Abstract
Epigenetic mechanisms play essential roles in determining distinct cell fates during the development of multicellular organisms. Histone proteins represent crucial epigenetic components that help specify cell identities. Previous work has demonstrated that during the asymmetric cell division of the Drosophila male germline stem cells (GSCs), histones H3 and H4 are asymmetrically inherited, such that preexisting (old) histones are segregated towards the self-renewing GSC whereas newly synthesized (new) histones are enriched towards the differentiating daughter cell. In order to further understand the molecular mechanisms underlying this striking phenomenon, two key questions must be answered: when and how old and new histones are differentially incorporated by sister chromatids, and how epigenetically distinct sister chromatids are specifically recognized and segregated. Here we discuss recent advances in our understanding of the molecular mechanisms and cellular bases underlying these fundamental and important biological processes responsible for generating two distinct cells through one cell division.
Keywords: Epigenetics, histones, germline, stem cells, asymmetric cell division, centromere
Opening remarks: an overview of asymmetric stem cell division in multicellular organisms
The processes of development and homeostasis in multicellular organisms require the specification of a wide variety of highly specialized cell types. In order to achieve this remarkable cellular diversity to create complex tissues and maintain their replenishing capacity, adult stem cells need to maintain a balance between the differentiated cell population and the stem cell population. For examples, skin [1] muscle [2], gut [3], blood [4] or testis [5, 6] in mice, as well as midgut [7, 8] and gonads [9, 10] in Drosophila have all been shown to have stem cell activities. Many stem cells achieve this by undergoing ACD (Asymmetric Cell Division), through which two daughter cells are produced, each with their own distinct cell fate (reviewed in [11–15]).
Previous studies have revealed a cohort of extrinsic and intrinsic mechanisms that help establish distinct daughter cell fates following ACD. From an extrinsic perspective, signaling molecules that act in the extracellular environment can help specify distinct cell fates. Many adult stem cells rely on a unique micro-environment known as the niche to provide extrinsic cues to maintain proper stem cell identity and activity (reviewed in [6, 16]). Niches support stem cell identity by secreting ligands, which bind to receptors at the neighboring cell and initiate a signaling cascade that promotes stem cell identity. Conversely, cells that leave the niche following ACD fail to receive the instructive cues provided by the niche, and therefore do not maintain stem cell identity but undergo differentiation to generate different cells types to maintain adult tissues. In this manner, extrinsic cues act within a short distance to help balance stem cell renewal with proper differentiation during homeostasis. In addition, asymmetric partitioning of key cell fate determining factors can provide an intrinsic mechanism to generate distinct cell fates (reviewed in [12]). Intrinsic mechanisms have been well described in a host of different systems, which often involve specifically localized proteins or RNAs, as well as polarized cell division. However, it remains unclear whether epigenetic mechanisms may intrinsically regulate cell fate decisions during ACD. Given the crucial roles epigenetic mechanisms play in regulating differential gene expression that defines distinct cell identities, it may serve as an important mechanism in determining cell fate. In this review, we will first go over previous work showing the phenomenon that sister chromatids can carry distinct epigenetic information followed by non-random segregation. We will then focus on recent findings that shed light on how this process is regulated in asymmetrically dividing stem cells. We will conclude by discussing other remaining questions, in the context of recent advances in their respective fields, with the hope to better understand how these mechanisms may help shape diverse epigenomes in multicellular organisms from a variety of different biological contexts.
Generating asymmetry among sister chromatids
Previously, a tag-exchange system was employed to selectively label old and new histones, a major epigenetic information carrier, in order to visualize their segregation patterns in the mitotic male germ cells of Drosophila melanogaster. Strikingly, during the ACD of germline stem cells (GSCs), histones H3 and H4 are asymmetrically inherited, such that a majority of old histones are segregated towards the stem daughter cell whereas new histones are enriched towards the differentiating daughter cell [17, 18]. This asymmetry is critical to the germline maintenance, as disruption of asymmetric histone inheritance leads to phenotypes ranging from cell death and stem cell loss to tumorigenesis. To understand the molecular machinery and cellular pathway(s) underlying this phenomenon, two fundamental processes involved in histone incorporation and segregation were investigated: DNA replication and mitosis [19, 20]. During DNA replication, histones are asymmetrically incorporated such that old H3 and H4 are preferentially incorporated to the leading strand, whereas new H3 and H4 are preferentially segregated towards the lagging strand. Furthermore, fork movement is highly coordinated, such that replication fork shows a high percentage of unidirectional or asymmetric bidirectional fork progression. Taken together, asymmetries in histone incorporation coupled with coordinated replication fork movement could generate sister chromatids each enriched with unique epigenetic information [18]. To understand how epigenetically distinct sister chromatids are recognized and segregated during mitosis, key players in sister chromatid segregation were investigated. These studies demonstrated a series of asymmetries in cis-elements at the sister chromatids such as sister centromeres as well as asymmetries in trans-factors such as the mitotic spindle. These components act together to distinguish epigenetically distinct sister chromatids and ensure their nonrandom segregation during ACD [21].
Part I: Different modes of replication-coupled nucleosome assembly across cell types
Across all organisms, the process of DNA replication allows the genetic information of a cell to be copied and transferred reliably to its daughter cells through many cell divisions [22–24]. For eukaryotic cells, genome replication is linked to the process of epigenome duplication [25–29]. The nucleosome, the basic unit of the eukaryotic epigenome, is composed of 147-base pairs of DNA wound around the histone octamer, an 8-membered complex consisting of two copies each of the histone proteins H2B, H2A, H3 and H4 [30–35]. During DNA replication, old nucleosomes ahead of the fork must be disassembled to allow for replication fork passage [36, 37]. Following disassembly, old histones are retained and deposited onto newly synthesized DNA strands in a process termed histone recycling [38]. New histones must be recruited and deposited to restore nucleosome density [39]. Together, the two processes of old histone recycling and new histone recruitment are referred to as the process of RCNA (Replication-Coupled Nucleosome Assembly) [40, 41].
Over the years of studying DNA replication and RCNA, different models have been proposed to explain how intrinsic differences in leading versus lagging strand synthesis could impact the process of histone recycling. A bias in old histone incorporation towards the leading strand seems logical, as the leading strand, by virtue of its continuous mode of synthesis, has a slight temporal advantage over the lagging strand in the competition for recycling old histones following the replication fork passage (Figure 1A). Electron microscopy studies have demonstrated that the leading strand does in fact ‘outpace’ the lagging strand during strand-synthesis, as evidenced by the fact that nucleosomes are incorporated ~225nm behind the advancing fork on the leading strand, compared to ~285nm on the sister lagging strand [37]. Despite these differences, previous studies investigating the distribution of old histones on newly synthesized sister chromatids have demonstrated a variety of patterns ranging from symmetric to asymmetric. Some studies have shown that old histones are equally distributed between sister chromatids following passage of the replication fork [36, 42, 43], whereas others have shown that old histone incorporation displays a clear strand preference. However, preferential deposition of old histones has been observed on both the leading strand [44–48] and the lagging strand [49], raising questions as to whether there is a consensus pattern in histone recycling, and, if not, what factors may act to regulate different histone deposition modes during RCNA. In this section of the review, recent findings from the Drosophila male germline will be discussed in the context of other recent advances.
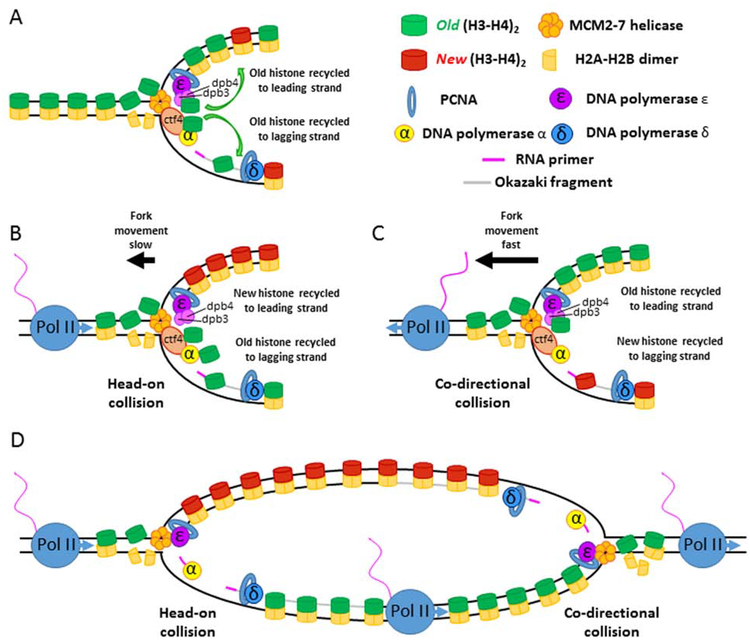
Figure 1: Conservative replication-coupled nucleosome assembly in the context of transcription.
(A) Old histone recycling pathways at the replication fork. Old histones are recycled to the lagging strand via interactions with MCM2-Ctf4-Pol α. Old histones are recycled to the leading strand via interactions with Dpb3 and Dpb4. (B) Histone inheritance in a case of head-on replication-transcription collisions. Head-on collisions slow the replication fork, allowing the MCM2-Ctf4-Polα histone recycling pathway to preferentially place old histones onto the lagging-strand. New histones, by default, populate the leading strand. (C) Histone inheritance in cases of co-directional replication-transcription collisions. Co-directional replication-transcription collisions do not slow replication fork, allowing the MCM2-Dpb3/Dpb4 histone recycling pathway to preferentially place old histones onto the leading-strand. New histones, by default, populate the lagging strand. (D) Conservative replication-coupled nucleosome assembly showing asymmetric histone inheritance at a bidirectional replication fork flanked by active transcription. Old histones are segregated towards one sister chromatid, whereas new histone are segregated towards the other. Transcription machinery interacts with both edges of the replication bubble, but in opposite orientations, thereby biasing histone inheritance to different strands (leading versus lagging), but the same sister chromatid.
1. Replication proteins regulate histone incorporation at the replication fork
Recent studies have revealed several key players involved in the process of RCNA. For instance, MCM2, an essential subunit of the MCM2–7 replicative helicase, was found to play a crucial role in retaining old histones displaced ahead of the advancing replication fork (Figure 1A) [50]. Loss of MCM2’s chaperone activity results in biased old histone recycling towards the leading strand. From these studies, it has been hypothesized that MCM2 functions at the fork to buffer the temporal differences between leading strand and lagging strand synthesis to allow old histones to be more evenly deposited onto the two sister chromatids [51]. Further studies have demonstrated that once retained by MCM2, old histones can be transferred to the lagging strand via an interaction with CTF4 and DNA polymerase α (Pol α) [52]. Additionally, Proliferating Cell Nuclear Antigen (PCNA), a molecule initially characterized as a processivity factor for DNA polymerases [53], was found to coordinate histone deposition at the replication fork [54, 55]. As CTF4, Pol α and PCNA are all enriched at the lagging strand [56], it has been hypothesized that this could serve as an axis to deposit old histones onto the lagging strand (Figure 1A) [55, 57].
Other studies have demonstrated that key replication proteins could also act as chaperones to facilitate deposition of old histones onto the leading strand. For example, DPB3 and DPB4, two subunits of the leading-strand DNA polymerase ε (Pol ε), have been shown to regulate deposition of old histones onto the leading strand (Figure 1A) [58]. Loss-of-function of either subunit compromises old histone deposition onto the leading strand, which results in a biased incorporation of old histones towards the lagging strand.
Interestingly, recent studies have suggested that transcription can also impact histone inheritance at the replication fork by modulating the rate of progression. Prior research has shown that head-on collisions (Figure 1B), where transcription direction is opposite to that of replication fork movement, are more likely to slow fork progression compared to co-directional progression, where transcription and replication are oriented towards the same direction (Figure 1C) [59]. Studies have further demonstrated that chromatin maturation rates vary according to these interactions. In case of co-directional replication and transcription, the leading strand matures faster compared to the lagging strand. Conversely, in cases of head-on collisions, the lagging strand matures faster [60]. While it remains unclear whether interactions between replication and transcription could bias old histone re-incorporation at the replication fork, a recent study suggests that slower rates of fork progression in case of head-on collisions could reduce the temporal difference between leading and lagging strand synthesis, thereby allowing the lagging strand to be more effective in recycling old histones (Figure 1B and 1D). Conversely, faster fork movement in the presence of co-directional replication with transcription could result in a more pronounced temporal delay of lagging strand synthesis, which may give leading strand advantages for re-incorporating old histones (Figures 1C and 1D) [61]. It is necessary to rigorously test these models in order to better understand the molecular and temporal factors responsible for old and new histone incorporation patterns at the replication fork.
2. Technology advances revealed distinct histone incorporation patterns in different biological systems
(1). Sequencing-based methods revealed distinct histone incorporation patterns at leading versus lagging strands in yeast and cell culture system:
Recent years have witnessed the development of a series of sequencing-based techniques to study replication-coupled histone inheritance patterns in different systems (Table 1). Two recent studies employed strand-specific sequencing methods to characterize old versus new histone distribution at newly synthesized DNA strands immediately after passage of the replication fork. In one study, Yu et al. utilized a method called Enrichment and Sequencing of Protein-Associated Nascent DNA (eSPAN) to map old versus new histone distribution at newly replicated DNA in the yeast Saccharomyces cerevisiae. This study showed a slight preference in old histone distribution towards the lagging strand [58]. In the another study, Petryk et al. used a similar method termed Sister Chromatids After Replication by DNA sequencing (SCAR-seq) to investigate histone incorporation patterns in cultured mouse embryonic stem cells (mESCs) and found a slight bias of old histone distribution towards the leading strand [51]. The different patterns of histone distribution in yeast versus mESCs raises interesting and important questions regarding the underlying mechanisms.
Table 1: Different technologies for studying Replication-Coupled Nucleosome Assembly (RCNA).
SuperResolved Chromatin Fibers (SRCF) [18]; Sister Chromatids After Replication by DNA sequencing (SCAR-seq) [51]; Enrichment and Sequencing of Protein Associated Nascent DNA (eSPAN) [58]; Mapping In Vivo Nascent Chromatin with EdU and Sequencing (MINCE-seq) Nascent Chromatin Avidin Pull-down (NChAP) [60]; Nascent Chromatin Occupancy Profiling (NCOP) [129].
| Technique | Sequence information | Single-molecule resolution | Unbiased versus candidate approach | High-throughput | Sister chromatid resolution |
|---|---|---|---|---|---|
| SRCF [18] | no | yes | candidate | no | yes |
| SCAR-seq [51] | yes | no | candidate | yes | yes |
| eSPAN [58] | yes | no | candidate | yes | yes |
| MINCE-seq [128] | yes | no | unbiased | yes | no |
| NChAP [60] | yes | no | unbiased | yes | yes |
| NCOP [129] | yes | no | unbiased | yes | no |
Several factors could contribute to these differences: Given the roles that replication components such as CTF4, DPB3 and DPB4 have in RCNA, it is possible that their relative abundance at the fork could bias histone incorporation towards the leading strand versus the lagging strand in different systems. Other proteins with crucial roles in DNA replication, such as FACT, RPA or the TONSL-MMS22L complex, have also been reported to regulate histone incorporation patterns [62–67]. These studies raise an intriguing possibility that the histone incorporation pattern at a specific gene region, in any given cell type, and at the particular development stage could depend on coordinated activities of these factors at the replication fork, which are themselves subject to dynamic developmental regulation of multicellular organisms [68, 69].
(2). Chromatin fiber combined with superresolution microscopy method allows visualization of distinct histone incorporation patterns in Drosophila:
By combining the chromatin fiber technique [70–72] with superresolution imaging, the SuperResolved Chromatin Fibers (SRCF; Table 1) method allows direct visualization of histone incorporation patterns at replicating regions in early-stage male germ cells of Drosophila [18]. Using this method, it was found that old and new H3 and H4 display asymmetric incorporation patterns at the replication fork, with a bias towards the leading strand. Conversely, old and new histone H2A showed largely symmetric incorporation pattern at the replicative regions. These distinction in histone distribution patterns suggest molecular specificities in the process of RCNA. Interestingly, previous studies have also demonstrated that different histones show distinct behaviors during DNA replication as well as during post-replication chromatin maturation. During replication, H3 and H4 are incorporated as (H3-H4)2 tetramers, whereas H2A and H2B are incorporated as dimers. Whereas one old (H3-H4)2 tetramer can only be inherited by one strand (leading or lagging), the presence of two old H2A-H2B dimers could allow for their reincorporation by both the leading and the lagging strands, thus ensuring a more symmetric inheritance pattern at the replication fork. Additionally, the relative stability of H2A-H2B dimers versus the (H3-H4)2 tetramer in the nucleosome structure could also account for their different distribution patterns observed on newly replicated chromatin fibers. Previous studies have shown that (H3-H4)2 tetramers are relatively stable in the nucleosome structure, while nucleosomal H2A-H2B dimers display a much more rapid exchange during post-replication chromatin maturation. A rapid exchange following replication fork passage could negate asymmetries in H2A-H2B inheritance initially established in the wake of the replication fork. More experiments are needed to better understand whether differences between H3-H4 and H2A-H2B first appear during the processes of RCNA or during chromatin maturation.
Notably, the distinct inheritance patterns observed for H3-H4 versus H2A-H2B on newly replicated chromatin fibers mirror the differences in global histone segregation patterns observed during ACD of GSCs, suggesting that distinct segregation patterns at the replication fork underlie distinct histone inheritance modes observed in mitotic and post-mitotic daughter cells[17, 18, 73]. Interestingly, histone inheritance patterns in the Drosophila male germline can vary based on the differentiation state of cells: GSCs have asymmetric inheritance while more differentiated germ cells show globally symmetric inheritance [17, 18], suggesting that the processes of RCNA are likely subject to cell-type-specific regulation during development. A key area for future research will be better understanding what factors are involved in regulating changes in histone inheritance observed in the Drosophila male germline.
(3). Chromatin fiber and DNA fiber methods track fork movement in cells from developing tissues:
Chromatin fiber or DNA fiber technology combined with a sequential nucleoside analog incorporation assay can be used to analyze replication fork progression patterns at single-molecule level. Using these methods, fibers isolated from the Drosophila testes showed a high incidence of unidirectional and asymmetric bidirectional replication fork movements. In contrast, chromatin fibers and DNA fibers isolated from the replicative Drosophila somatic cells showed largely symmetric bidirectional replication fork movement. Together with the strand bias between old and new H3, biased and coordinated fork movement could expand asymmetric H3 incorporation at individual forks to long-range asymmetry between sister chromatids [18]. On the contrary, the symmetric bidirectional replication fork movement mode could ensure both sister chromatids inherit similar amount of old histones. With the subsequent activities of epigenetic “readers” and “writers”, this process could ensure propagation of cells with similar “epigenetic memory”, as seen in yeast, cultured cells, or transit-amplifying cells in stem cell lineages.
Given their compatibility with small numbers of cells, these methods are highly adaptable to other biological systems, particularly cells isolated from their physiological environment during development. Together with the SRCF analysis of strand bias of different histones, these methods would allow for characterization of both fork movement and histone inheritance during the process of RCNA.
Coordinated fork movement has been reported in organisms ranging from bacteria to multicellular eukaryotes. In most cases, this coordination is required to avoid deleterious head-on collisions between replication progression and active transcription [74]. For example, robust transcription at the Drosophila rDNA locus during DNA replication requires coordinated forks to prevent genome instability associated with head-on replication-transcription collisions [75]. Similar patterns have also been reported for rDNA replication in other species [76, 77]. Regulated fork movement has also been reported in Saccromyces pombii, where fork movement is coordinated to ensure that the mating-type locus is replicated unidirectionally [78]. This coordination is developmentally relevant, as differences in leading versus lagging strand synthesis play a causal role during the process of mating-type switching in S. pombii.
In many cases, biased fork movement is regulated by replication components that bind in a sequence-specific manner. In the two examples above at replicating rDNA locus and at the mating-type locus, specific replication machinery proteins bind proximal to origins and impede the efficient bidirectional progression of the replication fork [79]. However, these physical blocking activities at specific DNA sequences are usually limited to a single genomic locus. Proteins that have been reported to regulate fork movement more globally often interact with the replisome. For example, SUppressor of Underreplicated Regions (SuUR) acts with Rif1 to slow fork movement in endoreplicating Drosophila cells to allow for selective amplification of the genome [80–82]. Recently, several components of the Polycomb group complex (PcG) have been reported to directly interact with advancing replication forks to regulate both the rate and the directionality of fork progression. Compromising activities of a PcG component Enhancer of zeste [E(z)] resulted in increased unidirectional fork movement in mouse embryonic fibroblasts [83]. Future studies will be needed to determine if components of PcG complex, or proteins such as SuUR and/or Rif1, function to regulate fork progression in Drosophila male GSCs in order to generate epigentically-distinct sister chromatids in preparation for ACD.
Part II: Sister chromatid recognition and segregation during asymmetric cell division
Even though old and new histones could be differentially incorporated during DNA replication, in order to achieve the asymmetric inheritance pattern, the mitotic machinery must have a mechanism to distinguish sister chromatids based on their histone composition. Many hypotheses have been proposed to explain non-random sister chromatid segregation. An “immortal DNA strand” hypothesis by John Cairn proposes that stem cells preferentially inherit older DNA strands to avoid replication-introduced errors [84]. However, this hypothesis has been challenged by many subsequent studies [85–90]. On the other hand, biased sister chromatid segregation has been reported in multiple systems, including a subpopulation of mouse muscle stem cells (i.e. satellite cells) [91], as well as mouse and human ESC systems [92, 93], where the DNA methyltransferase Dnmt3 has been shown to contribute to this process [93].
A previous study demonstrated that Drosophila male GSCs do not exhibit “immortal strand” phenomenon [85, 94]. However, sister chromatids of X and Y chromosomes display biased strand segregation while the second and the third autosomes show a co-segregation pattern without strand bias [95]. The putative Drosophila DNA methyltransferase Dnmt3 is required for biased sex chromosome segregation, suggesting potential involvement of epigenetic regulation [96]. The findings that old and new histone H3 and H4 segregate asymmetrically [17, 18] provoke the mechanisms underlying non-random sister chromatid segregation in asymmetrically dividing Drosophila male GSCs. Recently, an axis of cis-elements from sister chromatids and trans-factors from the mitotic machinery have been shown to coordinate in a spatiotemporally controlled manner to ensure non-random sister chromatid segregation [21].
1. Centromeres with epigenetic asymmetry between sister chromatids
Centromeres are specific chromosomal domains that recruit the kinetochore protein complex and constitute centromere-associated network protein complex, which act together to ensure proper attachment and accurate segregation of sister chromatids during mitosis. Centromere is epigenetically defined by a histone H3 variant called centromere protein-A (CENP-A) [97–99]. During mitosis, centromeres resolve into sister centromeres, with one centromere on each sister chromatid. Genetically and epigenetically sister centromeres are considered identical in a symmetrically dividing cell. Two hypotheses have been proposed for asymmetrically dividing cells, termed “strand-specific imprinting and selective chromatid segregation” and “silent sister chromatids” [100–103]. These hypotheses propose that epigenetic differences at the sister centromere region could guide preferential attachment and segregation of the sister chromatids to reach an asymmetric outcome. However, up to date no clear in vivo evidence in any multicellular organisms has been shown to support these hypotheses.
In unicellular organism yeast Saccharomyces cerevisiae, it has been shown that several kinetochore components segregate asymmetrically in a lineage-specific manner post-meiotically [104]. The authors suggested that this phenomenon could be related to lineage-defining asymmetry in multicellular organisms, such as in stem cell lineages. Recently, it has been shown that in asymmetrically dividing Drosophila Intestinal Stem Cells (ISCs), old CENP-A is preferentially retained in ISCs for stem cell activity [105]. Together, these observations suggest that centromeres could play an important role in determining cell fate.
Recently, studies using asymmetrically dividing Drosophila male GSCs showed asymmetrically segregated CENP-A, with approximately 1.4- to 1.5-fold more CENP-A towards the future GSC (Figure 2A) [21]. Interestingly, in prometaphase GSCs when sister centromeres are resolved, quantitative asymmetry is already detectable. The stronger centromere is subsequently attached by the microtubules (MTs) emanating from the mother centrosome towards the GSC side, in order to ensure the sister chromatid carrying it to be segregated into the future GSC (Figure 2A). Recently, a similar phenomenon has also been reported in Drosophila females GSC by Dattoli et al. [106]. It has also been shown that new CENP-A incorporation occurs in late G2 to early prophase in both male and female GSCs of Drosophila. In the male GSCs, using a photoconvertable CENP-A-Dendra2 to distinguish old and new CENP-A, it has been shown that both old and new CENP-A proteins are enriched at the sister chromatid segregated into GSCs. Together, these results suggest that the asymmetric sister centromere could drive non-random sister chromatid segregation.
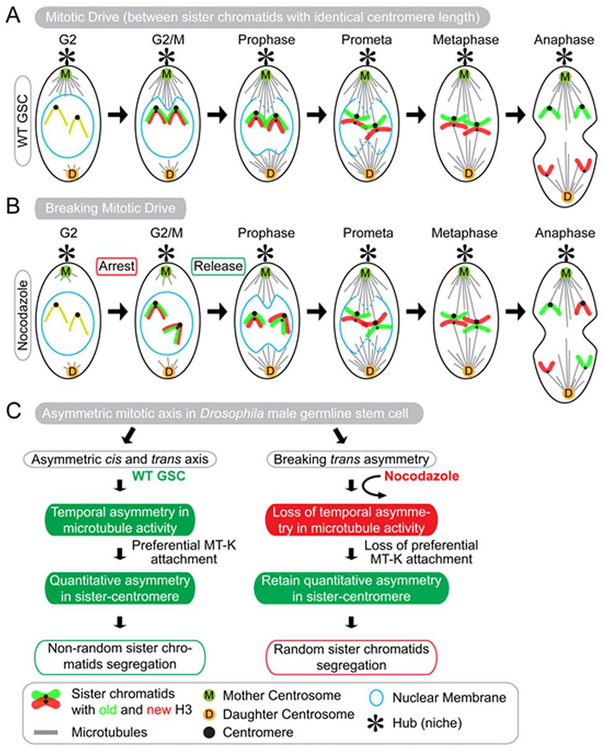
Figure 2: Schematic illustration of cis-asymmetry and trans-asymmetry, which coordinate to ensure nonrandom sister chromatids segregation in Drosophila male GSCs.
(A) Dynamic interactions between cis and trans factors at different stages during the cell cycle of Drosophila male GSCs. From mid-G2 to G2/M transition, microtubules from the mother centrosome actively interact with and break nuclear envelope locally at the stem cell side in early prophase. Microtubules emanated from the mother centrosome then interact with the centromeres and preferentially attach to the stronger centromere. At a later time point in prometaphase, microtubules from the daughter centrosome break the nuclear envelope at the differentiating daughter cell side and attach to the weaker centromere. (B) Breaking the trans-asymmetry using nocodazole (NZ) disrupts the temporal asymmetry in microtubule activity, which results in the loss of preferential sister chromatid attachment and randomizes their segregation in anaphase. (C) Summary of disrupting mitotic drive which leads to randomized sister chromatid segregation.
Additionally, studies in Caenorhabditis elegans with holocentric chromosomes have shown that CENP-A acts as a molecular “ruler” to determine chromosome length during early embryonic development. A linear relationship exists between the CENP-A amount and the chromosome length, with the shorter chromosomes contain less CENP-A compared to the longer chromosomes. Perturbation of the CENP-A levels caused by overexpression or partial knockdown results in alteration of the chromosome length and mitotic errors [107]. Studies in Saccharomyces cerevisiae have demonstrated that centromeres play a critical role in promoting chromosome condensation in cis by recruiting the kinases Aurora B and Bub1. Spreading of the condensation from the centromere to the chromosomal arms is facilitated by Shugoshin and histone deacetylase Hst2 [107]. Chromosomes lacking a centromere fail to condense and are arrest for mitotic progression. However, targeting Aurora B to a centromere-ablated chromosome or releasing Shugoshin from inhibition enhance chromosomal condensation and facilitate mitotic progression. These results indicate that centromeres act as a licensing locus and regulate the entire process of chromosomal condensation [108]. Although it remains unclear how asymmetric centromeres form between genetically identical sister chromatids in the Drosophila GSCs, considering the diverse nature of CENP-A/centromere, it is possible that centromeres act as a sensor/licensing locus to read the epigenetic differences between sister chromatids to scale up or down sister centromere sizes accordingly. In the future, it would be interesting to investigate these possibilities to understand the molecular mechanisms that establish asymmetric sister centromeres in Drosophila GSCs.
2. Asymmetric mitotic spindle facilitates non-random sister chromatids segregation
It has been shown that centrosomes are inherited with a non-random pattern in several systems [109–113]. The mother and daughter centrosomes are intrinsically distinct based on their age difference, which results in their differential activity as microtubule organizing center (MTOC) [109, 114–116]. In some systems, the mother centrosome has a higher MTOC activity and is inherited by the stem cells, such as in Drosophila male GSCs and mouse neural progenitor cells [109, 110]. In other systems, such as Drosophila neuroblasts and female GSCs, daughter centrosomes have a higher MTOC activity and are inherited by stem cells [111–113]. Even though stem cells could inherit either mother or daughter centrosome, the inheritance of the centrosome associated with higher MTOC activity seems to be conserved.
Recently it was shown that in Drosophila male GSCs MTs emanated from mother centrosome versus daughter centrosome are temporally asymmetric: The mother centrosome actively emanates MTs as early as in mid-G2 phase whereas the daughter centrosome only becomes active at the G2/M transition (Figure 2A, 2C) [21]. The more active MTs from the mother centrosome robustly interact with the nuclear envelope for a polarized nuclear envelope breakdown proximal to the stem cell side, followed by preferential attachment to the stronger sister centromere in early prophase GSCs. By contrast, the daughter centrosome emanates MTs later, leading to their attachment to the weaker sister centromere for segregating towards the differentiating daughter cell side. The spindle assembly checkpoint (SAC) functions. For example, SAC provides a feedback control that monitors the tension between the microtubule and the kinetochore-centromere-sister chromatid axis, in order to balance the force generated on the kinetochore and ensure bipolar spindle microtubule attachment [117–119]. An imbalance of the tension has been shown to activate a correction pathway against erroneous microtubules attachment. In GSCs, the differential attachment of microtubules to the asymmetric sister centromeres presumably generates an unequal force, which should activate SAC and arrest mitosis. The fact that GSCs can proceed with mitosis without being arrested suggest either a different tension sensing machinery or mechanisms allow for bypassing this in GSCs.
Interestingly, disruption of the temporal asymmetry of microtubules from mother versus daughter centrosomes by treatment with the microtubule depolymerizing drug nocodazole (NZ) leads to loss of their preferential attachment to sister chromatids, resulting in randomized segregation pattern of sister chromatids (Figure 2B, 2C). Under this condition, the asymmetry between individual sister centromeres is retained, suggesting that the temporal asymmetry of microtubule activity is critical for nonrandom sister chromatid recognition and segregation. Collectively, these results suggest a stem cell-specific ‘mitotic drive’ with two steps: The first step involves establishment of asymmetry at individual sister centromeres and the second step is to recognize sister centromere asymmetry by the mitotic machinery.
A ‘meiotic drive’ hypothesis has been proposed to understand how certain chromosomes could be retained in developing egg during meiosis I, in which the allele with a stronger kinetochore is retained by the oocyte while the allele with a weaker kinetochore is segregated to polar bodies for degeneration [120, 121]. In female mice, the stronger kinetochore often associates with the longer “selfish” centromere and has more affinity to the meiotic spindle, which itself has been asymmetrically modified due to polarized cellular signaling [122–125]. Additionally, it has been shown that a microtubule motor protein regulates ‘meiotic drive’ in maize, indicating a role of microtubules in selective attachment to centromeres [126, 127].
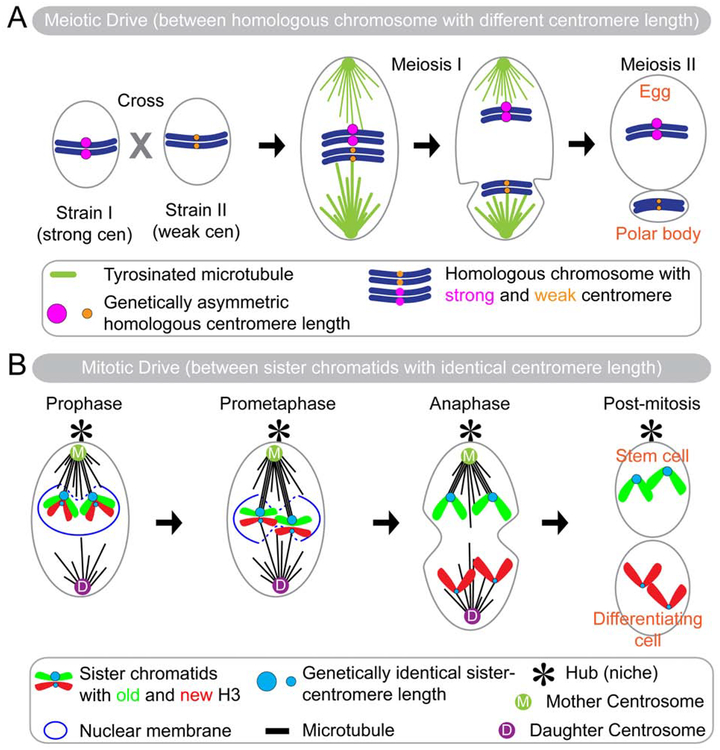
However, it remained unclear whether these asymmetric functions of centromere could act in mitosis and regulate sister chromatid segregation. Taken together results reported in Ranjan et al. and Dattoli et al., it seems that a similar mechanism also exists in mitosis. Nevertheless, the ‘mitotic drive’ has distinct features compared with the ‘meiotic drive’. In meiosis the centromere difference occurs between specific homologous chromosomes (Figure 3A), whereas in mitosis it occurs between genetically identical sister chromatids (Figure 3B). In meiosis, homologous chromosome has inherent asymmetry from two different mouse lines, one has a stronger centromere than the other one (Figure 3A). The stronger centromere has a greater number of minor satellite repeats, which provide more space for assembling CENP-A nucleosomes and subsequently kinetochore proteins. In contrast, in mitosis, sister centromeres have identical sequences in theory, therefore how they could assemble CENP-A asymmetrically remain a mystery and will need further studies. One possibility is that during replication of centromeric DNA sequences, old CENP-A is asymmetrically recycled like old H3, which may carry some post-translational modifications, in order to recruit more CENP-A chaperone CAL-1 to incorporate more new CENP-A during the subsequent G2 phase, prior to asymmetric mitosis.
Figure 3: Schematic illustration to compare the meiotic drive in mouse oogenesis and the mitotic drive in Drosophila male GSCs.
(A) A cross between two mouse strains, strain I with the strong centromere and strain II with the weak centromere, leads to meiotic drive phenomenon during meiosis I. The stronger centromere is preferentially retained in the future oocyte and the weaker centromere in the future polar bodies. Asymmetric tyrosination facilitates the preferential MT-K attachment: more tyrosinated microtubule destabilizes stronger centromere attachment and hence stabilizes attachment with weaker centromeres. Stronger centromere has longer α-satellite repeat and hence gets more space to incorporate more CENP-A. (B) Mitotic Drive occurs between sister-chromatids. Theoretically, sister chromatids have identical sequences at sister centromeres, but in GSCs CENP-A is incorporated differentially at sister centromeres through an unknown mechanism. The stronger centromere is stabilized by more microtubule emanated from the mother centrosome and segregates into the self-renewed stem cell. By contrast, the weaker centromere is attached by microtubule emanated from the daughter centrosome and segregates into differentiating daughter cell. This centromere driven nonrandom sister chromatid segregation is termed as “mitotic drive”.
Conclusions and perspectives
A long-standing question in developmental biology of any multicellular organisms is how distinct cell fates can be established and maintained. It has been shown for decades that cells carry the “epigenetic memory” to maintain their identities over many cell divisions. Asymmetric histone inheritance provides an elegant intrinsic mechanism that not only help cells maintain certain “memory”, but also take on distinct cell fates. In fact, this later role could be more relevant to challenges that cells always face in developing tissues in vivo compared to cells in culturing conditions, since very often the symmetric outcome expected from conventional mitosis needs to be altered for an asymmetric one.
Although the studies in Drosophila GSCs started to provide molecular insight, cellular bases and biological significance of non-random sister chromatid segregation, many questions remain. For example, understand the factors responsible for different RCNA patterns in stem cells versus progenitor cells represents a crucial area for future research. Given GSCs’ proximity to niche-secreted signaling molecules, it would be intriguing to explore how intrinsic histone inheritance pattern could respond to extrinsic signaling molecules. Moreover, outstanding questions remain, such as what molecular features differ between old and new histones including CENP-A; how differential inheritance may dictate distinct gene expression patterns and cell fates; and how this mode of asymmetry may change in accordance with the mode of stem cell division. Additionally, it will be important to investigate whether asymmetric histone inheritance is a common feature of asymmetrically dividing cells. It is highly likely that the extent of this asymmetry could be cell-type-specific. For example, it is likely that asymmetric histone inheritance occurs at gene regions that are differentially expressed between the two daughter cells. If the number of differentially expressed genes is large enough which cover broad genomic regions, asymmetric histone inheritance would appear global. However, if these differentially expressed genes are just a small subset which are applicable for many stem cell lineages, asymmetric histone inheritance might be local. Interestingly, in metazoan species, germ cell differentiation may represent one of the most drastic cellular differentiation pathways, given how morphologically and functionally mature gametes are distinct from progenitor germ cells. It would be intriguing to study histone inheritance patterns in the germline from other species, as well as at different stages of germ cell differentiation, since the asymmetric outcome could be required at distinct stages. For example, in the male germline this difference is likely established at the male germline stem cell asymmetric division stage, while in female this outcome is needed at the stage to specify oocyte. Characterizing histone establishment and inheritance patterns at specific gene loci and different developmental stages should help uncover the roles that epigenetic mechanisms have in generating the multitude of diverse cell types required to pattern and maintain a multicellular organism.
Outstanding Questions.
Are Polycomb group, SuUR and/or Rif1 proteins involved in regulating fork movement?
What molecular features of histones are involved in dictating their incorporation patterns at the replication fork?
Do MCM2-CTF4-Pol α and Dpb3/Dpb4 have equal affinity for old versus new histones?
How is CENP-A recruited asymmetrically at the sister centromeres?
What mechanisms guide temporal asymmetry of microtubule activity?
Is mitotic drive a common feature in asymmetrically dividing cells?
How to break symmetry to achieve an asymmetric outcome in multicellular organisms during development, homeostasis and regeneration?
Trend Box.
Non-random sister chromatid segregation has been proposed in asymmetrically dividing cell.
Sister chromatids has asymmetric epigenetic marks due to asymmetric incorporation of old versus new histones.
Differential epigenetic inheritance and differential gene expression has been proposed to regulate distinct cell fate.
Acknowledgements:
We thank X.C. lab members for insightful suggestions. Supported by NIH 5T32GM007231 and F31GM115149-01A1 (M.W.), NIGMS/NIH R01GM112008 and R35GM127075, the Howard Hughes Medical Institute, the David and Lucile Packard Foundation, and Johns Hopkins University startup funds (X.C.).
Glossary
- Replication fork
a structure defined as the transition from unreplicated dsDNA and newly separated and replicating single-stranded DNA (ssDNA), which serve as templates for synthesizing new DNA strands on both the continuously-synthesized leading strand and the discontinuously-synthesized lagging strand.
- Replication-coupled nucleosome assembly (RCNA)
The combined process of old histone recycling and new histone incorporation that occurs contemporaneously with the progression of the replication fork.
- Unidirectional DNA replication
Progression of the replication fork in only one direction as opposed to bi-directional replication progression.
- Asymmetric histone recycling
Biased recycling of old histones onto duplicating sister chromatids during replication fork progression.
- Asymmetric sister centromere
In general, the underlying DNA sequence of sister centromeres is identical in an organism and incorporates the same amount of the CENP-A nucleosome to form symmetric sister centromeres. In Drosophila male GSCs, however, one sister centromere has approximately 1.5-fold more CENP-A than the other, creating asymmetry between individual sister centromere pairs.
- Temporally asymmetric microtubule activity
Centrosomes are MTOCs where both centrosomes nucleate spindle MTs at almost the same time upon mitotic entry. In Drosophila male GSCs, the mother centrosome nucleates MTs earlier than the daughter centrosome.
- Mitotic drive
The stem cell mitotic drive occurs due to asymmetric strength between sister centromeres and temporal asymmetry in MT activity in Drosophila male GSCs. This leads to the retention of stronger sister centromeres in the self-renewed stem cell.
- Nonrandom sister chromatid segregation
The sister chromatid inheritance is thought to be a random process in most mitosis. However, under certain circumstances such as ACDs, sister chromatids show a nonrandom distribution, with certain chromatids preferentially segregated to one daughter cell.
Footnotes
Conflict of Interest
No conflict of interest.
References
- 1.Clayton E, et al. , A single type of progenitor cell maintains normal epidermis. Nature, 2007. 446(7132): p. 185–9. [DOI] [PubMed] [Google Scholar]
- 2.Dhawan J and Rando TA, Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol, 2005. 15(12): p. 666–73. [DOI] [PubMed] [Google Scholar]
- 3.Barker N, et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature, 2007. 449(7165): p. 1003–7. [DOI] [PubMed] [Google Scholar]
- 4.Crane GM, Jeffery E, and Morrison SJ, Adult haematopoietic stem cell niches. Nat Rev Immunol, 2017. 17(9): p. 573–590. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida S, Sukeno M, and Nabeshima Y, A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science, 2007. 317(5845): p. 1722–6. [DOI] [PubMed] [Google Scholar]
- 6.Morrison SJ and Spradling AC, Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell, 2008. 132(4): p. 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Micchelli CA and Perrimon N, Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature, 2006. 439(7075): p. 475–9. [DOI] [PubMed] [Google Scholar]
- 8.Ohlstein B and Spradling A, The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature, 2006. 439(7075): p. 470–4. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann R, Germline stem cells: origin and destiny. Cell Stem Cell, 2012. 10(6): p. 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller MT and Spradling AC, Male and female Drosophila germline stem cells: two versions of immortality. Science, 2007. 316(5823): p. 402–4. [DOI] [PubMed] [Google Scholar]
- 11.Knoblich JA, Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol, 2010. 11(12): p. 849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkei ZG and Yamashita YM, Emerging mechanisms of asymmetric stem cell division. J Cell Biol, 2018. 217(11): p. 3785–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clevers H, Stem cells, asymmetric division and cancer. Nat Genet, 2005. 37(10): p. 1027–8. [DOI] [PubMed] [Google Scholar]
- 14.Morrison SJ and Kimble J, Asymmetric and symmetric stem-cell divisions in development and cancer. Nature, 2006. 441(7097): p. 1068–74. [DOI] [PubMed] [Google Scholar]
- 15.Kahney EW, et al. , Symmetry from Asymmetry or Asymmetry from Symmetry? Cold Spring Harb Symp Quant Biol, 2017. 82: p. 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Losick VP, et al. , Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell, 2011. 21(1): p. 159–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tran V, et al. , Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science, 2012. 338(6107): p. 679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wooten M, et al. , Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat Struct Mol Biol, 2019. 26(8): p. 732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran V, Feng L, and Chen X, Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions. Chromosome Res, 2013. 21(3): p. 255–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie J, et al. , Breaking Symmetry - Asymmetric Histone Inheritance in Stem Cells. Trends Cell Biol, 2017. 27(7): p. 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranjan R, Snedeker J, and Chen X, Asymmetric centromere and dynamic microtubules coordinate to ensure biased sister chromatid segregation in Drosophila male germline stem cells. http://biorxiv.org/cgi/content/short/416446v2. Cell Stem Cell, in press, available online on Sep. 26, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgers PMJ and Kunkel TA, Eukaryotic DNA Replication Fork. Annu Rev Biochem, 2017. 86: p. 417–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell SP and Dutta A, DNA replication in eukaryotic cells. Annu Rev Biochem, 2002. 71: p. 333–74. [DOI] [PubMed] [Google Scholar]
- 24.DePamphilis ML, Review: nuclear structure and DNA replication. J Struct Biol, 2000. 129(2–3): p. 186–97. [DOI] [PubMed] [Google Scholar]
- 25.Alabert C, Jasencakova Z, and Groth A, Chromatin Replication and Histone Dynamics. Adv Exp Med Biol, 2017. 1042: p. 311–333. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran S, Ahmad K, and Henikoff S, Capitalizing on disaster: Establishing chromatin specificity behind the replication fork. Bioessays, 2017. 39(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller TC and Costa A, The architecture and function of the chromatin replication machinery. Curr Opin Struct Biol, 2017. 47: p. 9–16. [DOI] [PubMed] [Google Scholar]
- 28.Ehrenhofer-Murray AE, Kamakaka RT, and Rine J, A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics, 1999. 153(3): p. 1171–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alabert C and Groth A, Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol, 2012. 13(3): p. 153–67. [DOI] [PubMed] [Google Scholar]
- 30.Luger K, et al. , Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 1997. 389(6648): p. 251–60. [DOI] [PubMed] [Google Scholar]
- 31.Arents G, et al. , The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A, 1991. 88(22): p. 10148–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klug A, et al. , A low resolution structure for the histone core of the nucleosome. Nature, 1980. 287(5782): p. 509–16. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg RD and Thomas JO, Chromatin structure; oligomers of the histones. Science, 1974. 184(4139): p. 865–8. [DOI] [PubMed] [Google Scholar]
- 34.McGinty RK and Tan S, Nucleosome structure and function. Chem Rev, 2015. 115(6): p. 2255–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKnight SL and Miller OL Jr., Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell, 1977. 12(3): p. 795–804. [DOI] [PubMed] [Google Scholar]
- 36.Annunziato AT, Assembling chromatin: the long and winding road. Biochim Biophys Acta, 2013. 1819(3–4): p. 196–210. [DOI] [PubMed] [Google Scholar]
- 37.Sogo JM, et al. , Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol, 1986. 189(1): p. 189–204. [DOI] [PubMed] [Google Scholar]
- 38.Probst AV, Dunleavy E, and Almouzni G, Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol, 2009. 10(3): p. 192–206. [DOI] [PubMed] [Google Scholar]
- 39.Hammond CM, et al. , Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol, 2017. 18(3): p. 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snedeker J, Wooten M, and Chen X, The Inherent Asymmetry of DNA Replication. Annu Rev Cell Dev Biol, 2017. 33: p. 291–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worcel A, Han S, and Wong ML, Assembly of newly replicated chromatin. Cell, 1978. 15(3): p. 969–77. [DOI] [PubMed] [Google Scholar]
- 42.Jackson V and Chalkley R, A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell, 1981. 23(1): p. 121–34. [DOI] [PubMed] [Google Scholar]
- 43.Russev G and Hancock R, Assembly of new histones into nucleosomes and their distribution in replicating chromatin. Proc Natl Acad Sci U S A, 1982. 79(10): p. 3143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley D and Weintraub H, Conservative segregation of parental histones during replication in the presence of cycloheximide. Proc Natl Acad Sci U S A, 1979. 76(1): p. 328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seale RL, Studies on the mode of segregation of histone nu bodies during replication in HeLa cells. Cell, 1976. 9(3): p. 423–9. [DOI] [PubMed] [Google Scholar]
- 46.Weintraub H, Cooperative alignment of nu bodies during chromosome replication in the presence of cycloheximide. Cell, 1976. 9(3): p. 419–22. [DOI] [PubMed] [Google Scholar]
- 47.Seidman MM, Levine AJ, and Weintraub H, The asymmetric segregation of parental nucleosomes during chrosome replication. Cell, 1979. 18(2): p. 439–49. [DOI] [PubMed] [Google Scholar]
- 48.Leffak IM, Grainger R, and Weintraub H, Conservative assembly and segregation of nucleosomal histones. Cell, 1977. 12(3): p. 837–45. [DOI] [PubMed] [Google Scholar]
- 49.Roufa DJ and Marchionni MA, Nucleosome segregation at a defined mammalian chromosomal site. Proc Natl Acad Sci U S A, 1982. 79(6): p. 1810–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, et al. , A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol, 2015. 22(8): p. 618–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petryk N, et al. , MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science, 2018. 361(6409): p. 1389–1392. [DOI] [PubMed] [Google Scholar]
- 52.Roufail J, et al. , A novel integrative healing services approach for neurosurgery inpatients: Preliminary experiences and cost calculations. Interdiscip Neurosurg, 2018. 13: p. 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choe KN and Moldovan GL, Forging Ahead through Darkness: PCNA, Still the Principal Conductor at the Replication Fork. Mol Cell, 2017. 65(3): p. 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Shibahara K, and Stillman B, PCNA connects DNA replication to epigenetic inheritance in yeast. Nature, 2000. 408(6809): p. 221–5. [DOI] [PubMed] [Google Scholar]
- 55.Shibahara K and Stillman B, Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell, 1999. 96(4): p. 575–85. [DOI] [PubMed] [Google Scholar]
- 56.Yu C, et al. , Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell, 2014. 56(4): p. 551–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakano S, Stillman B, and Horvitz HR, CAF-1-Mediated Chromatin Assembly Generates a Bilateral Asymmetry in C. elegans Neuroanatomy. Cell, 2011. 147(7): p. 1525–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu C, et al. , A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science, 2018. 361(6409): p. 1386–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pomerantz RT and O’Donnell M, What happens when replication and transcription complexes collide? Cell Cycle, 2010. 9(13): p. 2537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasseur P, et al. , Dynamics of Nucleosome Positioning Maturation following Genomic Replication. Cell Rep, 2016. 16(10): p. 2651–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziane R, Camasses A, and Radman-Livaja M, Mechanics of DNA Replication and Transcription Guide the Asymmetric Distribution of RNAPol2 and New Nucleosomes on Replicated Daughter Genomes. bioRxiv, 2019: p. 553669. [Google Scholar]
- 62.Yang J, et al. , The Histone Chaperone FACT Contributes to DNA Replication-Coupled Nucleosome Assembly. Cell Rep, 2016. 14(5): p. 1128–1141. [DOI] [PubMed] [Google Scholar]
- 63.Han J, et al. , Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev, 2010. 24(14): p. 1485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kundu LR, et al. , Biphasic chromatin binding of histone chaperone FACT during eukaryotic chromatin DNA replication. Biochim Biophys Acta, 2011. 1813(6): p. 1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S, et al. , RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science, 2017. 355(6323): p. 415–420. [DOI] [PubMed] [Google Scholar]
- 66.Zhang H, et al. , RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol Cell, 2017. 65(2): p. 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campos EI, et al. , Analysis of the Histone H3.1 Interactome: A Suitable Chaperone for the Right Event. Mol Cell, 2015. 60(4): p. 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmad K and Henikoff S, No strand left behind. Science, 2018. 361(6409): p. 1311–1312. [DOI] [PubMed] [Google Scholar]
- 69.Serra-Cardona A and Zhang Z, Replication-Coupled Nucleosome Assembly in the Passage of Epigenetic Information and Cell Identity. Trends Biochem Sci, 2018. 43(2): p. 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cohen SM, et al. , DNA replication and the GINS complex: localization on extended chromatin fibers. Epigenetics Chromatin, 2009. 2(1): p. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahmad K and Henikoff S, Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A, 2002. 99 Suppl 4: p. 16477–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blower MD, Sullivan BA, and Karpen GH, Conserved Organization of Centromeric Chromatin in Flies and Humans. Developmental Cell, 2002. 2(3): p. 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie J, et al. , Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell, 2015. 163(4): p. 920–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rocha EP and Danchin A, Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res, 2003. 31(22): p. 6570–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sasaki T, et al. , Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol Cell Biol, 1999. 19(1): p. 547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rothstein R, Michel B, and Gangloff S, Replication fork pausing and recombination or “gimme a break”. Genes Dev, 2000. 14(1): p. 1–10. [PubMed] [Google Scholar]
- 77.Lebofsky R and Bensimon A, DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol Cell Biol, 2005. 25(15): p. 6789–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dalgaard JZ and Klar AJ, A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev, 2001. 15(16): p. 2060–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gadaleta MC and Noguchi E, Regulation of DNA Replication through Natural Impediments in the Eukaryotic Genome. Genes (Basel), 2017. 8(3). [Google Scholar]
- 80.Nordman JT, et al. , DNA copy-number control through inhibition of replication fork progression. Cell Rep, 2014. 9(3): p. 841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yarosh W and Spradling AC, Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev, 2014. 28(16): p. 1840–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munden A, et al. , Rif1 inhibits replication fork progression and controls DNA copy number in Drosophila. Elife, 2018. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piunti A, et al. , Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat Commun, 2014. 5: p. 3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cairns J, Mutation selection and the natural history of cancer. Nature, 1975. 255(5505): p. 197–200. [DOI] [PubMed] [Google Scholar]
- 85.Yadlapalli S, Cheng J, and Yamashita YM, Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J Cell Sci, 2011. 124(Pt 6): p. 933–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yadlapalli S and Yamashita YM, Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature, 2013. 498(7453): p. 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiel MJ, et al. , Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature, 2007. 449(7159): p. 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sotiropoulou PA, Candi A, and Blanpain C, The Majority of Multipotent Epidermal Stem Cells Do Not Protect Their Genome by Asymmetrical Chromosome Segregation. Stem Cells, 2008. 26(11): p. 2964–2973. [DOI] [PubMed] [Google Scholar]
- 89.Waghmare SK, et al. , Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. The EMBO Journal, 2008. 27(9): p. 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fei J-F and Huttner WB, Nonselective Sister Chromatid Segregation in Mouse Embryonic Neocortical Precursor Cells. Cerebral Cortex, 2009. 19(suppl_1): p. i49–i54. [DOI] [PubMed] [Google Scholar]
- 91.Rocheteau P, et al. , A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell, 2012. 148(1–2): p. 112–25. [DOI] [PubMed] [Google Scholar]
- 92.Conboy MJ, Karasov AO, and Rando TA, High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol, 2007. 5(5): p. e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elabd C, et al. , DNA methyltransferase-3-dependent nonrandom template segregation in differentiating embryonic stem cells. J Cell Biol, 2013. 203(1): p. 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yadlapalli S, Cheng J, and Yamashita YM, Reply to: Overlooked areas need attention for sound evaluation of DNA strand inheritance patterns in Drosophila male germline stem cells. J Cell Sci, 2011. 124: p. 4138–39. [DOI] [PubMed] [Google Scholar]
- 95.Yadlapalli S and Yamashita YM, Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature, 2013. 498(7453): p. 251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamashita YM, Nonrandom sister chromatid segregation of sex chromosomes in Drosophila male germline stem cells. Chromosome Res, 2013. 21(3): p. 243–54. [DOI] [PubMed] [Google Scholar]
- 97.Cheeseman IM and Desai A, Molecular architecture of the kinetochore–microtubule interface. Nature Reviews Molecular Cell Biology, 2008. 9(1): p. 33–46. [DOI] [PubMed] [Google Scholar]
- 98.Earnshaw W, et al. , Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. The Journal of Clinical Investigation, 1986. 77(2): p. 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Saitoh H, et al. , CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell, 1992. 70(1): p. 115–125. [DOI] [PubMed] [Google Scholar]
- 100.Klar AJ, A model for specification of the left-right axis in vertebrates. Trends in genetics: TIG, 1994. 10(11): p. 392–396. [DOI] [PubMed] [Google Scholar]
- 101.Klar AJS, Lessons learned from studies of fission yeast mating-type switching and silencing. Annual Review of Genetics, 2007. 41: p. 213–236. [DOI] [PubMed] [Google Scholar]
- 102.Kahney EW, et al. , Symmetry from Asymmetry or Asymmetry from Symmetry? Cold Spring Harbor Symposia on Quantitative Biology, 2017. 82: p. 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lansdorp PM, Immortal strands? Give me a break. Cell, 2007. 129(7): p. 1244–7. [DOI] [PubMed] [Google Scholar]
- 104.Thorpe PH, Bruno J, and Rothstein R, Kinetochore asymmetry defines a single yeast lineage. Proceedings of the National Academy of Sciences, 2009. 106(16): p. 6673–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.García Del Arco A, Edgar BA, and Erhardt S, In Vivo Analysis of Centromeric Proteins Reveals a Stem Cell-Specific Asymmetry and an Essential Role in Differentiated, Non-proliferating Cells. Cell Reports, 2018. 22(8): p. 1982–1993. [DOI] [PubMed] [Google Scholar]
- 106.Dattoli AA, et al. , CENP-A drives asymmetric cell division and maintains stem identity. bioRxiv, 2019: p. 631598. [Google Scholar]
- 107.Ladouceur AM, et al. , CENP-A and topoisomerase-II antagonistically affect chromosome length. The Journal of Cell Biology, 2017. 216(9): p. 2645–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kruitwagen T, et al. , Centromeres License the Mitotic Condensation of Yeast Chromosome Arms. Cell, 2018. 175(3): p. 780–795.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamashita YM, et al. , Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science (New York, N.Y.), 2007. 315(5811): p. 518–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X, et al. , Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature, 2009. 461(7266): p. 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Conduit PT and Raff JW, Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Current biology: CB, 2010. 20(24): p. 2187–2192. [DOI] [PubMed] [Google Scholar]
- 112.Salzmann V, et al. , Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Molecular Biology of the Cell, 2013. 25(2): p. 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Januschke J, et al. , Drosophila neuroblasts retain the daughter centrosome. Nature Communications, 2011. 2: p. 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pelletier L and Yamashita YM, Centrosome asymmetry and inheritance during animal development. Current Opinion in Cell Biology, 2012. 24(4): p. 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gasic I, Nerurkar P, and Meraldi P, Centrosome age regulates kinetochore–microtubule stability and biases chromosome mis-segregation. eLife, 2015. 4: p. e07909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Loncarek J and Bettencourt-Dias M, Building the right centriole for each cell type. The Journal of Cell Biology, 2018. 217(3): p. 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pinsky BA and Biggins S, The spindle checkpoint: tension versus attachment. Trends Cell Biol, 2005. 15(9): p. 486–93. [DOI] [PubMed] [Google Scholar]
- 118.King JM, Hays TS, and Nicklas RB, Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J Cell Biol, 2000. 151(4): p. 739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.King JM and Nicklas RB, Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J Cell Sci, 2000. 113 Pt 21: p. 3815–23. [DOI] [PubMed] [Google Scholar]
- 120.Pardo-Manuel de Villena F and Sapienza C, Female meiosis drives karyotypic evolution in mammals. Genetics, 2001. 159(3): p. 1179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pardo-Manuel de Villena F and Sapienza C, Nonrandom segregation during meiosis: the unfairness of females. Mamm Genome, 2001. 12(5): p. 331–9. [DOI] [PubMed] [Google Scholar]
- 122.Chmatal L, et al. , Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol, 2014. 24(19): p. 2295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Akera T, et al. , Spindle asymmetry drives non-Mendelian chromosome segregation. Science, 2017. 358(6363): p. 668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Iwata-Otsubo A, et al. , Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes that Drive in Female Meiosis. Curr Biol, 2017. 27(15): p. 2365–2373 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kursel LE and Malik HS, The cellular mechanisms and consequences of centromere drive. Curr Opin Cell Biol, 2018. 52: p. 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schroeder CM and Malik HS, Kindr Motors Drive in Meiosis. Cell, 2018. 173(4): p. 813–815. [DOI] [PubMed] [Google Scholar]
- 127.Dawe RK, et al. , A Kinesin-14 Motor Activates Neocentromeres to Promote Meiotic Drive in Maize. Cell, 2018. 173(4): p. 839–850 e18. [DOI] [PubMed] [Google Scholar]
- 128.Ramachandran S and Henikoff S, MINCE-Seq: Mapping In Vivo Nascent Chromatin with EdU and Sequencing. Methods Mol Biol, 2018. 1832: p. 159–168. [DOI] [PubMed] [Google Scholar]
- 129.Gutierrez MP, MacAlpine HK, and MacAlpine DM, Nascent chromatin occupancy profiling reveals locus- and factor-specific chromatin maturation dynamics behind the DNA replication fork. Genome Res, 2019. 29(7): p. 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]