Abstract
A growing number of studies indicate that host-species-specific and virus-strain-specific interactions of viral molecules with the host innate immune system play a pivotal role in determining virus host range and virulence. Because interacting proteins are likely constrained in their evolution, mutations that are selected for to improve virus replication in one species, may stochastically alter the ability of a viral antagonist to inhibit immune responses in hosts the virus has not yet encountered. Based on recent findings of host-species interactions of poxvirus, herpesvirus and influenza virus proteins, we propose a model for viral fitness and host range, which considers the full interactome between a specific host-species and a virus, resulting from the combination of all interactions, positive and negative, that influence whether a virus can productively infect a cell and cause disease in different hosts.
Keywords: host-pathogen interactions, PKR, poxvirus, herpesvirus, influenza virus, viral fitness interactome
Host Range of Viruses
Most viruses have a set of host species they are able to productively infect, a property known as host range (see Glossary), which can vary dramatically between different viruses. Whereas some viruses have very restricted host ranges and might only infect a single host species, other viruses have broad host ranges and infect multiple different host species. Extremely different host ranges can sometimes even be found among closely related viruses. For illustrative purposes, we focus primarily on a representative DNA virus family, Poxviridae, and influenza A viruses (IAV) as representatives of RNA viruses. Poxviruses include viruses which only infect one species such as variola virus, the causative agent of human smallpox, and cowpox viruses which infect at least several dozen different mammalian species [1]. While waterfowl are the natural hosts of IAV, these viruses can and do adapt to infect a broad range of host species, including other birds, pig and humans [2].
Obviously, if a virus cannot enter a cell, generally by binding a specific receptor molecule on the surface of the host cell to mediate virus entry, it will be unable to infect that cell. In general, poxviruses are not restricted by this entry step, as they promiscuously bind and enter cells from a variety of species. In contrast, IAV bind specific sialic acid-containing glycan receptors, and this interaction strongly influences viral host range [1] [3]. However, binding and entry are only one determinant of species specificity. Immediately after cell entry, viruses have to navigate additional processes, including appropriate intracellular trafficking, efficiently commandeering the host cell machinery including transcriptional and translational pathways, and evading a broad array of pattern recognition receptors and antiviral proteins, collectively known as host restriction factors. All of these steps are critical for the virus lifecycle, and disruption of any one of them can theoretically limit the host range of viruses.
This review focuses on host restriction factors, which are proteins that have evolved to recognize and restrict virus replication, often exerting their effect before the virus completes a single replication cycle. Some host restriction factors are most active against a particular virus family, for example TRIM5α primarily restricts retroviruses [4], while other host restriction factors are broadly active against many virus families, such as protein kinase R (PKR, eIF2aK2) [5]. These host restriction factors are engaged in an “evolutionary arms race” with the viral antagonists that viruses have acquired to evade the host immune response, and often display strong signatures of positive selection [6]. Over evolutionary time, different species have acquired modifications in the repertoire of host restriction factors that they encode. For example, chickens have lost the retinoic acid inducible gene I (RIG-I) [7], while TRIMCyp, a LINE-1 mediated fusion between TRIM5α and cyclophilin A, has evolved independently in some Old World and New World monkeys [8–11]. Therefore, in order to cross species-barriers and productively infect new hosts, viruses must be able to inhibit or evade unique sets of host restriction factors, the exact composition of which varies from species to species. The inability to inhibit one or more of these proteins during the course of infection is likely to reduce or completely eliminate viral fitness in a new host. Studies of host range are an active area of research for most medically important viruses. In this review, for the sake of brevity, we detail recent insights into the nature of host-virus interactions that govern species specificity of poxviruses, herpesviruses, and influenza viruses. These viruses have been the focus of substantial research investigating their host range, and in the case of poxviruses and IAV represent viruses with the capacity for broad host range, while herpesviruses tend to be more host-specific. Taken together, the research focused on host range determinants of these viruses highlights the fact that host restriction factors are under selection by a variety of different viral antagonists from multiple virus families, and that mutations that may evade inhibition by one viral antagonist may stochastically render the host susceptible to inhibition by unrelated viruses. These insights, we believe, are generalizable and therefore we propose the Viral Fitness Interactome (VFI) as a conceptual framework to incorporate the many interactions that regulate the ability of a virus to productively infect a given host species.
Implicit in this concept of the VFI is the realization that while reductive analyses of interactions between a single host antiviral protein and its cognate viral antagonist are useful for dissecting molecular determinants of the interaction, it is imperative to consider these interactions across a broad range of host species. Furthermore, unbiased deep mutational scanning methods are now available, enabling labs to systematically determine the impact of amino acid mutations across the entire gene of both host and virus proteins, to define specific residues that govern these interactions across different host species and different viral strains. The viral-host interactome is in delicate balance, and perturbations to single viral antagonist-host antiviral interactions can be sufficient to dramatically alter the outcome of viral infection.
Species-specific PKR inhibition in poxviruses and herpesviruses
Recent work that focused on the interaction of PKR with viral antagonists demonstrated that the interaction of viral proteins with the host innate immune system influences viral fitness and has important implications for cross-species transmission. PKR is an antiviral protein kinase found in vertebrates, which is activated by double-stranded (ds) RNA, produced during the replication cycle of most viruses. After binding to dsRNA, PKR is activated through dimerization and autophosphorylation. This activated PKR phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α), which results in both the general suppression of mRNA translation and the induction of antiviral genes, thereby inhibiting virus replication. [12]. To counteract the antiviral function of PKR, viruses have evolved a substantial number of inhibitors which target multiple steps in the PKR pathway [5, 13]. One of these inhibitors from poxviruses, called K3 in the prototypic poxvirus vaccinia virus (VACV), is homologous to the N-terminus of eIF2α. K3 acts as a pseudosubstrate inhibitor by binding to activated PKR, thereby precluding the interaction with eIF2 [14, 15]. K3, encoded by the gene K3L, was identified as a host range factor because it was shown to be important for VACV replication in Syrian hamster and mouse cells, but not in human cells [14, 16]. Studies of PKR-pseudosubstrate inhibitor interactions have led to seminal insights into the role that conflict-driven evolution plays in determining host range.
The dN/dS ratio, which measures the rate of accumulation of nonsynonymous mutations relative to synonymous mutations, has proven to be an effective tool to identify candidate antiviral factors and to define amino acid (aa) residues that are critical for their activity [6]. Interestingly, dN/dS and phylogenetic analyses demonstrated that the kinase domain of PKR evolved much faster in vertebrates than the kinase domains of related eIF2α kinases, which are activated by different stress stimuli [12, 17]. These studies also identified multiple positively selected sites throughout the PKR gene in primates and other vertebrate lineages. These positively selected sites were confirmed to be important determinants of host range because mutation of some of these sites in the kinase domain between PKR from different species changed the sensitivity of PKR to K3 inhibition [17, 18]. For example, gibbon PKR was sensitive to K3-mediated inhibition in a yeast assay, but substitution of two positively-selected amino acids in helix αG with two residues found in the more resistant human PKR increased the resistance of the chimeric PKR to K3-mediated inhibition. This increased resistance correlated with an approximately five-fold reduction in virus titer in cells infected with VACV lacking K3L in gibbon cells. In contrast, parental and K3L-deficent VACV strains replicated comparably well in human cells, consistent with human PKR being inhibited primarily by E3 instead of K3 [16, 18]. Similarly, human (largely resistant) and mouse (sensitive) PKR showed opposing sensitivities to K3 inhibition in cell-based reporter assays, which correlated with the importance of K3 for virus replication in cells from the respective species. The exchange of one amino acid in helix αG between human and mouse PKR partially reversed the sensitivities to K3 inhibition, highlighting the role that K3-PKR interactions play in host range [17].
Because VACV has been propagated in the lab over many decades, we lack detailed information about its original host and therefore cannot determine the ability of K3 to inhibit PKR from the original VACV reservoir species. However, recent studies of other poxviruses have addressed the interactions of K3 orthologs with PKR from the species that they naturally infect. For example, myxoma virus (MYXV), a leporipoxvirus that only infects rabbits and hares, encodes a K3 ortholog called M156 which has less than 30% amino acid identity with VACV K3 [19]. MYXV infection has a mortality rate approaching 100% for naïve European rabbits. The intentional release of MYXV virus into Australia and Europe provided one of the best-studied examples of population level host-virus co-evolution in a natural setting. Less than a decade after the release of MYXV the mortality rates dropped dramatically, apparently due in part to attenuation of the virus and in part to increased resistance of the rabbit hosts over time [20]. Consistent with MYXV host range, M156 inhibited PKR only inhibited European rabbit PKR but not PKR from humans, sheep or five different rodent species. Moreover, human PKR, but not rabbit PKR, was able to restrict MYXV replication in congenic cell lines [19]. Importantly, a naturally occurring variant of M156 found in more than 50% of Australian MYXV field isolates [21–23] rendered M156 unable to inhibit PKR as measured in complementary yeast and cell culture assays. This mutation also led to the attenuation of MYXV in vitro, raising the possibility that this mutation contributed to virus attenuation in the field [19].
A third distinct genus of poxviruses, capripoxvirus (CaPV), has also shown remarkable impacts of their K3 orthologs on virus host range. The CaPVs sheeppox (SPPV) and goatpox (GTPV) infect both sheep and goats, and cause high morbidity and mortality rates, while lumpy skin disease virus (LSDV) infects only cattle and normally causes lower morbidity and mortality rates [24]. K3 orthologs from these CaPVs show between 93 and 97% amino acid sequence identity to each other and 40% sequence identity in comparison to VACV K3. Inhibition assays showed that CaPV K3 orthologs strongly inhibited sheep and goat PKR, whereas mouse PKR was not inhibited, consistent with the CaPV host range. Surprisingly, human PKR was inhibited well even though CaPV does not infect humans, whereas PKR from LSDV-permissive cows was only weakly inhibited. In contrast, sheep, goat and human PKR were poorly inhibited by VACV K3, whereas cow and mouse PKR were strongly inhibited. As predicted from these data, VACV strains that expressed CaPV K3 orthologs as the only PKR inhibitor replicated well in sheep, goat and human cells, but replicated poorly in cow cells, whereas a virus containing only VACV K3 replicated to high titers only in cow cells [25]. It is noteworthy that CaPV K3 orthologs inhibited human PKR, which shares only 60% identity with sheep and goat PKR, more efficiently than they inhibited cow PKR, which shares 87% identity with sheep and goat PKR. This observation clearly demonstrates that neither the overall similarity nor the phylogenetic relatedness of PKR are necessarily good predictors of the sensitivity to inhibition by a particular viral antagonist (see Outstanding Questions). These studies of MYXV and CaPV K3 orthologs extend our knowledge about the importance of species-specific PKR inhibition for viral host range and raise interesting points. First, MYXV, SPPV, and GTPV K3 orthologs were highly effective at suppressing PKR from their natural host species. Cow PKR was the only exception, being only weakly inhibited by LSDV K3, yet this modest inhibition was still sufficient to drive some VACV replication in cow cells. It is possible that this low level of PKR inhibition might be sufficient to permit virus replication in cows infected with LSDV. Furthermore, this modest inhibition might contribute to the lesser disease severity in comparison to sheep and goats infected by SPPV or GTPV that more efficiently antagonize their native PKR. Second, whereas MYXV M156 only efficiently inhibited European rabbit PKR but not PKR from species which are resistant to MYXV infection, CaPV K3 homologs also efficiently inhibited PKR from humans, even though CaPV does not productively infect humans. This indicates that while PKR inhibition is essential for poxviruses to replicate, it is not the sole determinant for successful infection. It is noteworthy, however, that CaPVs, which clearly have not been subjected to evolutionary pressure in human cells, contain potent inhibitors of human PKR. Although PKR shows a remarkable flexibility to harbor mutations that modify sensitivity to pseudosubstrate-mediated inhibition, the requirement to maintain an interaction with its substrate eIF2α likely limits the number of possible variants [17, 18]. Because a limited number of sites broadly determine the ability of a pseudosubstrate inhibitor to interact with PKR, viruses may stochastically evolve the ability to inhibit PKR or other host restriction factors from species that the virus hasn’t encountered, simply as a function of the limited evolutionary options available to its target (Figure 1). Another example for species-specific inhibition of PKR can be found in herpesviruses. These viruses, in general, co-speciated with their mammalian hosts and perhaps as a consequence are often highly host-restricted [26]. For example, in nature human cytomegalovirus (HCMV) only replicates in humans, and in cell culture HCMV is restricted to human and chimpanzee derived cells [27]. Consistent with this host restriction, the HCMV PKR antagonist TRS1 (hTRS1) only inhibited human PKR [28]. In contrast, CMVs derived from rhesus macaques (Rh), African green monkeys (AGM), and squirrel monkeys (Sm) can productively infect human cells [29–31]. Surprisingly, in infection-based assays, only SmTRS1 inhibited human PKR, whereas all other primate CMV TRS1 orthologs derived from these viruses did not inhibit human PKR [28, 32]. The differential inhibition of human PKR by these various TRS1 orthologs is mediated by a single aa residue in the PKR αG helix at position 489. Mutation of this residue in human PKR to the AGM-encoded aa (F489S) rendered PKR resistant to inhibition by hTRS1 although it was still sensitive to SmTRS1. Interestingly, residue 489 is also a determinant of PKR susceptibility to VACV K3 [18, 33]. Despite identification of this key residue, it remains an open question how non-human primate CMVs are able to replicate in human cells, because human PKR is remarkably non-polymorphic and PKR inhibition is thought to be essential for CMV replication [32, 34].
Outstanding questions.
If phylogenetic relatedness is not a good predictor for host-virus interactions, will incorporating structure-based analyses yield more accurate predictions?
Why do many viruses expend energy and genomic space to encode multiple antagonists that target the same host-restriction factor?
How much does intraspecies resistance to a given virus vary between individuals? What is the spectrum of susceptibility within a population, and how does naturally occurring variation in host restriction factors influence this variation?
To what extent does the viral genetic background (strain or species) influence host-virus interactions? Similarly, how profoundly do proteomic differences in cells derived from the same genetic background influence host-virus interactions and tissue tropism?
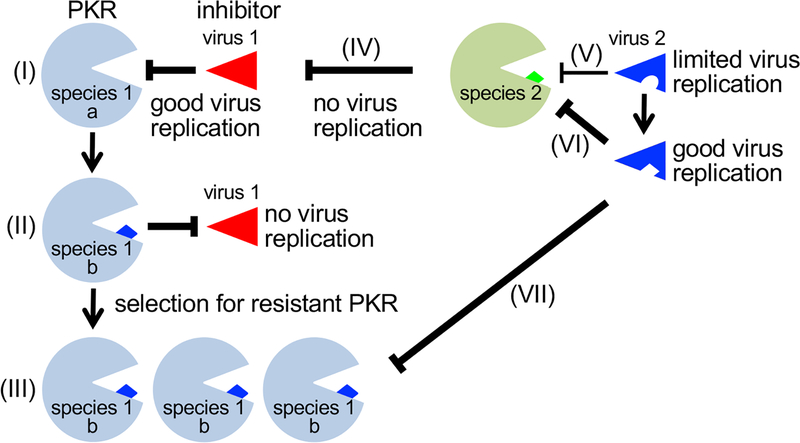
Figure 1. Consequences of host-virus molecular arms races for cross-species transmission.
(I) Good inhibition of an antiviral protein in species 1 (PKR allele a) by an inhibitor from virus 1 exerts selective pressure and leads to the selection for the resistant allele b, which was present in the population at low frequency (II). This variant cannot be inhibited by virus 1 and thus prevents virus infection. (III) The provided selective advantage leads to the fixation of this allele in the population. (IV) The inhibitor of virus 1 is unable to inhibit PKR from species 2, which precludes virus replication. (V) An inhibitor of virus 2 suboptimally inhibits PKR from species 2, which leads to limited virus replication, (VI) allowing the virus to evolve a better inhibitor, which results in good virus replication. (VII) By chance, the evolved inhibitor from virus 2, is also a good inhibitor for the PKR b allele from species 1, which enables virus 2 to gain a foothold in species 1 and results in successful cross-species transmission of the virus.
Although the critical residues governing species-specific interaction are unclear, RhTRS1 overexpression as a result of gene duplication can inhibit human PKR [35]. A similar elevated expression of RhTRS1 was detected during RhCMV infection of human fibroblasts, although the mechanism underlying this increase is not yet known [36]. More broadly, gain of function experiments such as those described in this section often demonstrate that improving the activity of a single viral antagonist can make the difference between infection and resistance. This observation suggests that the network of host-virus interactions is in relative balance but that perturbations of any one interaction may alter host susceptibility to a given virus, either positively or negatively.
Species-specific interaction of viral multifunctional proteins and importance for host range and virulence
While in the previous section we discussed the implications of PKR inhibition by viral proteins that mainly target a single host protein, and therefore present a more straightforward model for host-virus interactions, smaller viruses have often evolved multifunctional proteins that inhibit multiple host restriction factors. One such example is influenza A virus (IAV) nonstructural protein 1 (NS1), which is the only known dedicated host restriction factor antagonist encoded by this virus, although it is important to note that other IAV proteins, such as PB2 and PB1-F2 have also been reported to antagonize host antiviral factors [37–39].
IAV naturally infects waterfowl but often crosses species barriers to infect a wide range of hosts, including humans, pigs and horses. Ferrets have also proven to be a particularly useful laboratory model to study IAV transmission dynamics [40]. The ability of IAV to infect new host species is multifactorial, and studying adaptations that are necessary for efficient replication in new hosts remains an area of intense research. NS1 is the primary antagonist of the host interferon response, and therefore likely to be a key determinant of IAV host range. NS1 is encoded by the eighth segment of the IAV genome, along with two differentially spliced transcripts: the nuclear export protein (NEP), which is essential for virus replication, and an as yet uncharacterized non-structural protein, NS3. NS1 is comprised of a dsRNA binding domain, a variable length linker region, an effector domain, and a variable length C-terminal tail. Multiple studies have identified amino acid residues critical for NS1-mediated inhibition of several host antiviral proteins including RIG-I [41–45], PKR [46, 47], TRIM25 [48] and RNF135 (RIPLET) [49]. While many of these interactions have been shown to be virus strain-specific, much less is known about whether these NS1 mutations contribute to cross-species transmission events.
NS1 splice regulation influences IAV host range.
Although NS1 alters host splicing through interactions with CPSF30 [50] and also regulates the splice products of genome segment 7 [51], there are conflicting reports as to whether NS1 regulates its own splicing [52–56]. Nevertheless, changes in segment eight splicing efficiency have a substantial impact on virus replication in different species. Chua, et al., demonstrated that IAV uses an inefficient splice site as a “molecular timer” to coordinate viral replication in a variety of mammalian cells [57]. This splicing efficiency can vary dramatically between different virus strains [58]. Huang, et al. recently identified an exonic splicing enhancer (ESE) motif in the NEP message. A nucleotide G540A mutation in this ESE correlated with increased susceptibility of humans to this virus and accelerated the rate of NS1 accumulation. The increased amount of NS1 improved virus replication in mammalian cells and also maintained replication in avian cells [56]. Interestingly, other recent avian to human transmission events have also been correlated with G540A [59–62]. However, most avian influenza viruses appear to contain G540 even though A540 replicates efficiently in avian cells. It is unclear if this observation indicates a requirement for higher levels of NEP in avian hosts, or if a required “setpoint” level of splicing is selected for once the virus adapts to a host. Although not directly analogous, these changes in splicing efficiency may fill a similar evolutionary role to that of gene duplication in large DNA viruses [63], which results in overexpression of a given gene and thus improved replication via mass action effects without requiring genomic structural changes that are likely difficult or impossible in smaller RNA viruses.
Influenza NS1 species determinants.
Despite their reputation as a “mixing vessel”, pigs are no more susceptible to infection with IAV than other mammals [64]. However, they are more likely to be held in close proximity to waterfowl and poultry, and are therefore at an increased risk of exposure to avian influenza viruses. Understanding the viral determinants that mediate host adaptation in relevant species is therefore an important public health concern. While substantial work has been carried out to identify NS1 residues that govern interactions with various antiviral proteins, much less is known about how these polymorphisms influence viral fitness in new hosts. The amino acid change D74S, particularly in the context of two other mutations, P3S and R41K, permitted virus replication of an otherwise strictly avian virus (A/FPV/Rostock/34) in mammalian cell culture and in mouse infections, while still allowing efficient virus replication in avian cells [65]. Due to its multi-functional nature, attributing this improved replication to NS1 anti-immune functions alone is difficult, and the observed phenotype may be due to a combination of altered immune inhibition, and changes in viral splicing and polymerase activity [66–70].
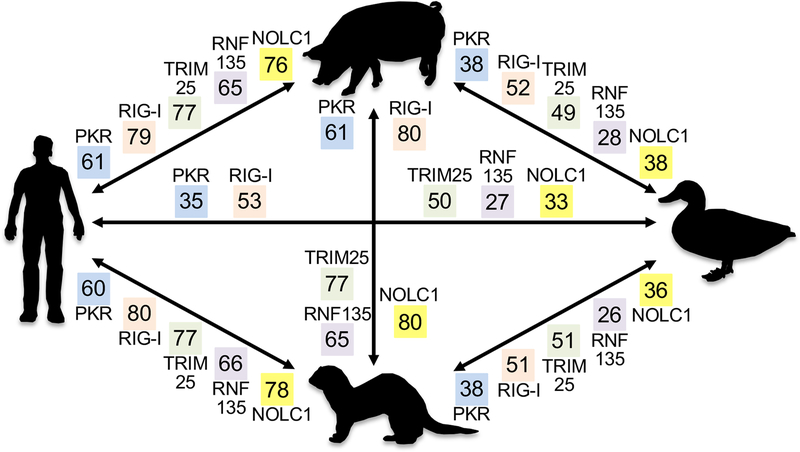
Understanding how these and other NS1 variations influence host-virus interactions in different species is critical, because NS1 has more than 60 interacting partners in the host proteome [71]. For example, NS1 interacts with nucleolar and coiled-body phosphoprotein 1 (NOLC1) and induces apoptosis in human-derived cells [72–74]. However, as shown in Figure 2, NOLC1, and many other NS1-interacting proteins, are very different between species. Therefore, it’s reasonable to postulate that many of these protein orthologs may have different interaction profiles between species, and differences in host-virus interactions with these orthologs are likely to influence host range. This variation demands additional scrutiny, as a recent report has demonstrated that RIG-I orthologs from a variety of mammals and birds exhibit remarkably different abilities to inhibit IAV replication [75].
Figure 2. Key host interaction partners of influenza A virus NS1 are highly divergent between relevant host species.
The amino acid identities between pig (Sus scrofa), waterfowl (mallard duck (Anas platyrhynchos) or goose (Anser anser domesticus)), ferret (Mustela putorius furo) and human (Homo sapiens) for PKR, RIG-I (DDX58), TRIM25, RNF135/RIPLET and NOLC1 are shown in colored boxes. For waterfowl, mallard duck proteins were analyzed, except for PKR (goose), because a high-quality mallard PKR sequence is not available. Sequence identities shown were determined from psi-blast searches and are very similar to sequence identities obtained from multiple sequence alignments. Accession numbers for PKR (eIF2aK2): NP_002750.1; NP_999484.1; XP_012918395.1 (first 39 aa missing from sequence); AXJ21467.1. RIG-I (DDX58): NP_055129.2; NP_998969.2; XP_004765417.2; BAO25514.1. TRIM25: NP_005073.2; XP_005657028.3; XP_012909123.1; XP_012948210.2. RNF135 (Riplet): NP_115698.3; XP_003131783.1; XP_004747119.1; XP_027327216.1 (contains a unique 97 aa N-terminal extension, which was excluded from analyses). NOLC1: NP_001271317.1; XP_005671467.1; XP_004749466.1; XP_027316116.1.
NS1 from the related influenza B virus, which naturally infects only humans and seals [76], was shown to inhibit ISG15-mediated virus inhibition in a species-specific manner, inhibiting human but not mouse ISG15, and shows approximately 100-fold higher binding affinity for human, relative to mouse, dog and cow ISG15 [77–80]. Consistent with this observation, ISG15-deficient mice were shown to be more susceptible to IBV infection, whereas ISG-deficient humans had no change in IBV susceptibility [81]. Further studies are warranted to determine what NS1 residues are necessary to bind ISG15 in a species-specific manner, and whether ISG15-NS1 interactions are a general determinant of host range.
Systematic screens of virus mutations in multiple hosts.
Currently, there has not been a systematic comparative survey of how mutations in NS1 alter virus replication in species that are important for the virus lifecycle. Systematic approaches will be necessary to define these critical mutations and their activity in a variety of host species. One approach, deep mutational scanning, has proven its utility in assessing the impact of mutations on the function of multiple IAV proteins [82–85], and has recently been used to identify mutations that permit avian influenza to replicate more efficiently in human cells [86]. These systematic, library-based approaches have the advantage that once a library is generated it is possible to screen it in cells derived from multiple species to define the breadth of species impacted by a given mutation in an unbiased manner. Testing these mutational libraries in multiple species is vital because the specific composition of host restriction factors that a virus must antagonize also varies from species to species as, for example, chickens do not express RIG-I [7]. While it is relatively straightforward to generate libraries of all single mutations, the number of molecules required to screen all combinations of multiple mutations, i.e. two or three mutations increases exponentially. Therefore, evaluating host-virus interactions mediated by multiple residues, or epistatic interactions within a viral antagonist is not currently feasible. However, in combination with cataloging naturally occurring mutations and laboratory based experimental evolution, deep mutational scanning will be a valuable tool to define the structural and genetic pathways that lead to increased fitness in a new host.
The other critical piece in understanding host range is defining how host proteins interact with their viral antagonists. We envision two complementary approaches to investigate host determinants of viral susceptibility. First, analyzing the activity of a panel of a particular host restriction factor derived from multiple hosts in a congenic background will enable fine molecular dissection of the critical residues that influence virus susceptibility. CRISPR-based knockout combined with targeted recombination such as a Flp/FRT site enables the construction of defined cell lines that can be used to analyze the activity of a given host restriction factor and to make direct comparisons between orthologs. Second, as our thesis is that the network of all host-virus interactions is what determines host range, it is important to define which host proteins contribute to viral susceptibility in cell types derived from multiple different species. A recent, elegant approach employed a CRISPR-based library targeting interferon stimulated genes to identify a small panel of IFN-induced host restriction factors capable of inhibiting the IFN-sensitive HIV-1 LAI strain. This observation demonstrates that a network of host restriction factors exerting modest inhibitory effects can together restrict virus replication [87].
Viral fitness interactome
Many, if not most, viral proteins that interact with host factors are multifunctional like IAV NS1. Examples include Ebola virus VP35 [88], flavivirus non-structural protein 5 [89] and the poxvirus E3L family [90]. For viruses that infect multiple species as IAV does, it can be assumed that many of the host proteins targeted by these viral antagonists vary between species similar to the differences shown in Figure 2. Therefore, these interactions are likely to show host-specific differences, particularly if their interacting partners vary at important interacting surfaces. It can be further assumed that differences in the expression levels of the host proteins, either between different species or different tissues, also lead to changes in the host-virus interactome. To account for the importance of host-specific differences in host-virus interactions for virus replication, we use the term Viral Fitness Interactome (VFI), which integrates the combinatory effects of all interactions between viral molecules with antiviral and pro-viral host factors (Figure 3). The VFI varies between different hosts and cell types depending on host and cell-type specific expression differences. It can be additionally assumed that different virus strains or species exhibit unique VFI profiles.
Figure 3. Host-specific Viral Fitness Interactome.
The Viral Fitness Interactome (VFI) results from the combinatory effects of all interactions between viral molecules with antiviral and pro-viral host factors. The VFI varies between different hosts and cell types. In this model, viral fitness (VF) of a single virus, as indicated by the size of the plus sign, is the result of interactions of a viral protein (VP) with the orthologs of four different host proteins (HP1–4). The relative strength of the interactions is indicated by the thickness of the lines connecting VP and HP. In species A, one strong interaction (with HP1, as indicated by the thickness of the line), one intermediate interaction (HP2) and two weaker interactions contribute to high VF. In species B, the interactions between VP and HPs are different, yet the combinatorial effects result in the same VF as found in species A. In species C, intermediate or weak interactions occur with HP1, 2 and 3, and there is no interaction with the antiviral HP4, resulting in low VF. In species D, the interactions between VP and HP1–3 are the same as species C; however, HP4 is not present, so the VF is therefore higher than in species C.
Concluding Remarks
We have made substantial strides in our understanding of how host-virus interactions drive evolution and influence host susceptibility. These interactions occur at all stages of the viral replication cycle, and constitute both pro- and antiviral interactions. The antiviral activity mediated by host restriction factors has provided several insights into host-virus interactions, and has important implications for virus host range. Recent work has demonstrated that neither phylogenetic relatedness nor sequence similarity are so far accurate predictors of host restriction factor susceptibility to a viral antagonist and highlights some limitations of animal models for human virus infections. Furthermore, these interactions appear to have at least some element of stochasticity, as restriction factors from some hosts are susceptible to inhibition by viral antagonists that cannot infect the host. Additionally, the network of possible host-virus interactions may vary between species, as host restriction factors are gained, lost, and modified over evolutionary time. Perturbations to any node in this network of interactions may alter susceptibility of a given host, either positively or negatively. Therefore, it is critical to consider the full viral fitness interactome to account for the combinatory effects of all interactions between viral molecules with antiviral and pro-viral host factors. Deep mutational scanning to sample all possible mutations, coupled with a directed effort to characterize viral antagonist activity against a much broader range of host restriction factor orthologs from multiple species may allow us to better define the available landscape of functional mutations. Defining the impact of these molecular interactions, viable mutations that change host-virus interactions, and how the individual interactions fit together as a network will enable us to predict, detect and act better against high risk viruses and will play an important role for maintaining and improving human and animal health.
Highlights.
The host range of a virus is governed by multiple molecular interactions, from receptor interaction, to coopting the cellular machinery and evading innate immune recognition. Failure of any one of these steps can reduce or eliminate viral replication in a given host.
Recent work in poxvirus-, influenza virus-, and herpesvirus-based systems has identified multiple evolutionary mechanisms for host adaptation. These studies have shown that these changes can have unpredictable effects on host-virus interactions and host susceptibility in both closely and distantly related species.
The Viral Fitness Interactome (VFI) results from the combinatory effects of all pro- and antiviral interactions, which ultimately determines host susceptibility to a virus. The VFI varies between different hosts, cell types and viruses.
Systematic, unbiased mutational analysis will be necessary to develop a comprehensive picture of the VFI.
Acknowledgments:
This work was supported by grant AI114851 (to S.R.) and AI135257 (to G.B.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Glossary
- Host range
The spectrum of host species that are susceptible to infection by a given pathogen.
- Host range factor
A viral gene product that is important for the infection of cells from some species but that is dispensable for the infection in cells from other species. Often targets antiviral host proteins.
- Host restriction factor
A diverse group of often interferon-stimulated genes that exert antiviral effects and restrict viral replication.
- PKR
Protein kinase R, is a host restriction factor, which is activated by dsRNA formed during virus infection. It exerts its antiviral function by inhibiting protein synthesis.
- Positive selection
The process that increases the frequency of advantageous traits in a population
- Pseudosubstrate inhibitor
An inhibitor that mimics the substrate of an enzyme and inhibits its activity
- Ortholog
A gene found in multiple species that evolved from a single ancestral gene through the process of speciation.
- RIG-I
Retinoic acid inducible gene I, is a host restriction factor, which is recognizes non-self RNA species and induces an interferon response.
- Viral antagonist
A viral molecule that has evolved to inhibit host factors. Often antiviral or host restriction factors.
References
- 1.Haller SL et al. (2014) Poxviruses and the evolution of host range and virulence. Infect Genet Evol 21, 15–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slemons RD et al. (1974) Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis 18 (1), 119–24. [PubMed] [Google Scholar]
- 3.Aytay S and Schulze IT (1991) Single amino acid substitutions in the hemagglutinin can alter the host range and receptor binding properties of H1 strains of influenza A virus. J Virol 65 (6), 3022–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grutter MG and Luban J (2012) TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr Opin Virol 2 (2), 142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dzananovic E et al. (2018) Viral proteins targeting host protein kinase R to evade an innate immune response: a mini review. Biotechnol Genet Eng Rev 34 (1), 33–59. [DOI] [PubMed] [Google Scholar]
- 6.Daugherty MD and Malik HS (2012) Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46, 677–700. [DOI] [PubMed] [Google Scholar]
- 7.Barber MR et al. (2010) Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A 107 (13), 5913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayah DM et al. (2004) Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430 (6999), 569–73. [DOI] [PubMed] [Google Scholar]
- 9.Brennan G et al. (2008) TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci U S A 105 (9), 3569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Virgen CA et al. (2008) Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci U S A 105 (9), 3563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson SJ et al. (2008) Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci U S A 105 (9), 3557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenburg S et al. (2016) Evolution of eIF2α Kinases: Adapting Translational Control to Diverse Stresses In Evolution of the Protein Synthesis Machinery and Its Regulation (Hernández G and Jagus R eds), pp. 235–260, Springer International Publishing. [Google Scholar]
- 13.Langland JO et al. (2006) Inhibition of PKR by RNA and DNA viruses. Virus Res 119 (1), 100–10. [DOI] [PubMed] [Google Scholar]
- 14.Beattie E et al. (1991) Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology 183 (1), 419–22. [DOI] [PubMed] [Google Scholar]
- 15.Dar AC and Sicheri F (2002) X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell 10 (2), 295–305. [DOI] [PubMed] [Google Scholar]
- 16.Langland JO and Jacobs BL (2002) The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299 (1), 133–41. [DOI] [PubMed] [Google Scholar]
- 17.Rothenburg S et al. (2009) Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol 16 (1), 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elde NC et al. (2009) Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 457 (7228), 485–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng C et al. (2016) Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc Natl Acad Sci U S A 113 (14), 3855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr PJ et al. (2015) Myxoma virus and the Leporipoxviruses: an evolutionary paradigm. Viruses 7 (3), 1020–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerr PJ et al. (2012) Evolutionary history and attenuation of myxoma virus on two continents. PLoS Pathog 8 (10), e1002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerr PJ et al. (2013) Comparative analysis of the complete genome sequence of the California MSW strain of myxoma virus reveals potential host adaptations. J Virol 87 (22), 12080–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr PJ et al. (2019) Punctuated Evolution of Myxoma Virus: Rapid and Disjunct Evolution of a Recent Viral Lineage in Australia. J Virol 93 (8). doi: 10.1128/JVI.01994-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuppurainen ESM et al. (2017) Review: Capripoxvirus Diseases: Current Status and Opportunities for Control. Transbound Emerg Dis 64 (3), 729–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park C et al. (2019) Species-specific inhibition of antiviral protein kinase R by capripoxviruses and vaccinia virus. Ann N Y Acad Sci 1438 (1), 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeoch DJ et al. (1995) Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol 247 (3), 443–58. [DOI] [PubMed] [Google Scholar]
- 27.Perot K et al. (1992) Primary chimpanzee skin fibroblast cells are fully permissive for human cytomegalovirus replication. J Gen Virol 73 (Pt 12), 3281–4. [DOI] [PubMed] [Google Scholar]
- 28.Child SJ et al. (2012) Species specificity of protein kinase r antagonism by cytomegalovirus TRS1 genes. J Virol 86 (7), 3880–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher DM et al. (1974) Persistent shedding of cytomegalovirus in the urine of healthy Rhesus monkeys. Proc Soc Exp Biol Med 145 (3), 794–801. [DOI] [PubMed] [Google Scholar]
- 30.Black PH et al. (1963) Isolation of a cytomegalovirus from African green monkey. Proc Soc Exp Biol Med 112, 601–5. [DOI] [PubMed] [Google Scholar]
- 31.Rangan SR and Chaiban J (1980) Isolation and characterization of a cytomegalovirus from the salivary gland of a squirrel monkey (Saimiri sciureus). Lab Anim Sci 30 (3), 532–40. [PubMed] [Google Scholar]
- 32.Carpentier KS and Geballe AP (2016) An Evolutionary View of the Arms Race between Protein Kinase R and Large DNA Viruses. J Virol 90 (7), 3280–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carpentier KS et al. (2016) A Single Amino Acid Dictates Protein Kinase R Susceptibility to Unrelated Viral Antagonists. PLoS Pathog 12 (10), e1005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marshall EE et al. (2009) Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol 83 (9), 4112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennan G et al. (2014) Adaptive gene amplification as an intermediate step in the expansion of virus host range. PLoS Pathog 10 (3), e1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Child SJ et al. (2018) Antagonism of the Protein Kinase R Pathway in Human Cells by Rhesus Cytomegalovirus. J Virol 92 (6). doi: 10.1128/JVI.01793-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber M et al. (2015) Influenza virus adaptation PB2–627K modulates nucleocapsid inhibition by the pathogen sensor RIG-I. Cell Host Microbe 17 (3), 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo SM et al. (2017) Inhibition of Avian Influenza A Virus Replication in Human Cells by Host Restriction Factor TUFM Is Correlated with Autophagy. MBio 8 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazel-Sanchez B et al. (2018) H5N1 Influenza A Virus PB1-F2 Relieves HAX-1-Mediated Restriction of Avian Virus Polymerase PA in Human Lung Cells. J Virol 92 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belser JA et al. (2011) The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4 (5), 575–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S et al. (2018) Identification of two residues within the NS1 of H7N9 influenza A virus that critically affect the protein stability and function. Vet Res 49 (1), 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jureka AS et al. (2015) Structural Basis for a Novel Interaction between the NS1 Protein Derived from the 1918 Influenza Virus and RIG-I. Structure 23 (11), 2001–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo Z et al. (2007) NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am J Respir Cell Mol Biol 36 (3), 263–9. [DOI] [PubMed] [Google Scholar]
- 44.Mibayashi M et al. (2007) Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81 (2), 514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Opitz B et al. (2007) IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol 9 (4), 930–8. [DOI] [PubMed] [Google Scholar]
- 46.Hatada E et al. (1999) Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol 73 (3), 2425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schierhorn KL et al. (2017) Influenza A Virus Virulence Depends on Two Amino Acids in the N-Terminal Domain of Its NS1 Protein To Facilitate Inhibition of the RNA-Dependent Protein Kinase PKR. J Virol 91 (10). doi: 10.1128/JVI.00198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gack MU et al. (2009) Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5 (5), 439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oshiumi H et al. (2009) Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem 284 (2), 807–17. [DOI] [PubMed] [Google Scholar]
- 50.Ayllon J and Garcia-Sastre A (2015) The NS1 protein: a multitasking virulence factor. Curr Top Microbiol Immunol 386, 73–107. [DOI] [PubMed] [Google Scholar]
- 51.Robb NC and Fodor E (2012) The accumulation of influenza A virus segment 7 spliced mRNAs is regulated by the NS1 protein. J Gen Virol 93 (Pt 1), 113–8. [DOI] [PubMed] [Google Scholar]
- 52.Robb NC et al. (2010) Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J Gen Virol 91 (Pt 9), 2331–40. [DOI] [PubMed] [Google Scholar]
- 53.Lu Y et al. (1994) The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev 8 (15), 1817–28. [DOI] [PubMed] [Google Scholar]
- 54.Fortes P et al. (1994) Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J 13 (3), 704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garaigorta U and Ortin J (2007) Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res 35 (14), 4573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang X et al. (2017) An NS-segment exonic splicing enhancer regulates influenza A virus replication in mammalian cells. Nat Commun 8, 14751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chua MA et al. (2013) Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep 3 (1), 23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Backstrom Winquist E et al. (2012) Inefficient splicing of segment 7 and 8 mRNAs is an inherent property of influenza virus A/Brevig Mission/1918/1 (H1N1) that causes elevated expression of NS1 protein. Virology 422 (1), 46–58. [DOI] [PubMed] [Google Scholar]
- 59.Pu J et al. (2015) Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A 112 (2), 548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma C et al. (2015) Emergence and evolution of H10 subtype influenza viruses in poultry in China. J Virol 89 (7), 3534–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu M et al. (2015) Genetic diversity of avian influenza A (H10N8) virus in live poultry markets and its association with human infections in China. Sci Rep 5, 7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bi Y et al. (2016) Genesis, Evolution and Prevalence of H5N6 Avian Influenza Viruses in China. Cell Host Microbe 20 (6), 810–821. [DOI] [PubMed] [Google Scholar]
- 63.Bayer A et al. (2018) Adaptation by copy number variation in monopartite viruses. Curr Opin Virol 33, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long JS et al. (2019) Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17 (2), 67–81. [DOI] [PubMed] [Google Scholar]
- 65.Kanrai P et al. (2016) Identification of specific residues in avian influenza A virus NS1 that enhance viral replication and pathogenicity in mammalian systems. J Gen Virol 97 (9), 2135–48. [DOI] [PubMed] [Google Scholar]
- 66.Robb NC et al. (2011) The influenza A virus NS1 protein interacts with the nucleoprotein of viral ribonucleoprotein complexes. J Virol 85 (10), 5228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen G et al. (2014) Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe 15 (4), 484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin L et al. (2012) Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J Virol 86 (4), 1942–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carrillo B et al. (2014) The influenza A virus protein NS1 displays structural polymorphism. J Virol 88 (8), 4113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hale BG (2014) Conformational plasticity of the influenza A virus NS1 protein. J Gen Virol 95 (Pt 10), 2099–105. [DOI] [PubMed] [Google Scholar]
- 71.Klemm C et al. (2018) Immunomodulatory Nonstructural Proteins of Influenza A Viruses. Trends Microbiol 26 (7), 624–636. [DOI] [PubMed] [Google Scholar]
- 72.Zhu C et al. (2013) Interaction of avian influenza virus NS1 protein and nucleolar and coiled-body phosphoprotein 1. Virus Genes 46 (2), 287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu CY et al. (2015) Identification of NS1 domains of avian H5N1 influenza virus which influence the interaction with the NOLC1 protein. Virus Genes 50 (2), 238–44. [DOI] [PubMed] [Google Scholar]
- 74.Zhu C et al. (2017) The interaction between NOLC1 and IAV NS1 protein promotes host cell apoptosis and reduces virus replication. Oncotarget 8 (55), 94519–94527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao Q et al. (2015) RIG-I from waterfowl and mammals differ in their abilities to induce antiviral responses against influenza A viruses. J Gen Virol 96 (Pt 2), 277–87. [DOI] [PubMed] [Google Scholar]
- 76.Osterhaus AD et al. (2000) Influenza B virus in seals. Science 288 (5468), 1051–3. [DOI] [PubMed] [Google Scholar]
- 77.Versteeg GA et al. (2010) Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J Virol 84 (10), 5423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sridharan H et al. (2010) Species specificity of the NS1 protein of influenza B virus: NS1 binds only human and non-human primate ubiquitin-like ISG15 proteins. J Biol Chem 285 (11), 7852–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao C et al. (2016) Influenza B virus non-structural protein 1 counteracts ISG15 antiviral activity by sequestering ISGylated viral proteins. Nat Commun 7, 12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang Y and Wang X (2018) Structural insights into the species preference of the influenza B virus NS1 protein in ISG15 binding. Protein Cell doi: 10.1007/s13238-018-0598-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Speer SD et al. (2016) ISG15 deficiency and increased viral resistance in humans but not mice. Nat Commun 7, 11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bloom JD (2014) An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol 31 (8), 1956–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doud MB and Bloom JD (2016) Accurate Measurement of the Effects of All Amino-Acid Mutations on Influenza Hemagglutinin. Viruses 8 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ashenberg O et al. (2017) Deep mutational scanning identifies sites in influenza nucleoprotein that affect viral inhibition by MxA. PLoS Pathog 13 (3), e1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du Y et al. (2018) Genome-wide identification of interferon-sensitive mutations enables influenza vaccine design. Science 359 (6373), 290–296. [DOI] [PubMed] [Google Scholar]
- 86.Soh YS et al. (2019) Comprehensive mapping of adaptation of the avian influenza polymerase protein PB2 to humans. Elife 8. doi: 10.7554/eLife.45079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.OhAinle M et al. (2018) A virus-packageable CRISPR screen identifies host factors mediating interferon inhibition of HIV. Elife 7. doi: 10.7554/eLife.39823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Basler CF et al. (2019) Virus and host interactions critical for filoviral RNA synthesis as therapeutic targets. Antiviral Res 162, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shah PS et al. (2018) Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 175 (7), 1931–1945 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perdiguero B and Esteban M (2009) The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res 29 (9), 581–98. [DOI] [PubMed] [Google Scholar]