Abstract
Background
22q11.2 copy number variants (CNVs) are among the most highly penetrant genetic risk variants for developmental neuropsychiatric disorders such as schizophrenia (SCZ) and autism spectrum disorder (ASD). However, the specific mechanisms through which they confer risk remain unclear.
Methods
Using a functional genomics approach, we integrated transcriptomic data from the developing human brain, genome-wide association findings for SCZ and ASD, protein interaction data, and gene expression signatures from SCZ and ASD post-mortem cortex to: 1) organize genes into the developmental cellular and molecular systems within which they operate; 2) identify neurodevelopmental processes associated with polygenic risk for SCZ and ASD across the allelic frequency spectrum; and 3) elucidate pathways and individual genes through which 22q11.2 CNVs may confer risk for each disorder.
Results
Polygenic risk for SCZ and ASD converged on partially overlapping neurodevelopmental modules involved in synaptic function and transcriptional regulation, with ASD risk variants additionally enriched for modules involved in neuronal differentiation during fetal development. The 22q11.2 locus formed a large protein network during development that disproportionately affected SCZ- and ASD-associated neurodevelopmental modules, including loading highly onto synaptic and gene regulatory pathways. SEPT5, PI4KA, and SNAP29 genes are candidate drivers of 22q11.2 synaptic pathology relevant to SCZ and ASD, and DGCR8 and HIRA are candidate drivers of disease-relevant alterations in gene regulation.
Conclusions
The current approach offers a powerful framework to identify neurodevelopmental processes affected by diverse risk variants for SCZ and ASD, and elucidate the mechanisms through which highly penetrant multi-gene CNVs contribute to disease risk.
Keywords: 22q11.2 copy number variants, schizophrenia, autism spectrum disorder, genetic risk, functional genomics, brain development
Introduction
Copy number variants (CNVs) at chromosome 22q11.2 are among the genetic variants most strongly associated with developmental neuropsychiatric disorders. The hemizygous deletion, 22q11.2 deletion syndrome (22q11DS), typically spans a gene-rich ~2.5 Mb region(1) and is the greatest known single genetic risk factor for schizophrenia (SCZ). Approximately 25% of 22q11DS patients develop a psychotic disorder by adulthood(2); conversely, ~0.3% of patients with SCZ have 22q11DS(3,4). Interestingly, the reciprocal duplication (22q11DupS) may protect against SCZ(4,5), whereas both 22q11DS and 22q11DupS confer risk for autism spectrum disorder (ASD;6). As psychiatry increasingly looks towards precision medicine approaches to therapeutic development, clarifying the mechanisms through which 22q11.2 CNVs confer risk for SCZ and ASD, and how these converge with risk from broader variants associated with each disorder, is a critical goal.
Our knowledge of the genetic architectures of both SCZ and ASD has advanced considerably in the last decade. It is now clear that SCZ and ASD are both highly polygenic and genetically heterogeneous disorders, with risk variants for each disorder spanning common (i.e., present in >1% of the population), rare (i.e., present in <1% of the population), and de novo variants (i.e., new mutations in offspring), in addition to CNVs(7). Despite this polygenicity, risk variants for SCZ and ASD each appear to converge on a subset of biological pathways. Neuronal and synaptic genes have been repeatedly implicated in SCZ, including the N-methyl-D-aspartate receptor (NMDAR) and activity-regulated cytoskeleton-associated protein (Arc) complexes, and targets of the fragile X mental retardation protein (FMRP;4,8–12). Similarly, neuronal and synaptic genes have been implicated in ASD, along with chromatin and transcription regulators, and FMRP targets(6,13–17). However, many genes in these gene-sets are developmentally regulated, and SCZ and ASD are both neurodevelopmental in origin. Indeed, brain development is tightly controlled through changes in transcription factor activity and chromatin condensation that alter gene expression patterns at precise times in development(18). These changes program the progression of stem cell proliferation, neuronal differentiation, and synaptogenesis during prenatal development, and myelination and synaptic refinement during postnatal development. Examining the convergence of SCZ and ASD risk variants on groups of developmentally co-expressed genes may therefore yield fundamental insights into the neurodevelopmental pathogenesis of each disorder (e.g., 16,17), and provide a powerful lens to understand how 22q11.2 CNVs confer risk for each disorder.
Here, we used network analyses of transcriptomic data from the developing human brain(18) to organize genes into the developmental cellular and molecular systems within which they operate(19). We then conducted a comprehensive examination of neurodevelopmental processes associated with risk variants for SCZ and ASD across the allelic frequency spectrum. Finally, we identified the brain developmental 22q11.2 protein network, and developed a framework leveraging this disease-informed lens to predict pathways and individual genes underlying 22q11.2 CNV-mediated risk for SCZ and ASD.
Methods and Materials
Defining Neurodevelopmental Co-Expression Modules
Modules of developmentally co-expressed genes were constructed using weighted gene co-expression network analysis (WGCNA) of BrainSpan data, the largest, publicly-available developmental brain transcriptome dataset derived from healthy human donors(18). Samples spanning the full period of brain development (i.e., ~6 post-conception weeks-30 years; 1061 samples; Table S1) were included. Briefly, correlations were computed for the expression of each gene with every other gene, and genes were clustered based on connectivity to identify modules of co-expressed genes. Modules were functionally annotated using: 1) brain cell-type markers(20); 2) gene ontology (GO) using g:Profiler(21); and 3) genes expressed in specific brain regions and developmental periods using the Specific Expression Analysis tool(22). The first principal component of each module (i.e., module eigengene) was used to summarize the developmental trajectory of each module.
Identifying Neurodevelopmental Modules Associated with SCZ and ASD
Summary statistics, gene-lists, and/or variant lists from genome-wide association studies of SCZ and ASD were compiled and tested for module enrichment using methods appropriate to each data type (see Table 1). Briefly, when summary statistics for common(9,12,23) and rare variants(24) were available, they were tested for enrichment using MAGMA(25). De novo mutations (DNMs) in SCZ(10,26–33) and ASD(34,35) were tested for enrichment using DenovolyzeR(36). Genes in SCZ-(4) and ASD-associated(15) CNV loci were tested for enrichment using logistic regression, controlling for gene length. Genes containing more deleterious ultra-rare variants in SCZ patients than controls(11); manually curated high-confidence ASD genes according to the SFARI database(37); and genes associated with ASD across rare, de novo, and copy number variants(15) were tested for enrichment using Fisher’s Exact tests with the GeneOverlap package(38).
Table 1.
Genome-wide SCZ and ASD Risk Variant Datasets Tested for BrainSpan Module Enrichment.
| Dataset | Reference | Variant Type | Description | Patient N | Control N | Analysis |
|---|---|---|---|---|---|---|
| SCZ | ||||||
| SCZ PGC-deCODE* | 12 | Common | PGC + deCODE meta-analysis | 36,989 | 113,075 | MAGMA |
| SCZ CLOZUK-PGC* | 9 | Common | CLOZUK + PGC meta-analysis | 40,675 | 64,643 | MAGMA |
| SCZ Richards | 24 | Rare | Rare variants (MAF<1%) | 5,585 | 8,103 | MAGMA |
| SCZ Genovese | 11 | Ultra-rare | 43 genes enriched for deleterious ultra-rare variants (p<0.01) | 4,877 | 6,242 | Fisher’s Exact Test |
| SCZ DNM | 10, 26–33 | De novo | Compilation of DNMs | 1,139 | NA | DenovolyzeR |
| SCZ CNV | 4 | CNV | 13 unique loci (FDR<0.05) | 21,094 | 20,227 | Logistic Regression |
| ASD | ||||||
| ASD iPSYCH-PGC | 23 | Common | iPSYCH + PGC meta-analysis | 18,381 | 27,969 | MAGMA |
| ASD SFARI | www.sfari.org | Gene list | 80 literature-curated genes (score 1 or 2) | NA | NA | Fisher’s Exact Test |
| ASD Sanders | 15 | Rare | 65 genes (FDR<0.1) | 3,982 | 1,911 | Fisher’s Exact Test |
| ASD DNM | 34, 35 | De novo | Compilation of DNMs | 4,424 | NA | DenovolyzeR |
| ASD CNV | 15 | CNV | 9 unique loci, +3 nested loci, in SSC and AGP cohorts (FDR<0.05) | 4,687 | 2,100 | Logistic Regression |
The PGC-deCODE meta-analysis includes 1,513 SCZ cases and 36,989 controls from the deCODE study not in the CLOZUK-PGC study and the SCZ CLOZUK-PGC meta-analysis includes 5,220 SCZ cases and 18,823 controls not in the SCZ PGC-deCODE study.
Characterizing the 22q11.2 Locus
To examine whether any BrainSpan module was over-represented among 22q11.2 genes, given the number of genes in each module, or whether SCZ- and ASD-associated modules were over-represented overall, logistic regression controlling for gene length was conducted for each module, and for all disease-associated modules together, respectively.
To characterize the total protein-coding gene content of the 22q11.2 locus relative to all regions of the genome, and per module, the rank of the 22q11.2 locus was compared to the empirical distribution from all equal-sized genomic regions, generated using the GenomicRanges package(39) in 250 kb sliding steps (i.e. 12,149 genome-wide ~2.57 Mb regions). Extreme-high rank values for the 22q11.2 locus relative to all genomic regions (PGENOMIC-REGION<0.05) were considered statistically significant. A Mann-Whitney U test also examined whether 22q11.2 ranks were higher for SCZ- and ASD-associated modules, overall, compared to non-associated modules.
Characterizing the 22qBD-PPI Network
Genes form protein-protein interaction (PPI) networks that fulfill critical biological roles, and perturbing protein networks is a common characteristic of disease-associated mutations(40). However, protein interactions depend on the tissue and temporal availability of each protein(41). We therefore identified and characterized the brain developmental PPI (BD-PPI) network for the 22q11.2 locus (i.e., 22qBD-PPI network) by first compiling a comprehensive catalogue of pair-wise protein interactions from the BioGRID(42) and InWeb3 PPI databases(43,44); PPI pairs were then thresholded for co-expression during at least one stage of brain development (Spearman’s ρ>0.7) to identify PPI likely to exist in the brain during development.
The 22qBD-PPI network was defined as its seed genes and corresponding BD-PPI pairs, and was first tested for over-representation in any BrainSpan modules, given the number of genes in each module, using Fisher’s Exact tests. We then examined the 22qBD-PPI network’s rank relative to the BD-PPI networks of 2 null distributions: 1) all 12,149 ~2.57 Mb regions of the genome, described above (PGENOMIC-REGION NEWORK); and 2) 10,000 random gene-sets, equal in number of seed BrainSpan genes to the 22q11.2 locus (PGENE-SET NETWORK). For each null distribution, extreme-high rank values (P<0.05) were considered statistically significant. Mann-Whitney U tests additionally examined whether 22qBD-PPI network ranks were higher overall for SCZ- and ASD-associated modules than for non-associated modules.
Prioritizing Disease-Relevant 22q11.2 Genes
To prioritize 22q11.2 genes likely to drive risk for SCZ and ASD, the number of genes in each individual 22q11.2 gene’s BD-PPI network overlapping disease-associated BrainSpan modules was compared to the null distribution of all BrainSpan genes’ BD-PPI networks. 22q11.2 genes with extreme-high BD-PPI network ranks (PGENE NETWORK<0.05) were considered enriched for disease-associated modules and functionally annotated using g:Profiler.
See Supplementary Methods for details on all statistical analyses.
Results
SCZ and ASD Risk Variants Converge on Partially Overlapping Neurodevelopmental Modules
Eighteen BrainSpan modules were identified across 17,216 genes (Figure S1A; Table S2) with distinct developmental expression trajectories, cell-type enrichment patterns (Figure S1B), and functional characteristics, capturing broad aspects of neurotypical development (see Supplementary Results for detailed biological characterization and validation of BrainSpan data and modules; Tables S3–S5; Figures. S1–S11).
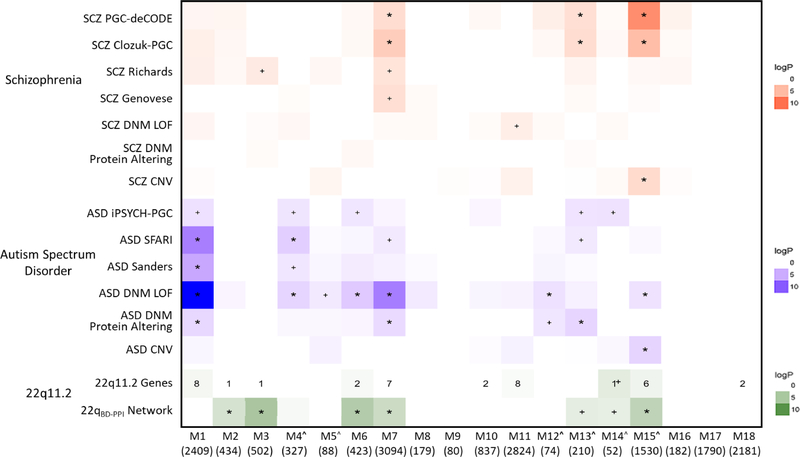
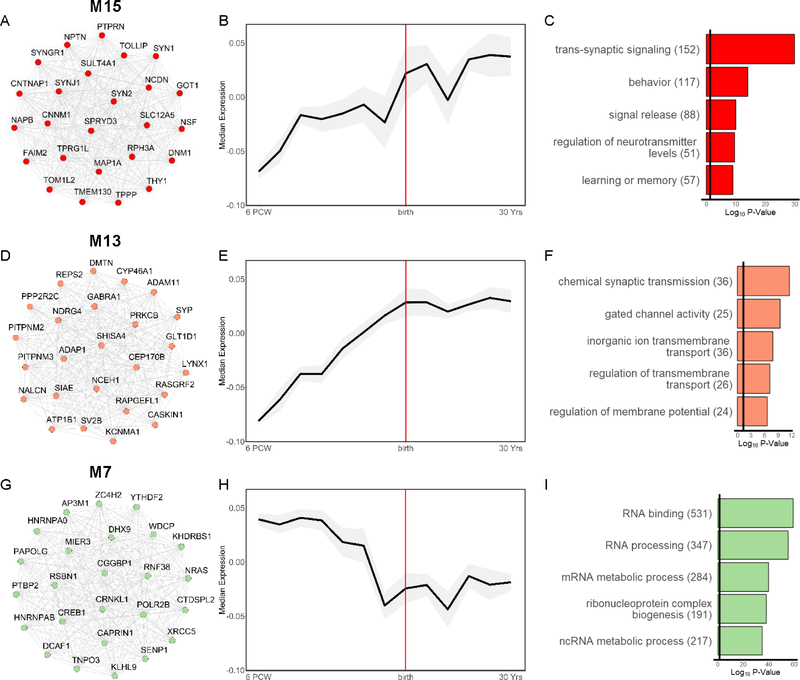
SCZ and ASD risk variant enrichment for BrainSpan modules is summarized in Figure 1. SCZ-associated variants converged on 3 modules. SCZ-associated common variants and CNVs were significantly over-represented in M15 (FDR-adjusted p<0.05, corrected for number of modules), a neuronal marker-enriched module that increased in expression through adolescence and was enriched for postnatal-expressed genes (Figures 2A–C, S3O). GO analysis indicated that M15 was involved in synaptic transmission, modulation of neurotransmission, and learning and memory. SCZ-associated common variants were additionally over-represented in M13, a neuronal module that increased in expression during fetal development, with relatively stable postnatal expression (Figure 2D–F). Consistent with this, M13 was enriched for genes expressed during late fetal and postnatal development (Figure S3M). M13 was enriched for synaptic transmission and regulation of the membrane potential, and its hub genes included ATP1B1 and KCNMA1, which encode components of the Na+/K+ pump and a voltage-gated K+ channel, respectively, that regulate the electrochemical gradient(45,46). Thus, M13 appears to be involved in regulating basic neuronal excitability. Finally, SCZ-associated common variants were significantly over-represented in M7, with rare variants nominally enriched. M7 was involved in RNA processing and binding, and its hub genes included POLR2B, which encodes a major subunit of RNA polymerase II, the central enzyme responsible for transcribing mRNA, and CREB1, which encodes the transcription factor CREB, known to regulate neuronal differentiation, survival, and plasticity(47; Figure 2G–I). M7 showed its highest expression during fetal development but was minimally enriched for genes associated with specific stages of development (Figure S3G). Thus, M7 appears to regulate gene expression and RNA processing across development.
Figure 1. Genetic Risk for SCZ and ASD Converge on Partially Overlapping and Partially Distinct Neurodevelopmental Modules.
BrainSpan module enrichment for SCZ (red) and ASD (blue) risk variant datasets, 22q11.2 genes (green), and the 22q11.2 brain-development PPI network (22qBD-PPI network; here shown tested for module enrichment using Fisher’s Exact tests). Log p values are indicated by color intensity for all datasets with enrichment odds ratios >1.0. To show potential convergence of enrichment across datasets, all enrichment values for p<0.05 are annotated (+p<0.05; *FDR corrected p<0.05 for number of modules). Numerical values denote number of 22q11.2 genes in each module. ^Module enriched for neuronal cell-type markers.
Figure 2. Characterization of BrainSpan Modules Associated with Both SCZ and ASD.
Left: top 25 hub genes, Middle: developmental expression trajectory (median ± median absolute deviation of module eigengene in grey shading), and Right: top 5 significantly enriched biological process and molecular function gene ontology (GO) terms for the SCZ- and ASD- associated modules: (A-C) M15, (D-F) M13, and (G-I) M7. Hub genes and GO terms are colored by module. Edges between hub genes denote positive correlations between genes.
Risk variants for ASD showed overlapping enrichment for SCZ-associated modules, as well as enrichment for 4 distinct modules (Figure 1). Thus, loss-of-function (LoF) DNMs in ASD were significantly over-represented in M7; protein-altering DNMs in ASD were over-represented in M13; and LoF DNMs and ASD-associated CNVs (many of which are also associated with SCZ; Table S4) were over-represented in M15.
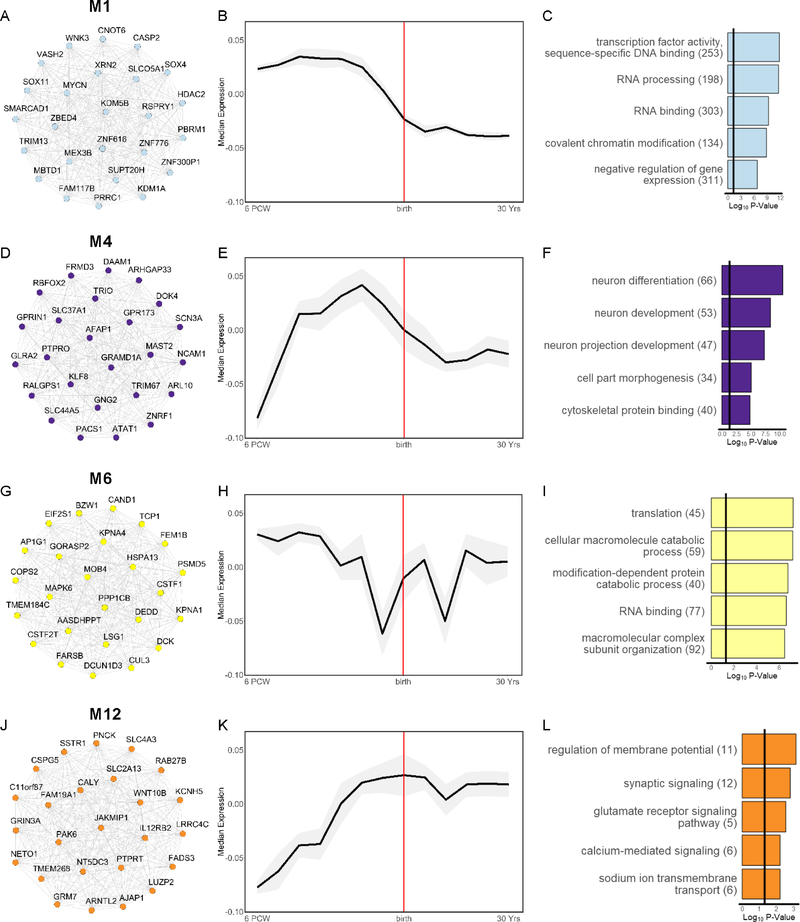
The additional modules significantly associated with ASD were M1, M4, M6, and M12. Thus, rare variants, broadly, and LoF and protein-altering DNMs in ASD were consistently over-represented in M1. M1 peaked in expression during fetal development and was highly enriched for genes with fetal-specific expression (Figures. 3A–C, S3A). M1 was involved in transcription, chromatin modification, and gene expression, and its hub genes included the transcription factors SOX11 and SOX4, which are critical for promoting neuronal differentiation(48). Thus, M1 appears to be important for regulating fetal gene expression patterns that direct neuronal differentiation. LoF DNMs and SFARI-defined ASD genes were additionally over-represented in M4. M4 was enriched for neuronal markers, peaked in expression during mid-fetal development, and was involved in neuron differentiation and projection development (Figure 3D–F). Consistent with this, M4 was enriched for genes with fetal-specific expression (Figure S3D), and its hub genes included DOK4, ARHGAP33, and TRIO, which are involved in axon and dendrite morphogenesis(49–51). LoF DNMs in ASD were also over-represented in M6, which showed maximal expression during fetal development but was not enriched for genes expressed in any specific developmental stage (Figure S3F). M6 was involved in translation and protein catabolism (Figure 3G–I). Finally, LoF DNMs in ASD were over-represented in M12, a neuronal module that increased in expression until plateauing in late fetal development, and was involved in regulating the membrane potential (Figures 3J–L, S3L). Interestingly, ASD-associated common variants were nominally over-represented across multiple modules significantly enriched for rare and/or de novo variants in ASD (M1, M4, M7, M15). Module enrichment for gene-sets implicated in prior genetic studies of SCZ or ASD were consistent with GO analyses (Figure S1C). See Figure S4 for summary of non-SCZ or ASD-associated modules.
Figure 3. Characterization of BrainSpan Modules Associated Only with ASD.
Left: top 25 hub genes, Middle: developmental expression trajectory (median ± median absolute deviation of module eigengene in grey shading), and Right: top 5 significantly enriched biological process and molecular function gene ontology (GO) terms for the ASD-associated modules: (A-C) M1, (D-F) M4, (G-I) M6, and (J-L) M12. Hub genes and GO terms are colored by module. Edges between hub genes denote positive correlations between genes.
As expected, LoF and protein-altering DNMs in control subjects were not over-represented in any SCZ- or ASD-associated modules, and no modules were significantly enriched for synonymous DNMs in ASD, SCZ, or controls, ps>0.05.
Thus, polygenic risk for SCZ and ASD each appear to coalesce on neurodevelopmental networks involved in postnatal neuronal excitability and synaptic transmission, and pan-developmental regulation of gene expression; however, risk variants for ASD additionally converge on networks involved in early neuronal differentiation and synapse formation.
The 22qBD-PPI Network Loads Highly onto SCZ- and ASD-Associated Modules
Forty-six protein-coding genes are contained in the ~2.57 Mb 22q11.2 locus, 38 of which were indexed in BrainSpan (Table 2; see Figure S12 for expression trajectory of each gene). Given the number of genes in each module, there were nominally more 22q11.2 genes than expected in M14 (i.e., 1 gene; SEPT5; p=0.02; Figure 1), although this did not survive FDR correction. Similarly, there were not significantly more 22q11.2 genes across all SCZ- and ASD-associated modules than expected, p>0.05. Relative to all regions of the genome, the protein-coding gene content of the overall 22q11.2 locus was in the 93.6 percentile, and among individual BrainSpan modules, 22q11.2 gene content was enriched for 3 modules, including the ASD-associated M1 module and the SCZ- and ASD-associated M15 module (PGENOMIC-REGION<.05; Table S6). However, the genomic-region ranks of the 22q11.2 locus were not significantly higher across all SCZ- and ASD-associated compared to non-associated modules, p>0.05.
Table 2. BrainSpan Module Membership of 22q11.2 Genes.
The 38 22q11.2 genes indexed in BrainSpan are annotated by module assignment, Kme score (i.e., correlation with parent module eigengene) and pLI score (i.e., loss-of-function function intolerance; bolded text indicates predicted pLI gene).
| Module | Gene | Definition | kME Score | pLI Score |
|---|---|---|---|---|
| M1 | ZNF74 | zinc finger protein 74 | 0.84 | 0.0004 |
| DGCR8 | DGCR8, microprocessor complex subunit | 0.76 | 0.9999 | |
| SLC25A1 | solute carrier family 25 member 1 | 0.75 | 0.6335 | |
| ARVCF | armadillo repeat gene deleted in velocardiofacial syndrome | 0.67 | 0.0000 | |
| CLTCL1 | clathrin heavy chain like 1 | 0.61 | 0.0000 | |
| KLHL22 | kelch like family member 22 | 0.56 | 0.7026 | |
| TMEM191A | transmembrane protein 191A | 0.18 | NA | |
| SCARF2 | scavenger receptor class F member 2 | 0.01 | NA | |
| M2 | CRKL | v-crk avian sarcoma virus CT10 oncogene homolog-like | 0.51 | 0.1635 |
| M3 | CDC45 | cell division cycle 45 | 0.78 | 0.0691 |
| M6 | MRPL40 | mitochondrial ribosomal protein L40 | 0.78 | 0.0201 |
| ESS2 | DiGeorge syndrome critical region gene 14 | 0.75 | 0.0000 | |
| M7 | UFD1 | ubiquitin recognition factor in ER associated degradation 1 | 0.80 | 0.9974 |
| SNAP29 | synaptosome associated protein 29 | 0.75 | 0.1517 | |
| HIRA | histone cell cycle regulator | 0.64 | 1.0000 | |
| RTL10 | retrotransposon Gag like 10 | 0.55 | 0.0000 | |
| TRMT2A | tRNA methyltransferase 2 homolog A | 0.35 | 0.0000 | |
| THAP7 | THAP domain containing 7 | 0.21 | 0.0293 | |
| DGCR2 | DiGeorge syndrome critical region gene 2 | 0.17 | 0.0011 | |
| M10 | AIFM3 | apoptosis-inducing factor, mitochondrionassociated, 3 | 0.94 | 0.0000 |
| P2RX6 | purinergic receptor P2X, ligand-gated ion channel, 6 | 0.80 | 0.0000 | |
| M11 | PRODH | proline dehydrogenase (oxidase) 1 | 0.86 | 0.0000 |
| ZDHHC8 | zinc finger, DHHC-type containing 8 | 0.76 | 0.9890 | |
| COMT | catechol-O-methyltransferase | 0.63 | 0.0008 | |
| TXNRD2 | thioredoxin reductase 2 | 0.59 | 0.0000 | |
| SERPIND1 | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | 0.42 | 0.0000 | |
| C22orf39 | chromosome 22 open reading frame 39 | 0.24 | 0.0000 | |
| DGCR6L | DiGeorge syndrome critical region gene 6-like | 0.17 | 0.1286 | |
| GSC2 | goosecoid homeobox 2 | 0.15 | 0.0372 | |
| M14^ | SEPT5 | septin 5 | 0.80 | 0.9146 |
| M15^ | PI4KA | phosphatidylinositol 4-kinase alpha | 0.88 | 0.0008 |
| TANGO2 | transport and golgi organization 2 homolog | 0.82 | 0.0000 | |
| SLC7A4 | solute carrier family 7 member 4 | 0.72 | 0.0001 | |
| RTN4R | reticulon 4 receptor | 0.57 | 0.5264 | |
| MED15 | mediator complex subunit 15 | 0.38 | 0.9610 | |
| LZTR1 | leucine zipper like transcription regulator 1 | 0.25 | 0.0000 | |
| M18 | CLDN5 | claudin 5 | 0.43 | 0.7405 |
| TSSK2 | testis-specific serine kinase 2 | 0.38 | 0.0042 | |
Module enriched for neuronal cell-type markers.
We next characterized the 22q11.2 brain developmental PPI network (22qBD-PPI network), given that perturbation of protein networks is a common characteristic of mutations associated with many diseases(40). The 22q11.2 locus formed a large BD-PPI network, consisting of 239 genes from 38 seed genes (Figure S13). Given the number of genes in each module, the 22qBD-PPI network was significantly over-represented in the SCZ- and/or ASD-associated modules M6, M7, and M15, as well as M2 and M3 (FDR p<0.05; Figure 1).
Some BrainSpan modules consist of genes that are more highly connected than other modules (ANOVA p<2.0×10−16; Table S7), and highly connected genes are more likely to be found in the BD-PPI network of any random set of genes. This may be biologically meaningful, with gene networks that are highly connected during brain development potentially more susceptible to perturbation. Nevertheless, to better understand the 22qBD-PPI network, we next characterized the connectivity of the 22qBD-PPI network relative to all regions of the genome, and to random gene-sets derived from an equal number of seed genes.
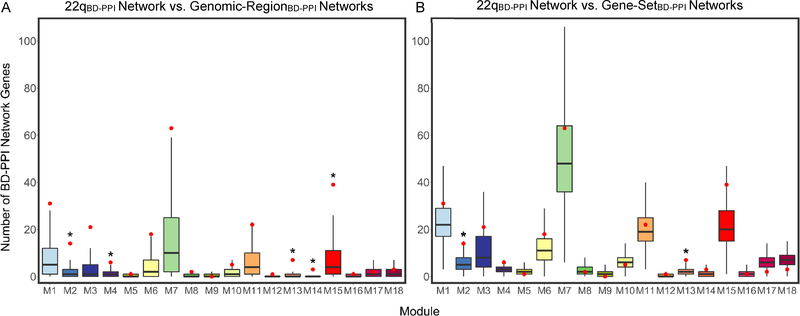
Relative to all genomic regions, the 22qBD-PPI network was significantly enriched for 5 modules, (PGENOMIC-REGION NETWORK<0.05), including the SCZ- and/or ASD-associated neuronal modules, M4, M13, and M15 (Figure 4A, Table S6). Further, the 22qBD-PPI network ranks relative to all Genomic-RegionBD-PPI networks were significantly higher for SCZ- and/or ASD-associated modules, overall, compared to non-SCZ- or ASD-associated modules, p=0.03.
Figure 4. Characterizing the 22qBD-PPI Network Relative to Genomic-Region and Gene-Set BD-PPI Networks.
Null distributions for number of BD-PPI network genes per module across all: (A) ~2.57 Mb genomic regions (i.e., 12,149 Genomic-RegionBD-PPI networks) and (B) random 38 seed gene gene-sets (i.e., 10,000 Gene-SetBD-PPI networks), derived to characterize the 22q11.2 locus relative to all regions of the genome, and the connectivity of 22q11.2 genes within brain development protein networks, respectively. Boxplots mark the median number of BD-PPI network genes per module, with lower and upper hinges corresponding to the 25th and 75th quartiles, respectively, and the whiskers extending to 1.5 x interquartile range. The number of 22qBD-PPI network genes in each module are marked in red. *22qBD-PPI network P<0.05 relative to null Genomic-RegionBD-PPI or Gene-SetBD-PPI network distribution.
Relative to random sets of 38 seed genes, the 22qBD-PPI network remained significantly enriched for the SCZ- and ASD-associated M13 module, as well as the non-associated M2 module (PGENE-SET NETWORK<0.05; Figure 4B; Table S6). The 22qBD-PPI network ranks relative to Gene-SetBD-PPI networks were also higher overall for SCZ- and/or ASD-associated modules than for non-associated modules, p=0.03.
This suggests that 22q11.2 CNVs confer risk for SCZ and ASD by not only spanning a genomic region that is rich in protein-coding genes, but by spanning genes that form protein networks in the developing brain that are highly and disproportionately connected to broader networks involved in SCZ and ASD pathogenesis.
SEPT5, PI4KA, and SNAP29 are Candidate Drivers of Synaptic Pathology, and DGCR8 and HIRA are Candidate Drivers of Dysregulated Gene Expression
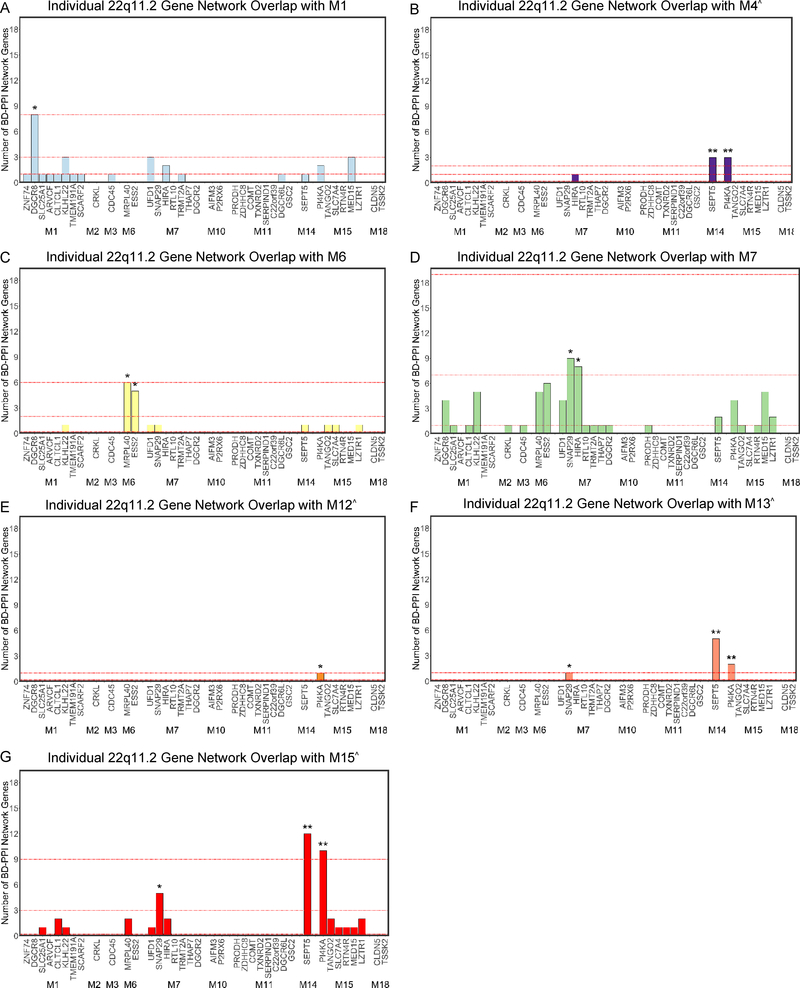
To identify specific genes driving 22qBD-PPI network loading onto SCZ- and ASD-associated modules, we next examined the rank of each 22q11.2 gene’s BD-PPI network per module, relative to all BrainSpan genes. The SEPT5 and PI4KA BD-PPI networks were enriched for 3 neuronal modules associated with SCZ and/or ASD: M4, M13, and M15 (PGENE NETWORK<0.05; Figure 5); PI4KA was additionally enriched for the ASD-associated M12 module. SNAP29’s BD-PPI network was also enriched for M13 and M15, as well as M7 (PGENE NETWORK<0.05). Indeed, the individual SEPT5, PI4KA, and SNAP29 networks were each enriched for cell parts and/or biological processes related to synaptic function (Figure 6A–F). HIRA’s BD-PPI network was also enriched for M7 (Figure 5D), and was enriched for genes involved in chromatin organization and gene expression (Figure 6G–H). Additionally, DGCR8’s BD-PPI network was enriched for M1 (Figure 5A), and was enriched for RNA binding and processing (Figure 6I–J). Finally, MRPL40 and ESS2’s BD-PPI networks were enriched for M6 (Figure 5C); the ESS2 network was not enriched for any GO terms, while MRPL40’s network was enriched for ribosomal components and mitochondrial elongation (Figure 6K–M).
Figure 5. Prioritizing Individual 22q11.2 Genes Based on BD-PPI Network Overlap with Each SCZ- and/or ASD-Associated BrainSpan Module.
Number of genes in each individual 22q11.2 gene’s BD-PPI network belonging to the: (A) M1, (B) M4, (C) M6, (D) M7, (E) M12, (F) M13, and (G) M15 SCZ- and/or ASD-associated modules. In each plot, 22q11.2 genes are shown grouped by module membership along the x-axis. The top, second, and third red dotted lines in each plot mark the number of BD-PPI network genes nearest to the 99th, 95th, and 75th percentile (when above 0) of all BrainSpan genes for each module. Bar plots are colored by module color. *PGENE NETWORK<0.05 and **PGENE NETWORK<0.01 for each module, relative to the BD-PPI networks of all BrainSpan genes. ^Module enriched for neuronal cell-type markers.
Figure 6. Characterization of BD-PPI Networks for 22q11.2 Genes Prioritized to Drive Risk for SCZ and/or ASD.
Left: BD-PPI network genes and Right: significantly enriched GO terms (up to top 5 terms, where applicable) for the BD-PPI networks of: (A-B) SEPT5, (C-D) PI4KA, (E-F) SNAP29, (G-H) HIRA, (I-J) DGCR8, (K-L) MRPL40, and (M) ESS2. Individual gene nodes in each network are colored by BrainSpan module assignment. GO plots are colored by module membership of the 22q11.2 gene of interest.
Support for Leveraging BD-PPI Networks to Understand 22q11.2 CNV-Mediated Risk for SCZ and ASD
Investigation of gene expression signatures in SCZ and ASD post-mortem cortex(43) indicated that nearly all BrainSpan modules associated with genetic risk for SCZ and ASD were enriched for down-regulated genes in each disorder (Figure S14A); the 22qBD-PPI network was also enriched for down-regulated genes in SCZ and ASD, given the size of the 22qBD-PPI network and each gene expression gene-set (ps<0.05). Relative to all BrainSpan genes, the SEPT5 and PI4KA BD-PPI networks ranked highly for overlap with down-regulated genes in both SCZ and ASD (PGENE NETWORK<0.05), with the SNAP29 and KLHL22 BD-PPI networks additionally enriched for overlap with down-regulated genes in ASD (Figure S15).
Exploratory analyses of broader CNVs and genes associated with SCZ and ASD confirmed that high BD-PPI network loading onto SCZ- and ASD-associated BrainSpan modules and down-regulated genes in SCZ or ASD cortex is a common characteristic of disease-associated CNVs (Figure S16) and genes (Figure S17). Similarly, using more inclusive co-expression thresholds to define the BD-PPI networks of each gene highlighted similar 22q11.2 genes as likely drivers of SCZ and ASD risk (Figures S18, S19). Further, using a machine learning approach to classify SCZ- and ASD-associated genes based on BD-PPI network ranks prioritized similar 22q11.2 genes (Tables S2, S12). Finally, study bias could not account for our prioritization of 22q11.2 genes likely to drive disease risk (Figure S20). Together, these results support the utility of BD-PPI networks for identifying neurodevelopmental mechanisms and specific genes through which multi-gene CNVs confer risk for SCZ and ASD. See Supplementary Results for details.
Discussion
Recent genetic discoveries in SCZ and ASD have brought renewed focus to understanding the mechanisms through which genetic variation confers risk for these disorders at both the polygenic and individual variant level. Here, using a developmental functional genomics approach, we identified overlapping and distinct neurodevelopmental processes associated with SCZ and ASD risk, and then leveraged this disease-informed lens to elucidate mechanisms underlying disease risk in highly penetrant multi-gene CNVs. Our analyses confirmed the capacity of 22q11.2 CNVs to disrupt disease-relevant neurodevelopmental modules and highlighted both well-studied (e.g., DGCR8 and HIRA) and relatively understudied genes (e.g., SEPT5 and PI4KA) as candidate 22q11.2 genes likely to drive disease risk.
Prior studies using transcriptomic data from the developing brain to understand how risk variants contribute to disease largely focused on rare variants in ASD. These studies suggested that ASD risk variants preferentially disrupt neuronal development and transcriptional and chromatin regulation during fetal development(16,17,35). One early study of DNMs in SCZ also implicated disruptions in neurogenesis and transcriptional regulation(26); however, this was a relatively small study. Here, by creating co-expression networks from brain samples spanning the full period of development and testing an expanded set of risk variants for SCZ and ASD simultaneously, we confirmed prior enrichment patterns in ASD and identified co-expression modules with distinct enrichment patterns for ASD versus SCZ. We found robust enrichment in SCZ for a neuronal module that increased in expression across development and was involved broadly in postnatal synaptic transmission and refinement of neurotransmission; a neuronal module involved in regulating neuronal excitability that stabilized in expression around birth; and a module involved in regulating transcription and splicing across development. ASD risk variants showed overlapping enrichment for these modules, as well as robust enrichment for distinct modules involved in neuronal differentiation, and in regulating chromatin organization and gene transcription during fetal development. While our findings thus confirm prior findings implicating neuronal and synaptic genes in both SCZ and ASD(7,52), risk variants for ASD appear to partially diverge from those for SCZ by loading highly onto gene networks that orchestrate neuronal differentiation during fetal development. It is tempting to suggest that the higher loading of ASD risk variants onto early neuronal differentiation networks may be a key mechanism underlying the earlier onset of ASD relative to SCZ; however, experimental validation is necessary to confirm this.
By integrating BrainSpan co-expression and PPI data, we identified protein networks likely to be strongly affected by 22q11.2 CNVs during brain development. One prior study focused on 16p11.2 CNVs integrated PPI and BrainSpan data and found that ‘neuropsychiatric’ CNV networks were enriched for varying developmental periods, with the overall 22q11.2 network showing particular enrichment during childhood(41). Here, using a disease-informed framework, we focused on understanding how 22q11.2 CNVs confer risk for SCZ and ASD, more specifically. We found that 22q11.2 genes formed a large BD-PPI network that: 1) loaded highly onto multiple SCZ- and ASD-associated modules, relative to all regions of the genome; 2) was more highly connected to genes in SCZ- and/or ASD-associated modules than genes in non-disease associated modules; and 3) overlapped with genes that are down-regulated in SCZ and ASD cortex, with overlapping genes involved in neuronal and synaptic function. Thus, 22q11.2 CNVs may confer risk for SCZ and ASD, at least partially, by perturbing synaptic networks that modulate neuronal excitability and signaling. Consistent with this, 22q11DS mouse models show altered neuronal excitability, activity, and dendritic spine stability(53,54).
A primary advantage of our approach was our ability to systematically survey 22q11.2 genes to prioritize genes likely to contribute disease risk. Our analyses consistently highlighted SEPT5 and PI4KA as candidate drivers of synaptic pathology relevant to SCZ and ASD, and DGCR8 as a candidate driver of dysregulated gene expression that may be most relevant to ASD. SNAP29 and HIRA were also prioritized as contributing to SCZ and ASD. Notably, both SEPT5 and SNAP29 have been found to negatively modulate neurotransmitter release. SEPT5 encodes the filamentous protein septin 5, which has been found to associate with synaptic vesicles to create a physical barrier that inhibits vesicle fusion with the membrane, and thereby, inhibits neurotransmitter release(55,56). Similarly, SNAP29 encodes a SNARE protein that competes with α-SNAP for binding to SNARE complexes after membrane fusion, thus reducing SNARE protein recycling and synaptic vesicle availability(57). Conversely, altered PI4KA expression may contribute to disease risk by perturbing the proteomic composition of the plasma membrane (PM). PI4KA encodes the kinase PI4KIIIα, which is involved in synthesizing the phosphoinositide, phosphatidylinositol 4,5-biphosphate (PI(4,5)P2), that recruits and regulates PM proteins and thereby governs diverse processes including vesicular trafficking, ion transport, and actin cytoskeletal dynamics(58). PI4KA depletion was found to disrupt PI(4,5)P2-associated protein localization, cellular responses to stimulation(59,60), and synapse maintenance(61). Our prioritization of PI4KA is consistent with prior associations of PI4KA polymorphisms in SCZ(62,63,64), and with growing evidence that PI4KA may play an important role in synaptic function(60,61).
DGCR8 is a core component of the microprocessor complex involved in processing miRNAs, which regulates gene expression at the protein level by binding target mRNAs to silence their translation(65). HIRA is a histone chaperone that influences gene expression by facilitating the deposition of the non-canonical histone variant, H3.3, into chromatin(66,67). Given these gene regulatory functions, DGCR8 and HIRA have been previously hypothesized to play important roles in 22q11.2 CNV phenotypes(67,68). Here, using our disease-informed functional genomic lens, we provide independent, unbiased evidence for prioritizing DGCR8 and HIRA in 22q11.2 CNV-mediated risk for neurodevelopmental disorders.
Some caveats to the current study should be noted. First, in our analyses testing SCZ and ASD risk variants for module enrichment, some studies shared overlapping samples. As the genetic architectures of SCZ and ASD include many variants with small effects, collaborative cross-study analyses are increasingly common to facilitate power. We utilized independent datasets where possible, and focused on modules showing consistent disease association. Second, generating BD-PPI networks identifies protein networks putatively affected by 22q11.2 CNVs, but cannot tell us the functional output of decreased versus increased expression of each gene. We therefore cannot determine how deletions versus duplications affect these networks, nor quantify the full consequences of mutations in genes whose actions involve regulating the expression of other genes. Similarly, while using individual gene BD-PPI network rankings is useful for prioritizing 22q11.2 genes predicted to be highly connected within disease-associated modules, experimental validation is necessary to confirm the contribution of specific 22q11.2 genes to disease-relevant phenotypes (e.g., see 1,69). As high-throughput functional assays become increasingly feasible, quantifying the transcriptomic and proteomic consequences of altered expression of each 22q11.2 gene will likely yield further insights into 22q11.2 CNV-mediated disease risk. Nevertheless, identifying convergent neurodevelopmental networks affected by polygenic risk for SCZ and ASD, and systematically prioritizing 22q11.2 genes that load highly onto these networks, offers a critical step towards understanding the mechanisms through which 22q11.2 CNVs confer risk for SCZ and ASD.
Supplementary Material
KEY RESOURCES TABLE.
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Deposited Data; Public Database | BrainSpan microarray data | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE25219 | RRID:SCR_004219 | NA |
| Deposited Data; Public Database | dbVar CNV database | https://www.ncbi.nlm.nih.gov/dbvar/ | RRID:SCR_003219 | NA |
| Deposited Data; Public Database | BioGrid PPI 3.4 database | https://downloads.thebioarid.ora/BioGRID | RRID:SCR_007393 | NA |
| Deposited Data; Public Database | InWeb3 PPI database | https://doi.org/10.1371/journal.pgen.1001273 | NA | NA |
| Software; Algorithm | g:Profiler (rev 1741, built 2017-10-19) | https://biit.cs.ut.ee/gprofiler/gost | RRID:SCR_006809) | NA |
| Software; Algorithm | Specific Expression Analysis Tool | http://genetics.wustl.edu/jdlab/csea-tool-2/ | NA | NA |
| Software; Algorithm | Ensembl Variant Effect Predictor version 90 | http://grch37.ensembl.org/Homo_sapiens/Tools/VEP | RRID:SCR_002344 | NA |
Acknowledgements
The authors thank Gerhard Helleman for statistical advice and Elizabeth Ruzzo for feedback and advice on analyses. This work was supported by grants from the National Institutes of Mental Health (R01MH085953 to CEB, R01MH100900 to CEB, T32MH096682 to JKF, K08MH11877 to JKF). This manuscript has been posted on the preprint server bioRxiv.
Footnotes
Conflicts of Interest. The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guna A, Butcher NJ, Bassett AS (2015): Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J Neurodev Disord. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van L, Boot E, Bassett AS (2017): Update on the 22q11.2 deletion syndrome and its relevance to schizophrenia. Curr Opin Psychiatry. 30: 191–196. [DOI] [PubMed] [Google Scholar]
- 3.Kirov G, Rees E, Walters JTR, Escott-Price V, Georgieva L, Richards AL, et al. (2014): The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry. 75: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. (2017): Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 49: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees E, Kirov G, Sanders A, Walters JTR, Chambert KD, Shi J, et al. (2014): Evidence that duplications of 22q11.2 protect against schizophrenia. Mol Psychiatry. 19: 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. (2011): Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 70: 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandal MJ, Leppa V, Won H, Parikshak NN, Geschwind DH (2016): The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci. 19: 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pocklington AJ, Rees E, Walters JTR, Han J, Kavanagh DH, Chambert KD, et al. (2015): Novel Findings from CNVs Implicate Inhibitory and Excitatory Signaling Complexes in Schizophrenia. Neuron. 86: 1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. (2018): Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. (2014): De novo mutations in schizophrenia implicate synaptic networks. Nature. 506: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, et al. (2016): Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 19: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014): Biological insights from 108 schizophrenia-associated genetic loci. Nature. 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. (2014): The contribution of de novo coding mutations to autism spectrum disorder. Nature. 515: 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. (2014): Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 515: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. (2015): Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 87: 1215–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, et al. (2013): Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 155: 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. (2013): Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 155: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. (2011): Spatio-temporal transcriptome of the human brain. Nature. 478: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geschwind DH, Konopka G (2009): Neuroscience in the era of functional genomics and systems biology. Nature. 461: 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, et al. (2017): A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst. 4: 60–72.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reimand J, Kull M, Peterson H, Hansen J, Vilo J (2007): g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35: W193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dougherty JD, Schmidt EF, Nakajima M, Heintz N (2010): Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic Acids Res. 38: 4218–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grove J, Ripke S, Als TD, Mattheisen M, Walters R, Won H, et al. (2017, November 25): Common risk variants identified in autism spectrum disorder. bioRxiv. [Google Scholar]
- 24.Richards AL, Leonenko G, Walters JT, Kavanagh DH, Rees EG, Evans A, et al. (2016): Exome arrays capture polygenic rare variant contributions to schizophrenia. Hum Mol Genet. 25: 1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015): MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 11: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, et al. (2013): Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 154: 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambalavanan A, Girard SL, Ahn K, Zhou S, Dionne-Laporte A, Spiegelman D, et al. (2016): De novo variants in sporadic cases of childhood onset schizophrenia. Eur J Hum Genet. 24: 944–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guipponi M, Santoni FA, Setola V, Gehrig C, Rotharmel M, Cuenca M, et al. (2014): Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One. 9: e112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, et al. (2012): De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 44: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Li M, Yang Z, Hu X, Wu H-M, Ni P, et al. (2015): Increased co-expression of genes harboring the damaging de novo mutations in Chinese schizophrenic patients during prenatal development. Sci Rep. 5: 18209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, et al. (2014): De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 19: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, et al. (2011): Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 43: 860–863. [DOI] [PubMed] [Google Scholar]
- 33.Takata A, Xu B, Ionita-Laza I, Roos JL, Gogos JA, Karayiorgou M (2014): Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron. 82: 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosmicki JA, Samocha KE, Howrigan DP, Sanders SJ, Slowikowski K, Lek M, et al. (2017): Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 49: 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takata A, Miyake N, Tsurusaki Y, Fukai R, Miyatake S, Koshimizu E, et al. (2018): Integrative Analyses of De Novo Mutations Provide Deeper Biological Insights into Autism Spectrum Disorder. Cell Rep. 22: 734–747. [DOI] [PubMed] [Google Scholar]
- 36.Ware JS, Samocha KE, Homsy J, Daly MJ (2015): Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr Protoc Hum Genet. 87: 7.251–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons Foundation Autism Research Initiative (n.d.): SFARI Gene. Retrieved November 28, 2017, from https://gene.sfari.org/.
- 38.Shen L, Sinai M (2013): GeneOverlap: Test and visualize gene overlaps. R package version. 1.
- 39.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al. (2013): Software for computing and annotating genomic ranges. PLoS Comput Biol. 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, et al. (2015): Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 161: 647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, et al. (2015): Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 85: 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatr-Aryamontri A, Breitkreutz B-J, Oughtred R, Boucher L, Heinicke S, Chen D, et al. (2015): The BioGRID interaction database: 2015 update. Nucleic Acids Res. 43: D470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lage K, Karlberg EO, Størling ZM, Olason PI, Pedersen AG, Rigina O, et al. (2007): A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 25: 309–316. [DOI] [PubMed] [Google Scholar]
- 44.Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. (2011): Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 7: e1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Langhans SA (2015): Transcriptional regulators of Na,K-ATPase subunits. Front Cell Dev Biol. 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montgomery JR, Meredith AL (2012): Genetic activation of BK currents in vivo generates bidirectional effects on neuronal excitability. Proc Natl Acad Sci U S A. 109: 18997–19002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ortega-Martínez S (2015): A new perspective on the role of the CREB family of transcription factors in memory consolidation via adult hippocampal neurogenesis. Front Mol Neurosci. 8: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J (2006): The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 20: 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchida M, Enomoto A, Fukuda T, Kurokawa K, Maeda K, Kodama Y, et al. (2006): Dok-4 regulates GDNF-dependent neurite outgrowth through downstream activation of Rap1 and mitogen-activated protein kinase. J Cell Sci. 119: 3067–3077. [DOI] [PubMed] [Google Scholar]
- 50.Schuster S, Rivalan M, Strauss U, Stoenica L, Trimbuch T, Rademacher N, et al. (2015): NOMA-GAP/ARHGAP33 regulates synapse development and autistic-like behavior in the mouse. Mol Psychiatry. 20: 1120–1131. [DOI] [PubMed] [Google Scholar]
- 51.Peng Y-J, He W-Q, Tang J, Tao T, Chen C, Gao Y-Q, et al. (2010): Trio is a key guanine nucleotide exchange factor coordinating regulation of the migration and morphogenesis of granule cells in the developing cerebellum. J Biol Chem. 285: 24834–24844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forsyth JK, Lewis DA (2017): Mapping the Consequences of Impaired Synaptic Plasticity in Schizophrenia through Development: An Integrative Model for Diverse Clinical Features. Trends Cogn Sci. 21: 760–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fénelon K, Xu B, Lai CS, Mukai J, Markx S, Stark KL, et al. (2013): The pattern of cortical dysfunction in a mouse model of a schizophrenia-related microdeletion. J Neurosci. 33: 14825–14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Z, Williams DJ, Xu B, Gogos JA (2018): Altered function and maturation of primary cortical neurons from a 22q11.2 deletion mouse model of schizophrenia. Transl Psychiatry. 8: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beites CL, Xie H, Bowser R, Trimble WS (1999): The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 2: 434–439. [DOI] [PubMed] [Google Scholar]
- 56.Beites CL, Campbell KA, Trimble WS (2005): The septin Sept5/CDCrel-1 competes with alpha-SNAP for binding to the SNARE complex. Biochem J. 385: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su Q, Mochida S, Tian JH, Mehta R, Sheng ZH (2001): SNAP-29: a general SNARE protein that inhibits SNARE disassembly and is implicated in synaptic transmission. Proc Natl Acad Sci U S A. 98: 14038–14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falkenburger BH, Jensen JB, Dickson EJ, Suh B-C, Hille B (2010): Phosphoinositides: lipid regulators of membrane proteins. J Physiol. 588: 3179–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, et al. (2012): PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 199: 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Balakrishnan SS, Basu U, Shinde D, Thakur R, Jaiswal M, Raghu P (2018): Regulation of PI4P levels by PI4KIIIα during G-protein-coupled PLC signaling in Drosophila photoreceptors. J Cell Sci. 131. doi: 10.1242/jcs.217257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bulat V, Rast M, Pielage J (2014): Presynaptic CK2 promotes synapse organization and stability by targeting Ankyrin2. J Cell Biol. 204: 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jungerius BJ, Hoogendoorn MLC, Bakker SC, Van’t Slot R, Bardoel AF, Ophoff RA, et al. (2008): An association screen of myelin-related genes implicates the chromosome 22q11 PIK4CA gene in schizophrenia. Mol Psychiatry. 13: 1060–1068. [DOI] [PubMed] [Google Scholar]
- 63.Kaur H, Jajodia A, Grover S, Baghel R, Jain S, Kukreti R (2014): Synergistic association of PI4KA and GRM3 genetic polymorphisms with poor antipsychotic response in south Indian schizophrenia patients with low severity of illness. Am J Med Genet B Neuropsychiatr Genet. 165B: 635–646. [DOI] [PubMed] [Google Scholar]
- 64.Vorstman JAS, Chow EW, Ophoff RA, van Engeland H, Beemer FA, Kahn RS, et al. (2009): Association of the PIK4CA schizophrenia-susceptibility gene in adults with the 22q11.2 deletion syndrome. Am J Med Genet B Neuropsychiatr Genet. 150B: 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajman M, Schratt G (2017): MicroRNAs in neural development: from master regulators to fine-tuners. Development. 144: 2310–2322. [DOI] [PubMed] [Google Scholar]
- 66.Chen W-Y, Shih H-T, Liu K-Y, Shih Z-S, Chen L-K, Tsai T-H, et al. (2015): Intellectual disability-associated dBRWD3 regulates gene expression through inhibition of HIRA/YEM-mediated chromatin deposition of histone H3.3. EMBO Rep. 16: 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valenzuela N, Fan Q, Fa’ak F, Soibam B, Nagandla H, Liu Y, et al. (2016): Cardiomyocyte-specific conditional knockout of the histone chaperone HIRA in mice results in hypertrophy, sarcolemmal damage and focal replacement fibrosis. Dis Model Mech. 9: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fénelon K, Mukai J, Xu B, Hsu P-K, Drew LJ, Karayiorgou M, et al. (2011): Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 108: 4447–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hiroi N (2018): Critical reappraisal of mechanistic links of copy number variants to dimensional constructs of neuropsychiatric disorders in mouse models. Psychiatry Clin Neurosci. 72: 301–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.