyl isocyano group was conjugated to the lysine amino acid to yield ε-N-2-isocyanoisobutyryl-lysine (NCibK, 1, Figure 1). In addition, we also designed another analogue containing a 3-isocyanopropyl-1-carbamoyl (ICPrc) group, ε-N-isocyano-lysine (NCK, 2, Figure 1). We hypothesized that NCK could be chemically decaged by tetrazines via a [4 + 1]-cycloaddition reaction to yield a wild-type lysine residue, thus providing a new bioorthogonal cleavage mechanism for protein activation. We envision that the site-specific introduction of the NCibK or NCK ncAAs into proteins will enable precision protein labeling and chemical activation, respectively.
For the synthesis of NCK and NCibK ncAAs, isocyanide moieties were prepared by dehydrating N-formamide with phosphorus oxychloride. NCK was prepared by coupling the p-nitrophenyl carbonate-functionalized isocyanide to lysine with a 68% yield (Supporting Information). Instead of forming the isocyanide prior to coupling to lysine, NCibK was prepared by first incorporating an N-formyl aminoisobutyryl group into lysine, followed by converting this moiety to an isocyanoisobutyryl group in the presence of phosphorus oxychloride.
Bioorthogonal Reaction between NCibK and Tetrazines.
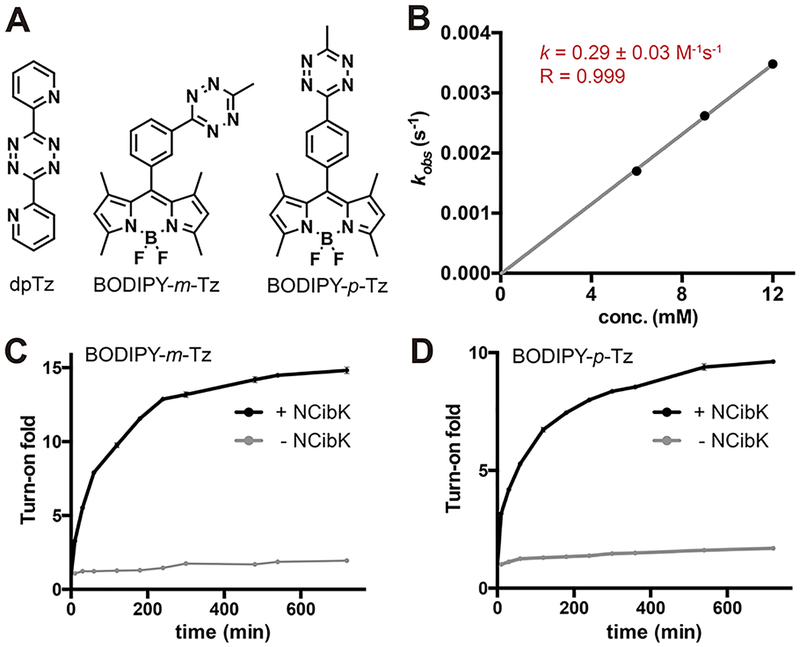
To determine tetrazine reactivity with the isocyano group, the rate constant for the reaction between NCibK and 3,6-di-2-pyridyl-1,2,4,5-tetrazine (dpTz) in phosphate-buffered saline (PBS) at 25 °C was obtained by using UV-vis spectroscopy (Figure 2A). By following the increase in pyrazole product absorbance at 430 nm upon reaction of NCibK with a 10–20-fold excess of dpTz, we determined the rate constant as 0.29 ± 0.03 M−1 s−1, which is comparable to that for the [3 + 2]-cycloaddition reaction between dibenzocyclooctyne (DIBO) and benzyl azide (0.057 ± 0.001 M−1 s−1, Figure 2B).44
Figure 2.
Characterization of reactions between NCibK and tetrazines. (A) Structures of tetrazine derivatives used in this study.(B) Kinetics of NCibK with dpTz resulted in a rate constant of k =0.29 ± 0.03 M−1 s−1. (C) Fluorogenic turn-on reaction between NCibK and BODIPY-m-Tz. (D) Fluorogenic turn-on reaction between NCibK and BODIPY-p-Tz.
Tetrazine-fluorophore conjugates, including BODIPY-tetrazine (Tz), are a unique class of turn-on probes in which the tetrazine moiety serves as both a fluorescence quencher and a bioorthogonal handle.25,28,45–48 Upon reaction with dienophiles, tetrazine is no longer able to quench the fluorophore, resulting in enhanced fluorescence of fluorophore conjugates. To examine the ability of isocyanides to activate the fluorescence of tetrazine-caged fluorophores, we incubated different BODIPY-Tz conjugates with NCibK. We found that the reaction between NCibK and BODIPY-m-Tz, the best-known turn-on probe for TCO, resulted in a 14.8-fold increase in fluorescence (Figure 2C).25 The reaction of NCibK with BODIPY-p-Tz, a probe with lower activation efficiency for TCO, yielded a 9.6-fold activation (Figure 2D). Due to their good signal-to-noise ratios, we anticipate that these isocyanide-mediated fluorogenic activations, like those between strained dienophiles and tetrazine-fluorophore conjugates, will result in numerous applications in fluorescence imaging.
Genetic Incorporation of Isocyanide-Containing Amino Acids in E. coli.
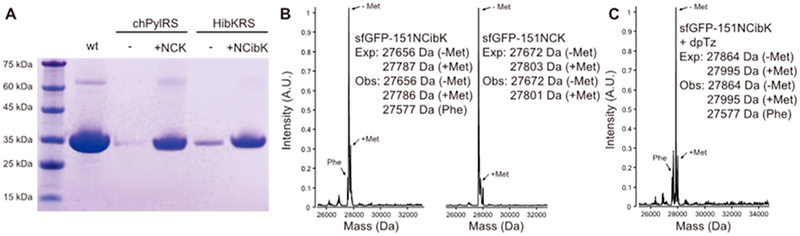
To genetically incorporate NCibK and NCK into proteins, the pyrrolysyl-tRNA synthetase (PylRS)/ pair, which typically encodes ncAAs bearing a lysine moiety and the ε-N-carbonyl group of pyrrolysine, was used.2,3,49–51 We screened a collection of Methanosarcina barkeri PylRS mutants that are able to genetically incorporate ncAAs with structures similar to NCibK and NCK.14,52–59 The activities of these PylRS mutants were determined based on their ability to incorporate NCibK or NCK into superfolder green fluorescence protein modified with a C-terminal hexahistidine-tag (sfGFP, encoded on plasmid pET22b-T5-sfGFP*) and containing an amber codon at position 151. Escherichia coli BL21 (DE3) cells containing the pET22b-T5-sfGFP* plasmid were independently transformed with each of the PylRS mutants, encoded on plasmid pUltra-PylRS.60,61 A quantitative fluorescence assay was carried out in LB medium in the presence or absence of 1 mM NCibK or NCK. We identified a PylRS mutant (HibK-1) containing two amino acid substitutions (Cys313Thr and Tyr349Phe) that exhibited an increase in fluorescence in the presence of NCibK, suggesting successful expression of full-length NCibK-containing sfGFP (Figure S1).57 Several PylRS mutants were found to be capable of NCK incorporation, based on observed increases in fluorescence of sfGFP. By comparison, a chimeric PylRS mutant exhibited the largest increases in sfGFP expression level (Figure S1). The chimeric PylRS-IPYE mutant (chPylRS-IPYE), comprised of residues 1–149 of M. barkeri PylRS, residues 185–454 of Methanosarcina mazei PylRS, and four mutations (Val31Ile, Thr56Pro, His62Tyr, and Ala100Glu), was obtained via phage-assisted continuous evolution for improved enzymatic efficiency without loss of substrate specificity.59 To investigate the efficiency and fidelity of NCibK and NCK incorporation, HibK-1 and chPylRS-IPYE mutants were used to express sfGFP in LB medium in the presence or absence of 1 mM NCibK and NCK, respectively. SDS-PAGE analysis of Ni-NTA purified sfGFP mutants revealed that full-length sfGFPs were expressed only for the two combinations of HibK-1 enzyme plus 1 mM NCibK or chPylRS-IPYE enzyme in the presence of 1 mM NCK (Figure 3A). Yields of sfGFP mutants substituted with NCibK and NCK and expressed in LB medium were 8.46 and 10.93 mg L−1, respectively, comparing with 90 mg L−1 for wild-type sfGFP. ESI-MS analysis confirmed the site-specific incorporation of NCibK and NCK into GFP (Figure 3B). A minor peak corresponding to the misincorporation of phenylalanine at 151 position of GFP was observed for the purified sfGFP-151NCibK. Thus, the expression of sfGFP-151NCibK was also carried out in M9-glucose minimal medium to obtain a homogeneous protein (Figure S2).
Figure 3.
Incorporation of NCK and NCibK into sfGFP proteins in E. coli. (A) Expression of sfGFP mutants in the presence or absence (−) of 1 mM NCK or NCibK, analyzed by SDS-PAGE. (B) ESI-MS analysis of sfGFP mutants containing NCibK or NCK. Two desired peaks were observed for each sample. One is the full-length sfGFP mutant, and the other one is the full-length sfGFP mutant without N-terminal Met. The 27577 Da peak corresponds to the misincorporation of Phe at 151 position of sfGFP. (C) ESI-MS analysis of NCibK-containing sfGFP mutant reacted with dpTz. The 27995 and 27864 Da peaks correspond to the conjugated products with and without N-terminal Met, respectively.
Site-Specific Labeling of Proteins with NCibK.
To demonstrate that a tetrazine-functionalized probe can site-specifically react with isocyano groups genetically inserted into proteins, we reacted purified NCibK-containing sfGFP with 1000 equiv of dpTz in PBS buffer at RT. ESI-MS analysis confirmed the quantitative labeling of NCibK-containing sfGFP with the tetrazine moiety after 2 h (Figure 3C). Furthermore, no hydrolysis of the conjugated product was detected by ESI-MS after storage in PBS buffer for one month, suggesting good stability of the NCibK-tetrazine conjugate under physiological conditions (Figure S3). To verify that our isocyanide-based strategy represents a general approach to protein labeling, we substituted NCibK for the Lys99 residue of myoglobin expressed in E. coli. This was accomplished using a pBad-myo-K99* plasmid containing a pBad promoter-driven myoglobin gene with an amber codon at a permissive site (Lys99TAG).62 Myoglobin expression was carried out using E. coli BL21 (DE3) cells cotransformed with the plasmids pBadmyo-K99* and pUltra-HibK-1 in 2×YT medium. SDS-PAGE analysis revealed that full-length myoglobin was expressed only in the presence of 1 mM NCibK (Figure S4). ESI-MS analysis of myoglobin containing the Lys99NCibK substitution revealed an observed mass of 18449 Da, in agreement with the calculated mass (Figure S5). To demonstrate that NCibK-containing myoglobin can site-specifically react with tetrazine moieties, we incubated myoglobin containing the Lys99NCibK with 1000 equiv of dpTz in PBS buffer (pH 7.8) at RT. ESIMS analysis confirmed the successful labeling of NCibK-containing myoglobin with the dpTz within 2 h (Figure S5C). Next, we demonstrated that NCibK-containing myoglobin could be site-specifically labeled with BODIPY-p-Tz. SDS-PAGE and in-gel fluorescence analyses confirmed that only myoglobin containing NCibK was labeled with BODIPY-p-Tz; that is, no labeling was detected in wild-type myoglobin (Figure S4).
Genetic Incorporation of Isocyanide-Containing Amino Acids in Mammalian Cells.
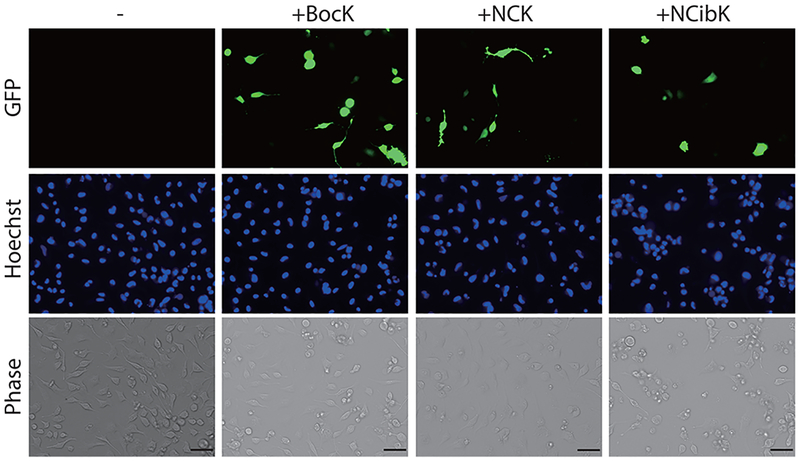
PylRS/tRNAPylCUA pairs evolved in E. coli have been previously shown to be functional in mammalian cells.53,57,60 To evaluate the incorporation of NCK and NCibK into proteins in mammalian cells, we constructed a plasmid (pAcBac1-chPylRS) containing four copies of U6 promoter-driven M. barkeri pyrrolysyltRNACUA, as well as a chPylRS gene expressed from a CMV promoter.63–65 To determine the incorporation efficiency of NCK and NCibK into mammalian proteins, the pAcBac1-chPylRS plasmid was co-transfected into HeLa cells along with the pAcBac1-Pyl-EGFP* plasmid harboring an EGFP gene with an amber codon at a permissive site (Tyr39). To judge the relative incorporation efficiency, ε-N-Boc-lysine (BocK), the best-known substrate for chPylRS, was used as a positive control. Thirty-six hours after transfection, increased levels of EGFP fluorescence were observed only in cultures containing one of the three ncAAs, suggesting that, like BocK, NCK and NCibK can be genetically incorporated in site-specific fashion into EGFP in mammalian cells (Figure 4 and S6).
Figure 4.
Incorporation of BocK, NCK, and NCibK into EGFPY40TAG in mammalian cells analyzed by fluorescence microscopy in the presence or absence (−) of ncAAs. Scale bar = 50 μm.
Fluorescence microscopy also revealed that NCK, in particular, was incorporated without obvious toxicity at a level similar to that of BocK incorporation (Figure 4 and S7).
Tetrazine-Mediated Activation of Isocyano-Caged Proteins.
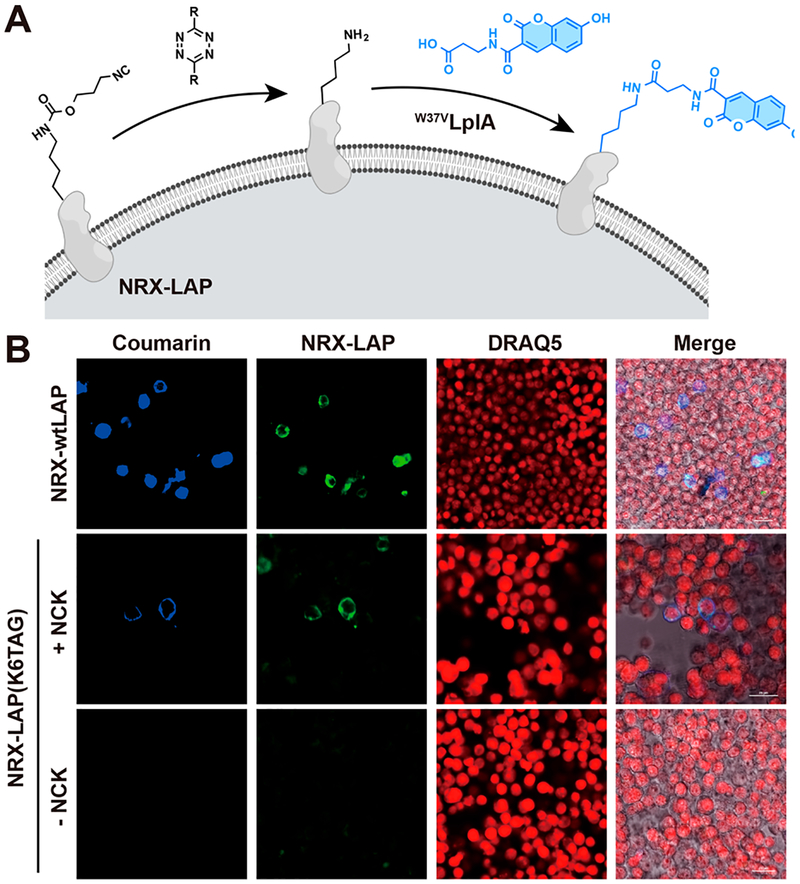
With NCK incorporation systems in hand for both bacterial and mammalian cells, we investigated if the isocyano group can be used as a reversible caging tool for protein activation in vitro as well as in living cells. A sfGFP mutant, containing NCK at residue 151, was purified from E. coli and used as a model protein. To determine whether NCK can be decaged to lysine via [4 + 1]-cycloaddition followed by a β-elimination reaction, NCK-containing sfGFP was incubated with 1 mM dpTz in PBS buffer. ESI-MS analysis of the NCK-containing sfGFP before and after dpTz-mediated decaging revealed a 112 Da shift of the main peak, corresponding to removal of the isocyano caging group (Figure S8). In addition to the major elimination products, we also observed reaction intermediates involved in the decaging process (Figure S8). To demonstrate if this isocyanide and tetrazine-mediated decaging strategy is also effective in mammalian cells, we site-specifically incorporated NCK into a lipoic acid ligase-acceptor peptide (LAP) tag. This tag is used in studies of probe incorporation mediated by enzymes (PRIME, Figure 5A).66,67 The LAP tag is a 13-amino acid peptide in which a critical lysine residue is able to covalently link to carboxylic acid residues in the presence of lipoic acid ligase (LplA) and adenosine triphosphate (ATP).
Figure 5.
Tetrazine-mediated activation of isocyano-caged lipoic acid ligase-acceptor peptide (LAP) tag in living cells. (A) The reaction scheme for tetrazine-mediated activation of isocyano-caged proteins. NCK was site-specifically incorporated into the LAP-tagged neurexin1β (NRX-LAP) to block the critical lysine residue. The NCK-modified LAP tag was then decaged by tetrazine treatment, followed by reaction with coumarin fluorophore catalyzed by lipoic acid ligase (LplA). (B) HEK293T cells expressing NRX-LAP or NRXLAP-TAG with or without NCK were mixed with 1 mM dpTz in PBS for 20 min, followed by the incubation with 1 mM pyrrolidine for 8 h. After PRIME labeling (blue) and cell fixation, 0.1 μg/mL anti-Myc antibody and 7.5 μg/mL FITC-conjugated secondary antibody (green) were added to check the expression level of NRX-LAP. Nucleus staining was carried out with 5 μM DRAQ5 (red) before confocal imaging. Scale bars = 25 μm.
To demonstrate that NCK can be used to cage the lysine residue present in the LAP tag, we chose LAP-tagged neurexin1β, a synaptic adhesion protein that reacts with carboxylic acid-functionalized coumarin to enable neurexin1β imaging in live cells.68 We found that human embryonic kidney (HEK) 293T cells expressing wild-type LAP-tagged neurexin1β could be stained with coumarin fluorophore in the presence of 10 μM LplA and 1 mM ATP (Figure 5B). Since coumarin labeling of LAP-tagged neurexin1β depends on the critical Lys6 residue within the LAP tag, we generated LAP-tagged neurexin1β-Lys6NCK by substituting NCK at this lysine residue to block LAP conjugation activity. As expected, labeling with coumarin was only observed for cells expressing LAP-tagged neurexin1β-Lys6TAG in the presence of both 1 mM NCK and dpTz for decaging (Figure 5B and S9). Removal of either NCK in the media or the dpTz-mediated decaging step will lead to no significant coumarin fluorescence, demonstrating that isocyano-caged lysine can be chemically restored to lysine by tetrazine treatment in living cells.
DISCUSSION
In summary, we have accomplished the genetic incorporation of isocyanide-containing ncAAs into proteins in both bacterial and mammalian cells. Furthermore, we have demonstrated the utility of this methodology for bioorthogonal protein labeling and bioorthogonal protein decaging via a [4 + 1]-cycloaddition reaction with tetrazine probes. Compared to currently used bioorthogonal reactivating partners for tetrazines, the isocyano group has a smaller size, along with better stability and water-solubility, thus minimizing the disruption of protein structures after incorporation. Furthermore, isocyanide-tetrazine [4 + 1] cycloaddition reaction is compatible with the strain-promoted azide alkyne cycloaddition reactions. Thus, the addition of isocyanide to genetic code will enable the multifunctionalization of proteins in a site-specific manner. The ease of synthesis, versatility of reactivity, and biocompatibility of isocyanides render them as attractive bioorthogonal handles for biological and biomedical applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. P. G. Schultz for kindly providing the plasmids pET22b-Tf-sfGFP151TAG and pBad-myo99TAG, and Prof. A. Y. Ting for kindly providing the plasmids pNICE-LAP-neurexin-1beta, pNICE-LAP-neurexin-1beta-TAG, and pYFJ16-Lpla(W37V). This work was supported by the Cancer Prevention Research Institute of Texas (CPRIT RR170014), NIH (R35-GM133706), the Robert A. Welch Foundation (C-1970), the John S. Dunn Foundation Collaborative Research Award, and the Hamill Innovation Award.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.9b00678.
Additional experimental methods, screening of PylRS mutants for NCibK and NCK incorporation, ESI-MS analysis of isolated sfGFP-NCibK, stability of sfGFPNCibK conjugate with dpTz, incorporation of NCibK into myoglobin, incorporation of NCK into EGFP, incorporation of BocK or NCK into EGFP-Y40TAG, time-dependent study of tetrazine-mediated sfGFP-NCK decaging, and tetrazine-mediated LAP tag activation in living cells (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Prescher JA, and Bertozzi CR (2005) Chemistry in Living Systems. Nat. Chem. Biol 1, 13. [DOI] [PubMed] [Google Scholar]
- (2).Liu CC, and Schultz PG (2010) Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem 79, 413–444. [DOI] [PubMed] [Google Scholar]
- (3).Xiao H, and Schultz PG (2016) At the Interface of Chemical and Biological Synthesis: An Expanded Genetic Code. Cold Spring Harbor Perspect. Biol 8, No. a023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lang K, and Chin JW (2014) Cellular Incorporation of Unnatural Amino Acids and Bioorthogonal Labeling of Proteins. Chem. Rev 114, 4764–4806. [DOI] [PubMed] [Google Scholar]
- (5).Jing C, and Cornish VW (2011) Chemical Tags for Labeling Proteins Inside Living Cells. Acc. Chem. Res 44, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang L (2017) Genetically Encoding New Bioreactivity. New Biotechnol. 38, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Chin JW (2017) Expanding and Reprogramming the Genetic Code. Nature 550, 53–60. [DOI] [PubMed] [Google Scholar]
- (8).Wang Q, Parrish AR, and Wang L (2009) Expanding the Genetic Code for Biological Studies. Chem. Biol 16, 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Sletten EM, and Bertozzi CR (2009) Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem., Int. Ed 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Patterson DM, Nazarova LA, and Prescher JA (2014) Finding the Right (Bioorthogonal) Chemistry. ACS Chem. Biol 9, 592–605. [DOI] [PubMed] [Google Scholar]
- (11).Shih H-W, Kamber DN, and Prescher JA (2014) Building Better Bioorthogonal Reactions. Curr. Opin. Chem. Biol 21, 103–111. [DOI] [PubMed] [Google Scholar]
- (12).Lim RKV, and Lin Q (2010) Bioorthogonal Chemistry: Recent Progress and Future Directions. Chem. Commun. (Cambridge,U. K.) 46, 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Seitchik JL, Peeler JC, Taylor MT, Blackman ML, Rhoads TW, Cooley RB, Refakis C, Fox JM, and Mehl RA (2012) Genetically Encoded Tetrazine Amino Acid Directs Rapid Site-Specific in Vivo Bioorthogonal Ligation with Trans-Cyclooctenes. J. Am. Chem. Soc 134, 2898–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, and Chin JW (2012) Genetic Encoding of Bicyclononynes and Trans -Cyclooctenes for Site-Specific Protein Labeling in Vitro and in Live Mammalian Cells via Rapid Fluorogenic Diels-Alder Reactions. J. Am. Chem. Soc 134, 10317–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, and Chin JW (2012) Genetically Encoded Norbornene Directs Site-Specific Cellular Protein Labelling via a Rapid Bioorthogonal Reaction. Nat. Chem 4, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ramil CP, Dong M, An P, Lewandowski TM, Yu Z, Miller LJ, and Lin Q (2017) Spirohexene-Tetrazine Ligation Enables Bioorthogonal Labeling of Class B G Protein-Coupled Receptors in Live Cells. J. Am. Chem. Soc 139, 13376–13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Blizzard RJ, Backus DR, Brown W, Bazewicz CG, Li Y, and Mehl RA (2015) Ideal Bioorthogonal Reactions Using A Site-Specifically Encoded Tetrazine Amino Acid. J. Am. Chem. Soc 137, 10044–10047. [DOI] [PubMed] [Google Scholar]
- (18).Liu L, Liu Y, Zhang G, Ge Y, Fan X, Lin F, Wang J, Zheng H, Xie X, Zeng X, and Chen PR (2018) Genetically Encoded Chemical Decaging in Living Bacteria. Biochemistry 57, 446–450. [DOI] [PubMed] [Google Scholar]
- (19).Fan X, Ge Y, Lin F, Yang Y, Zhang G, Ngai WSC, Lin Z, Zheng S, Wang J, Zhao J, Li J, and Chen PR (2016) Optimized Tetrazine Derivatives for Rapid Bioorthogonal Decaging in Living Cells. Angew. Chem., Int. Ed 55, 14046–14050. [DOI] [PubMed] [Google Scholar]
- (20).Zhang G, Li J, Xie R, Fan X, Liu Y, Zheng S, Ge Y, and Chen PR (2016) Bioorthogonal Chemical Activation of Kinases in Living Systems. ACS Cent. Sci 2, 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Li J, Jia S, and Chen PR (2014) Diels-Alder Reaction-Triggered Bioorthogonal Protein Decaging in Living Cells. Nat. Chem. Biol 10, 1003–1005. [DOI] [PubMed] [Google Scholar]
- (22).Zhao J, Liu Y, Lin F, Wang W, Yang S, Ge Y, and Chen PR (2019) Bioorthogonal Engineering of Bacterial Effectors for Spatial-Temporal Modulation of Cell Signaling. ACS Cent. Sci 5, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Liu DS, Tangpeerachaikul A, Selvaraj R, Taylor MT, Fox JM, and Ting AY (2012) Diels-Alder Cycloaddition for Fluorophore Targeting to Specific Proteins inside Living Cells. J. Am. Chem. Soc 134, 792–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Uttamapinant C, Howe JD, Lang K, Beránek V, Davis L, Mahesh M, Barry NP, and Chin JW (2015) Genetic Code Expansion Enables Live-Cell and Super-Resolution Imaging of Site-Specifically Labeled Cellular Proteins. J. Am. Chem. Soc 137, 4602–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Carlson JCT, Meimetis LG, Hilderbrand SA, and Weissleder R (2013) BODIPY-Tetrazine Derivatives as Superbright Bioorthogonal Turn-on Probes. Angew. Chem., Int. Ed 52, 6917–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Meimetis LG, Carlson JCT, Giedt RJ, Kohler RH, and Weissleder R (2014) Ultrafluorogenic Coumarin-Tetrazine Probes for Real-Time Biological Imaging. Angew. Chem., Int. Ed 53, 7531–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lee Y, Cho W, Sung J, Kim E, and Park SB (2018) Monochromophoric Design Strategy for Tetrazine-Based Colorful Bioorthogonal Probes with a Single Fluorescent Core Skeleton. J. Am. Chem. Soc 140, 974–983. [DOI] [PubMed] [Google Scholar]
- (28).Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, and Weissleder R (2010) Bioorthogonal Turn-On Probes for Imaging Small Molecules Inside Living Cells. Angew. Chem., Int. Ed 49, 2869–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Devaraj NK, Thurber GM, Keliher EJ, Marinelli B, and Weissleder R (2012) Reactive Polymer Enables Efficient in Vivo Bioorthogonal Chemistry. Proc. Natl. Acad. Sci. U. S. A 109, 4762–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liu K, Enns B, Evans B, Wang N, Shang X, Sittiwong W, Dussault PH, and Guo J (2017) A Genetically Encoded Cyclobutene Probe for Labelling of Live Cells. Chem. Commun. (Cambridge, U. K.) 53, 10604–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Doemling A (2006) Recent Developments in Isocyanide-Based Multicomponent Reactions in Applied Chemistry. Chem. Rev 106, 17–89. [DOI] [PubMed] [Google Scholar]
- (32).Kaur T, Wadhwa P, Bagchi S, and Sharma A (2016) Isocyanide Based [4 + 1] Cycloaddition Reactions: An Indispensable Tool in Multi-Component Reactions (MCRs). Chem. Commun. (Cambridge, U. K.) 52, 6958–6976. [DOI] [PubMed] [Google Scholar]
- (33).Giustiniano M, Basso A, Mercalli V, Massarotti A, Novellino E, Tron GC, and Zhu J (2017) To Each His Own: Isonitriles for All Flavors. Functionalized Isocyanides as Valuable Tools in Organic Synthesis. Chem. Soc. Rev 46, 1295–1357. [DOI] [PubMed] [Google Scholar]
- (34).Fusetani N (2004) Biofouling and Antifouling. Nat. Prod. Rep 21, 94–104. [DOI] [PubMed] [Google Scholar]
- (35).Garson MJ, and Simpson JS (2004) Marine Isocyanides and Related Natural Products–Structure, Biosynthesis and Ecology. Nat. Prod. Rep 21, 164–179. [DOI] [PubMed] [Google Scholar]
- (36).Ziegler T, Gerling S, and Lang M (2000) Preparation of Bioconjugates through an Ugi Reaction This Work Was Financially Supported by the Fonds Der Chemischen Industrie and Aventis Research & Technologies, Frankfurt Am Main. Angew. Chem., Int. Ed 39, 2109–2112. [DOI] [PubMed] [Google Scholar]
- (37).Stöckmann H, Neves AA, Stairs S, Brindle KM, and Leeper FJ (2011) Exploring Isonitrile-Based Click Chemistry for Ligation with Biomolecules. Org. Biomol. Chem 9, 7303–7305. [DOI] [PubMed] [Google Scholar]
- (38).Stairs S, Neves AA, Stöckmann H, Wainman YA, Ireland-Zecchini H, Brindle KM, and Leeper FJ (2013) Metabolic Glycan Imaging by Isonitrile-Tetrazine Click Chemistry. ChemBioChem 14, 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wainman YA, Neves AA, Stairs S, Stöckmann H, Ireland-Zecchini H, Brindle KM, and Leeper FJ (2013) Dual-Sugar Imaging Using Isonitrile and Azido-Based Click Chemistries. Org. Biomol. Chem 11, 7297–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tu J, Xu M, Parvez S, Peterson RT, and Franzini RM (2018) Bioorthogonal Removal of 3-Isocyanopropyl Groups Enables the Controlled Release of Fluorophores and Drugs in Vivo. J. Am. Chem. Soc 140, 8410–8414. [DOI] [PubMed] [Google Scholar]
- (41).Imming P, Mohr R, Müller E, Overheu W, and Seitz G (1982) [4 + 1]Cycloaddition of Isocyanides to 1,2,4,5-Tetrazines: A Novel Synthesis of Pyrazole. Angew. Chem., Int. Ed. Engl 21, 284–284. [Google Scholar]
- (42).Leslie AK, Li D, and Koide K (2011) Amine-Promoted β-Elimination of a β-Aryloxy Aldehyde for Fluorogenic Chemodosimeters. J. Org. Chem 76, 6860–6865. [DOI] [PubMed] [Google Scholar]
- (43).Tu J, Svatunek D, Parvez S, Liu AC, Levandowski BJ, Eckvahl HJ, Peterson RT, Houk KN, and Franzini RM (2019) Stable, Reactive, and Orthogonal Tetrazines: Dispersion Forces Promote the Cycloaddition with Isonitriles. Angew. Chem., Int. Ed 58, 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ning X, Guo J, Wolfert MA, and Boons G-J (2008) Visualizing Metabolically Labeled Glycoconjugates of Living Cells by Copper-Free and Fast Huisgen Cycloadditions. Angew. Chem., Int. Ed 47, 2253–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wieczorek A, Werther P, Euchner J, and Wombacher R (2017) Green- to Far-Red-Emitting Fluorogenic Tetrazine Probes − Synthetic Access and No-Wash Protein Imaging inside Living Cells. Chem. Sci 8, 1506–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).de Moliner F, Kielland N, Lavilla R, and Vendrell M (2017) Modern Synthetic Avenues for the Preparation of Functional Fluorophores. Angew. Chem., Int. Ed 56, 3758–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Brown SP, and Smith AB (2015) Peptide/Protein Stapling and Unstapling: Introduction of s -Tetrazine, Photochemical Release, and Regeneration of the Peptide/Protein. J. Am. Chem. Soc 137, 4034–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Tucker MJ, Courter JR, Chen J, Atasoylu O, Smith AB, and Hochstrasser RM (2010) Tetrazine Phototriggers: Probes for Peptide Dynamics. Angew. Chem., Int. Ed 49, 3612–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Wang L, Xie J, and Schultz PG (2006) Expanding the Genetic Code. Annu. Rev. Biophys. Biomol. Struct 35, 225–249. [DOI] [PubMed] [Google Scholar]
- (50).Chin JW (2014) Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu. Rev. Biochem 83, 379–408. [DOI] [PubMed] [Google Scholar]
- (51).Wan W, Tharp JM, and Liu WR (2014) Pyrrolysyl-TRNA Synthetase: An Ordinary Enzyme but an Outstanding Genetic Code Expansion Tool. Biochim. Biophys. Acta, Proteins Proteomics 1844, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Chatterjee A, Sun SB, Furman JL, Xiao H, and Schultz PG (2013) A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia Coli. Biochemistry 52, 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kim CH, Kang M, Kim HJ, Chatterjee A, and Schultz PG (2012) Site-Specific Incorporation of ε-N-Crotonyllysine into Histones. Angew. Chem., Int. Ed 51, 7246–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Ai H, Shen W, Sagi A, Chen PR, and Schultz PG (2011) Probing Protein-Protein Interactions with a Genetically Encoded Photo-Crosslinking Amino Acid. ChemBioChem 12, 1854–1857. [DOI] [PubMed] [Google Scholar]
- (55).Nguyen DP, Elliott T, Holt M, Muir TW, and Chin JW (2011) Genetically Encoded 1,2-Aminothiols Facilitate Rapid and Site-Specific Protein Labeling via a Bio-Orthogonal Cyanobenzothiazole Condensation. J. Am. Chem. Soc 133, 11418–11421. [DOI] [PubMed] [Google Scholar]
- (56).Furman JL, Kang M, Choi S, Cao Y, Wold ED, Sun SB, Smider VV, Schultz PG, and Kim CH (2014) A Genetically Encoded Aza-Michael Acceptor for Covalent Cross-Linking of Protein-Receptor Complexes. J. Am. Chem. Soc 136, 8411–8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Xiao H, Xuan W, Shao S, Liu T, and Schultz PG (2015) Genetic Incorporation of ε-N-2-Hydroxyisobutyryl-Lysine into Recombinant Histones. ACS Chem. Biol 10, 1599–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Elsässer SJ, Ernst RJ, Walker OS, and Chin JW (2016) Genetic Code Expansion in Stable Cell Lines Enables Encoded Chromatin Modification. Nat. Methods 13, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Bryson DI, Fan C, Guo L-T, Miller C, Söll D, and Liu DR (2017) Continuous Directed Evolution of Aminoacyl-TRNA Synthetases. Nat. Chem. Biol 13, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Xiao H, Peters FB, Yang P-Y, Reed S, Chittuluru JR, and Schultz PG (2014) Genetic Incorporation of Histidine Derivatives Using an Engineered Pyrrolysyl-TRNA Synthetase. ACS Chem. Biol 9, 1092–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Chen Y, Loredo A, Gordon A, Tang J, Yu C, Ordonez J, and Xiao H (2018) A Noncanonical Amino Acid-Based Relay System for Site-Specific Protein Labeling. Chem. Commun. (Cambridge, U. K.) 54, 7187–7190. [DOI] [PubMed] [Google Scholar]
- (62).Young TS, Ahmad I, Yin JA, and Schultz PG (2010) An Enhanced System for Unnatural Amino Acid Mutagenesis in E. Coli. J. Mol. Biol 395, 361–374. [DOI] [PubMed] [Google Scholar]
- (63).Xiao H, Chatterjee A, Choi S, Bajjuri KM, Sinha SC, and Schultz PG (2013) Genetic Incorporation of Multiple Unnatural Amino Acids into Proteins in Mammalian Cells. Angew. Chem., Int. Ed 52, 14080–14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Chatterjee A, Xiao H, Bollong M, Ai H-W, and Schultz PG (2013) Efficient Viral Delivery System for Unnatural Amino Acid Mutagenesis in Mammalian Cells. Proc. Natl. Acad. Sci. U. S. A 110, 11803–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee K-F, Slesinger PA, and Wang L (2007) Genetically Encoding Unnatural Amino Acids for Cellular and Neuronal Studies. Nat. Neurosci 10, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Uttamapinant C, White KA, Baruah H, Thompson S, Fernández-Suárez M, Puthenveetil S, and Ting AY (2010) A Fluorophore Ligase for Site-Specific Protein Labeling inside Living Cells. Proc. Natl. Acad. Sci. U. S. A 107, 10914–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Liu DS, Nivón LG, Richter F, Goldman PJ, Deerinck TJ, Yao JZ, Richardson D, Phipps WS, Ye AZ, Ellisman MH, et al. (2014) Computational Design of a Red Fluorophore Ligase for Site-Specific Protein Labeling in Living Cells. Proc. Natl. Acad. Sci. U. S. A 111, E4551–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Liu DS, Phipps WS, Loh KH, Howarth M, and Ting AY (2012) Quantum Dot Targeting with Lipoic Acid Ligase and HaloTag for Single-Molecule Imaging on Living Cells. ACS Nano 6, 11080–11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.